Summary
While horizontal basal cells (HBCs) make minor contributions to olfactory epithelium (OE) regeneration during homeostatic conditions, they possess a potent, latent capacity to activate and subsequently regenerate the OE following severe injury. Activation requires, and is mediated by, the downregulation of the transcription factor (TF) TP63. In this paper, we describe the cellular processes that drive the nascent stages of HBC activation. The compound phorbol 12-myristate 13-acetate (PMA) induces a rapid loss in TP63 protein and rapid enrichment of HOPX and the nuclear translocation of RELA, previously identified as components of HBC activation. Using bulk RNA sequencing (RNA-seq), we find that PMA-treated HBCs pass through various stages of activation identifiable by transcriptional regulatory signatures that mimic stages identified in vivo. These temporal stages are associated with varying degrees of engraftment and differentiation potential in transplantation assays. Together, these data show that our in vitro HBC activation system models physiologically relevant features of in vivo HBC activation and identifies new candidates for mechanistic testing.
Highlights
-
•
Treating HBCs with PMA induces the rapid degradation of TP63 protein
-
•
PMA-treated HBCs express previously identified in vivo activation genes
-
•
Analysis of PMA-treated HBC gene networks reveals candidate activation genes
-
•
PMA treatment unlocks HBC engraftment and partial differentiation potential
Barrios-Camacho et al. develop an in vitro model of nascent horizontal basal cells (HBCs) activation. Transcriptomic profiling of HBCs at various stages of activation reveals discrete gene regulatory networks also seen in the in vivo injury context. Despite initiating activation gene signatures, HBCs can regress and re-establish dormancy markers. Further, activated HBCs differ in engraftment and differentiation responses when transplanted.
Introduction
The only neurons in the human body that directly interface the external environment are housed in the olfactory epithelium (OE), a specialized neuroepithelium lining a portion of the nasal cavity (Carr et al., 2001). In response to continuous exposure and injury, the OE has evolved a remarkable and nearly life-long capacity to regenerate its neuronal and non-neuronal constituents. The extent of the regeneration makes it unique by comparison to the few other sites in the nervous system where adult neurogenesis takes place (Schwob et al., 2017). Crucial to this process is the activity of two stem cell populations, both of which possess multipotent capacity within the context of epithelial regeneration (Carter et al., 2004; Jang et al., 2003; Schwob et al., 2017). Globose basal cells (GBCs) are a constitutively proliferating and differentiating stem cell pool (Schwob et al., 2017). Horizontal basal cells (HBCs), on the other hand, are usually dormant in the uninjured OE but activate in response to severe injury (Fletcher et al., 2011, 2017; Gadye et al., 2017; Herrick et al., 2017; Leung et al., 2007; Schnittke et al., 2015) with the rare spontaneous differentiation in the absence of acute injury (Herrick et al., 2017; Iwai et al., 2008; Packard et al., 2011). In the anosmic, aged OE, focal sites of aneuronal OE, and respiratory metaplasia can be observed (Child et al., 2018; Fitzek et al., 2022; Holbrook et al., 2005). In both pathological tissue states, the GBC population is lost, while the HBC population fails to activate and regenerate the neuronal compartment (Child et al., 2018; Fitzek et al., 2022).Consequently, understanding the cohort of regulators that facilitate the HBC transition out of dormancy post-lesion would facilitate the clinical use of HBCs for the treatment of anosmia.
Previous studies have shown that the loss of the transcription factor (TF) Tp63 is necessary and sufficient to release HBCs from dormancy and into active multipotency in vivo (Schnittke et al., 2015; Schwob, 2005, 2017; Fletcher et al., 2011; Packard et al., 2011). TP63 loss and the subsequent activation of HBCs can be achieved in vivo with selective ablation of the sustentacular (Sus) cell population, whereas the death of neurons following olfactory bulbectomy is insufficient to release HBCs from dormancy on its own although the threshold for HBC activation is lowered (Herrick et al., 2017). In addition to mediating HBC differentiation, TP63 is critical for their development, such that HBCs fail to form in TP63 knockout (KO) mice at postnatal day 0 (Packard et al., 2011). Similarly, Notch1 KO lowers the threshold for HBC activation in the uninjured OE thus implicating Notch signaling in the maintenance of HBC dormancy during homeostatic tissue states (Herrick et al., 2017). Additionally, HBC-specific deletion of the nuclear factor κB (NF-κB) transcriptional effector Rela blunts regeneration of the OE after methimazole (MTZ) lesion (Chen et al., 2017). Lastly, while HBCs can return to dormancy following injury, expression of Hopx is selectively enriched in a subpopulation of differentiation-committed HBCs (Gadye et al., 2017).
A common limitation in understanding HBC activation is poor temporal resolution of TP63 loss in vivo, which in turn confounds the characterization of subsequent transcriptomic changes. Additionally, while lineage-tracing studies have been performed on injury-activated HBCs in vivo, these studies examine events occurring 24 h or more after lesion (Gadye et al., 2017) or Tp63 KO by HBC-specific genetic recombination (Fletcher et al., 2011). While previous studies from our lab found that sustained treatment of cultured HBCs with retinoic acid (RA) eventually induces the loss of Tp63, treatment times were extensive (Peterson et al., 2019). Therefore, there is an urgent need to develop an in vitro HBC system that facilitates the temporal coupling of TP63 decline with changes in gene expression to better understand the cellular processes underlying nascent HBC activation. To that end, we present a new in vitro platform for achieving physiologic and reversible TP63 loss within 12 h, allowing us to probe nascent HBC activation with fine temporal resolution. This model recapitulates a myriad of acute molecular processes and developmental programs that govern in vivo HBC-mediated regeneration of the OE. Using this model, we identify the precise window during which TP63 undergoes precipitous decline. Additionally, we find a transient and robust nuclear translocation of the NF-κB effector RELA preceding precipitous TP63 loss. This activation model unlocks engraftment potential and generates a heterogeneous HBC population that can either self-renew or transition away from an HBC state, consistent with post-lesion behavior of HBCs in situ following injury. Finally, we study the transcriptional regulatory signatures throughout the acute activation process and identify candidates that may be responsible for fate-determining events in nascent HBCs.
Results
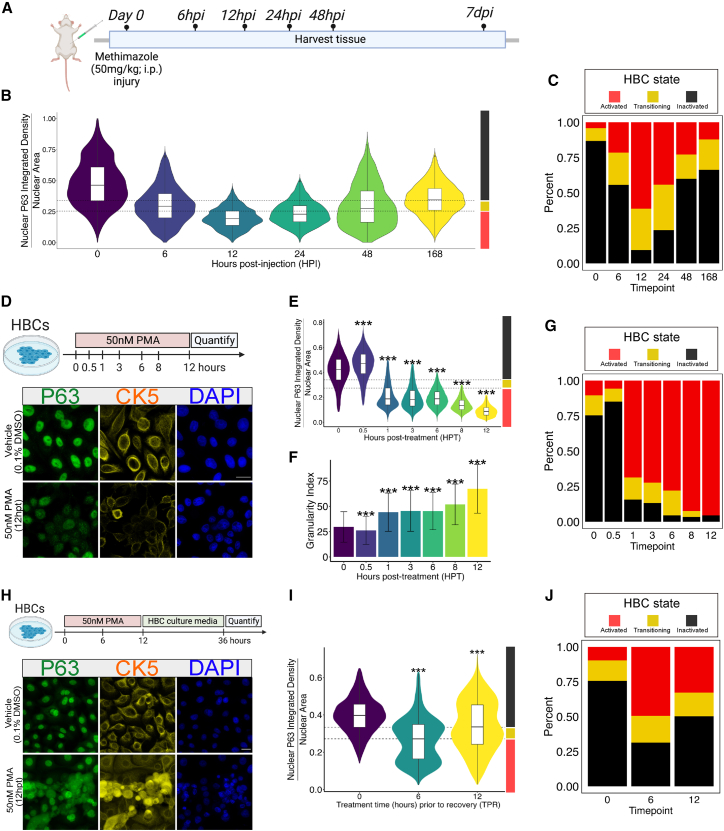
While previous analyses of in vivo lesioned HBCs have revealed near-absent expression of TP63 at 18 h post methyl bromide (MeBr)(Schnittke et al., 2015), the kinetics of acute TP63 loss following injury are unknown but critical to the evaluation of any in vitro activation model. We mapped the in situ kinetics of TP63 loss during the acute phase of OE lesion following administration of MTZ in wild-type mice. To do so, we coupled confocal microscopy with high-throughput CellProfiler-mediated image analysis to quantify nuclear TP63 immunofluorescence (IF) levels over time in 18,557 HBCs (McQuin et al., 2018). TP63 levels were assessed on a per-cell nucleus basis at 0, 6, 12, 24, and 48 h post MTZ injection (hours post injection; HPI) and 7 days post injection (DPI), at which time the OE has partially regenerated (Figure 1A). We observed a high degree of HBC heterogeneity during the acute activation phase, with some HBCs demonstrating rapid responsiveness to injury (indicated by rapid TP63 loss) and others demonstrating a degree of resistance (preservation of near-baseline TP63 expression or minimal decline) (Figure 1B). This observed heterogeneity prompted us to classify our HBCs into three populations on the basis of TP63 IF labeling (Figure 1C; for IF thresholds see Table S2). These results suggest HBCs distribute across a spectrum of TP63 expression across all OE states, including unlesioned tissue. Injury induces compositional changes; at 12 HPI, 95% of HBCs are in an activated or transitioning state, while retaining a small (5%) dormant fraction (see Table S2). Lastly, analysis of recovering tissue (7DPI) indicates that HBC composition has not returned to that of the unlesioned condition, despite re-establishing a sizable resting population (time point 0 = 74.9% resting, 14.3% transitioning, 10.6% activated vs. time point 168 HPT = 51.6% resting, 25.1% transitioning, 23.1% activated). Together, these data show that HBC activation is not a synchronous process and that multi-stage HBCs compose the OE across all tissue contexts. Most importantly, we observe that overall TP63 expression in vivo reaches a nadir at 12 h post MTZ injection.
