Abstract
While multiple factors impact disease, artificial intelligence (AI) studies in medicine often use small, non-diverse patient cohorts due to data sharing and privacy issues. Federated learning (FL) has emerged as a solution, enabling training across hospitals without direct data sharing. Here, we present FL-PedBrain, an FL platform for pediatric posterior fossa brain tumors, and evaluate its performance on a diverse, realistic, multi-center cohort. Pediatric brain tumors were targeted due to the scarcity of such datasets, even in tertiary care hospitals. Our platform orchestrates federated training for joint tumor classification and segmentation across 19 international sites. FL-PedBrain exhibits less than a 1.5% decrease in classification and a 3% reduction in segmentation performance compared to centralized data training. FL boosts segmentation performance by 20 to 30% on three external, out-of-network sites. Finally, we explore the sources of data heterogeneity and examine FL robustness in real-world scenarios with data imbalances.
Subject terms: Paediatric research, Computational science, CNS cancer, Machine learning, Cancer imaging
Federated learning (FL) has emerged as a potential solution to train machine learning models in multiple clinical datasets while preserving patient privacy. Here, the authors develop an MRI-based FL platform for pediatric posterior fossa brain tumors—FL-PedBrain—and evaluate it on a diverse multi-center cohort.
Introduction
AI has created untapped opportunities for accelerating precision in medicine, including transformations in medical imaging that offer improved efficiency and enhanced disease diagnosis, therapy planning, and surveillance. To date, even with relatively small datasets, studies have shown promise of AI across imaging modalities, clinical subspecialties, and organ domains, such as disease delineation1–4, diagnosis5–9, outcomes10,11, and underlying genomics12,13, some of which surpass human performance.
Ultimately, “big data” is a key to the success of AI in medicine. For this reason, many AI investigations have focused on relatively common adult diseases pooled from one or a few large centers, e.g., breast or lung cancer, pneumonias, heart disease, intracranial hemorrhage, or acute ischemic strokes. However, many rare or pediatric diseases with data scattered across hospitals currently do not benefit from the advancements in AI. Even CheXNet, a chest radiograph dataset with >100,000 annotations5 and considered “large” among medical datasets dwarfs in comparison to non-medical ImageNet, which contains >14 million annotated images14. Current AI models on cross-sectional imaging, e.g., MRI or CT—often considered the clinical workhorse—are trained on significantly fewer datasets, raising questions of AI reliability and generalization and thereby further exacerbating a general lag in AI adoption in healthcare.
Medical data is not scarce, however. Large tomes exist across the world in the form of electronic health records and imaging archives and are fertile grounds for large-scale AI developments. Unfortunately, barriers to data sharing across institutions—while necessary for patient privacy—have impeded progress in AI for healthcare. FL has emerged as one potential solution that enables model training across multiple, decentralized datasets, without direct patient data sharing15. It offers better privacy and local data autonomy while facilitating learning from a distributed data source in which diverse factors contribute to disease phenotypes and their outcomes. From such a network, AI can learn complex relationships and potentially uncover new clinical perspectives.
Recent FL works in medicine have shown feasibility16–19 but with limited scope, e.g., few participating FL sites or small range of classes or datasets that are limited in diversity and size. Prior FL investigations20–22 have examined segmentation of adult gliomas that typically arise in the supratentorial brain. Children, however, present with more diverse brain tumor pathologies, the majority occurring in the posterior fossa (PF) spaces that include the brainstem and the cerebellum.
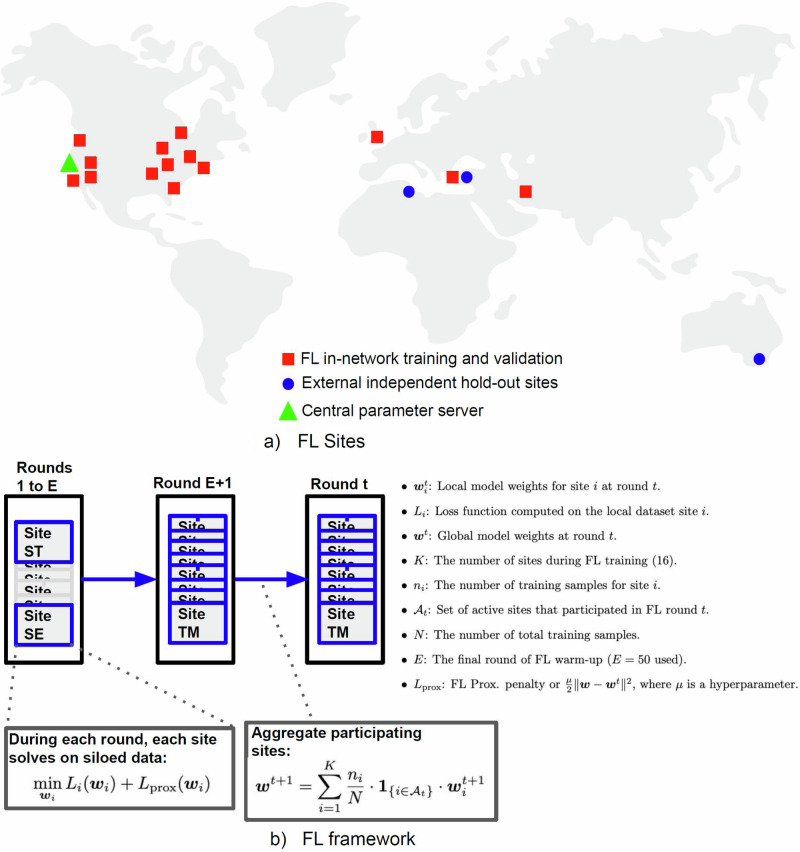
In this work, we present an end-to-end, MRI-based FL platform for PF tumors, FL-PedBrain, on a large international pediatric dataset of 19 institutions from North America, Europe, West Asia, North Africa, and Australia (Fig. 1). We targeted pediatric tumors, given both their pathologic diversity and general scarcity even within subspecialty pediatric hospitals. Hence, a successful FL platform could uniquely benefit this data-sparse, yet vulnerable population.
Fig. 1. Federated Learning Platform.
