Abstract
Purpose:
Classic Hodgkin lymphoma (cHL) is a B-cell lymphoma that occurs primarily in young adults and, less frequently, in elderly individuals. A hallmark of cHL is the exceptional scarcity (1%–5%) of the malignant Hodgkin Reed–Sternberg (HRS) cells within a network of nonmalignant immune cells. Molecular determinants governing the relationship between HRS cells and their proximal microenvironment remain largely unknown.
Experimental Design:
We performed spatially resolved multiplexed protein imaging and transcriptomic sequencing to characterize HRS cell states, cellular neighborhoods, and gene expression signatures of 23.6 million cells from 36 newly diagnosed Epstein–Barr virus (EBV)-positive and EBV-negative cHL tumors.
Results:
We show that MHC-I expression on HRS cells is associated with immune-inflamed neighborhoods containing CD8+ T cells, MHC-II+ macrophages, and immune checkpoint expression (i.e., PD1 and VISTA). We identified spatial clustering of HRS cells, consistent with the syncytial variant of cHL, and its association with T-cell–excluded neighborhoods in a subset of EBV-negative tumors. Finally, a subset of both EBV-positive and EBV-negative tumors contained regulatory T-cell–high neighborhoods harboring HRS cells with augmented proliferative capacity.
Conclusions:
Our study links HRS cell properties with distinct immunophenotypes and potential immune escape mechanisms in cHL.
Translational Relevance.
Classic Hodgkin lymphoma (cHL) is a B-cell lymphoma that occurs primarily in young adults and, less frequently, in elderly individuals. Cure rates with radiation therapy and multiagent chemotherapy exceed 90%. However, treatment is associated with a life-long risk of secondary malignancies, cardiac dysfunction, and other complications. Immune-directed therapies with antibody–drug conjugates and immune checkpoint inhibitors have been shown to be effective as second-line therapies, but further improvements will require a deeper understanding of the tumor-immune architecture of cHL. We performed an in situ analysis of malignant Hodgkin Reed–Sternberg cells and their proximal immune microenvironment in newly diagnosed and previously untreated cHL. We identified differences in the tumor microenvironment of Epstein–Barr virus (EBV)-positive and EBV-negative cHL that may be relevant for future immunotherapy approaches if confirmed in larger patient cohorts.
Introduction
Hodgkin Reed–Sternberg (HRS) cells, the malignant cells in classic Hodgkin lymphoma (cHL), constitute only 1% of cells in the tumor mass and are surrounded by an extraordinarily heterogeneous tumor microenvironment (TME; refs. 1, 2). The rarity of HRS cells and their inability to grow in culture (3) has posed a considerable technical challenge in characterizing the molecular pathogenesis of cHL. The application of single-cell technologies to study cHL has enabled the identification of specific immune cell populations within the cHL TME, including PD1+ T helper 1 (Th1)-polarized T effector cells (4), PD1− CD4+ regulatory T cells (Treg; ref. 4), and immunosuppressive LAG3+ T cells (5), and recently, identification of cell–cell interactions, including PDL1+ tumor-associated macrophages with PD1+ CD4+ helper T cells (6) and CTLA4+ T cells with CD86+ (CTLA4 ligand) HRS cells and tumor-associated macrophages (7, 8). Most recently, sequencing of circulating tumor DNA in patients with cHL has revealed two distinct cHL genomic subtypes (9). Much of this work has been accomplished through the ex-vivo characterization of isolated cells or tumor cell products, and thus, spatial interactions between HRS cells and their proximal immune microenvironment remain incompletely understood.
Approximately 30% of cHL cases occur in the setting of prior Epstein–Barr virus (EBV), a virus that infects nearly 95% of the world’s population. However, only a small proportion of these individuals will develop an EBV-associated malignancy, including cHL (10–13). Essentially all malignant cells in EBV-positive cancers express EBV-related genes, including Epstein–Barr nuclear antigen 1, latent membrane protein (LMP) 1 and 2, EBV-encoded small RNAs (EBER), and EBV BART-region micro RNAs (14). Interestingly, EBV not only potently transforms B cells, but it also hyperactivates the cellular immune response more than any other tumor-associated virus. This perhaps explains why only a very small number of EBV-infected individuals develop EBV-associated cancers (15), raising the question of how tumor cells survive within a particularly hostile TME.
Several molecular mechanisms may enable HRS cells to escape lethal attacks from surrounding cytotoxic CD8+ T cells and natural killer (NK) cells, including loss of MHC class I (MHC-I) expression, expression of cell surface molecules that impair CD8+ T-cell or NK-cell function (e.g., PDL1 and CD95L), attraction of immunosuppressive Tregs and macrophages, and secretion of immunosuppressive molecules (16). To better understand the relationship between HRS cells and their neighboring immune cells, we generated an integrated dataset of multiplexed protein imaging and transcriptomic data, spanning over 23 million cells from 36 newly diagnosed cHLs, including both EBV-positive and EBV-negative cHL.
Materials and Methods
Patients and tissue
This study includes newly diagnosed specimens from patients with cHL who were admitted to Memorial Sloan Kettering Cancer Center (MSKCC) from 2012 to 2017 (n = 36; Supplementary Table S1). All patients signed statements of informed consent under protocols approved by the MSKCC Institutional Review Board (IRB; IRB number 21-269). The study was conducted in accordance with the Declaration of Helsinki. Informed written consent was obtained from each participant. All tumors were surgically resected and immediately formalin-fixed, paraffin-embedded (FFPE) with standard tissue processing in the MSKCC surgical pathology lab (Clinical Laboratory Improvement Amendments accredited). FFPE blocks were maintained in the MSKCC Department of Pathology temperature-controlled storage units. Adjacent FFPE tissue sections were freshly cut for hematoxylin and eosin (H&E; one section, 5 microns), multiplexed immunofluorescence (mpIF; one section, 3 microns), IHC (three sections, 5 microns), and NanoString (five sections, 10 microns). H&Es were reviewed by a board-certified pathologist (Ahmet Dogan). EBV status for each patient was determined through EBER in situ hybridization and IHC for latent membrane protein 1 (LMP1). The normal human tissue microarrays for antibody validation were processed and consented to as described above.
H&E staining
H&E staining was performed using the Ventana Symphony automated H&E stainer with standard clinical protocol. Tissue sections were baked for 1 hour at 60°C, hydrated, stained with hematoxylin (Leica catalog #3801560), stained with bluing reagent (Leica catalog #3802918), stained with eosin counterstain (Leica catalog #3801600), rinsed, dehydrated, and coverslipped.
Multiplexed immunofluorescence: antibody conjugation, staining, and data acquisition
Selection of primary antibody clones for the mpIF panel of 29 proteins; conjugation of primary antibodies to Cy2, Cy3, or Cy5 Bis NHS Ester dyes (GE catalog #PA22000, PA13000, or PA25000, respectively); validation of the conjugated primary antibody; and testing of epitope stability to alkaline H2O2-based signal inactivation on normal human tissue microarrays was performed using previously described methods (Supplementary Fig. S1; ref. 17). CD15 and Pax5 were not included for HRS cell identification due to poor antibody conjugation. The antibody clones, dye/protein ratios, and mpIF staining concentrations are listed in Supplementary Table S2. mpIF (Cell Dive) was performed for 30 out of 36 tumors based on tissue availability using previously described methods (17). A normal human tissue microarray containing tonsil, placenta, colon, skin, and spleen (at least one positive and one negative control for each of the 29 proteins) was included on each cHL mpIF slide to assess the quality and specificity of each marker for each slide. Fields of view (FOV) were placed evenly throughout the tumor tissue to capture intratumor heterogeneity. The order of the markers in the Cell Dive panel was determined based on epitope stability to hydrogen peroxide signal deactivation. Tissue sections underwent 14 cycles of background imaging, staining, imaging, and signal inactivation. Images were acquired using the Cytell Cell Imaging System (Cytiva, Issaquah, WA). Image App software was used for image acquisition and registration (using DAPI). An acquired background image following each cycle of dye inactivation was used to subtract autofluorescence from the subsequent stain round.
Multiplexed immunofluorescence analysis
Image analysis
Indica Labs’ HALO Image Analysis software (RRID:SCR_018350) was used for image visualization, low-level image annotation, cell segmentation, and marker thresholding. For each FOV, images for the 29 markers and DAPI were stacked. Markers with technical issues or unspecific staining (based on comparison to the on-slide normal human tissue microarray) were excluded from analysis for all FOVs of the sample. Regions within each marker channel of all FOVs containing high-intensity artifacts (e.g., folded tissue and dust spots) were annotated for downstream exclusion. A custom nuclear segmentation algorithm using HALO Image Analysis software was pretrained using 10 manually annotated cHL FOVs each containing approximately 10,000 cells to overcome the difficulty of cell segmentation in this high-density tumor type. Nuclear segmentation quality was assessed and optimized in a minimum of two FOVs from each sample (apart from HL_17, which had one FOV). Manual thresholds for marker positivity were set for each marker in each sample on the mean cytoplasm (for cytoplasmic markers) or mean nucleus (for nuclear markers) pixel intensity in reference to marker expression on the on-slide normal human tissue microarray and again assessed and optimized in a minimum of two FOVs from each sample. Cells from additional FOVs were visually assessed for samples with larger numbers of FOVs. In each sample, a random sampling of cells across different regions of the tissue defined as positive and negative for each marker was examined to ensure that the thresholds were representative.