Figure 1.
PMA treatment leads to TP63 degradation in vitro but is not deterministic
(A) Schematic illustration of MTZ injury time course.
(B) An integrated violin and boxplot representing the Nuclear P63 integrated density/nuclear area quantification results of each time point (n = 2).
(C) Bar plot showing proportional composition of HBC state at each time point.
(D) Schematic illustration of the PMA-mediated HBC activation assay (top). Representative sum projected z stack confocal 63x images of vehicle-treated and 12h-long 50 nm-treated HBCs (below).
(E) An integrated violin and boxplot representing the Nuclear P63 integrated density/nuclear area quantification results of each time point (n = 3).
(F) An integrated violin and boxplot representing the Granularity I output from CellProfiler.
(G) Bar plot showing proportional composition of HBC state at each time point post-PMA treatment.
(H) Schematic illustration of the PMA-mediated HBC recovery assay (top). Representative sum projected z stack confocal 63x images of vehicle-treated and 12h-long 50 nm-treated HBCs (bottom).
(I) An integrated violin and boxplot representing the Nuclear P63 integrated density/nuclear area quantification results of each time point (n = 3).
(J) Bar plot showing proportional composition of HBC state at each time point post-PMA treatment and recovery. For (B, E, and I), dashed lines represent TP63 fluorescence expression level thresholds corresponding to HBC state. Right y axis color blocks on (B, E, and I) correspond to these states, shown in (C, G, and J). Extreme outliers were removed utilizing the Tukey method (1.5 x IQR) (E and I). For (E, F, and I) statistical significance was determined by one-way ANOVA followed by Dunnett’s test. For (E, F, and I): ∗p < 0.05,∗∗p < 0.001,∗∗∗p < 0.0001 (E and I). For (F), mean and standard deviation is reported. Scale bars: 20 μm (D and H). See Table S2 for information about sample sizes and number of cells per condition.
Treatment with PMA induces activation in vitro through the rapid loss of TP63
We set out to develop an in vitro assay capable of modeling and probing the underlying molecular mechanisms of nascent HBC activation (Figure 1D, top). To induce activation, we utilized phorbol 12-myristate 13-acetate (PMA), which is an activator of numerous signaling pathways previously implicated in HBC activation, such as the NF-κB pathway (Holden et al., 2008). Following treatment of in vitro HBCs with 50 nM PMA, we quantified nuclear TP63 expression with previously described CellProfiler-mediated approach. We quantified cells at 6 time points over a 12-h period to assess the kinetics and extent of nuclear TP63 loss. Mirroring the heterogeneity demonstrated by in vivo HBCs following OE injury, results from this in vitro experiment showed triphasic change in nuclear TP63 IF over the 12-h time course. Thirty minutes after PMA treatment, we observed a 10% increase of nuclear TP63 protein compared to vehicle, possibly reflecting homeostatic mechanisms protecting TP63 pools (Figure 1E). However, levels rapidly fall between 30 min and 1-h to approximately 52% of vehicle-treated cells. Levels then appeared to stabilize between 1 and 6 HPT before undergoing a second phase of reduction between 6 and 12 HPT. At the end of the 12-h treatment period, HBCs lost 78% of their nuclear TP63 IF. Additionally, cytoplasmic TP63 does not increase as nuclear signal decreases; indeed, PMA induces a whole-cell loss of TP63 (Figure S1). Visual inspection of high-magnification images (Figure 1D, bottom) shows the emergence of a granular, punctate staining pattern of TP63 in both the nucleus and cytoplasm, likely representing sites of TP63 sequestration and degradation. This change in IF texture was quantified using the MeasureGranularity module in CellProfiler, which revealed linearly increasing levels of punctate TP63 protein (Figure 1F). While most cells rapidly responded to treatment, a minor fraction of HBCs at 8 and 12 HPT maintained near-baseline TP63 IF levels and thus appeared resistant to PMA treatment (Figure 1G). Critically, we found that if treated for 24 h, HBCs detached from the plate, thus rendering extended incubation impractical.
In the acute post-injury environment in vivo, the HBC population bifurcates into cells that are fated to either differentiate or return to dormancy. Having established that PMA induces TP63 loss, we assessed physiologic plasticity in the HBCs activated in vitro by testing whether baseline TP63 levels are restored after PMA treatment. Accordingly, we treated HBCs with PMA for either 6 or 12 h before washing out the drug and allowing cells to recover in maintenance media for 36 h (Figure 1H, top). Results from this experiment showed that HBCs can resynthesize TP63 following acute PMA treatment, with HBCs experiencing a net loss of only 13% of TP63 protein after 12 h of PMA treatment and recovery (Figure 1I). Interestingly, we observed a bifurcation in the population when cells were treated with PMA for 6 h and then transitioned back to maintenance media. Under this paradigm, roughly half of the cells re-established vehicle-treated TP63 levels whereas the other half continued to lose TP63 in a manner consistent with activation (Figure 1J). Additionally, we observed a three-dimensional reorganization in the culture. Whereas vehicle-treated HBCs form a stereotyped cobblestone monolayer, cultures that were transiently activated contained regions of piled-up HBCs (Figure 1H, bottom). Together, these results suggest that a transient activation event in this system may be leveraged to study physiologic subpopulations of HBCs with characteristics that mirror those in the acute post-injury setting in vivo.
Bulk RNA sequencing of PMA-treated HBCs reveals transcriptional networks involved in in vitro activation
Having established a model system for nascent HBC activation, we assessed transcriptomic changes during this process using bulk RNA-seq of vehicle-treated HBCs and two populations of PMA-treated HBCs (6 HPT and 12 HPT). Principal-component analysis indicates that 92% of the variance in the data is explained by exposure to PMA, while 6% of the variance is explained by PMA treatment length (6 h vs. 12 h) (Figure S2A). Analysis of differentially expressed genes (DEGs; treatment/vehicle) in the PMA-treated conditions revealed a core PMA-induced treatment signature, irrespective of length of treatment (n = 6,458 DEGs) (Figure S2B). We evaluated the gene ontology (GO) networks that were either up- or downregulated in our 6 HPT/vehicle- and 12 HPT/vehicle-specific signatures (6 HPT = 1,388 genes; 12 HPT = 1,271 genes) as well as the GO networks induced by PMA treatment alone (n = 6,458). We utilized Metascape to identify significantly enriched pathways across these six contexts (Zhou et al., 2019). At 6 HPT, the ontology networks “transcriptional machinery,” “NF-κB signaling,” and “mRNA modifications” are among those upregulated (Figures S2C and S2D). In contrast, this analysis revealed that the networks for “membrane trafficking,” “mitochondrial transport,” and “the regulation of cell-matrix adhesion” were downregulated (Figures S2C and S2D). At 12 HPT, “MAP2K signaling,” “cytoskeletal remodeling,” and “axon development” are a set of upregulated categories (Figures S1E and S1F). In contrast, we found downregulated signatures involving “cell cycle,” “ncRNA processing,” and “RNA polymerase III transcription” (Figures S2E and S2F). In addition to the time point-specific ontology analysis, we also interrogated the ontology networks specifically induced by PMA, irrespective of length of treatment (Figures S2G and S2H). In sum, we found that treating HBCs with PMA upregulated various morphogenic, developmental, and differentiation ontology categories, while simultaneously downregulating cell cycle, DNA replication, and cell division categories. These results, paired with the time point-specific ontology changes, show that distinct metabolic and signaling pathways are recruited as the transcriptome shifts across the activation landscape.