Participating sites (a) and FL procedure (b). a North America: Stanford Children’s Hospital (ST—Palo Alto, California), Seattle Children’s Hospital (SE—Seattle, Washington), Phoenix Children’s Hospital (PH—Phoenix, Arizona), Primary Children’s Hospital (UT—Salt Lake City, Utah), Children’s Hospital Orange County (CH—Orange County, California), Dayton Children’s Hospital (DY—Dayton, Ohio), Indiana University Riley Children’s (IN—Indianapolis, Indiana), Lurie Children’s Hospital of Chicago (CG—Chicago, Illinois), NYU Langone Medical Center (NY—New York City, New York), Children’s Hospital of Philadelphia (CP—Philadelphia, Pennsylvania), Duke Children’s Hospital (DU—Durham, North Carolina), Boston Children’s Hospital (BO—Boston, Massachusetts), Toronto Sick Kids Hospital (TO—Toronto, Canada); Europe: Great Ormand Street Hospital (GO—London, United Kingdom),Tepecik Health Sciences (TK—Izmir, Turkey), Koç University (KC—Istanbul, Turkey); North Africa: Centre International Carthage Médical (TU—Monastir, Tunisia); West Asia: Tehran University of Medical Sciences (TM—Tehran, Iran); Australia: The Children’s Hospital at Westmead (AU—Sydney, Australia). b Our FL framework incorporates FL warm-up on the largest sites and proximal regularization to learn on heterogeneous sites, but we report the best results with μ = 0.
We examine a heterogeneous group of pediatric PF tumors with diverse clinical outcomes, dependent on tumor pathology or genomics, surgical resection margins, or their candidacy for new drug therapies. We conducted tumor classification and segmentation jointly, as success of such tasks prior to surgery can directly impact precision in surgical margins, radiation targets, and alter other therapy strategies that aim cure with minimal risks. Specifically, we orchestrated FL training with real data from 19 participating sites from five continents and compared its efficacy against the traditional pooled data approach, i.e., centralized data sharing (CDS). We investigated real-life scenarios where some sites provide missing or imbalanced data. Finally, we explored the underlying sources of data heterogeneity, such as variations in image quality, or site-specific tumor features.
Results
Study cohort
A total of 1468 unique PF tumor subjects (mean age 7.6 years; 48% females) were included, comprising 596 MB, 210 EP, 335 PA, and 327 DIPG. Table 1 summarizes the demographic information and the tumor pathologies distributed across the 19 institutions.
Table 1.
Demographic table
| Site ID | Total subjects | EP* | DIPG* | MB* | PA* | EP** | DIPG** | MB** | PA** |
|---|---|---|---|---|---|---|---|---|---|
| TM | 13 | 0 (N/A) | 0 (N/A) | 7 (67%) | 6 (71%) | N/A | N/A | 70.6 | 106.2 |
| PH | 55 | 12 (42%) | 14 (50%) | 15 (60%) | 14 (47%) | 77.8 | 105.5 | 94 | 76.9 |
| TO | 92 | 26 (61%) | 0 (N/A) | 60 (64%) | 6 (100%) | 60.2 | N/A | 92.2 | 98.9 |
| UT | 129 | 17 (75%) | 18 (60%) | 42 (68%) | 52 (42%) | 30.1 | 91.5 | 85.7 | 104.7 |
| DU | 24 | 0 (N/A) | 0 (N/A) | 24 (64%) | 0 (N/A) | N/A | N/A | 66.5 | N/A |
| CP | 96 | 20 (N/A) | 0 (N/A) | 43 (N/A) | 33 (N/A) | N/A | N/A | N/A | N/A |
| IN | 118 | 5 (38%) | 21 (52%) | 63 (77%) | 29 (50%) | 58.4 | 75.8 | 93.7 | 102.2 |
| ST | 328 | 39 (57%) | 84 (51%) | 93 (66%) | 112 (48%) | 175.35 | 98.3 | 107.1 | 108.2 |
| SE | 241 | 42 (71%) | 44 (39%) | 113 (58%) | 42 (51%) | 49.6 | 83 | 84.2 | 96.7 |
| CG | 150 | 10 (50%) | 75 (49%) | 54 (66%) | 11 (38%) | 83.9 | 96.9 | 88.8 | 105.6 |
| NY | 26 | 7 (75%) | 10 (64%) | 9 (67%) | 0 (N/A) | 94 | 92.7 | 159.2 | N/A |
| CH | 14 | 0 (N/A) | 4 (50%) | 3 (50%) | 7 (60%) | N/A | 120 | 85 | 42.8 |
| GO | 78 | 23 (48%) | 14 (21%) | 27 (48%) | 14 (36%) | 64.9 | 82.2 | 70.9 | 83.2 |
| BO | 19 | 0 (N/A) | 0 (N/A) | 19 (40%) | 0 (N/A) | N/A | N/A | 100.4 | N/A |
| KC | 3 | 1 (100%) | 0 (N/A) | 2 (71%) | 0 (100%) | 144 | 60 | 109.7 | 24 |
| DY | 28 | 4 (0%) | 5 (60%) | 13 (92%) | 6 (50%) | 58 | 104.1 | 99.1 | 50.6 |
| TK | 16 | 2 (100%) | 4 (50%) | 7 (45%) | 3 (71%) | 327.6 | 127.8 | 198.2 | 130.3 |
| AU | 32 | 0 (N/A) | 32 (34%) | 0 (N/A) | 0 (N/A) | N/A | 83.3 | N/A | N/A |
| TU | 6 | 2 (100%) | 2 (50%) | 2 (50%) | 0 (N/A) | 217 | 70 | 105.6 | N/A |
*Number and percentage of males.
**Mean ages for each tumor type per site (EP ependymoma, DIPG diffuse intrinsic pontine glioma, MB medulloblastoma, PA pilocytic astrocytoma).
N/A Information not available.
Classification
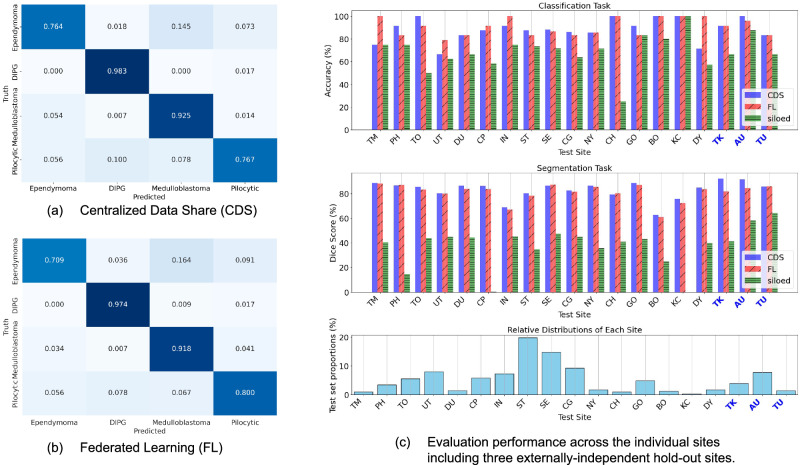
Table 2 summarizes the model performances, comparing FL and CDS. FL achieved classification performance on par with CDS, without a statistically discernible difference. We present FL and CDS confusion matrices summarizing the classification performance on the four tumor pathologies (MB, EP, PA, DIPG) in Fig. 2a, b. Figure 2c also illustrates per site, overall accuracies. Compared to either FL and CDS, Siloed training significantly underperforms and shows large performance variance across the sites (Fig. 2c).