Cell loss computation
DAPI images from each cycle were processed with intensity normalization by histogram matching. The sum of the squared differences was used to generate a pixel-level bit mask image indicating areas of cell loss/drift between the first DAPI image and each subsequent DAPI image from all cycles. This bit mask and cell coordinates were used to calculate a loss/drift percentage for each cell in every cycle of imaging. Cells with greater than 10% loss/drift of pixels were flagged for removal from the dataset, and the cell loss percentage for each sample was calculated.
Initial data processing
Each cell was assigned a unique ID. Cells in regions with artifacts, within the 20-micron border region of each FOV, or with greater than 10% loss/drift of pixels were removed from all analyses. For all pairs of cells within a 50-micron radius, the pairwise distance between the cells’ cellular centroids (determined following nuclear segmentation) was calculated for downstream spatial analyses. Intensity values for each marker were normalized at the sample level by dividing the intensities for each marker by the value of the threshold. Intensity values below the threshold (less than 1) were set to 1. The log of the intensity values for each marker was divided by the value of the 97.5 percentile computed across each FOV.
Cell type assignment
Cells were assigned to a cell type using a two-step method. First, positive and negative combinations of cell identity markers were used to label cell types, which are hierarchically grouped by “Category” (Tumor vs. Immune), “Cell type,” and “Cell subtype” (Supplementary Table S3). Second, for recovery of HRS cells, recovery of unknown cells that did not fit a cell type definition, reassignment of T cell/null phenotype cells, and reassignment of CD4+/CD8+ T cells that occurred at higher than biologically expected percentages, a cell reassignment hierarchy was designed (https://github.com/mskcc/cHL-spatial-profiling/blob/main/reassignment_rules.md). For the rank comparison, we first computed the rank for each marker based on its single-cell intensities and then normalized to 0 to 1 using NormRank = (Rank + 1)/(nCells + 2). Next, in the case in which the pair of markers of interest were both positive (e.g., CD4+/CD8+ T cells), we compared the ranks and switched the marker with the lower rank value to negative. In the case in which the pair of markers of interest were both negative (e.g., T cell/null phenotype), we compared the ranks of CD4 and CD8 and switched the marker with the higher rank to positive. Lastly, in the case in which we wanted to preserve a subset of the cell type being reassigned (e.g., we expect 5% of the T cells to be CD4+/CD8+ T cells), we selected the cells to reassign by computing the log odds ratio (OR) of the normalized ranks and selected the cells with the largest difference in their log OR for reassignment/marker positivity “flipping.” Following cell type labeling, because our mpIF panel was not sufficiently extensive for labeling every immune cell type in the body, 4.7% of total cells were labeled “other leukocyte” based on positivity for only CD45. Additionally, 5.6% of total cells were labeled “negative” based on positivity for no cell identity markers. Again, our mpIF panel does not include the full range of stromal markers explaining this “negative” population. Finally, because of the difficulty in cell segmentation of the lymph node given the high density of cells, 17.7% of cells were labeled “unknown” based on positivity for markers signifying at least two cell lineages.
Dimensionality reduction
Based on marker data availability in the greatest number of FOVs, the normalized intensities of 23 out of the 29 markers were used in the Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) embedding (ICOS, B7H3, VISTA, CD40, CD40L, and MHC-I were excluded). The cells containing information on these 23 markers represent 24 out of 30 patients. A weighted-down sampling of cell subtypes was performed using the function sample_n from the package dplyr. The weight for each cell is 1/sqrt (n_type) where n_type is the count for the cell subtype. A total of 1,276,712 cells were used in the total cell UMAP embedding and 200,012 cells were used in the HRS cell UMAP embedding.
Statistical analysis
Fractions of each cell state (Supplementary Table S4) were transformed to log odds at the FOV level and R function wilcox.test () with the default two-sided option was used to compute the significance of the differences and effect sizes as log ORs. P values were adjusted for multiple testing with Bonferroni adjustment separately for immune cell states and tumor cell states. A cell state fraction was considered statistically significant if the adjusted P value was less than 0.05 and biologically significant if the median fraction difference between the two groups was greater than 0.1, the minimum cell state (numerator) count was 300 cells, and the minimum population (denominator) count was 1,000 cells. For spatially restricted statistics, fractions were computed at the FOV level for cells within 30 microns of the cell of interest (e.g., HRS cell).
Hodgkin Reed–Sternberg cell neighborhoods
Immune cells were grouped by being in a specific HRS neighborhood (e.g., syncytial HRS cell neighborhood) based on neighborhoods (30-micron radius) having a minimum of 95% HRS cells of that specific category (e.g., 95% of HRS cells in a syncytial HRS cell neighborhood are HRS cells in an aggregate). The specific HRS neighborhoods characterized were EBV-positive, EBV-negative, syncytial, non-syncytial, and HRS cells positive and/or negative for the following combinations: B2M+/MHC-I+, B2M−/MHC-I−, B2M+/MHC-I+/MHC-II+/PDL1+, and B2M−/MHC-I−/MHC-II−/PDL1−.
Hodgkin Reed–Sternberg cell spatial topology classification
The Delaunay triangulation was run on all cellular coordinates in each FOV using the function triangulate from package RTriangle. Two HRS cells were considered neighbors either if they shared an edge in the triangulation graph or if there was at least one path with just one non-HRS cell between them. We then constructed a new graph connecting neighboring HRS cells as just defined. This graph was then split into its connected components using the function components from the package igraph. This resulted in disjoint sub-graphs of connected HRS cells (a minimum of two HRS cells per connected component). In total, there were 40,680 HRS connected components of which the majority contained <20 HRS cells (specifically 39,611 or 97.2% of the connected components). Given this, we then defined an HRS aggregate to be any sub-graph of 20 or more HRS cells.
Coefficient of variation
The coefficient of variation (CoV) was used as a measure of intratumor heterogeneity. First, the fraction of HRS cells in each FOV f in state s () was computed.
Next, the CoV of this fraction () for each patient p and state s was computed by taking mean () and standard deviation () of the FOVs in patient p.
Note: The number of FOVs in patient p is denoted by .
IHC staining and analysis
IHC for B2M, MHC-I, and MHC-II was performed for all tumors using an automated staining system (Leica Bond RX) and previously described methods (17). The same primary antibody clones used for mpIF were used for IHC (Supplementary Table S2). The staining was scored exclusively in HRS cells as positive, cytoplasmic, or negative for B2M and MHC-I and positive or negative for MHC-II. Slides were scored blindly by a board-certified hematopathologist (Ahmet Dogan).
NanoString targeted RNA sequencing and analysis
NanoString was performed for all but four tumors, which were excluded due to low RNA quantity post-extraction. RNA extraction was performed on the entire FFPE tissue section. FFPE sections were deparaffinized and RNA extracted using previously described methods (17). A minimum of 100 ng of RNA per sample was used to measure the expression of 750 immune-related genes and 20 internal reference genes (PanCancer IO 360 gene expression panel) using NanoString Technologies’ nCounter platform. Normalization using the internal reference genes was performed using nSolver. The deconvolution of immune cell subtypes from their gene expression profiles was performed using CIBERSORT (18). Differential expression analysis was run using the DESeq2 (RRID:SCR_015687) Bioconductor (RRID:SCR_006442) package. The overlap of differentially expressed genes was computed against Hallmark, Reactome, and KEGG (RRID:SCR_012773) gene sets from the Molecular Signatures Database (MSigDB). Gene sets are abbreviated in figures and the text as the following:
-
1
Antigen processing/presentation: KEGG_Antigen_processing_and_presentation
-
2
Interaction lymphoid/non-lymphoid: REACTOME_Immunoregulatory_interactions_between_a_lymphoid_and_a_non_lymphoid_cell
-
3
NK cell–mediated toxicity: KEGG_Natural_killer_cell_mediated_cytotoxicity
-
4
Interferon alpha/beta/gamma: union of
-
a.
HALLMARK_Interferon_gamma_response
-
b.
HALLMARK_Interferon_alpha_response
-
c.
REACTOME_Interferon_signaling
-
d.
REACTOME_Interferon_gamma_signaling
-
e.
REACTOME_Interferon_alpha_beta_signaling
-
a.