PMA treatment of cultured HBCs induces in vivo activation genes
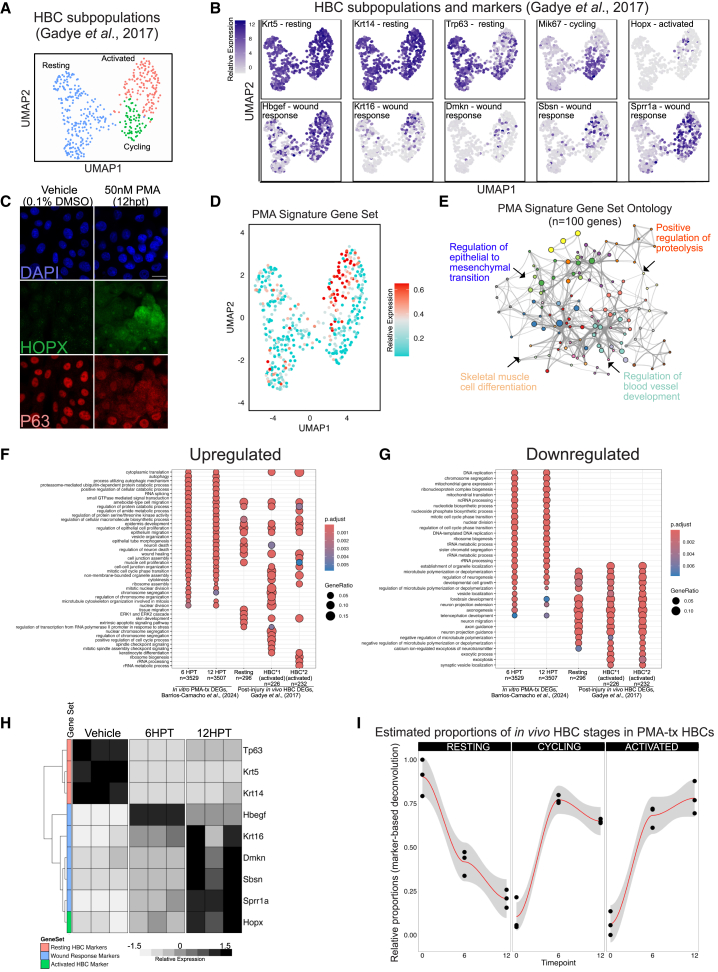
We next compared the gene expression of HBCs activated via PMA in vitro and HBCs activating natively under injury conditions in vivo. Given the potential wide-ranging effects of PMA on cultured cells, we assessed transcriptomic similarity using specific established marker genes and by analyzing the expression of entire gene sets. For our in vivo reference, we re-analyzed a single-cell dataset of resting and activated HBCs from the injured, regenerating wild-type mouse OE harvested at multiple time points after MTZ injury (Gadye et al., 2017). We clustered Krt5(+) HBCs from the Gadye dataset into three groups based on their expression of three marker genes: dormancy regulator Tp63, proliferation marker Mki67, and activation marker Hopx. Hopx was additionally chosen in part because of its known role in cellular differentiation in other tissues (Hng et al., 2020; Palpant et al., 2017). These markers identify three developmentally sequential HBC subpopulations: Tp63(+) resting HBCs, Mki67(+) transitioning/cycling HBCs, and Hopx(+) fully activated HBCs (Figure 2A). A set of ten candidate genes that were highlighted by Gadye as key during the regeneration process in vivo were mapped onto the re-analyzed, native HBC UMAP (Uniform Manifold Approximation and Projection) to highlight the gene list’s correlation with HBC status in vivo, confirming the clustering into three subpopulations (Figure 2B). In keeping with the in vivo analysis, PMA-activated HBCs demonstrated, by immunostaining, an inverse correlation between increasing HOPX protein and decreasing TP63 protein at 12 HPT (Figure 2C). This result is corroborated by prior work demonstrating enrichment of HOPX in human keratinocytes following PMA treatment (Yang et al., 2010). In contrast with the results in vivo, HOPX labeled a very high percentage of the PMA-activated HBCs. Nonetheless, PMA treatment does induce a specific state highly analogous to in vivo injury-activated HBCs.
Figure 2.
PMA-treated HBCs correspond to in vivo HBCs state
(A) Clustering of HBC subpopulations found in the regenerating olfactory epithelium (Gadye et al., 2017) as described in experimental procedures.
(B) UMAP expression plot of marker genes differentiating the HBC subpopulations in vivo (from Gadye dataset): resting-HBC markers – Krt5, Krt14; dormant-HBC marker – Tp63; cycling-HBC marker – Mki67; activated HBC – Hopx; wound response markers – Hbgef, Krt16, Dmskn, Sbsn, Sprr1a.
(C) Representative immunostaining for TP63 and HOPX, an activated HBC marker previously identified. Cells were treated for 12 h with PMA before staining. Scale bar: 20 μm.
(D) Relative signature expression in HBCs. Quantity reflects the collective expression of the gene set relative to a randomized control set of the same size (see Tirosh et al., 2016.). The “PMA signature” comprises the top 100 differentially upregulated genes from the highly concordant combined 6 HPT and 12 HPT in vitro datasets (see Figure S2). Only Sprr1a is common between the PMA and in vivo (n = 100) gene sets. Cyan indicates low signature expression; red indicates high signature expression.
(E) Gene ontology network visualizing and summarizing the Biological Process terms that are enriched in the 100-gene PMA signature.
(F) clusterProfiler dotplot of GO Biological Process ontology comparing upregulated DEGs identified in the current manuscript to those identified in Gadye et al. (2017).
(G) clusterProfiler dotplot of GO Biological Process ontology comparing downregulated DEGs identified in the current manuscript to those identified in Gadye et al. (2017).
(H) Heatmap visualizing the expression of HBC markers and wound response genes detected in PMA-treated in vitro HBCs.
(I) Marker-based deconvolution of the bulk RNA-seq samples based on the single-cell gene expression patterns of resting, cycling, and activated HBCs in vivo. This approach relies on the raw single-cell data to generate markers segregating HBC states and then estimating their respective proportion in PMA-treated HBCs. The y axis defines the proportion of PMA-treated HBCs estimated to be either “resting,” “cycling,” or “activated.” For (F) and (G) FDR-adjusted p value <0.01. See NIHMS917517-supplement-5 for Gadye et al., gene lists. Please note that in vitro DEGs were converted to their mouse orthologs before enrichment analysis. Ontology categories were simplified utilizing the Wang method, with a similarity cutoff of 0.7.
Using the module detection function of Seurat (Tirosh et al., 2016), we plotted the collective expression of the top 100 DEGs induced by PMA treatment, including both 6 HPT and 12 HPT sets given the high degree of concordance between the sets (Figure S2). Expression of this signature was highly elevated in the same in vivo-activated HBC cluster (Figure 2D), reaffirming transcriptomic similarity between PMA-activated HBCs in vitro and injury-activated HBCs in vivo. Importantly, while the PMA-induced gene set was enriched for signaling, development, and differentiation GO terms (Figure 2E), only one gene overlapped with the previously published wound-response gene set (Sprr1a), thus indicating broader transcriptomic similarity between the two activated HBC populations.
We examined the overrepresented ontology categories across five distinct HBC populations to compare changes between in vitro and in vivo activation more comprehensively: PMA-treated HBCs in vitro (6 HPT and 12 HPT) and in vivo post-injury HBCs (resting, HBC∗1, and HBC∗2, the latter two representing the two stages of activated HBCs) (Gadye et al., 2017). All five HBC populations show upregulation of various epithelial remodeling categories. In addition, all five HBC populations show downregulation of developmental, microtubule polymerization, and neuron projection categories. PMA-treated HBCs, irrespective of length of treatment, could be defined by overlapping upregulated and downregulated ontology categories. Interestingly, only three categories differed between the length of PMA treatment: “regulation of chromosome organization,” “vesicle localization”, and “axonogenesis.” In contrast, in vivo HBCs could be defined by discrete patterns of upregulated, but not downregulated, ontology categories. HBC∗1, for example, show uniquely upregulated cell-cycle gene categories justifying their identification as proliferating HBCs, as we have done in this manuscript. Lastly, we found that both in vitro and in vivo HBCs depleted roughly equivalent categories. Interestingly, we found that only resting HBCs induced the ERK1/ERK2 cascade (Figure 2F).