Table 2.
Classification accuracies for CDS and FL on all validation sets
| Metric | Centralized training (CDS) | Federated learning (FL) |
|---|---|---|
| Accuracy | 0.8922 | 0.8799 |
| F1 Score | 0.8766 | 0.8561 |
Fig. 2. Performance of FL on the validation sites compared to CDS and siloed training using the ST site model.
Confusion matrices (a, b) for the classification task and per site performance (c). The hospitals TK, AU, and TU are external and independent hold-out sites. Source data are provided as a Source Data file. Source data are provided as a Source Data file.
Segmentation
As shown in Tables 3 and 4, FL achieves an overall segmentation performance that approaches CDS on both, the 16 validation datasets and the 3 hold-out test sites. Compared to either FL or CDS, Siloed training underperforms by >20%, a performance drop that is greater for segmentation than for classification (Fig. 2c). Both, FL and CDS, yielded the best segmentation performance on DIPG tumors, whereas performance on MB was lower than the other tumors (Table 3). Within the tumor subgroups, FL matched that of CDS performance on MB and PA tumor (no t-statistic difference), while FL slightly underperformed compared to CDS on EP and DIPG. Such variations might suggest heterogeneity in tumor voxel volume between the sites (see section on Heterogeneity). While the mean Dice Similarity Coefficient (DSC) on the validation sets were congruent for both FL and CDS, FL exhibited slightly higher variability, i.e., greater standard deviation, suggesting underlying differences in the model behavior.
Table 3.
Segmentation Statistics for CDS and FL on Validation Sets
| Metric | DSCs for CDS | DSCs for FL | IoU score for CDS | IoU score for FL |
|---|---|---|---|---|
| EP | 0.8378 ± 0.0842 | 0.8044 ± 0.1051 | 0.7291 ± 0.1144 | 0.6848 ± 0.1380 |
| DIPG | 0.8866 ± 0.0544 | 0.8622 ± 0.0861 | 0.8003 ± 0.0826 | 0.7663 ± 0.1129 |
| MB | 0.7782 ± 0.2275 | 0.7800 ± 0.2151 | 0.6782 ± 0.2261 | 0.6775 ± 0.2207 |
| PA | 0.8233 ± 0.2126 | 0.8194 ± 0.2087 | 0.7386 ± 0.2204 | 0.7312 ± 0.2139 |
The table compares the segmentation performance between CDS and FL on validation sets. A two-sided t-test for the DSC distributions with t-statistic (degrees of freedom), p value, effect size, and the 95% confidence interval for each class: 1) EP: 4.874 (54), 9.998e-06, 0.5, [0.01, 0.04], 2) DIPG: 3.511 (115), 0.0006, 0.37, [0.01,0.04], MB: −0.284 (146), 0.7771, 0.06, [−0.01,0.02], PA: 0.513 (89), 0.6090, 0.22, [0.00, 0.02].
Table 4.
Segmentation DSC performance on the three independent hold-out sites
| Sites | CDS-model on Holdouts | FL-model Holdouts | Siloed model on Holdouts |
|---|---|---|---|
| TU | 0.862 ± 0.076 | 0.860 ± 0.079 | 0.410 ± 0.32 |
| TK | 0.920 ± 0.023 | 0.818 ± 0.082 | 0.58 ± 0.264 |
| AU | 0.914 ± 0.043 | 0.845 ± 0.127 | 0.642 ± 0.134 |
These are sites that did not participate in the training, validation, and model development.
Supplementary Table 1 presents the classification and segmentation results for the 16 independently trained models, each using its respective site-specific dataset. The outcomes suggest subpar performance across the board, attributable to the limited size of individual datasets. Notably, models from sites UT and CP showed the highest segmentation DSCs, reaching 0.57. However, models from five sites did not converge.