-
5
Hepatitis B virus (HBV) infection: WIELAND_UP_BY_HBV_INFECTION (19)
-
6
Epithelial–mesenchymal transition: HALLMARK_Epithelial_mesenchymal_transition
Code availability
All code and detailed computational methods are publicly available at https://github.com/mskcc/cHL-spatial-profiling.
Data availability
All data supporting the findings of this study are publicly available at https://zenodo.org/records/10659311.
Results
Patient cohort and study design
We assembled a retrospective case series encompassing 36 patients with newly diagnosed cHL. All patients signed statements of informed consent under protocols approved by the MSKCC IRB. Most tumors were histologically classified as nodular sclerosis (NS), the most common subtype of cHL (Fig. 1A; Supplementary Table S1). Seven of 36 tumors expressed EBER RNAs and LMP1 protein. As expected, a larger fraction of EBV-positive tumors (3/7), compared only with 1/29 EBV-negative tumors, were of the mixed cellularity (MC) morphologic variant.
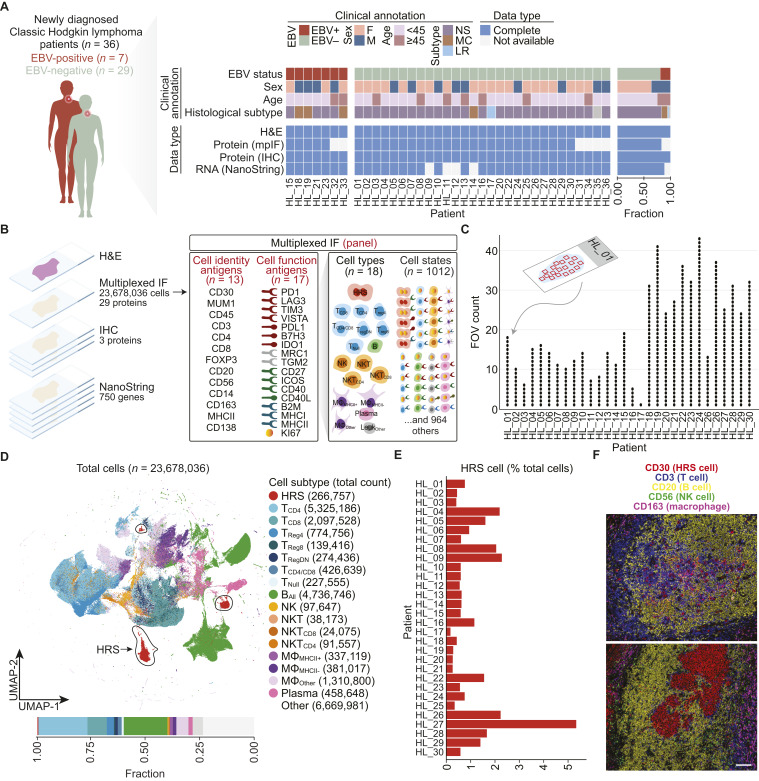
Figure 1.
Multidimensional molecular profiling of newly diagnosed cHL. A, Clinical characteristics of patients with cHL and their data availability. Tumors are grouped by EBV status. Gray (clinical annotation) indicates unknown characteristics. The bar graph (right) indicates the cohort-level summary. B, Study design. Derivation of cell types and cell states from cell identity and cell function antigens using mpIF. For cell type abbreviations, see Supplementary Table S3. C, Summary of FOV counts per patient. D, UMAP of all cells (n = 1,276,712 subsampled from 23,678,036) profiled by mpIF colored by cell subtype. HRS clusters are circled in the UMAP. Bar plot indicating the percentage of each cell type over total cells. E, Bar plot indicating the percentage of HRS cells over total cells in each patient. F, Representative mpIF FOVs overlaying five markers delineating major cell types from 2 patients (HL_15 FOV 14, HL_21 FOV 5). Scale bar, 100 microns. (A, Adapted from images by GraphicsRF/stock.adobe.com and wowow/stock.adobe.com). F, female; LR, lymphocyte rich; M, male.
We performed mpIF on 30 patient samples based on tissue availability, examining at single-cell resolution the co-expression of 29 proteins, of which 13 were cell identity–related and 17 were cell function–related (Fig. 1B; Supplementary Table S2).
For each protein, we validated the primary antibody clone, epitope stability to hydrogen peroxide signal inactivation, and conjugated primary antibody on normal human tissue microarrays (Supplementary Fig. S1; “Materials and Methods”). Additionally, each mpIF slide contained an adjacent section of the normal human tissue microarray for further quality control (“Materials and Methods”).
HALO Image Analysis software and a pretrained segmentation algorithm on 10 manually annotated cHL images each containing an average of 10,000 cells were used for cell segmentation (“Materials and Methods”).
Marker expression was categorized as positive and negative using per-marker thresholds and intensities for positive markers were normalized (“Materials and Methods”). We used a minimum of six different protein markers to identify each cell type. Cell type curation was based on the combination of positive cell staining for some markers and negative staining for other markers. For example, a CD8+ T cell was defined by positive staining for both CD8 and CD3 and the absence of staining with antibodies against CD4, FOXP3, CD20, CD56, CD30, and MUM1. We labeled 18 distinct cell types, including HRS cells and subpopulations of T cells, B cells, NK cells, macrophages, monocytes, and plasma cells (Fig. 1B; Supplementary Table S3; “Materials and Methods”).
Using combinations of cell types and positive and negative cell function antigens, we were able to label 1,012 unique cell states (Fig. 1B; Supplementary Table S4).
We examined an average of 20 FOVs per patient (median: 16 FOVs, range 1–43; Fig. 1C; Supplementary Table S5) with the total tumor area ranging from 6.4 to 153.3 mm2 (mean: 63.1 mm2, median: 34.6 mm2). Our method enabled us to examine a much larger tumor area than is typically examined in multiplexed imaging studies using tissue microarray cores.
Following the exclusion of a small percentage of cells that were displaced during mpIF staining (Supplementary Fig. S2; Supplementary Table S5), we determined in situ protein expression for 23,678,036 single cells within 587 high-dimensional FOVs (Supplementary Fig. S3).
To assess the global expression of functional markers and patient specificity of cell states, we used dimensionality reduction to visualize the mpIF data. The major cell lineages were well resolved in the UMAP (Fig. 1D) with intermixing of immune cells from all patients (Supplementary Fig. S4A). The percentage of HRS cells in each tumor ranged from 0.1% to 5.2%, with an overall percentage of about 1% HRS cells across all examined tumors (266,757/23,678,036 cells; Fig. 1E). HRS cells were embedded within an immune-rich TME with T cells and B cells being most abundant (Fig. 1D and F; Supplementary Table S6). We detected very rare populations of CD8+ regulatory T cells (<1% of the total cell population) and confirmed their existence in the mpIF images (Supplementary Fig. S4B). We observed extensive heterogeneity present in the composition of immune cell populations between both different tumors (Supplementary Fig. S4C) and within different FOVs of the same tumor (Supplementary Fig. S4D).
A subset of “other” cells could not be resolved into cell types and were classified as either “other leukocyte” based on positivity for only CD45, “negative” based on negativity for all cell identity markers, or “unknown” based on positivity for several cell identity markers of multiple lineages (Supplementary Table S6; “Materials and Methods”). The former category included cell types that could not be identified using our preselected panel of markers (e.g., eosinophils, mast cells, and granulocytes) and the latter category included cells that could not be unambiguously separated from other cells within a densely packed stroma.
Among co-inhibitory and co-stimulatory checkpoint receptors, CD27 and PD1 expression was limited to T cells and B cells, TIM3 was ubiquitously expressed in all cell types, and LAG3 and CD40L expression was rare (Supplementary Fig. S4E). Variability in PDL1 expression within the HRS cell population explained the separation of HRS cells into multiple clusters (Fig. 1D). We found a proliferating (Ki67+) subset within each cell population. Among antigen presentation proteins, beta-2 microglobulin (B2M) and MHC-I were expressed in all leukocytes.
Characterization of HRS cell states
The large number of cells in our cohort (>23 million) and the large tumor area captured by our spatial profiling platform provided an opportunity to characterize the protein expression of HRS cells, subsequently referred to as “HRS cell states,” and the spatial distribution of HRS cell states within their native TME.
The UMAP projection of all HRS cells showed widespread expression of B2M, MHC-I, MHC-II, CD40, TIM3, and PDL1, whereas expression of other cell function markers was restricted to smaller subsets of HRS cells (Fig. 2A). Although patient-specific clustering of immune cells was not observed in the UMAP projection, we did identify patient-specific clustering of HRS cells (Supplementary Fig. S5A).