In light of our comparison of in vivo data and the in vitro activation signature, we performed the reciprocal approach, analyzing bulk RNA-seq data from the in vitro HBCs to determine whether known gene expression changes in the post-lesion environment were recapitulated by PMA activation. We observed that PMA treatment caused a decrease in Tp63, Krt5, and Krt14 expression concurrent with unanimous upregulation of wound-response signature genes and Hopx in vitro (Figure 2H). Next, we wanted to predict the abundances of these in vivo HBC subtypes within our bulk transcriptomes. Using the BisqueRNA R package (Jew et al., 2020) and Gadye reference dataset, we observed a decrease in the predicted proportions of resting HBCs after PMA activation, along with an increase in the predicted proportion of cycling and activated HBCs (Figure 2I). Collectively, these analyses indicate that there is a bidirectional association between the gene expressions of in vitro- and in vivo-activated HBCs with regards to individual marker genes and activation-associated gene sets. In sum, the similarities between PMA activation in vitro and injury-induced activation in vivo are substantial, though not absolute.
Activating HBCs in vitro with PMA induces stage-specific developmental regulons
Having demonstrated a correspondence between activation by injury in vivo and activation by PMA treatment in vitro, we examined the transcriptional networks that are differentially regulated as activation proceeds in response to PMA. Previous analyses of the stages of HBC activation in vivo have identified TFs that regulate aspects of HBC dormancy, activation, and differentiation. To wit, TP63 is known to regulate HBC dormancy (Schwob et al., 2017), RELA seems to participate in the activation process (Chen et al., 2017), and HOPX is suggested to play a role in the regulation of HBC differentiation (Gadye et al., 2017). To investigate the roles of these and other TFs in the regulation of PMA-activated HBCs, we performed differential expression analysis with DESeq2 to identify TFs (n = 249) that are differentially expressed across the activation time points. As reported by the authors, TFs alone could not distinguish between HBC states in their single-cell RNA-seq dataset (Gadye et al., 2017). Consequently, we looked instead at the target genes induced by the TFs in our regulon analysis to determine if these could serve as a proxy for the activation process in vivo.
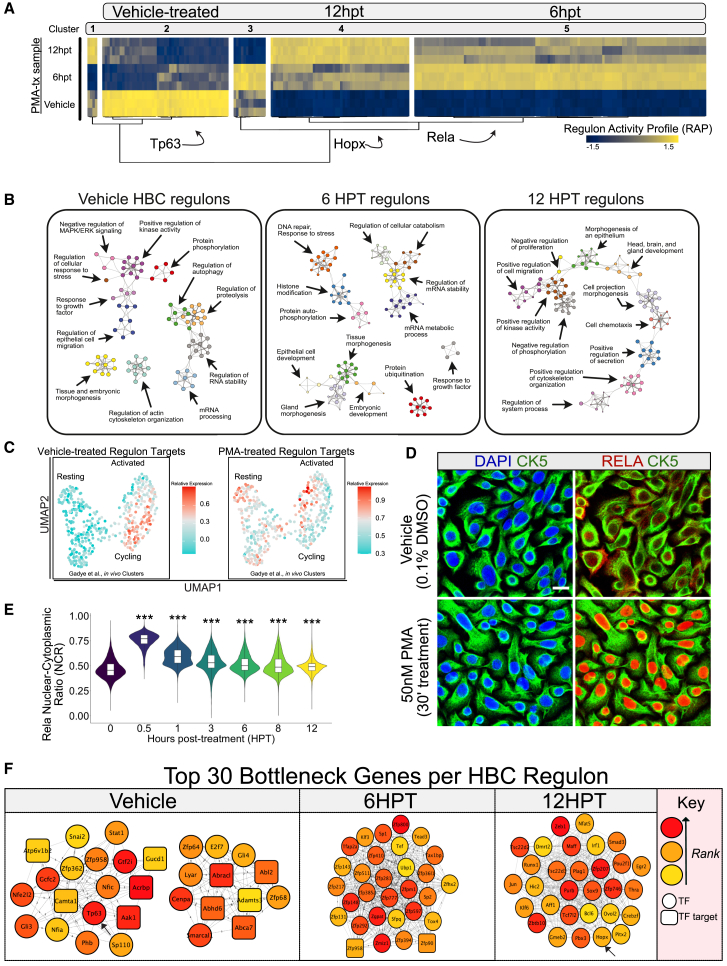
Accordingly, we analyzed the sets of TFs and their proposed targets (collectively “regulons”) according to gene co-expression analysis utilizing the R package RTN (version 1.2.1) (Castro et al., 2016; Corces et al., 2018; Fletcher et al., 2013) (Figure 3A). Our analysis revealed that the 249 TFs were associated with 4,736 proposed target genes (Figure 3A). The collective expression of the TFs and their proposed target genes (regulon activity profiles, RAPs) clustered into 5 distinct signatures which changed dynamically across PMA treatment. The three most prominent regulon clusters corresponded to the vehicle-treated state, early activation (6 HPT), and extended activation (12 HPT).
Figure 3.
Regulon analysis of PMA-treated HBCs reveals distinct networks underlying activation
(A) A heatmap representing the regulon activity profiles exported from RTN. Each vertical line represents an individual regulon (TF + target genes). Clustering is generated using Ward’s minimum variance method with Pearson correlations used as the distance used for clustering.
(B) Metascape enrichment of regulon clusters.
(C) Regulon target gene abundances from Figure 3A mapped onto the single-cell HBC data from Figure 2A for both vehicle-treated and PMA-treated transcription factors. Red indicates high expression and cyan indicates low expression.
(D) Representative immunofluorescence detailing RelA translocation 30′ after PMA treatment. Scale bar: 20 μm.
(E) An integrated violin and boxplot representing the nuclear-cytoplasmic index of RelA fluorescence over a 12-h long PMA time course (n = 3). One-way ANOVA followed by Dunnett’s Test.
(F) The top 30 bottleneck genes identified per regulon cluster. Upper left legend indicates rank of genes, as well as whether a gene is a transcription factor (circle) or a transcription factor target (square). The arrows at vehicle and at 12 HPT indicate Tp63 and Hopx, respectively.
We queried each cluster for TFs previously characterized in HBCs (Figure 3A). As expected, we identified Tp63 among the resting-state regulons, which suggests a transcriptional network important for the maintenance of inactivated HBCs and serves as an important positive control for our analytic approach. The 6 HPT regulons included RelA, which participates in the process of activation (Chen et al., 2017). The 12 HPT regulons included Hopx (Gadye et al., 2017) suggesting that this cluster may represent gene expression changes important for HBC differentiation. A role in differentiation was supported by ontology analysis of the TFs and their target genes (Figure 3B): the vehicle-treated regulons were associated with tissue and embryonic morphogenesis, growth factor response, and actin reorganization; the early-activation regulons were associated with DNA repair, protein ubiquitination, and morphogenesis; and the extended-activation regulons were associated with developmental processes and morphogenesis (Figure 3B). We also mapped the target genes in our regulon analysis onto the reanalyzed single-cell analysis (see Figure 2A for the reanalyzed clusters) to determine if the target gene sets track the activation process in vivo (Figure 3C). The target genes regulated by vehicle-treated TFs were enriched in the partially activated/cycling in vivo population (Krt5+/Tp63MED/Mki67+/Hopx-) (Figure 3C, left). The correspondence with cell-cycle progression is consistent with our previous report that primary HBC cultures allow expansion of HBCs by proliferation without differentiation (Peterson et al., 2019), and thus HBCs in the absence of PMA are closer to the partially activated/cycling in vivo population. In contrast, we found that that our collapsed PMA target genes were most strongly enriched in the more fully activated in vivo HBC population (Krt5+/Tp63none/Mki67-) encompassing those HBCs in vivo that express Hopx (Figure 3C, right). Given the in vivo findings implicating RelA as a crucial effector of HBC activation (Chen et al., 2017) and the present findings in our study which mapped RelA regulon activity to early stages of activation (Figure 3A), we sought to determine whether PMA treatment also induced RelA nuclear translocation during the 12-h long PMA-treatment protocol (Figures 3D and 3E). We observed robust nuclear translocation of RELA as soon as 30 min after PMA treatment (Figures 3D and 3E) which slowly tapered toward baseline levels nuclear-translocation index (NCI) over the remaining protocol time (Figure 3E). Notably, RELA translocation precedes the decline in nuclear TP63 staining (Figure 1B).