Visualizations and quality of FL training
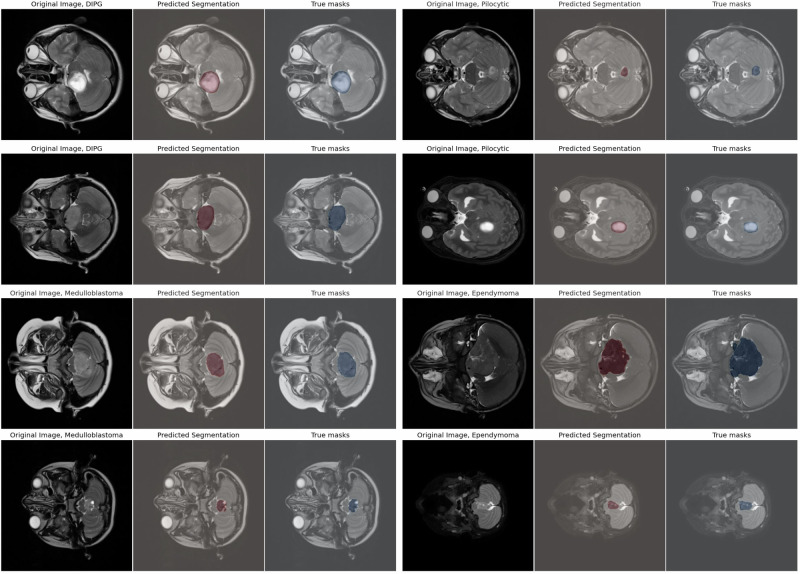
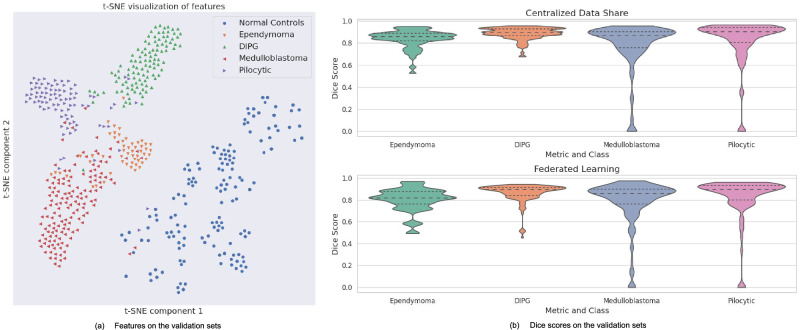
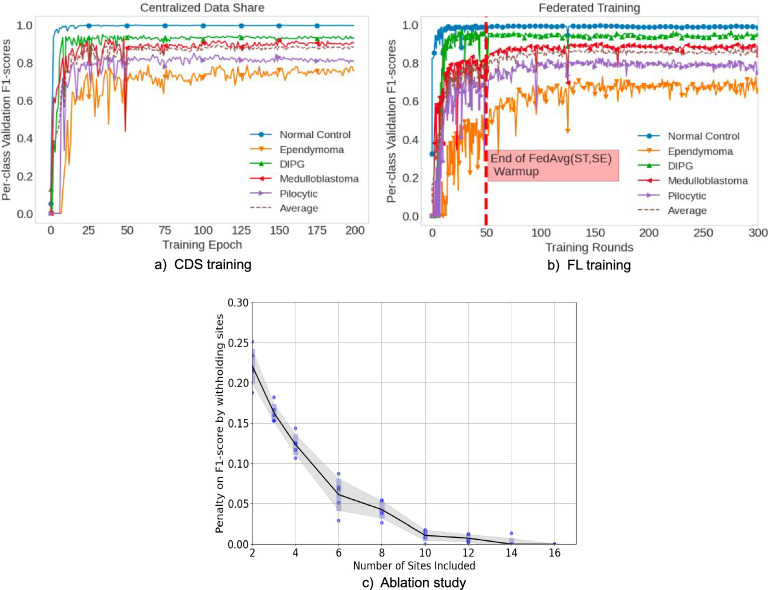
Figure 3 illustrates sample segmentation outputs from FL-PedBrain compared to the ground-truth segmentations. We also present a t-SNE23 visualization of projected embedded features from FL-PedBrain classification model from the validation set (Fig. 4a). Note unique tumor features that are also distinct from normal pediatric brains. A corresponding violin plot of all per-example Dice scores (DSC) in the 16-site population is also shown in Fig. 4b. Finally, Fig. 5 illustrates convergence during training, comparing CDS to FL. As expected, and consistent with expected observation15, CDS requires fewer learning updates to converge.
Fig. 3. Sample FL segmentation predictions compared to ground-truth segmentations.
Sample predictions of the FL-trained model compared to ground-truth segmentations across various tumor types sampled at different depth regions of the brain. The Source model is provided in the GitHub link.
Fig. 4. Classification and Segmentation Results.
Visualization of the FL-trained model features (a) and DSCs (Dice scores) (b). The violin plot displays the median, quartiles, and minimum and maximum values of the distribution. Source data are provided as a Source Data file.
Fig. 5. FL Training Runs.
Training runs comparing CDS (a) to FL (b), and ablation study showing impact of participating sites in the network (c). Runs using CDS (a) and FL (b) show fast convergence for both the classification and segmentation tasks. A federated warm-up was performed on the two largest sites first. These runs (c) show the influence of adding sites into the FL training network on the worst-case class prediction (Ependymoma) measured at 100 FL rounds, 150, 200, 250, and 300. The error bars represent 1 std. deviation of variation among five independent FL runs. Source data are provided as a Source Data file.
Heterogeneity
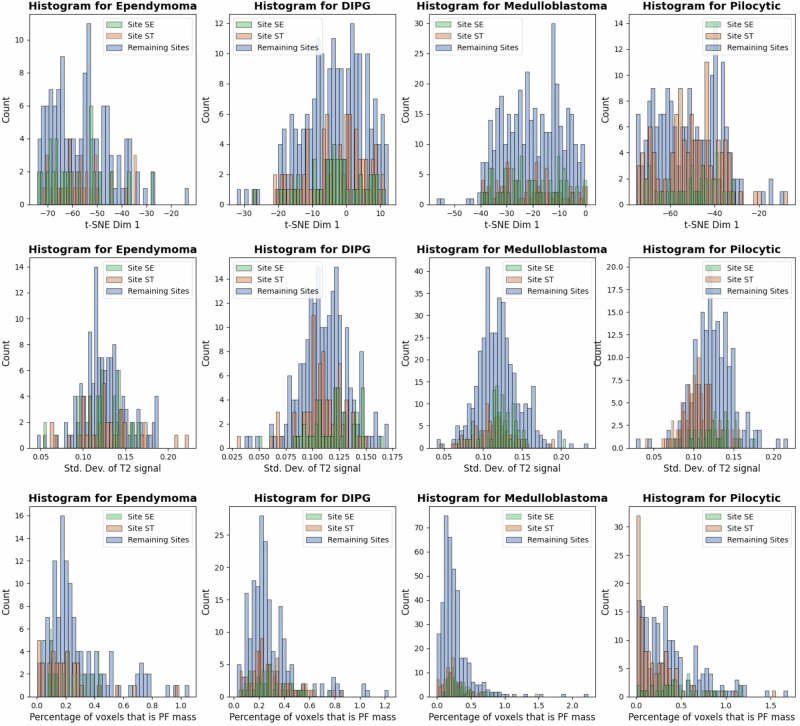
Since data heterogeneity is a key consideration in AI studies, we examined various sources of data heterogeneity. One notable factor was the significant class imbalances across the participating sites, both in the sample sizes and the pathologic subgroups, with some sites completely missing certain tumor types, as shown in Table 1. Interestingly, we observed differences in T2-MRI pixel variance in Fig. 6, especially for DIPG and PA, possibly reflecting larger variances in solid, hemorrhagic, necrotic, or cystic components, or other tumoral habitats unique to astrocytomas. We also found significant variation in relative tumor volumes across sites. Despite such sources of data heterogeneity—including extreme class imbalances—we found no evidence such factors impacted FL convergence.
Fig. 6. Visualization of heterogeneity.
Differences in T2-MRI pixel variance, especially for DIPG and Pilocytic, possibly reflecting larger variances in solid, hemorrhagic, necrotic, or cystic components. Significant variation in tumor volumes across sites were found. Source data are provided as a Source Data file.