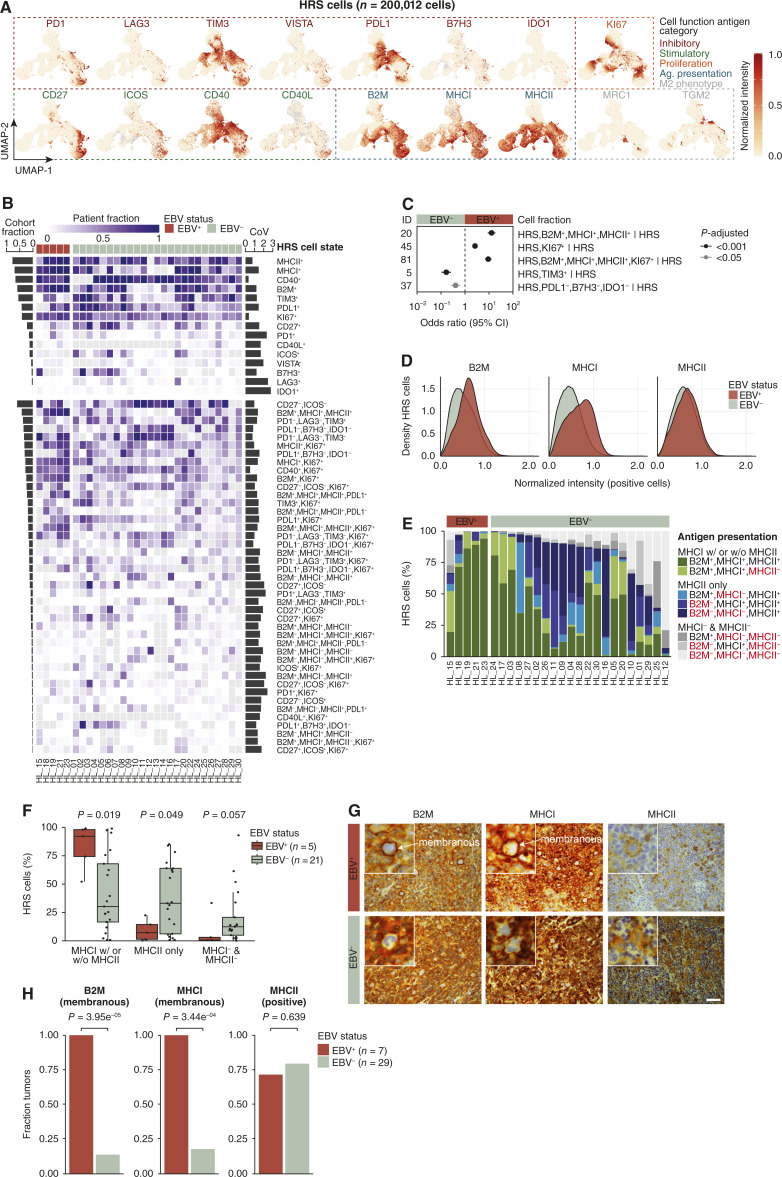
Figure 2.
Characterization of HRS cell states and their spatial heterogeneity. A, UMAP of HRS cells profiled by multiplexed immunofluorescence colored by normalized intensity of cell function antigens. B, Heatmap indicating the fraction of HRS cell states (over total HRS cells) at the patient level grouped by EBV status and ordered by decreasing cohort fraction depicted by the bar graph (left). Cell states with a minimum cohort fraction of 3% are included (except IDO1+). Gray indicates missing data. The bar graph (right) indicates the maximum CoV for each cell state for patients with a minimum of five FOVs (n = 29). See also Supplementary Table S7. C, Forest plot indicating effect size and 95% CI of statistically and biologically significant (see “Materials and Methods”) HRS cell fractions colored by P-adjusted (two-sided Wilcoxon test adjusted by Bonferroni correction) in the comparison of EBV-positive (n = 150 FOVs) and EBV-negative (n = 437 FOVs) patients. For exact P values, see Supplementary Table S8. D, Density plot indicating normalized intensity of B2M, MHC-I, and MHC-II. E, Stacked bar plot indicating percent of HRS cells positive for all combinations of B2M, MHC-I, and MHC-II. MHC-I with or without MHC-II refers to HRS cells positive for only MHC-I or both MHC-I and MHC-II. Patients with data for all three markers (n = 26) are shown. F, Box plot indicating the percent of HRS cells in three antigen presentation machinery states (two-sided Wilcoxon test). Each point represents one patient (box plot represents minimum, first quartile, median, third quartile, and maximum). G, B2M, MHC-I, and MHC-II IHC of an EBV-positive (HL_33) and EBV-negative (HL_14) tumor. Scale bar, 100 microns. H, Bar graph indicating the fraction of tumors expressing B2M, MHC-I, and MHC-II on HRS cells based on pathologist quantification of IHC staining (two-sided Fisher’s exact test). Ag, antigen; CI, confidence interval.
In total, we characterized 92 HRS cell states based on positivity for different combinations of 15 cell function markers. More than one-third of the 92 HRS cell states were very rare, each representing less than 3% of the HRS cells in the entire cohort, of uncertain biological significance, and likely only detectable due to the large number of cells profiled in our study (Supplementary Fig. S5B; Supplementary Table S7). Even some of the more common HRS cell states (e.g., MHC-II+ HRS cells) showed considerable heterogeneity between tumors (i.e., inter-patient heterogeneity; Fig. 2B).
The extraordinary depth of spatial profiling in our study provided an opportunity to characterize intratumoral heterogeneity in established cancer immunotherapy biomarkers such as PDL1 and MHC-I. For the subset of tumors for which were able to image a minimum of five FOVs (n = 29), we examined the spatial distribution of HRS cell states within each tumor (i.e., intra-patient heterogeneity). Some HRS cell states were distributed very unevenly throughout the tumor (i.e., high intratumor heterogeneity), whereas other HRS cell states were distributed very evenly (i.e., low intratumor heterogeneity; Supplementary Fig. S5C). For example, the fraction of MHC-II+ HRS cells in patient HL_01 ranged from 7.5% to 89.7% across 18 FOVs whereas the fraction of CD40+ HRS cells in patient HL_04 exhibited a much tighter range of 93.2% to 99.9% across 15 FOVs. Using CoV as a metric for intratumor heterogeneity (Fig. 2B; Supplementary Fig. S5B and S5D), we found CD40+ and Ki67+ HRS cells to be least spatially variable and IDO1+/Ki67+ and IDO1+ HRS cells to be most spatially variable within tumors (Supplementary Fig. S5E). PDL1, a common immunotherapy biomarker, exhibited variable intratumor heterogeneity among patients (Fig. 2B).
HRS cells in EBV-positive cHL maintain expression of MHC-I
Previous studies have reported decreased or absent expression of B2M and MHC-I in the majority of patients with cHL (up to 79%; ref. 20). Given that our initial assessment of HRS cell states demonstrated preserved MHC-I expression in most HRS cells from EBV-positive cHLs (Fig. 2B), we compared the co-expression of B2M, MHC-I, and MHC-II and all other HRS cell states across all HRS cells in EBV-positive and EBV-negative cHLs in our dataset.
Compared with HRS cells from EBV-negative cHLs, HRS cells from EBV-positive tumors exhibited increased Ki67 positivity, increased PDL1 positivity, and decreased TIM3 positivity (Fig. 2C; Supplementary Table S8). Most strikingly, HRS cells from EBV-positive tumors exhibited increased “triple-positive” expression of B2M+/MHC-I+/MHC-II+ [OR, 12.8; CI 8.86–18.5; P-adjusted 1.57E-29]. Even within the subgroup of B2M+/MHC-I+/MHC-II+ HRS cells, HRS cells from EBV-positive cHL tumors showed higher levels of B2M, MHC-I, and MHC-II protein expression (Fig. 2D).
We also examined various combinations of B2M, MHC-I, and MHC-II marker positivity at single-cell resolution in each tumor. HRS cells from EBV-positive tumors more frequently expressed MHC-I or both MHC-I and MHC-II, whereas HRS cells from EBV-negative tumors, in contrast, expressed only MHC-II or showed loss of both MHC-I and MHC-II antigen presentation machinery (Fig. 2E and F).
We performed immunohistochemical staining of our cHL cohort with antibodies against B2M, MHC-I, and MHC-II as an orthogonal approach for measuring these proteins. This further allowed us to closely evaluate the membranous expression of these markers in HRS cells, a feature that is not discernable using mpIF. Quantification by a hematopathologist blinded to the results of our mpIF analysis showed that all EBV-positive tumors, compared with less than 25% of EBV-negative tumors, exhibited membranous positivity for B2M and MHC-I in HRS cells (Fig. 2G and H). We did not observe distinct phenotypic features of the HRS cell states that lacked MHC-I expression.
Immune inflamed immunotype in MHC-I–positive cHL
Given the prominent role of MHC-I in regulating adaptive immunity (21) and the apparent loss of MHC-I expression in HRS cells from EBV-negative cHLs, we expected considerable differences between the proximal TME of HRS cells in EBV-positive versus EBV-negative cHLs. To further examine this question, we quantified cell states within a 30-micron radius of HRS cells, referred to as “HRS neighborhoods.” We specifically focused all neighborhood analyses on a 30-micron radius to restrict the number of cells within any given neighborhood to a maximum of 10 cells (if lymphocytes) across the diameter of the neighborhood given the overcrowded cellular landscape in cHL.