Topology analyses of regulon communities reveal bottleneck genes relevant for HBC activation
Transcriptional networks, like those identified in the regulon analysis, exist to shape cell states. Our regulon clusters highlight orchestrated sets of genes that mold a cell’s transcriptional potential for differentiation. To extend our analysis, we were interested in assessing the topological features of our regulon networks. Assessing the topology of biological networks offers the possibility of identifying individual genes whose presence in the network is important for maintaining other biological relationships. We utilized Cytoscape’s (3.8.2) cytoHubba (Chin et al., 2014) plug-in to identify the top 30 bottleneck genes in our regulon clusters (Figure 3F).
Inspection of the top 30 bottleneck genes identified previously validated regulators. As expected, we identified Tp63 as a bottleneck gene in our vehicle-treated HBC cluster, whose RAP decreases dramatically following PMA activation (Figure 3A). Given the master regulator role of Tp63 in HBC activation, its identification as a bottleneck gene (Figure 3F, left) suggests that the other 29 genes are relevant for inhibiting activation or promoting dormancy. Other bottleneck genes found in this group include Stat1, Gli3, and Nfe2l2 (Figure 3F, left). Analysis of our 12 HPT cluster identified Hopx as a top 30 bottleneck gene (Figure 3F, right). Hopx steadily increases as a function of PMA-mediated activation (Figure 3A), reaching a maximum at 12 HPT, implying that the regulon clusters observed at 12 HPT are important for sustaining activation and/or differentiation. Other genes in this group include Zeb1, Jun, Sox9, and Egr2 (Figure 3F, right). Our 6 HPT cluster represents RAPs which reach maxima at 6 HPT but then return to baseline at 12 HPT (Figure 3A). The phasic nature of the 6 HPT RAPs suggests that these genes might be functioning analogously to immediate-early genes (IEGs) (Kim et al., 2018; Minatohara et al., 2015; Okuno, 2011). Genes in this list include Tox4, Tead3, Klf3, and Tef (Figure 3F, middle).
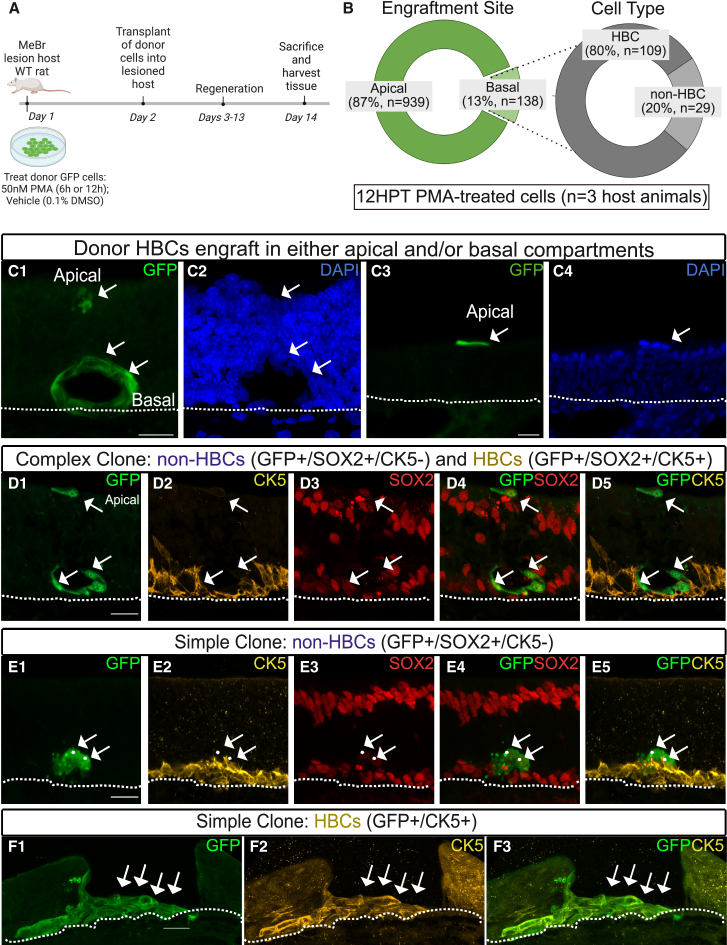
Transplantation of PMA-activated HBCs into the OE of lesioned host mice
The results from our PMA recovery assay prompted us to consider whether PMA activation resulted in a deterministic commitment to differentiation, or whether the recovery we observed reflected a resistance to activation. To specifically test this question, we carried out transplantation assays of HBCs at the same two stages of activation (6 HPT and 12 HPT) (Figure 4A). Utilizing HBCs derived from pan-GFP-expressing transgenic rats, we transplanted vehicle-, 6 HPT-, and 12 HPT-HBCs into MeBr-lesioned host rats. The OE was harvested 12 days after transplantation to assess engraftment and the composition of GFP-positive clones (Figures 4A and 4B).
Figure 4.
PMA treatment unlocks engraftment potential in 12 HPT-HBCs
(A) Schematic illustration of transplantation assay.
(B) Quantification of 12 HPT PMA-treated GFP+ HBCs (n = 1077) based on engraftment location: apical (n = 939 total cells) or basal (n = 138 total cells). n = 3 host rats. Quantification of 12H PMA-treated basal cells (n = 138) based on progeny outcome (HBC or non-HBC).
(C1–C4) Representative clones highlighting GFP engraftment sites (apical and/or basal). Scale bars: 20 μm.
(D1–D5) Representative complex clone comprising HBCs (GFP+/SOX2+/CK5+) and GBCs (GFP+, SOX2+, CK5-). Note representative unengrafted HBC atop the apical cell layer. Scale bar: 20 μm.
(E1–E5) Representative simple clone comprising non-HBCs (GFP+, SOX2+, CK5-). Scale bar: 20 μm.
(F1–F3) Representative oversized simple clone highlighting a swathe of engrafted HBCs. Note missing apical structures. Scale bar: 20 μm.
Vehicle-treated HBCs failed to engraft and differentiate, a result consistent with previous results showing that donor HBCs need to be activated to successfully engraft (Chen et al., 2004; Peterson et al., 2019; Schnittke et al., 2015). Likewise, we found that 6 HPT failed to engraft—a transplantation outcome indistinguishable from vehicle-treated cells. In contrast, we found that 12 h of PMA treatment induced engraftment but led to outcomes that differed from previous transplants of in vivo-activated HBCs (Schnittke et al., 2015) or prolonged RA-treated cultured HBCs (Peterson et al., 2019). After 12 h of PMA treatment, HBC transplantation led to two distinct outcomes: first, 87% of GFP + cells (n = 1,077 total) were seen resting at the apex of the regenerated tissue (Figure 4C3). These apical GFP + cells were often found as single cells but sometimes appeared as doublets (Figure 4C3). The remaining 13% of GFP + cells (n = 138) engrafted deeper in the basal compartment of the OE, a location normally populated by HBCs and GBCs (Figure 4C1). To identify these cells, we immunostained sections containing GFP + clones for CK5 and SOX2. Cells expressing CK5 and SOX2 were classified as HBCs, while cells expressing SOX2 without CK5 were classified as non-HBCs. Engrafted HBCs also display the elongated lens-like morphology and appear tightly adherent to the basal lamina both of which are highly typical of HBCs in situ. Based on this marker panel, 80% of the engrafted basal cells expressed SOX2(+)/CK5(+) and thus were classified as HBCs whereas 20% were SOX2(+)/CK5(−) and thus phenotypically not HBCs (Figure 4D). Additional features of the engrafted cells are worthy of note. The GFP+ cells tended to form pits near the base of the epithelium, identified by the lack of DAPI+ cells and interruptions in the dense cellularity of the epithelium (Figures 4C1, 4D1, and 4F1). Additionally, although most of the clones were simple, we did identify scattered complex clones, some of which contained both SOX2(+)/CK5(+) and SOX2(+)/CK5(−) cells (Figures 4D1–D5). Collectively, the transcriptional regulatory signatures and in vivo engraftment data suggest that treatment of cultured HBCs with PMA for 12 h unlocks developmental programs sufficient to drive engraftment and differentiation when transplanted into the regenerating OE, although the usual behavior of activated HBCs is not fully actuated.