Impact of FL Warm-up
FL across just two of the largest centers achieved >70% classification accuracy and segmentation DSC, except for the EP cases. By adding in the remaining sites, performance was significantly enhanced, as shown by Fig. 5c. We found that Federated Warm-up was important; without warm-up, training times were up to 10 times longer and overall performance lower, especially for EP classification and segmentation.
Better performance with more active FL sites
In our study, we assess the impact of site activity on FL performance by conducting an ablation experiment. This experiment measures the FL system’s performance relative to the quantity of active training sites, as depicted in Fig. 5c. We rerun the full FL experiment by integrating more sites into the training process (x axis), prioritizing those with larger datasets. The performance evaluation is based on the F1 score—specifically, the classification accuracy of the label that performs the poorest. Our findings indicate a positive correlation between the number of active sites and the F1 score: as more sites participate in the FL network, the F1 score improves, eventually equaling the peak score achieved when all available sites are active.
Future challenges and practical implementation
FL-PedBrain introduces logistical challenges, communication overhead, model synchronization, and computational demands.
Communication and logistical challenges
In FL, every participating hospital must regularly exchange model updates—specifically, the model weights after each FL training round. For our classification-segmentation model, this equates to transmitting ~125 MB of model weights per round. This culminates in a data transfer of ~74 GB per hospital for each training session with 200 rounds. Training the largest dataset for 1 epoch consumes ~3–4 minutes on a V100 GPU, and the time to then transfer all 16 models from each hospital to the central parameter server (coordinating hospital) in Fig. 1a is roughly 7 minutes at 1 MB/s internet upload rate, assuming that the central server’s download rate is much faster than 1 MB/s. This equates to about 10 minutes per round (1 epoch per round) and 2000 minutes to ship one trained model. Although CDS only requires a one-time collection of 200–1000 GB of DICOMs, FL offers benefits by removing the need for data use agreements and the need for deidentification, which can take a long time to establish and verify. Finally, FL provides advantages such as continuous quality control and oversight from each of the sites’ technical model builders. The provided figures are rough estimates; actual performance will vary as hospitals differ in computing power, communication standards, and data transfer speeds. Asynchronous Federated Learning (FL) is particularly beneficial in environments where hospitals exhibit diversity not just in data but also in computational and networking resources. Recent methods for heterogeneous FL24 can potentially alleviate communication and compute overheads.
Need for on-site technical expertise
Additionally, having both clinical and AI experts per site would greatly enhance and streamline the FL workflow, enabling them to (1) inspect the training and evaluation data for any obvious imaging artifacts or integrity of diagnosis and (2) monitor the training process as the model evolves. We intend our FL framework not to be used just for static datasets like in the CDS case but rather as a bedrock for active learning on growing datasets. Therefore, human integration into the FL pipeline is a very promising future direction.
Discussion
We present an FL system for pediatric cancer, FL-PedBrain, specifically targeting PF tumors. While brain tumors represent the most common solid neoplasm of childhood, they remain sparse compared to adult tumors, dispersed across pediatric or subspecialty centers. Thus, a successful collaborative platform that enables large-scale AI learning across institutions could uniquely benefit this population. Here, we capitalize on a large and diverse brain MRI dataset of pediatric PF tumors to date from 19 global institutions and present and evaluate an FL design that jointly conducts tumor pathology prediction and segmentation, optimized for this relatively data-sparse population.
Overall, we found robust generalization of FL-PedBrain across all sites, including the three external holdouts. Compared to CDS that uses pooled data from all sites, FL deviates by less than 1.5% in the classification and only 3% in the segmentation performances, with no statistical difference between CDS and FL on classification and slightly lower segmentation performance of FL on two of the four tumor groups. On the other hand, Siloed training—or training confined to a local site—performs ~20% worse compared to either FL or CDS, highlighting the risks of AI generalization and brittle models.
Prior FL studies on brain tumors have exclusively focused on segmentation of adult gliomas20–22. In this work, we trained the classifier and segmentation jointly. Unlike prior FL studies that employed extensive image manipulations, e.g., skull-stripping and rigid atlas-based brain co-registration20–22, we used real-life, raw MRI data that included brain tissue, skull, scalp, and head sizes of all ages, so that FL-PedBrain could be used in an end-to-end clinical deployment. Despite the heterogeneous dataset (infant to adult head sizes and diverse tumor pathologies beyond gliomas, e.g., embryonal and glial tumor cells of origin) and not requiring image manipulation prior to FL training, FL-PedBrain, performed segmentation on par with prior adult studies. FL-PedBrain also outperformed (F1 scores of 0.877 and 0.856 for CDS and FL, respectively) a prior pilot study9 that used pooled data for PF tumor prediction (F1 score of 0.800). Recent advances in FL strategies19,25–28 tackle learning on heterogeneous data and environments. Federated Proximal learning (FedProx25) is an adjustment to Federated Averaging that can accommodate for model drift. One important ingredient is the proximal weight penalty to ensure that the local updates do not stray too far from the global model, thereby making the training process more robust to data heterogeneity among clients. We have found that Federated Averaging (µ = 0) achieves higher and more consistent segmentation performance across the 19 sites on average compared to other advanced strategies such as weight transfer, exploiting synthetic data, and knowledge distillation26.
Moreover, we presented the Federated Warm-up method to combat challenges of severe non-uniform distributions of sample sizes across the FL network. This allows the training process to learn from the sites with the largest data samples for a few federated rounds. Thereafter, the learning proceeds to all the sites, including the ones with missing classes.
FL-PedBrain jointly classifies and segments brain tumors, which addresses several clinical needs. First, a more precise, pre-surgical knowledge of the PF tumor pathology could impact therapy. For example, less aggressive, safer resection margins may be desirable for more radio-sensitive MB compared to EP. Patient counseling and therapeutic strategy may vastly differ for non-resectable DIPG versus less aggressive but “infiltrative”-appearing PA tumors that can mimic DIPG. Second, since PF tumors often plague critical brain regions such as the brainstem, more precise tumor localization via segmentation that ports into surgical navigation can also optimize maximal resection for cure (e.g., EP or PA tumors) while minimizing risks. It may also enhance radiation targets and offer efficiency in radiomics or other quantitative tumor analytics.
Currently, manual maximal, linear measurements (x, y, z dimensions) are used to calculate tumor growth or regression. While useful, these are crude metrics for tumor tissues that are asymmetric or irregular, and also prone to interobserver variability. Thus, FL-PedBrain could be used to more reliably calculate tumor volumes across serial imaging. Segmentation masks generated from FL-PedBrain can also be plugged into a radiomics pipeline for tumor genomics4 or enhanced risk-stratification10 and potentially clarify patient candidacy for various individualized therapies.
