Abstract
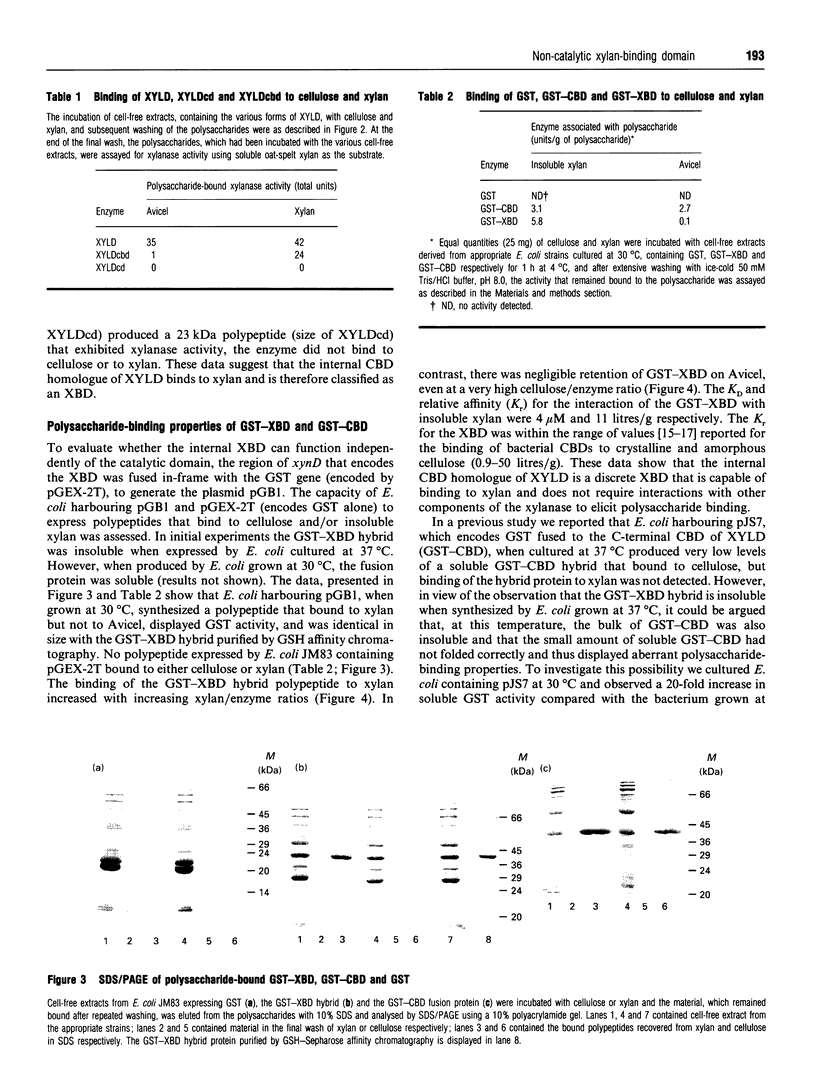
Xylanase D (XYLD) from Cellulomonas fimi contains a C-terminal cellulose-binding domain (CBD) and an internal domain that exhibits 65% sequence identity with the C-terminal CBD. Full-length XYLD binds to both cellulose and xylan. Deletion of the C-terminal CBD from XYLD abolishes the capacity of the enzyme to bind to cellulose, although the truncated xylanase retains its xylan-binding properties. A derivative of XYLD lacking both the C-terminal CBD and the internal CBD homologue did not bind to either cellulose or xylan. A fusion protein consisting of the XYLD internal CBD homologue linked to the C-terminus of glutathione S-transferase (GST) bound to xylan, but not to cellulose, while GST bound to neither of the polysaccharides. The Km and specific activity of full-length XYLD and truncated derivatives of the enzyme lacking the C-terminal CBD (XYLDcbd), and both the CBD and the internal CBD homologue (XYLDcd), were determined with soluble and insoluble xylan as the substrates. The data showed that the specific activities of the three enzymes were similar for both substrates, as were the Km values for soluble substrate. However, the Km values of XYLD and XYLDcbd for insoluble xylan were significantly lower than the Km of XYLDcd. Overall, these data indicate that the internal CBD homologue in XYLD constitutes a discrete xylan-binding domain which influences the affinity of the enzyme for insoluble xylan but does not directly affect the catalytic activity of the xylanase. The rationale for the evolution of this domain is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Béguin P., Aubert J. P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994 Jan;13(1):25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Coutinho J. B., Gilkes N. R., Warren R. A., Kilburn D. G., Miller R. C., Jr The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992 May;6(9):1243–1252. doi: 10.1111/j.1365-2958.1992.tb01563.x. [DOI] [PubMed] [Google Scholar]
- Din N., Forsythe I. J., Burtnick L. D., Gilkes N. R., Miller R. C., Jr, Warren R. A., Kilburn D. G. The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol Microbiol. 1994 Feb;11(4):747–755. doi: 10.1111/j.1365-2958.1994.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. M., Wood T. M., Williamson G., Faulds C., Hazlewood G. P., Black G. W., Gilbert H. J. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993 Sep 1;294(Pt 2):349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghangas G. S., Hu Y. J., Wilson D. B. Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J Bacteriol. 1989 Jun;171(6):2963–2969. doi: 10.1128/jb.171.6.2963-2969.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974 Nov 25;249(22):7130–7139. [PubMed] [Google Scholar]
- Hazlewood G. P., Davidson K., Laurie J. I., Romaniec M. P., Gilbert H. J. Cloning and sequencing of the celA gene encoding endoglucanase A of Butyrivibrio fibrisolvens strain A46. J Gen Microbiol. 1990 Oct;136(10):2089–2097. doi: 10.1099/00221287-136-10-2089. [DOI] [PubMed] [Google Scholar]
- Irwin D., Jung E. D., Wilson D. B. Characterization and sequence of a Thermomonospora fusca xylanase. Appl Environ Microbiol. 1994 Mar;60(3):763–770. doi: 10.1128/aem.60.3.763-770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M., Röhrig H., Schmidt J., Wieneke U., Schell J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Millward-Sadler S. J., Poole D. M., Henrissat B., Hazlewood G. P., Clarke J. H., Gilbert H. J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994 Jan;11(2):375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Sedmak J. J., Grossberg S. E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977 May 1;79(1-2):544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]