Discussion
We describe a robust primary in vitro system that allows us to characterize and experimentally manipulate HBCs across various stages of activation. PMA treatment induces the rapid loss of TP63 protein in most HBCs, while also revealing treatment-resistant HBCs. PMA recovery assays demonstrate that TP63 loss does not irreversibly drive HBCs toward differentiation, as evidenced by the robust re-synthesis of TP63 after removal of PMA. Complementary bioinformatic analysis of bulk RNA-seq data demonstrates that PMA-treated HBCs share many transcriptomic features with published single-cell RNA-seq of in vivo HBCs following lesion (Gadye et al., 2017). Furthermore, topology analysis of regulon networks reveals novel candidate regulators of HBC activation for future study. Lastly, in vivo transplantation experiments demonstrate that only sustained activated (12 HPT) can engraft and partially differentiate, albeit in a manner that is atypical by comparison with previous transplant HBC experiments (Chen et al., 2004; Peterson et al., 2019; Schnittke et al., 2015). Taken together, these data support the robustness of this PMA-mediated activation paradigm, while also revealing novel features of HBC activation.
PMA treatment leads to the robust, rapid loss of TP63 protein
HBC activation is predicated on the loss of TP63 protein. While TP63 regulation in other systems has been widely explored (Gu et al., 2006; Shen et al., 2023) and engages the ubiquitin-proteasome system (for complete review: Li and Xiao, 2014), the mechanisms by which TP63 is regulated in HBCs are largely unknown. However, we do know that neuronal depletion is insufficient cause for diminution of TP63 levels (Herrick et al., 2017). Thus, understanding mechanisms regulating TP63 stability is of critical clinical relevance, as directing TP63 loss in HBCs is a potential first step toward OE reconstitution after pathological regression of the tissue (Schwob et al., 2017).
Our data also reveal the presence of treatment- and injury-resistant HBCs as has also been reported in other publications from our lab and others (Haglin et al., 2020; Schnittke et al., 2015). These cells either retain or resynthesize TP63 rapidly. Additionally, our work reveals that the kinetics of TP63 loss in HBCs are not entirely uniform either in situ or in our culture system, suggesting that HBCs collapse these signals asynchronously. Whether there is a correlation between TP63 loss and fate determination, that is, whether HBCs that are more susceptible to TP63 loss execute a particular lineage outcome, is unknown. However, other papers from the lab have shown that lineage outcomes are influenced by the status of the Notch pathways and of RAC1 signaling in HBCs (Louie et al., 2023; 2024). Our data corroborate the idea that HBCs might need additional inputs to enact an expansive differentiation response following TP63 loss.
PMA-treated HBCs possess limited ability to differentiate after transplantation
OE transplantation experiments can be used to probe progenitor cell capacity experimentally. However, they are inefficient, with very low rates of analyzable cells (in this study: 1,077 analyzed cells/3 million infused cells), in no small part because the nasal cavity remains a voluminous open space from which infusates drain quickly. Despite this technical limitation, results obtained here following HBC transplantation suggest that PMA stimulation capacitated HBCs toward engraftment. However, by itself PMA treatment was unable to accomplish full activation to a truly pluripotent state since even after 12 h PMA treatment HBCs demonstrated only limited differentiation. This response to PMA stands in stark contrast to the behavior of uninjured HBCs observed in other HBC-transplantation assays. Our lab previously demonstrated that uninjured HBCs do not engraft (Chen et al., 2004; Peterson et al., 2019; Schnittke et al., 2015) and HBCs subjected to sustained RA treatment (14 days in vitro; Peterson et al., 2019) or harvested 18 h post-MeBr lesion (Schnittke et al., 2015) were collectively capable of forming all OE cell types. Thus, cells treated with PMA for 12 h are functionally distinct from uninjured HBCs when assayed by transplantation. However, the results also indicate that any additional contextual cues in the lesioned OE are insufficient to release the full potential of HBCs activated in this manner to generate Sus cells, neurons, or Bowman’s glands. The inability to drive large-scale regeneration efforts implies that PMA-induced signals instead cast HBCs into a plastic state and that more robust differentiation requires additional signals that we have not yet been able to replicate completely. Interestingly, our lab has shown that, while uninjured GBCs exhibit multipotency after transplantation, injury engenders them with even greater plasticity (Lin et al., 2017). That both cell types exhibit a similar property—plasticity acquired after injury—emphasizes the robustness of the PMA paradigm as it allows this property to be experimentally accessible in a rapid, robust fashion.
Topological analysis of regulons
Our work also presents a pipeline that pairs regulon analysis with topological analyses of these networks to comprehensively characterize and identify regulators for further mechanistic testing. Previous analyses of TF networks have highlighted the notion that the relationship between a TF and its targets provide a better tissue/cell-specific signature than the expression of TFs alone(Sonawane et al., 2017). We applied this perspective to our transcriptomic datasets and, to identify novel regulators, used topological analyses prioritizing bottleneck genes, which are thought to be essential for determining cell state (Gibney et al., 2013; Yu et al., 2007). Not only did this complementary analysis reveal novel candidates but it also encompassed candidates that had been previously identified in other experimental systems, emphasizing the robustness of this approach. For example, Nfe2l2, identified in the dormancy cluster, has been shown to synergistically cooperate with Tp63 in proliferating keratinocytes to sustain proliferation and promote differentiation (Kurinna et al., 2021), suggesting that Tp63 likely coordinates HBC dormancy through engagement with other TFs.
In summary, we have leveraged our previously established primary in vitro HBC culture model (Peterson et al., 2019) to establish a physiologically relevant platform to study nascent HBC activation.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James Schwob (jim.schwob@tufts.edu).
Materials availability
This study did not generate any unique reagents.
Data and code availability
Raw files, counts, and normalized counts from our bulk RNA-seq can be accessed under GEO: GSE215797. Single-cell RNA-seq data used in this analysis were accessed from GEO, GEO: GSE99251. All code used to generate the data in this manuscript can be accessed at https://github.com/barriosc. The accession number for the PMA time course dataset reported in this paper is Mendeley Data V1: https://doi.org/10.17632/54tkpr2zdv.1.
Animals
Animals were maintained on ad libitum standard rodent chow and water in an AALAC (Association for Assessment and Accreditation of Laboratory Animal Care)-accredited vivarium operating under a 12-h light/dark cycle. All procedures used on animals were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine. Eight 10-week-old Sprague-Dawley (Taconic NTac:SD) rats were used to generate primary wild-type HBC lines; these were maintained from a founder breeder pair. Pan-GFP-expressing transgenic rats (SD-Tg(ACT-EGFP) CZ-004OOsb strain) were bred and maintained in-house and used to generate pan-GFP HBC lines (Jang et al., 2008; Sasaki et al., 2001).
Tissue harvest and preparation for cryosectioning
Animals were deeply anesthetized with single, lethal intraperitoneal (i.p.) injection of triple cocktail of acepromazine (1.25 mg/kg), ketamine (37.5 mg/kg), and xylazine (7.5 mg/kg). Animals were perfused intracardially with 10 mL of 1X PBS followed by 35 mL of ice-cold formalin. The OE was dissected and then incubated with 30 mL of formalin for 2 h at room temperature (RT) and under vacuum. Tissue was immersed in a decalcifying solution of saturated EDTA for 48 h at 4°C, before being cryoprotected in 30% sucrose-PBS solution for 24 h. Tissue was embedded in OCT (Optimal Cutteing Temperature) compound, snap frozen in liquid nitrogen, and stored at −80°C before being sectioned on a Leica cryostat. OE samples were sectioned coronally at 10–20 μm of thickness, mounted on Superfrost Plus glass slides (Fisher Scientific, #12-550-150), and stored at −20°C. Rat OE was similarly processed, apart from the formalin perfusion volume (200 mL).
OE injury assays
Mice were subject to a single i.p. injection of 75 mg/kg MTZ (Sigma-Aldrich, #046KO705) dissolved in 1X sterile PBS (Gibco, #10010–023) (Haglin et al., 2020; Lin et al., 2017). 7-to-8-week-old rats were subjected to MeBr exposure via passive inhalation of 330–345 ppm MeBr mixed with pure air for 6 h (Schwob et al., 1995).
Transplantation
Passage 12–14 rat GFP cells were utilized as donor cells. Host rats were subjected to 330–345 ppm MeBr lesion for 6 h, with the transplants taking place 20 h after the termination of the MeBr exposure. Donor cells were treated with 50 nM PMA (6 h or 12 h) or 0.1% DMSO for vehicle control. Cells were grown to 90% confluency on 100 mm dishes in triplicate. 8-to-9-week-old littermate male rats were used as host animals. At the time of transplantation, cells were washed 3X with PBS and treated with Accutase at RT until detaching from the plate. After detachment, cells were spun down and resuspended in 150 μL DMEM/F12 and kept on ice. A tracheotomy was performed, and cells were infused into one naris. Animals were injected with 2 mL of saline and allowed to recover overnight. For detailed information, please see the supplemental experimental procedures.