We recognize several limitations of this work. First, the MRI scans originated from various MRI hardware across different sites, each employing unique protocols, leading to disparities in image quality. For example, we found site-to-site variations in T2-MRI image intensities. Second, differences in clinical practices and culture or site-specific barriers to MRI may impact tumor features at the time of diagnosis. For example, sites that use MRI to screen children at high risk of brain tumors, e.g., patients with specific genetic syndromes, might find tumors at an earlier phase versus under-resourced communities that catch tumors at a later period, when the patient finally decompensates. Alternatively, a particular subspecialty center may attract more complex or advanced tumors due to referral patterns. For example, we found that some sites tend to host larger size tumors, e.g., PA tumors, compared to others. Class imbalance within TK and AU sites likely contribute to a slightly lower classification performance. For segmentation task, where the scores are calculated per pixel across the entire 256 × 256 × 64 head volume, we observe similar performance between FL and CDS.
Nevertheless, we incorporated such heterogeneous conditions to properly investigate FL feasibility in a real-life setting. Real-world datasets are generally non-IID (non-independent and identically distributed) and can thus impact the final FL performance compared to baseline CDS. Here, we highlight multiple sources of heterogeneity inherent in our data: (1) imbalanced number of samples per site; (2) age and sex differences across sites (Table 2); (3) imbalanced or missing tumor classes on few sites of the federated network; (4) site-specific variations in the MRI signal intensities; and (5) site-specific variations in the tumor sizes. More sophisticated FL techniques such as Federated Proximal techniques and variations25 might improve training convergence with imbalanced classes. However, we have not found improvement using such methods. Despite such sources of data heterogeneity, we show robustness of FL-PedBrain as shown by the training convergence graph (Fig. 5b) with an FL performance that not only closely matches CDS, but also offers the advantages of AI training without data sharing across the sites within the FL network. Lastly, while the study does not account for human inter-reader variability, our segmentation masks reflect a consensus-based ground truth validated by six experts in the field.
In conclusion, we presented and evaluated a federated platform for pediatric brain tumors that is privacy-preserving and does not require sharing of data and showed its feasibility on a heterogeneous tumor pathology and diverse MRI dataset from 19 geographic centers. We emphasize the potential of FL in accelerating large-scale, clinically translatable AI for pediatric datasets and other heterogeneous, privacy-preserving data.
Next steps will include a study on the prospective deployment of real-time FL-PedBrain at local hospitals, requiring no additional data processing to enhance clinical usability. The methodology and results of this work lay the groundwork for future applications of FL in radiology and beyond, towards collaborative, efficient, and ethical AI-driven developments.
The key results are as follows:
FL-PedBrain, a platform for an MRI-based FL, performs on-par with the traditional CDS AI method for the concurrent classification and segmentation of pediatric posterior fossa brain tumors. Both, FL and CDS, approaches yield 20 to 30% higher performance improvements in segmentation compared to siloed learning from localized, limited data sources.
Heterogeneity is inherent in real-world medical image data and can be quantitatively described by class imbalances, MRI signal intensities, and even tumor sizes across different centers.
Despite data heterogeneity, FL-PedBrain achieves high generalization performance across 19 sites across the world.
Methods
This multi-center, retrospective study underwent approval by the Stanford University institutional review board (IRB) and execution of data use agreements across the participating sites, with a waiver of consent/assent (IRB No. 51059: Deep Learning Analysis of Radiologic imaging). Nineteen institutions from North America, Europe, West Asia, North Africa, and Australia participated in the study (Fig. 1a). Waiver of consent was granted by the IRB for the following reasons: (1) As a retrospective study, the research involves no more than minimal risk to the participants as the materials involved (data, documents, records) have already been collected and precautions will be taken to ensure confidentiality, (2) the waiver will not adversely affect the rights and welfare of the participants as there are procedures in place that protect confidentiality, and (3) the information learned during the study will not affect the treatment or clinical outcome of the participants.
The inclusion criteria were: patients who presented with a new, treatment-naive PF tumor; had pathologic confirmation for any of the following benign or malignant tumors: medulloblastoma (MB); ependymoma (EP); pilocytic astrocytoma (PA); and in the case of diffuse intrinsic pontine glioma (DIPG), MRI and/or biopsy-based diagnosis; obtained pre-treatment brain MRI that included axial T2-weighted imaging (T2-MRI). Subjects were excluded if the imaging was non-diagnostic due to severe motion degradation or other artifacts. Table 1 summarizes cohort demographics and site-specific tumor pathology.
Tumor segmentation was performed on axial T2-MRI by an expert board-certified, pediatric neuroradiologist (KY, >15 years’ experience), followed by a consensus agreement among three pediatric neuroradiologists (AJ, JW, MK) and two pediatric neurosurgeons (SC, RL). Segmentation was performed over the whole tumor, inclusive of cystic, hemorrhagic, or necrotic components within the tumor niche. T2-MRI was selected as it is most frequently acquired on routine MRI protocols; is embedded within pre-surgical navigation; and most reliably identifies the tumor margins regardless of enhancement, hence, recommended for pediatric glioma assessment29.
MRI acquisition
MRI of the brain was obtained using either 1.5 or 3 T MRI systems. The following vendors were employed across sites: GE Healthcare, Waukesha, WI; Siemens Healthineers, Erlangen, Germany; Philips Healthcare, Andover, MA; and Toshiba Canon Medical Systems USA Inc., Tustin, CA. The T2-weighted MRI (T2-MRI) sequence parameters were: T2 TSE clear/sense, T2 FSE, T2 propeller, T2 blade, T2 drive sense (TR/TE 2475.6-9622.24/80-146.048); slice thickness 1–5 mm with 0.5 or 1 mm skip; matrix ranges of 224–1024 × 256–1024.
Study design
Dataset distribution
Of the 19 sites, 16 sites were selected to participate in the model training and validation; the remaining three sites served as independent, external hold-out sites. A dataset from a database of normal pediatric brain MRI (N = 1667 from ST site) was used for pretraining. Within each of the 16 sites that participated in model training and validation, 75% of the MRI data was used in the training set; the remaining 25% was used as hold-out validation sets. Sample collection on sex and/or gender were not considered for sample selection.
Statistics and reproducibility
No statistical method was used to predetermine sample sizes of the training, validation, and external, independent validation sites. All data collected from the 19 sites were used. The training runs showed minor variations in convergence for different random seeds.