Compared with HRS neighborhoods in EBV-negative cHL, HRS neighborhoods in EBV-positive cHL contained increased CD8+ T cells (OR, 2.77; P-adjusted 1.24E-21), increased MHC-II+ macrophages (OR, 3.39; P-adjusted 4.98E-13), and decreased CD4+ helper T cells (OR, 0.403; P-adjusted 4.45E-26; Fig. 3A; Supplementary Table S8). Additionally, a larger fraction of CD8+ T cells in HRS neighborhoods in EBV-positive cHL were proliferating (Ki67+).
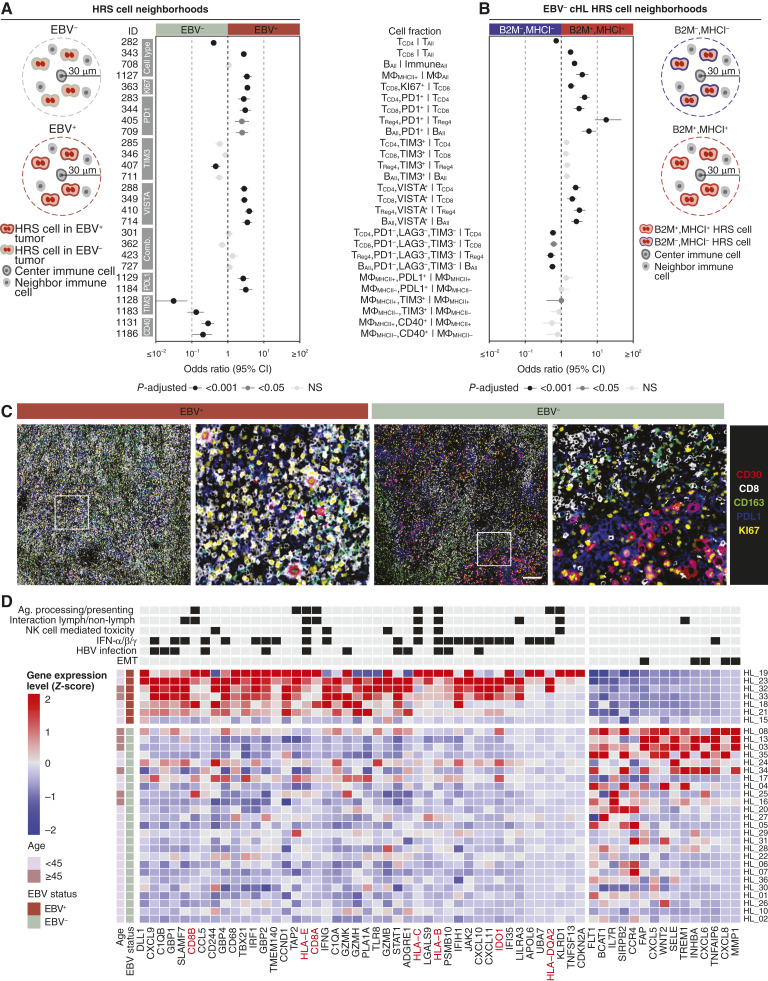
Figure 3.
EBV-positive cHL exhibits an immune-inflamed immunotype. A, Analysis approach to defining EBV-positive and EBV-negative HRS neighborhoods. The forest plot shows the effect size and 95% CI of immune cell fractions colored by P-adjusted (two-sided Wilcoxon test adjusted by Bonferroni correction) in the comparison of EBV-positive (n = 150 FOVs) and EBV-negative (n = 437 FOVs) HRS neighborhoods. For cell type abbreviations, see Supplementary Table S3. For exact P values, see Supplementary Table S8. B, Analysis approach to defining B2M/MHC-I positive and negative HRS cell neighborhoods. The forest plot shows the effect size and 95% CI of immune cell fractions colored by P-adjusted in the comparison of B2M+/MHC-I+ and B2M−/MHC-I− HRS neighborhoods in EBV-negative tumors. For exact P values, see Supplementary Table S10. C, Representative mpIF FOVs overlaying five markers from an EBV-positive (HL_19 FOV 3) and an EBV-negative tumor (HL_28 FOV 13). Scale bar, 500 microns. D, Heatmap indicating the scaled RNA expression values for differentially expressed genes (P-adjusted < 0.01) in EBV-positive (n = 7) vs. EBV-negative (n = 25) tumors sorted by fraction of samples with a Z-score in the same direction within each EBV group. Genes in red are in the mpIF panel. The annotated pathways represent pathways with the highest count of differentially expressed genes. See also Supplementary Table S11. EMT, epithelial–mesenchymal transition; IFN, interferon; Lymph, lymphoid.
In terms of immune checkpoint expression within each immune cell type, HRS neighborhoods in EBV-positive cHL contained increased fractions of (i) PD1+ and VISTA+ CD4+ helper T cells, CD8+ T cells, Tregs, and B cells (Supplementary Fig. S6A–S6D), and (ii) PDL1+ macrophages (MHC-II+ and MHC-II− macrophages; Fig. 3A). HRS neighborhoods in EBV-negative cHLs contained increased fractions of (i) TIM3+ Tregs, MHC-II+ macrophages, and MHC-II− macrophages, and (ii) CD40+ macrophages (MHC-II+ and MHC-II− macrophages; Fig. 3A).
Within the subgroup of EBV-positive cHLs, MC cHLs contained increased CD8+ T cells and decreased NK cells compared with NS cHLs (Supplementary Fig. S7; Supplementary Table S9). MC cHLs also harbored increased fractions of PD1+ CD4+ helper T cells, CD8+ T cells, B cells, NK, NKT, and CD8+ NKT cells, whereas NS cHLs harbored increased fractions of ICOS+ CD4+ helper T cells and CD4+ T regs and proliferating CD4+ T regs.
Our results thus far indicated a strong association between EBV status, expression of MHC-I in HRS cells, and the immune-inflamed immunotype. However, we also identified MHC-I–positive HRS cells in EBV-negative cHLs (Fig. 2E), which provided an opportunity to compare the relationship between MHC-I expression on HRS cells and HRS neighborhoods within the subgroup of EBV-negative cHLs. We observed a striking similarity between the neighborhoods of MHC-I–positive HRS cells in EBV-negative cHLs (Fig. 3B; Supplementary Table S10) and the neighborhoods of EBV-positive HRS cells (Fig. 3A). This suggests that MHC-I–positive HRS cells engender an inflamed tumor architecture even in the absence of EBV positivity.
We also compared the neighborhoods of HRS cells that were “quadruple-positive” for B2M+/MHC-I+/MHC-II+/PDL1+ with the neighborhoods of HRS cells that were “quadruple-negative” for all four markers in EBV-negative tumors (Supplementary Fig. S8; Supplementary Table S10). We again found increased CD8+ T cells, B cells, and MHC-II+ macrophages and decreased CD4+ helper T cells in “quadruple-positive” HRS neighborhoods, indicating that positivity for B2M/MHC-I is driving these differences in immune cell populations. Likewise, we found increased PD1+ and VISTA+ CD4+ helper T cells, CD8+ T cells, Tregs, and B cells, and decreased “triple-negative” PD1−/LAG3−/TIM3− CD4+ helper T cells, CD8+ T cells, Tregs, and B cells in the “quadruple-positive” HRS neighborhood. Unlike the B2M+/MHC-I+ HRS neighborhood (Fig. 3B), the “quadruple-positive” HRS neighborhood harbored increased fractions of TIM3+ CD4+ helper T cells, CD8+ T cells, Tregs, and B cells, and PDL1+ macrophages (both MHC-II+ macrophages and MHC-II− macrophages; Supplementary Fig. S8), indicating that positivity for B2M/MHC-I alone is not responsible for these immunophenotypes. Taken together, these neighborhood analyses suggest that expression of B2M/MHC-I, MHC-II, and PDL1 on HRS cells have independent and synergistic effects on their cellular neighborhoods.
Our data thus far demonstrated that HRS cells in EBV-positive cHL maintain expression of antigen presentation pathway proteins (B2M, MHC-I, and MHC-II) and are surrounded by activated CD8+ T cells and macrophages (Fig. 3C). To further characterize differences in the TME of EBV-positive and EBV-negative cHL, we next examined our transcriptomic data, specifically the expression of 750 immune-related genes in immediately adjacent tissue sections of the same tumors (Fig. 1B). One hundred and eleven genes were differentially expressed in EBV-positive and EBV-negative tumors (Fig. 3D; Supplementary Fig. S9A; Supplementary Table S11).