Immunohistochemistry
Slides were washed with 1X PBS before baking on a plate warmer (65°C) for 1 h. All pre-treatments and staining conditions took place in a humidified chamber. Tissue sections were pretreated with a 5-min incubation of 3% H2O2 in MeOH and 10 min of antigen retrieval (0.01 M pH 6.0 citrate buffer) in a commercial food steamer. Sections were then subjected to blocking with block buffer (10% normal donkey serum, 5% nonfat dry milk, 4% BSA, 0.1% Triton X-100 in 1X PBS) for one 1 h at RT. Primary antibodies were prepared in block buffer and incubated for either 1 h at RT or 16 h overnight at 4°C. Antibody concentrations can be found in Table S1.
In vitro model of HBCs and PMA activation
HBCs were cultured using our established primary culture HBC system (Peterson et al., 2019) with slight modifications to the antibiotic regimen. Instead of utilizing penicillin-streptomycin, we used Primocin (100 μg/mL final concentration) and anti-mycotic/antibiotic (1X final concentration, Gibco #15240062). For PMA-mediated activation, HBCs were treated with 50 nM PMA (Sigma-Aldrich #P8139) resuspended in their maintenance media for up to 12 h. Furthermore, all cells were grown on 35 mm poly-d-lysine/laminin-coated plasticware or glass chamber slides.
Immunocytochemistry
Upon the end of indicated treatment lengths, cells were washed with 1X PBS three times for 5 min. Cells were then fixed with 10% formalin for 15 min at RT. After three 1X PBS, 5-min washes, cells were permeabilized with ice-cold methanol on ice. Cells were then blocked for 1 h at RT utilizing blocking buffer. Primary antibodies were prepared in blocking buffer and incubated for either 1 h at RT or 16 h overnight at 4°C. Antibody concentrations can be found in Table S1.
Confocal microscopy
All samples, either tissue sections or in vitro HBCs, were imagined on a Zeiss LSM800 confocal microscope utilizing the multi-track mode. For the PMA-induced TP63 loss time course, the highest TP63 fluorescence Z-level was manually selected for each sample before creating a 5 × 5 mosaic imaging grid. All other in vitro experiments were imaged by creating a 2 × 2 mosaic grid with a multi-step (5–8 step) z stack, before being maximally projected in Fiji prior to analysis. All representative image samples underwent a z stack imaging approach utilizing Zeiss software recommendations before being maximally projected in Fiji. Images were background corrected utilizing the rolling ball algorithm in ImageJ. Representative images were prepared in Fiji before being assembled in Affinity Designer. Figure schemas were created with BioRender.com.
CellProfiler for quantitative image analysis
CellProfiler was used to quantify fluorescent images following preprocessing of images as described in these methods. CellProfiler pipelines for all quantitative imaging experiments can be retrieved from the aforementioned GitHub account link.
Bulk RNA-seq
Following indicated treatment lengths, cultured HBCs were prepared for bulk RNA paired-end (150 base pair) sequencing on an Illumina NovaSeq 6000 sequencer. For detailed information, please see the supplemental experimental procedures.
Single-cell RNA-seq analysis
Single-cell RNA-seq count data from Gadye et al. (2017) was accessed at GSE99251. Pre-processed and filtered expression data were analyzed with Seurat according to their vignette (Satija et al., 2015). Activated, resting, and cycling HBC subpopulations were identified. Iterated subclustering was used to isolate HBC populations from other cell types in the dataset including GBCs, Sus cells, and neuronal populations. To refine our HBC subpopulations, we used the following marker conditions: resting state, Tp63(+)/Mki67(−)/Hopx(−); partially activated, proliferating state, Tp63MED/Mki67(+)/Hopx(−); activated state, Tp63LOW/NEG/Mki67(−)/Hopx(+). For detailed methods on iterated subclustering, please see supplemental experimental procedures.
Preprocessing and quality control of bulk RNA-seq samples and differential gene expression (DGE)
Samples were aligned to the rat genome (rn6, version 2.7.4a) utilizing STAR(Dobin et al., 2013) (version 2.7.5b) with the following settings: “—quantMode GeneCounts.” Samples were normalized by using the estimateSIzeFactors function from DEseq2(Love et al., 2014). DGE analysis was carried out utilizing the “results” function of DESeq2. The alpha was set to 0.05.
Topological analyses of dynamically changing regulon networks and bottleneck gene identification
Regulon analysis was carried out with the R package RTN (v. 2.12.1). A list of rat TFs was accessed from the Animal Transcription Factor Database (Hu et al., 2019) (v. 3.0), cross-referenced with our normalized matrix counts for each sample, and 249 TFs were inputted. Standard package recommendations were followed, except for the deletion of the bootstrapping step due to limited sample size. Identified RAPs were exported directly into Cytoscape (Shannon et al., 2003) (v. 3.8.2) utilizing the R package RCy3 (v. 2.8.1) for further analysis. The five RAP clusters underwent further topological analysis to identify bottleneck genes utilizing the Cytoscape (v. 3.0) package CytoHubba (Chin et al., 2014) (v. 0.1).
Statistics and data visualization
Statistical analyses were carried out in R/RStudio (R version 4.0.2- 4.3.2, RStudio version 1.3.1056-2023.09.0+463). Visualization was performed in ggplot2 (versions 3.3.3-3.5.1), unless otherwise indicated in the figure legends. For groups with multiple comparisons, a one-way ANOVA followed by Dunnett’s test was calculated, relative to untreated or vehicle condition. Information about sample sizes and other statistical tests can be found in their respective figure legends. Sample size information for the in vitro model can be found in Table S2. Significance levels are demarcated by asterisks, which are as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Acknowledgments
The authors wish to thank Po Kwok-Tse for her outstanding technical contributions to this project. The authors also wish to thank Drs. Greg Bashaw and Brian Lin for their critical feedback on this manuscript. Funded by NIH grants R01 DC017869 (J.E.S), F30 DC017354 (M.J.Z), F30 DC018450 (J.D.L).
Author contributions
C.M.B.-C., conceptualization; C.M.B.-C., data curation; C.M.B.-C., M.J.Z., and J.D.L., formal analysis; J.E.S., funding acquisition; C.M.B.-C., J.D.L., W.J., and M.J.Z., investigation; C.M.B.-C., J.D.L., and M.J.Z., methodology; J.E.S., supervision; C.M.B.-C., writing – original draft; C.M.B.-C., J.D.L., M.J.Z., and J.E.S., writing – review and editing, All authors contributed to the article and approved the submitted version.
Declaration of interests
J.E.S. is a co-founder of Rhino Therapeutics, Inc.
Published: July 25, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.06.011.