Data preprocessing
Each site must possess the small but important knowledge to manage consistent data preprocessing, a task that, under CDS, would typically be centralized by a trusted party. To streamline preprocessing, we have minimized any complex preprocessing steps (e.g., brain registration to a common atlas or skull-stripping). Preprocessing only includes: (1) normalization of each 3D image to a simple 0–255 intensity range and (2) volume extraction of 64 congruent axial slices of 256 × 256. These preprocessing steps are executed via an automated script applied to the DICOM data across all 19 sites. The number of 64 slices was chosen such that it can handle virtually all of the variations of the individual sites’ T2 sequence parameters (e.g., TSE, FSE, Propeller, etc.) with a large range of slice thicknesses (e.g.,1–5 mm) based on site-specific scanner technology and protocols. Therefore, our FL system can accommodate a large range of sequence parameters and axial slices. While normal pediatric MRI data of the pediatric brain were not required in the FL experiments, we observed that it could help retrain the model to identify the geometry and spatial locations of the pediatric brain across all ages, i.e., infants to adult head sizes of teenagers. The normal dataset (N = 1667) was shared and distributed amongst the participating sites for both CDS and FL approaches. However, the normal cohort was not used in the validation or the hold-out test sets.
Federated model development and evaluation
We developed a 3D model that jointly performs tumor pathology prediction (MB, EP, PA, DIPG, normal) and segmentation masks using FL (Fig. 1b). In the CDS approach, we combined the datasets from all 16 sites into a single pool, on which we trained the model. We also examined a Siloed model trained using the training and validation data from a single site only (Site ST, which hosted the largest single institution dataset), which was then evaluated on the 16 hold-out validation sets and 3 external independent sites. In contrast, the FL strategy used a method known as Federated Averaging15. Within this framework, the 16 sites did not share data. Instead, they only share information via model parameters learned on each site-specific data.
Each FL round began with local model training at the individual sites, after which each site transmitted the learned weights back to a central server. Here, the model weights from each of the 16 sites were averaged, creating a unified, global set of weights. These weights were then distributed back to each site to initiate the next FL round, where local training resumed. This iterative process, alternating between local training and centralized averaging, continued through many FL rounds. Eventually, the finalized global model underwent evaluation across the 16 validation sets and three hold-out test sets, its performance reflecting the collaborative—yet segregated—approach that characterizes the FL paradigm.
We modified the conventional FL strategy by creating a “warm-up” phase for the initial model, called Federated Warm-up, which enabled an efficient FL training to hasten convergence, given the large disparity in data distributions that underlie the 16 participating centers. The FL training consists of two stages enabling efficient learning: an initial 50 rounds of Federated Averaging on the ST and SE sites followed by 150 additional rounds of Federated Averaging across all 16 sites. A convergence plot that illustrates this Federated Averaging “warm-up” is shown in Fig. 5.
We employed a 3D-UNet architecture, incorporating a Kinetics-pretrained encoder that was initially trained on large-scale video data30. The 3D architecture allowed for processing 64 slices of high-resolution planes per datum, necessitating substantial GPU memory to manage large batch sizes. For the CDS training, 200 epochs were conducted with a combined loss function of Cross-Entropy and Dice Score Loss, utilizing Adam Optimization with a learning rate of 0.0001. This combined loss function facilitated the learning of both classification and segmentation predictions.
For classification performance, we calculated model raw accuracies and F1 scores. For segmentation, we utilized the same model as in the classification task to calculate the DSCs. The DSC determines the overlap between the predicted and ground-truth segmentations and thus offers insights into the quality of segmentation. We also conducted a two-sided t-test on the DSCs and compared the performance between CDS and FL. The distribution of predictions is approximately normally distributed due to the large test sample sizes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We extend our gratitude to Raju and Bala Vegesna for their generous support of this work. We appreciate Amazon Web Services for their computing support. We also are extremely thankful to Brendan Kelly for his feedback on our manuscript. We thank Jonathan Duh for his contribution. M.Z. was funded by the National Institutes of Health (5T32CA009695-27). S.H.C. was supported by the Kathryn S.R. Lowry Endowed Chair to the University of Utah, Department of Neurosurgery. All research at GOSH NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR GOSH Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. V.R. was supported by the Canadian Institutes for Health Research, the Canadian Cancer Society Emerging Scholars Award, the Garron Family Cancer Centre, and the C.R. Younger Foundation. MW and NDF were supported by the Canadian Institutes of Health Research (Project Grant 462169). K.W.Y. was supported by the American Brain Tumor Association (DG1800019). L.P. was supported by Chambers-Okamura Faculty Scholarship in Pediatric Neurosurgery, the Shurl and Kay Curci Foundation, and Stanford Maternal & Child Health Research Institute.
Author contributions
All authors read and proofread the manuscript. E.H.L., K.W.Y. and L.M.P. conceived the study conception and design, and E.H.L. and K.W.Y conducted the experiments and analyzed the findings. Material preparation and data collection were performed by E.H.L., K.W.Y., L.M.P., M.H., J.W., M.K., J.M., G.F., O.C., U.R., M.Z., M.W.W., R.G., S.T., S.P., H.D., E.A., M.M., K.A.S., C.J.C., H.L., A.E., A.R., K.M., K.A., M.S., A.V., O.O., B.E.W., T.P., E.M.T., C.Y.H., A.J., J.C., V.R., S.H.C., G.A.G., S.S.W., M.E.M., R.M.L., M.W., N.D.F., N.A.V., J.H.M. and K.W.Y.
Peer review
Peer review information
Nature Communications thanks Hamid Tizhoosh and the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The MRI data in this study have been deposited in the Stanford Research Database at 10.25740/bf070wx6289. The dataset used to develop the model consisting of approximately 1200 patients with pediatric brain tumors is included and organized by site. The remaining patients used as the test sets will be included in the near future as we have plans to organize federated learning challenges. The test set data, along with pediatric brain normal, can be requested by the reader by emailing the authors. Furthermore, clinical factors and notes, including sex and age for each data sample, will also be provided in the near future. Please see the project link for any future updates. Source data are provided with this paper.
Code availability
We share the FL codebase related to our pediatric brain tumor machine-learning algorithm as part of this journal submission to foster future validation and research in this domain, as well as for other tumor types. The project link is https://github.com/edhlee/FL-PedBrain, which contains a video and interactive web-based demo. This link also contains data and any updates.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Laura M. Prolo, Kristen W. Yeom
Contributor Information
Edward H. Lee, Email: edward.heesung.lee@gmail.com
Laura M. Prolo, Email: lmprolo@stanford.edu
Kristen W. Yeom, Email: kyeom@stanford.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51172-5.