Among the differentially expressed genes, 22/111 are commonly used to distinguish mature human hematopoietic populations (so-called LM22 genes) in a computational method for quantifying cell fractions from bulk tissue gene expression profiles (CIBERSORT; ref. 18) and represented CD4 memory resting, CD8, Macrophage M1, and dendritic cell activated cells (Supplementary Fig. S9B and S9C). The association of EBV positivity with increased CD8+ T cells and MHC-II+ “M1” macrophages, as inferred using CIBERSORT, was consistent with our mpIF findings (Supplementary Fig. S9D).
The most consistently upregulated genes in EBV-positive cHL included: (i) the gene encoding delta-like canonical Notch ligand 1 (DLL1); (ii) all three CXCR3 receptor ligands (CXCL9, CXCL10, and CXCL11), which mediate the recruitment of CD8+ T cells, Th1 cells, and NK cells into tumors (22); (iii) genes encoding the complement component 1q (C1QA and C1QB); (iv) SLAMF7, which regulates effector function of NK cells (23); (v) antiviral genes IFIH1 and TLR8; (vi) several interferon-inducible GTPases (GBP1, GBP2, and GBP4; ref. 24); and (vii) IFNγ itself, which is known to activate the JAK/STAT signaling pathway and is typically secreted by CD8+ T cells, CD4+ Th1 cells, and NK cells (Fig. 3D). Genes that were downregulated in EBV-positive cHL included CXCL6 and CXCL8, which are chemo-attractants for neutrophilic granulocytes and interact with the chemokine receptors CXCR1 and CXCR2.
At the pathway level, upregulated genes in EBV-positive tumors associated with gene sets related to antigen processing and presentation, the interaction between a lymphoid and non-lymphoid cell, NK cell–mediated toxicity, IFNα/IFNβ/IFNγ signaling, and HBV infection (Fig. 3D; ref. 19). In EBV-negative tumors, several of the upregulated genes overlapped with an epithelial–mesenchymal transition gene set. Interestingly, patients over the age of 45 had a more pronounced EBV-negative–like transcriptome.
Identification of additional immune escape mechanisms in EBV-negative cHL
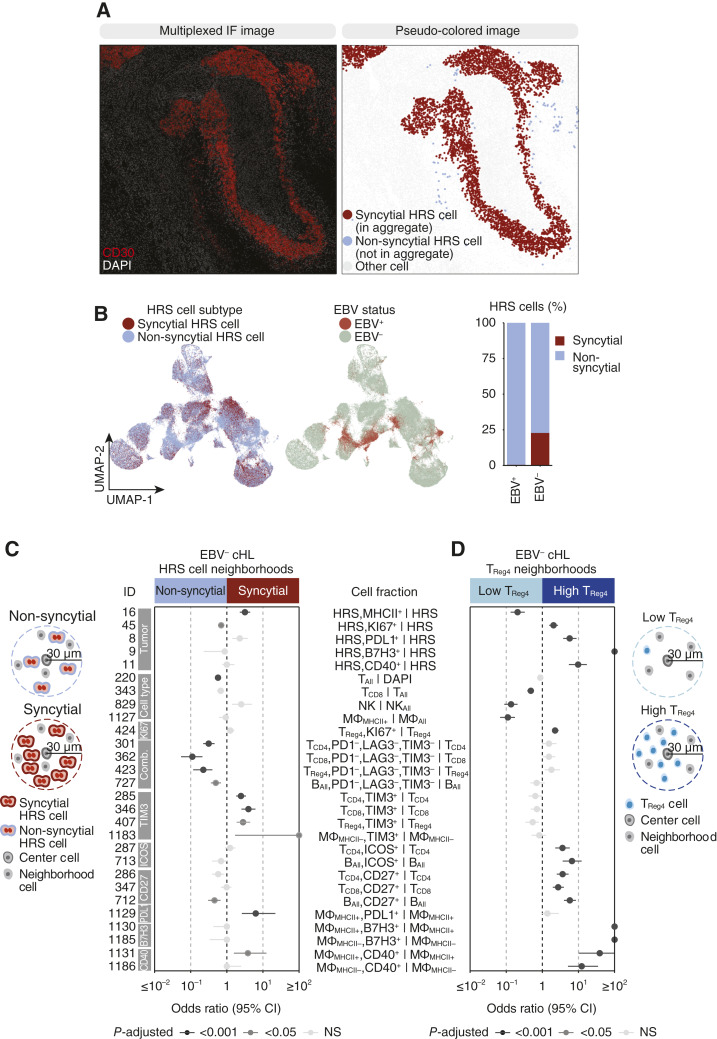
Pathologists have described a “syncytial variant” (1) of cHL in which HRS cells form cellular islands. To understand the relationship between this histologic variant and EBV, we first quantified HRS cellular islands by constructing graphs from the segmented mpIF images (Supplementary Fig. S10A) and identifying HRS cellular islands or “syncytial HRS cells” with a minimum of 20 neighboring HRS cells (Fig. 4A; Supplementary Fig. S10B; Supplementary Table S12; “Materials and Methods”). We observed near-perfect overlap between non-syncytial HRS cells and EBV-positive tumors in the UMAP projection (Fig. 4B) and a significantly larger fraction of syncytial HRS cells (i.e., HRS cells in cellular islands) in EBV-negative tumors compared with EBV-positive tumors (Supplementary Fig. S10C). EBV-positive tumors rarely contained syncytial HRS aggregates (Supplementary Fig. S10D), and the few EBV-positive HRS aggregates were all smaller than 30 HRS cells compared with the HRS aggregate size in EBV-negative tumors, which had an upper range of more than 1,000 HRS cells (Supplementary Fig. S10E). This indicates that syncytial cHL predominately occurs in EBV-negative tumors. To identify contributors of syncytial cHL or the effect of syncytial HRS cells on the TME, we characterized the cellular neighborhood of syncytial HRS cells versus non-syncytial HRS cells in EBV-negative tumors (Fig. 4C; Supplementary Table S10). Among HRS cell states, syncytial HRS neighborhoods harbored increased MHC-II+ HRS cells and decreased Ki67+ HRS cells. Among immune cell states, syncytial HRS neighborhoods were enriched for CD4+ helper T cells, CD8+ T cells, Tregs, and MHC-II− macrophages positive for TIM3 and MHC-II+ macrophages positive for PDL1, whereas non-syncytial HRS neighborhoods were enriched for naïve CD4+ helper T cells, CD8+ T cells, Tregs, and B cells “triple-negative” for PD1−/LAG3−/TIM3−. Most notably, non-syncytial HRS neighborhoods contained increased fractions of T cells indicating non-syncytial cHL exemplifies a T-cell infiltrated tumor, whereas syncytial cHL exemplifies a T-cell excluded tumor.
Figure 4.
Determinants of spatial neighborhoods in EBV-negative cHL. A, FOV showing two patterns of HRS cell spatial arrangement. B, UMAP of HRS cells colored by HRS cell subtype and EBV status. Stacked bar plot indicates the percent of syncytial and non-syncytial HRS cells by EBV status. C, Analysis approach to defining syncytial and non-syncytial HRS cell neighborhoods. The forest plot shows the effect size and 95% CI of cell fractions colored by P-adjusted in the comparison of syncytial and non-syncytial HRS neighborhoods in EBV-negative tumors. D, Analysis approach to defining CD4+ Treg high and low neighborhoods. The forest plot shows the effect size and 95% CI cell fractions colored by P-adjusted in the comparison of CD4+ Treg high and CD4+ Treg low neighborhoods in EBV-negative tumors. For cell type abbreviations, see Supplementary Table S3. For exact P-values, see Supplementary Table S10.
cHL often exhibits an HRS/lymphocyte “rosette” characteristic where HRS cells are in close contact with clusters of T cells, mainly composed of CD4+ helper T cells and immunosuppressive Tregs (3). To investigate the role of Tregs in cHL, we first explored whether their abundance, measured by the percentage of Tregs over immune cells, differs across FOVs or tumors. We found a considerable range of nearly 0 to more than 30% in the fraction of Tregs over immune cells across all FOVs in the cohort (Supplementary Fig. S11A). At the patient level, we found spatial heterogeneity in Treg abundance within some tumors (Supplementary Fig. S11B). Specifically, some tumors had low Treg abundance throughout (e.g., HL_27), others contained a mix of low and high Treg abundance FOVs (e.g., HL_19, HL_13), and three tumors (all EBV-negative) contained exclusively high Treg abundance FOVs. To determine the effect of Tregs on HRS cells and other cells in the TME, we compared high-abundance Treg cellular neighborhoods to low-abundance Treg cellular neighborhoods (Fig. 4D; Supplementary Table S10). Treg high neighborhoods contained increased proliferating HRS cells and HRS cells positive for PDL1, B7H3, and CD40, whereas Treg low neighborhoods contained increased MHC-II+ HRS cells. Among cells in the TME, Treg low neighborhoods contained increased fractions of CD8+ T cells, NK cells, and MHC-II+ macrophages, whereas Treg high neighborhoods contained increased proliferating Tregs and T and B cells positive for ICOS and CD27. Interestingly, Treg high neighborhoods harbored increased fractions of MHC-II+ and MHC-II− macrophages positive for B7H3 and CD40, mimicking expression patterns of HRS cells in these neighborhoods.