Supplemental information
References
- Carr V.M., Menco B.P., Yankova M.P., Morimoto R.I., Farbman A.I. Odorants as cell-type specific activators of a heat shock response in the rat olfactory mucosa. J. Comp. Neurol. 2001;432:425–439. doi: 10.1002/cne.1112. [DOI] [PubMed] [Google Scholar]
- Carter L.A., MacDonald J.L., Roskams A.J. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J. Neurosci. 2004;24:5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M.A.A., de Santiago I., Campbell T.M., Vaughn C., Hickey T.E., Ross E., Tilley W.D., Markowetz F., Ponder B.A.J., Meyer K.B. Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 2016;48:12–21. doi: 10.1038/ng.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Reed R.R., Lane A.P. Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc. Natl. Acad. Sci. USA. 2017;114:8089–8094. doi: 10.1073/pnas.1620664114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Fang H., Schwob J.E. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J. Comp. Neurol. 2004;469:457–474. doi: 10.1002/cne.11031. [DOI] [PubMed] [Google Scholar]
- Child K.M., Herrick D.B., Schwob J.E., Holbrook E.H., Jang W. The neuroregenerative capacity of olfactory stem cells is not limitless: Implications for aging. J. Neurosci. 2018;38:6806–6824. doi: 10.1523/JNEUROSCI.3261-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.-H., Chen S.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362 doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzek M., Patel P.K., Solomon P.D., Lin B., Hummel T., Schwob J.E., Holbrook E.H. Integrated age-related immunohistological changes occur in human olfactory epithelium and olfactory bulb. J. Comp. Neurol. 2022;530:2154–2175. doi: 10.1002/cne.25325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M.N.C., Castro M.A.A., Wang X., de Santiago I., O’Reilly M., Chin S.F., Rueda O.M., Caldas C., Ponder B.A.J., Markowetz F., Meyer K.B. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 2013;4 doi: 10.1038/ncomms3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher R.B., Das D., Gadye L., Street K.N., Baudhuin A., Wagner A., Cole M.B., Flores Q., Choi Y.G., Yosef N., et al. Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell Stem Cell. 2017;20:817–830.e8. doi: 10.1016/j.stem.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher R.B., Prasol M.S., Estrada J., Baudhuin A., Vranizan K., Choi Y.G., Ngai J. p63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72:748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadye L., Das D., Sanchez M.A., Street K., Baudhuin A., Wagner A., Cole M.B., Choi Y.G., Yosef N., Purdom E., et al. Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell. 2017;21:775–790.e9. doi: 10.1016/j.stem.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney P.A., Lu C., Caudy A.A., Hess D.C., Botstein D. Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proc. Natl. Acad. Sci. USA. 2013;110:E4393–E4402. doi: 10.1073/pnas.1318100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Lundqvist E.N., Coates P.J., Thurfjell N., Wettersand E., Nylander K. Dysregulation of TAp63 mRNA and protein levels in psoriasis. J. Invest. Dermatol. 2006;126:137–141. doi: 10.1038/sj.jid.5700010. [DOI] [PubMed] [Google Scholar]
- Haglin S., Berghard A., Bohm S. Increased retinoic acid catabolism in olfactory sensory neurons activates dormant tissue-specific stem cells and accelerates age-related metaplasia. J. Neurosci. 2020;40:4116–4129. doi: 10.1523/JNEUROSCI.2468-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D.B., Lin B., Peterson J., Schnittke N., Schwob J.E. Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc. Natl. Acad. Sci. USA. 2017;114:E5589–E5598. doi: 10.1073/pnas.1701333114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hng C.H., Camp E., Anderson P., Breen J., Zannettino A., Gronthos S. HOPX regulates bone marrow-derived mesenchymal stromal cell fate determination via suppression of adipogenic gene pathways. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-68261-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook E.H., Leopold D.A., Schwob J.E. Abnormalities of axon growth in human olfactory mucosa. Laryngoscope. 2005;115:2144–2154. doi: 10.1097/01.MLG.0000181493.83661.CE. [DOI] [PubMed] [Google Scholar]
- Holden N.S., Squires P.E., Kaur M., Bland R., Jones C.E., Newton R. Phorbol ester-stimulated NF-kappaB-dependent transcription: roles for isoforms of novel protein kinase C. Cell. Signal. 2008;20:1338–1348. doi: 10.1016/j.cellsig.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Hu H., Miao Y.-R., Jia L.-H., Yu Q.-Y., Zhang Q., Guo A.-Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai N., Zhou Z., Roop D.R., Behringer R.R. Horizontal Basal Cells Are Multipotent Progenitors in Normal and Injured Adult Olfactory Epithelium. Stem Cell. 2008;26:1298–1306. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W., Lambropoulos J., Woo J.K., Peluso C.E., Schwob J.E. Maintaining epitheliopoietic potency when culturing olfactory progenitors. Exp. Neurol. 2008;214:25–36. doi: 10.1016/j.expneurol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W., Youngentob S.L., Schwob J.E. Globose basal cells are required for reconstitution of olfactory epithelium after methyl bromide lesion. J. Comp. Neurol. 2003;460:123–140. doi: 10.1002/cne.10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H., Um J.W. Synapse development organized by neuronal activity-regulated immediate-early genes. Exp. Mol. Med. 2018;50:1–7. doi: 10.1038/s12276-018-0025-1. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinna S., Seltmann K., Bachmann A.L., Schwendimann A., Thiagarajan L., Hennig P., Beer H.D., Mollo M.R., Missero C., Werner S. Interaction of the NRF2 and p63 transcription factors promotes keratinocyte proliferation in the epidermis. Nucleic Acids Res. 2021;49:3748–3763. doi: 10.1093/nar/gkab167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C.T., Coulombe P.A., Reed R.R. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Li C., Xiao Z.X. Regulation of p63 protein stability via ubiquitin-proteasome pathway. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Coleman J.H., Peterson J.N., Zunitch M.J., Jang W., Herrick D.B., Schwob J.E. Injury Induces Endogenous Reprogramming and Dedifferentiation of Neuronal Progenitors to Multipotency. Cell Stem Cell. 2017;21:761–774.e5. doi: 10.1016/j.stem.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie J.D., Barrios-Camacho C.M., Bromberg B.H., Hintschich C.A., Schwob J.E. Spatiotemporal dynamics exhibited by horizontal basal cells reveal a pro-neurogenic pathway during injury-induced olfactory epithelium regeneration. iScience. 2024;27 doi: 10.1016/j.isci.2024.109600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie J.D., Bromberg B.H., Zunitch M.J., Schwob J.E. Horizontal basal cells self-govern their neurogenic potential during injury-induced regeneration of the olfactory epithelium. Development. 2023;150 doi: 10.1242/dev.201552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minatohara K., Akiyoshi M., Okuno H. Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front. Mol. Neurosci. 2015;8:78. doi: 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno H. Regulation and function of immediate-early genes in the brain: Beyond neuronal activity markers. Neurosci. Res. 2011;69:175–186. doi: 10.1016/j.neures.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Packard A., Schnittke N., Romano R.-A., Sinha S., Schwob J.E. DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J. Neurosci. 2011;31:8748–8759. doi: 10.1523/JNEUROSCI.0681-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant N.J., Wang Y., Hadland B., Zaunbrecher R.J., Redd M., Jones D., Pabon L., Jain R., Epstein J., Ruzzo W.L., et al. Chromatin and Transcriptional Analysis of Mesoderm Progenitor Cells Identifies HOPX as a Regulator of Primitive Hematopoiesis. Cell Rep. 2017;20:1597–1608. doi: 10.1016/j.celrep.2017.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J., Lin B., Barrios-Camacho C.M., Herrick D.B., Holbrook E.H., Jang W., Coleman J.H., Schwob J.E. Activating a Reserve Neural Stem Cell Population In Vitro Enables Engraftment and Multipotency after Transplantation. Stem Cell Rep. 2019;12:680–695. doi: 10.1016/j.stemcr.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Honmou O., Akiyama Y., Uede T., Hashi K., Kocsis J.D. Transplantation of an acutely isolated bone marrow fraction repairs demyelinated adult rat spinal cord axons. Glia. 2001;35:26–34. doi: 10.1002/glia.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N., Herrick D.B., Lin B., Peterson J., Coleman J.H., Packard A.I., Jang W., Schwob J.E. Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc. Natl. Acad. Sci. USA. 2015;112:E5068–E5077. doi: 10.1073/pnas.1512272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J.E. Restoring olfaction: a view from the olfactory epithelium. Chem. Senses. 2005;30:i131–i132. doi: 10.1093/chemse/bjh149. [DOI] [PubMed] [Google Scholar]
- Schwob J.E., Jang W., Holbrook E.H., Lin B., Herrick D.B., Peterson J.N., Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol. 2017;525:1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J.E., Youngentob S.L., Mezza R.C. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J. Comp. Neurol. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Wang H., Zhang L. TP63 Functions as a Tumor Suppressor Regulated by GAS5/miR-221-3p Signaling Axis in Human Non-Small Cell Lung Cancer Cells. Cancer Manag. Res. 2023;15:217–231. doi: 10.2147/CMAR.S387781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane A.R., Platig J., Fagny M., Chen C.-Y., Paulson J.N., Lopes-Ramos C.M., DeMeo D.L., Quackenbush J., Glass K., Kuijjer M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017;21:1077–1088. doi: 10.1016/j.celrep.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.M., Sim S.M., Kim H.Y., Park G.T. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur. J. Cell Biol. 2010;89:537–546. doi: 10.1016/j.ejcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Yu H., Kim P.M., Sprecher E., Trifonov V., Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw files, counts, and normalized counts from our bulk RNA-seq can be accessed under GEO: GSE215797. Single-cell RNA-seq data used in this analysis were accessed from GEO, GEO: GSE99251. All code used to generate the data in this manuscript can be accessed at https://github.com/barriosc. The accession number for the PMA time course dataset reported in this paper is Mendeley Data V1: https://doi.org/10.17632/54tkpr2zdv.1.