References
- 1.Mukherjee, P. et al. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat. Mach. Intell.2, 274–282 (2020). 10.1038/s42256-020-0173-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imag.34, 1993–2024 (2015). [DOI] [PMC free article] [PubMed]
- 3.Park, A. et al. Deep learning–assisted diagnosis of cerebral aneurysms using the HeadXNet model. JAMA Netw. Open2, e195600 (2019). 10.1001/jamanetworkopen.2019.5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, M. et al. MRI radiogenomics of pediatric medulloblastoma: a multicenter study. Radiology304, 406–416 (2022). 10.1148/radiol.212137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajpurkar, P. et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. https://arxiv.org/abs/1711.05225 (2017).
- 6.Lee, E. H. et al. Federated learning on heterogeneous data using chest CT. https://arxiv.org/abs/2303.13567 (2023).
- 7.Lee, E. H. et al. Deep covid detect: an international experience on Covid-19 lung detection and prognosis using chest CT. NPJ Digit. Med.4, 11 (2021). 10.1038/s41746-020-00369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, K. et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell181, 1423–1433 (2020). 10.1016/j.cell.2020.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quon, J. L. et al. Artificial intelligence for automatic cerebral ventricle segmentation and volume calculation: a clinical tool for the evaluation of pediatric hydrocephalus. J. Neurosurg. Pediatr.27, 131–138 (2020). 10.3171/2020.6.PEDS20251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam, L. T. et al. MRI-based radiomics for prognosis of pediatric diffuse intrinsic pontine glioma: an international study. Neuro-oncology advances 3, no. 1. 2021). [DOI] [PMC free article] [PubMed]
- 11.Kelly, B. et al. DEEP MOVEMENT: Deep learning of movie files for management of endovascular thrombectomy. Eur. Radiol.33, 5728–5739 (2023). 10.1007/s00330-023-09478-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, M. et al. Machine-learning approach to differentiation of benign and malignant peripheral nerve sheath tumors: a multicenter study. Neurosurgery89, 509 (2021). 10.1093/neuros/nyab212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, M. et al. Radiomic signatures of posterior fossa ependymoma: molecular subgroups and risk profiles. Neuro-Oncol.24, 986–994 (2022). 10.1093/neuonc/noab272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, J. et al. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, pp. 248–255. 2009).
- 15.McMahan, B. et al. Communication-efficient learning of deep networks from decentralized data. In: Artificial intelligence and statistics. PMLR. https://arxiv.org/abs/1602.05629 (2017).
- 16.Choudhury, O., Park, Y., Salonidis, T., Gkoulalas-Divanis, A. & Sylla, I. Predicting adverse drug reactions on distributed health data using federated learning. AMIA Annu. Symp. Proc.2019, 313 (2019). [PMC free article] [PubMed] [Google Scholar]
- 17.Yeganeh Y., Farshad A., Navab N., Albarqouni S. Inverse distance aggregation for federated learning with non-iid data. In: Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning: Second MICCAI Workshop, DART 2020, and First MICCAI Workshop, DCL 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings 2, pp. 150–159. Springer International Publishing (2020).
- 18.Feki, I., Ammar, S., Kessentini, Y. & Muhammad, K. Federated learning for COVID-19 screening from chest X-ray images. Appl. Soft Comput.106, 107330 (2021). 10.1016/j.asoc.2021.107330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, B. et al. Robustly federated learning model for identifying high-risk patients with postoperative gastric cancer recurrence. Nat. Commun.15, 742 (2024). 10.1038/s41467-024-44946-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheller, M. J. et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep.10, 12598 (2020). 10.1038/s41598-020-69250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pati, S. et al. Federated learning enables big data for rare cancer boundary detection. Nat. Commun.13, 7346 (2022). 10.1038/s41467-022-33407-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo, G. et al. Influence of data distribution on federated learning performance in tumor segmentation. Radiol. Artif. Intell.5, e220082 (2023). 10.1148/ryai.220082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maaten, L. V. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res.9, 11 (2008). [Google Scholar]
- 24.Kalra, S., Wen, J., Cresswell, J. C., Volkovs, M. & Tizhoosh, H. R. Decentralized federated learning through proxy model sharing. Nat. Commun.14, 2899 (2023). 10.1038/s41467-023-38569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, T. et al. Federated optimization in heterogeneous networks. Proc. Mach. Learn. Syst.2, 429–450 (2020). [Google Scholar]
- 26.Ye, M. et al. Heterogeneous federated learning: State-of-the-art and research challenges. ACM Comput. Surv.56, 1–44 (2023). [Google Scholar]
- 27.Shao, J., Wu, F. & Zhang, J. Selective knowledge sharing for privacy-preserving federated distillation without a good teacher. Nat. Commun.15, 349 (2024). 10.1038/s41467-023-44383-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahimi, M. M. et al. EvoFed: leveraging evolutionary strategies for communication-efficient federated learning. Advances in Neural Information Processing Systems, 36, https://arxiv.org/abs/2311.07485 (2024).
- 29.Erker, C. et al. Response assessment in paediatric high-grade glioma: recommendations from the response assessment in pediatric neuro-oncology (RAPNO) working group. Lancet Oncol.21, 317–329 (2020). 10.1016/S1470-2045(20)30173-X [DOI] [PubMed] [Google Scholar]
- 30.Carreira, J., and Zisserman, A. Quo vadis, action recognition? a new model and the kinetics dataset. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6299–6308 (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MRI data in this study have been deposited in the Stanford Research Database at 10.25740/bf070wx6289. The dataset used to develop the model consisting of approximately 1200 patients with pediatric brain tumors is included and organized by site. The remaining patients used as the test sets will be included in the near future as we have plans to organize federated learning challenges. The test set data, along with pediatric brain normal, can be requested by the reader by emailing the authors. Furthermore, clinical factors and notes, including sex and age for each data sample, will also be provided in the near future. Please see the project link for any future updates. Source data are provided with this paper.
We share the FL codebase related to our pediatric brain tumor machine-learning algorithm as part of this journal submission to foster future validation and research in this domain, as well as for other tumor types. The project link is https://github.com/edhlee/FL-PedBrain, which contains a video and interactive web-based demo. This link also contains data and any updates.