Discussion
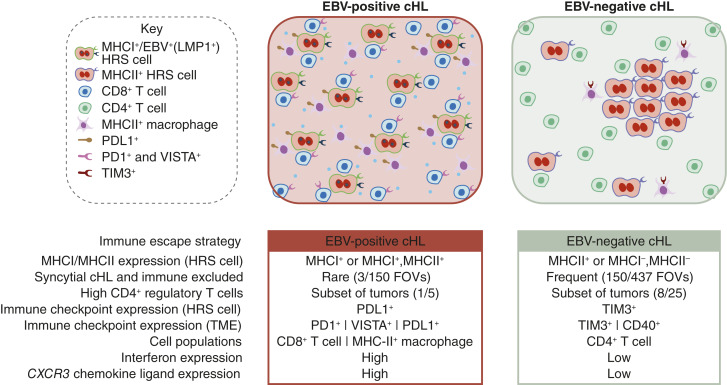
Our study represents a detailed in situ analysis of HRS cells and their proximal immune microenvironment in newly diagnosed and previously untreated cHL. We show that HRS cells in EBV-positive cHL consistently express MHC-I (or both MHC-I and MHC-II), reside in neighborhoods containing activated (PD1+ and VISTA+) T cells and B cells, shield themselves from immune attack through expression of PDL1 and habitation in a niche of PDL1+ macrophages, and proliferate in a cytokine milieu that is characterized by upregulation of IFNγ and CXCR3 receptor ligands (CXCL9, CXCL10, and CXCL11) and downregulation of CXCR1/CXCR2 receptor ligands (CXCL6 and CXCL8; Fig. 5). These data are consistent with experimental models of EBV infection, which have shown that LMP1, an EBV protein expressed during the EBV “latency program” of cHL, upregulates antigen presentation, regulates the expression of various co-stimulatory ligands, and induces potent T-cell responses that include not only CD8+ T cells but also CD4+ helper T cells (25). Functional studies in experimental models are required to provide mechanistic details of how EBV licenses the coexistence of MHC-positive cells with locally activated CD8+ T cells. In EBV-negative cHL, HRS cells seemed to use multiple different mechanisms of immune escape including downregulation of MHC-I, formation of a syncytial architecture, and attraction of Treg high cellular neighborhoods. Further work is required to identify molecular determinants driving these differences.
Figure 5.
A model of HRS cells and their TME in EBV-positive and EBV-negative cHL. Distinguishing features of EBV-positive and EBV-negative tumors. An FOV is characterized as syncytial cHL if it contains a minimum of one HRS aggregate.
Previous studies have used IHC or flow-based approaches to examine the relationship between EBV status and individual protein members of the antigen presentation machinery, in particular B2M, MHC-I, and MHC-II (4, 20, 26). Our study expands this prior work by characterizing the co-expression of all three proteins (B2M, MHC-I, and MHC-II) at single-cell resolution in a large number of HRS cells and by linking distinct co-expression patterns to specific HRS tumor neighborhoods. We confirm prior findings that EBV-positive cHLs harbor MHC-I expressing HRS cells and CD8+ T cells, and we also show that HRS neighborhoods in EBV-positive cHLs harbor increased fractions of CD8+ T cells and activated (PD1+ and VISTA+) T cells and B cells. Most importantly, we identified similar neighborhoods surrounding B2M+/MHC-I+ HRS cells in EBV-negative cHL, suggesting that HRS cell expression of MHC-I, and not EBV positivity, is responsible for these TME differences.
Overall, our data suggest that there is a fundamental difference between EBV-positive and EBV-negative cHL. Although our study was limited by a lack of genomic profiling of the HRS cells, our findings are consistent with a recently published study describing two genomic subtypes of cHL, H1 and H2, characterized by mutations in NF-κB, JAK-STAT, and PI3K pathways, or TP53 and KMT2D, respectively (9). The H2 subtype was enriched for EBV-positive tumors and exhibited increased CD8+ T cells and upregulation of T-cell activation genes, consistent with our data. Furthermore, we found an overlap between our list of EBV-related differentially expressed genes and the genomic subtype-related differentially expressed genes, specifically upregulation of TBX21 in EBV-positive H2 cHL and CCR4 and CXCL1 in EBV-negative H1 cHL. These parallels between EBV and genomic subtypes further support EBV-related cHL as a distinct subtype of the disease.
An important goal of our work was to develop an integrated approach using multiplex protein imaging and transcriptomics to evaluate the TME in routinely collected clinical cancer biospecimens. Our mpIF platform allowed us to examine large areas of each tumor and generate a single-cell proteomic dataset that exceeds prior studies (5, 7) by several orders of magnitude (>23 million cells). The size of our dataset enabled the characterization of otherwise rare HRS cells and their cellular neighborhoods. The large number of cell states in our dataset is a reflection of the very large number of cells that were profiled in our study and of our goal to provide an unbiased evaluation of protein co-expression patterns without preconceived cell states. Many of these states were observed in only a small fraction of cells and their biological significance warrants further study and validation using an independent method such as flow cytometry. It is unlikely that these patterns of protein co-expression or cell states can be attributed to technical issues or batch artifacts as we included replicates of a normal human tissue microarray on each tumor slide to serve as a positive staining control and minimize batch artifacts. The additional collection of gene expression data from immediately adjacent tumor sections allowed us to further interrogate functional differences between these tumors, develop a multidimensional portrait of the cHL tumor-immune architecture, and link our findings to prior studies focusing on protein (4, 6, 7, 26, 27) or gene expression analyses (28). The methods presented here provide a framework for future unbiased evaluation of spatial neighborhoods with the goal of identifying specific defects within the cancer immunity cycle (29).
Our study provides new insights into the architecture of the TME of cHL but is largely descriptive. Further studies are needed to validate the functional consequences for disease biology and potential implications for immunotherapy approaches. Clinical responses in cHL to antibodies against the PD1/PDL1 signaling axis, as defined by progression-free survival, have been associated with HRS cell expression of MHC-II (30) and a peripheral blood immune signature consistent with the expansion of clonally diverse CD4+ helper T cells (31). Our findings that HRS cells in EBV-positive cHL express not only MHC-II but also MHC-I and are surrounded by PD1+ immune effector cells and PDL1+ macrophages raise the question of whether EBV status might be a predictor of clinical response to PD1/PDL1 blockade. Interestingly, recent studies have reported encouraging responses to immune checkpoint blockade in patients with EBV-positive metastatic gastric cancer and non-Hodgkin lymphoma (32, 33).
Supplementary Material
Supplementary Figure S1. Validation of mpIF antibody panel.
Supplementary Figure S2. Tracking of cell loss across staining cycles.
Supplementary Figure S3. FOV placement for mpIF.
Supplementary Figure S4. cHL tumors exhibit heterogeneity in immune cell composition and states.
Supplementary Figure S5. Heterogeneity in HRS cell states and their spatial distribution.
Supplementary Figure S6. EBV-positive tumors exhibit increased expression of VISTA.
Supplementary Figure S7. EBV-positive MC tumors contain a T cell rich activated TME.
Supplementary Figure S8. B2M/MHCI/MHCII/PDL1-related HRS cell neighborhoods.
Supplementary Figure S9. Gene expression landscape of EBV-related cHL.
Supplementary Figure S10. Quantification of HRS cell patterns of spatial organization.
Supplementary Figure S11. Intertumoral heterogeneity in regulatory T cell abundance.
Supplementary Table S1. Patient characteristics.
Supplementary Table S2. mpIF panel and antibody information.
Supplementary Table S3. Cell type definitions.
Supplementary Table S4. List of cell states.
Supplementary Table S5. Cell count, cell loss, and field of view count for each patient.
Supplementary Table S6. Cell type counts for each patient.
Supplementary Table S7. Summary of HRS cell states for each patient.
Supplementary Table S8. Comparison of EBV-positive and EBV-negative cHL.
Supplementary Table S9. Comparison of EBV-positive NS and EBV-positive MC cHL.
Supplementary Table S10. Comparison of EBV-negative neighborhoods.
Supplementary Table S11. Differential expression of genes and cell types in EBV-positive and EBV-negative cHL.
Supplementary Table S12. HRS cell aggregates.
Acknowledgments
We acknowledge the use of the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748; all authors) and Cycle for Survival (I.K. Mellinghoff). Funding for this project was provided by the National Institutes of Health grant R35 NS105109 04 (I.K. Mellinghoff), Geoffrey Beene Cancer Research Center (I.K. Mellinghoff), Cycle for Survival (I.K. Mellinghoff), and National Institutes of Health/National Cancer Institute grant P30 CA008748 (all authors). M. Pourmaleki was also funded through the National Institutes of Health grant F31 CA271778 01.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
M. Roshal reports personal fees from Auron, grants from AstraZeneca, and grants and nonfinancial support from Genentech outside the submitted work. S.P. Shah reports grants from AstraZeneca and Bristol Myers Squibb during the conduct of the study. N.D. Socci reports grants from NIH during the conduct of the study. A. Dogan reports grants from Roche and AstraZeneca outside the submitted work. I.K. Mellinghoff reports grants from General Electric, Lilly, and Roche; grants and other support from Amgen, Kazia Therapeutics, Servier, and Agios; and other support from AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
M. Pourmaleki: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing. C.J. Jones: Conceptualization, resources, formal analysis, supervision, investigation, methodology, writing–original draft, writing–review and editing. S.D. Mellinghoff: Formal analysis, investigation. B.D. Greenstein: Formal analysis, investigation. P. Kumar: Resources, data curation, formal analysis, investigation. M. Foronda: Resources, data curation, investigation, writing–review and editing. D.A. Navarrete: Formal analysis, investigation, writing–review and editing. C. Campos: Formal analysis, supervision, investigation. M. Roshal: Resources, formal analysis, supervision, investigation. N. Schultz: Resources, data curation, supervision. S.P. Shah: Data curation, supervision. A. Schietinger: Data curation, supervision. N.D. Socci: Data curation, formal analysis, supervision, investigation, methodology. T.J. Hollmann: Resources, data curation, formal analysis, supervision, investigation, methodology. A. Dogan: Resources, data curation, formal analysis, supervision. I.K. Mellinghoff: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Connors JM, Cozen W, Steidl C, Carbone A, Hoppe RT, Flechtner HH, et al. Hodgkin lymphoma. Nat Rev Dis Primer 2020;6:61. [DOI] [PubMed] [Google Scholar]
- 2. Brice P, de Kerviler E, Friedberg JW. Classical Hodgkin lymphoma. Lancet 2021;398:1518–27. [DOI] [PubMed] [Google Scholar]
- 3. Wein F, Küppers R. The role of T cells in the microenvironment of Hodgkin lymphoma. J Leukoc Biol 2016;99:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cader FZ, Schackmann RCJ, Hu X, Wienand K, Redd R, Chapuy B, et al. Mass cytometry of Hodgkin lymphoma reveals a CD4+ regulatory T-cell–rich and exhausted T-effector microenvironment. Blood 2018;132:825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aoki T, Chong LC, Takata K, Milne K, Hav M, Colombo A, et al. Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov 2020;10:406–21. [DOI] [PubMed] [Google Scholar]
- 6. Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017;130:2420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patel SS, Weirather JL, Lipschitz M, Lako A, Chen PH, Griffin GK, et al. The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4–positive T cells that are PD-1–negative. Blood 2019;134:2059–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aoki T, Steidl C. Novel insights into Hodgkin lymphoma biology by single-cell analysis. Blood 2023;141:1791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alig SK, Shahrokh Esfahani M, Garofalo A, Li MY, Rossi C, Flerlage T, et al. Distinct Hodgkin lymphoma subtypes defined by noninvasive genomic profiling. Nature 2024;11:778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cohen JI, Fauci AS, Varmus H, Nabel GJ. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med 2011;3:107fs7–107fs7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res 2004;10:803–21. [DOI] [PubMed] [Google Scholar]
- 12. Young LS, Rickinson AB. Epstein-barr virus: 40 years on. Nat Rev Cancer 2004;4:757–68. [DOI] [PubMed] [Google Scholar]
- 13. Cohen JI. Epstein-Barr virus infection. N Engl J Med 2000;343:481–92. [DOI] [PubMed] [Google Scholar]
- 14. Farrell PJ. Epstein-Barr virus and cancer. Annu Rev Pathol 2019;14:29–53. [DOI] [PubMed] [Google Scholar]
- 15. Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 2016;16:789–802. [DOI] [PubMed] [Google Scholar]
- 16. Weniger MA, Küppers R. Molecular biology of Hodgkin lymphoma. Leukemia 2021;35:968–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pourmaleki M, Jones CJ, Ariyan CE, Zeng Z, Pirun M, Navarrete DA, et al. Tumor MHC class I expression associates with intralesional IL2 response in melanoma. Cancer Immunol Res 2022;10:303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 2004;101:6669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roemer MGM, Advani RH, Redd RA, Pinkus GS, Natkunam Y, Ligon AH, et al. Classical Hodgkin lymphoma with reduced β2M/MHC class I expression is associated with inferior outcome independent of 9p24.1 status. Cancer Immunol Res 2016;4:910–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol 2013;31:443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gutierrez-Guerrero A, Mancilla-Herrera I, Maravillas-Montero JL, Martinez-Duncker I, Veillette A, Cruz-Munoz ME. SLAMF7 selectively favors degranulation to promote cytotoxicity in human NK cells. Eur J Immunol 2022;52:62–74. [DOI] [PubMed] [Google Scholar]
- 24. Tretina K, Park ES, Maminska A, MacMicking JD. Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J Exp Med 2019;216:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang B, Choi IK. Facts and hopes in the relationship of EBV with cancer immunity and immunotherapy. Clin Cancer Res 2022;28:4363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oudejans J, Jiwa N, Kummer J, Horstman A, Vos W, Baak J, et al. Analysis of major histocompatibility complex class I expression on Reed-Sternberg cells in relation to the cytotoxic T-cell response in Epstein-Barr virus-positive and -negative Hodgkin’s disease. Blood 1996;87:3844–51. [PubMed] [Google Scholar]
- 27. Antel K, Chetty D, Oosthuizen J, Mohamed Z, Van der Vyver L, Verburgh E. CD68-positive tumour associated macrophages, PD-L1 expression, and EBV latent infection in a high HIV-prevalent South African cohort of Hodgkin lymphoma patients. Pathology 2021;53:628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chetaille B, Bertucci F, Finetti P, Esterni B, Stamatoullas A, Picquenot JM, et al. Molecular profiling of classical Hodgkin lymphoma tissues uncovers variations in the tumor microenvironment and correlations with EBV infection and outcome. Blood 2009;113:2765–3775. [DOI] [PubMed] [Google Scholar]
- 29. Mellman I, Chen DS, Powles T, Turley SJ. The cancer-immunity cycle: indication, genotype, and immunotype. Immunity 2023;56:2188–205. [DOI] [PubMed] [Google Scholar]
- 30. Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol 2018;36:942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cader FZ, Hu X, Goh WL, Wienand K, Ouyang J, Mandato E, et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat Med 2020;26:1468–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. [DOI] [PubMed] [Google Scholar]
- 33. Kim SJ, Hyeon J, Cho I, Ko YH, Kim WS. Comparison of efficacy of pembrolizumab between Epstein-Barr virus‒positive and ‒negative relapsed or refractory non-Hodgkin lymphomas. Cancer Res Treat 2019;51:611–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Validation of mpIF antibody panel.
Supplementary Figure S2. Tracking of cell loss across staining cycles.
Supplementary Figure S3. FOV placement for mpIF.
Supplementary Figure S4. cHL tumors exhibit heterogeneity in immune cell composition and states.
Supplementary Figure S5. Heterogeneity in HRS cell states and their spatial distribution.
Supplementary Figure S6. EBV-positive tumors exhibit increased expression of VISTA.
Supplementary Figure S7. EBV-positive MC tumors contain a T cell rich activated TME.
Supplementary Figure S8. B2M/MHCI/MHCII/PDL1-related HRS cell neighborhoods.
Supplementary Figure S9. Gene expression landscape of EBV-related cHL.
Supplementary Figure S10. Quantification of HRS cell patterns of spatial organization.
Supplementary Figure S11. Intertumoral heterogeneity in regulatory T cell abundance.
Supplementary Table S1. Patient characteristics.
Supplementary Table S2. mpIF panel and antibody information.
Supplementary Table S3. Cell type definitions.
Supplementary Table S4. List of cell states.
Supplementary Table S5. Cell count, cell loss, and field of view count for each patient.
Supplementary Table S6. Cell type counts for each patient.
Supplementary Table S7. Summary of HRS cell states for each patient.
Supplementary Table S8. Comparison of EBV-positive and EBV-negative cHL.
Supplementary Table S9. Comparison of EBV-positive NS and EBV-positive MC cHL.
Supplementary Table S10. Comparison of EBV-negative neighborhoods.
Supplementary Table S11. Differential expression of genes and cell types in EBV-positive and EBV-negative cHL.
Supplementary Table S12. HRS cell aggregates.
Data Availability Statement
All data supporting the findings of this study are publicly available at https://zenodo.org/records/10659311.