ABSTRACT
Many tissue-specific adult stem cell lineages maintain a balance between proliferation and differentiation. Here, we study how the H3K4me3 methyltransferase Set1 regulates early-stage male germ cells in Drosophila. Early-stage germline-specific knockdown of Set1 results in temporally progressive defects, arising as germ cell loss and developing into overpopulated early-stage germ cells. These germline defects also impact the niche architecture and cyst stem cell lineage non-cell-autonomously. Additionally, wild-type Set1, but not the catalytically inactive Set1, rescues the Set1 knockdown phenotypes, highlighting the functional importance of the methyltransferase activity of Set1. Further, RNA-sequencing experiments reveal key signaling pathway components, such as the JAK-STAT pathway gene Stat92E and the BMP pathway gene Mad, which are upregulated upon Set1 knockdown. Genetic interaction assays support the functional relationships between Set1 and JAK-STAT or BMP pathways, as both Stat92E and Mad mutations suppress the Set1 knockdown phenotypes. These findings enhance our understanding of the balance between proliferation and differentiation in an adult stem cell lineage. The phenotype of germ cell loss followed by over-proliferation when inhibiting a histone methyltransferase also raises concerns about using their inhibitors in cancer therapy.
Keywords: Germline stem cell, Maintenance, Differentiation, Chromatin, Histone modification, Epigenetic, Signaling pathway, Gene expression
Summary: Set1 maintains germline activity by ensuring proper JAK-STAT and BMP signaling in the Drosophila testis.
INTRODUCTION
In multicellular organisms, homeostasis and regeneration of many tissues largely depend on adult stem cells. These endogenous stem cells often undergo asymmetric cell divisions to allow both the stem cell to maintain its own population through self-renewal and the differentiation process to replace cells lost under physiological and pathological conditions (Kahney et al., 2019; Knoblich, 2008; Morrison and Spradling, 2008; Venkei and Yamashita, 2018). Disruption of these processes can potentially result in the misregulation of stem cell activity, leading to cancer or tissue degeneration (Clevers, 2005; Knoblich, 2010; Morrison and Kimble, 2006; Zion et al., 2020). There are two main modes of dysregulation in adult stem cell lineages that can result in the unrestrained cell proliferation that underlies tumorigenesis: constraints of normal stem cell expansion may be compromised or inactivated. This situation could occur if the dependence of stem cells on the niche is disrupted, resulting in a niche-independent overpopulation of stem cells or enhancement of stem cell activities. Alternatively, the transit-amplifying cells could fail to exit proliferation and enter the terminal differentiation program (Clarke and Fuller, 2006; Mukherjee et al., 2015; Sell, 2010; Zhang and Hsu, 2017).
Drosophila spermatogenesis is an excellent model system in which to study stem cell proliferation and differentiation (Davies and Fuller, 2008; Fuller, 1993; Gleason et al., 2018). Spermatogenesis is initiated with asymmetric division of the germline stem cell (GSC) to produce a self-renewed GSC and a gonialblast (Fig. 1A). The gonialblast undergoes four rounds of transit-amplifying mitotic divisions as spermatogonial cells. After mitosis, the 16 spermatogonial cells enter meiosis with a prolonged G2 phase as the primary spermatocytes. In addition to the GSC-derived germline lineage, the testis also has at least two somatic cell populations: the hub cells and the cyst cells (de Cuevas and Matunis, 2011). The hub cells are post-mitotic and they support the GSCs and the cyst stem cells (CySCs) (Kiger et al., 2001; Leatherman, 2013; Leatherman and Dinardo, 2010; Losick et al., 2011; Tran et al., 2000; Yamashita et al., 2003). The CySCs give rise to the cyst cells that support the germ cells throughout spermatogenesis (Leatherman and Dinardo, 2008; Lim and Fuller, 2012). The CySC also undergoes an asymmetric cell division to produce a self-renewed CySC and a cyst cell, which never divides again (Cheng et al., 2011). Two cyst cells encapsulate the germ cells as they divide and differentiate (Fig. 1A) (Fuller and Spradling, 2007; Matunis et al., 2012; Spradling et al., 2011).
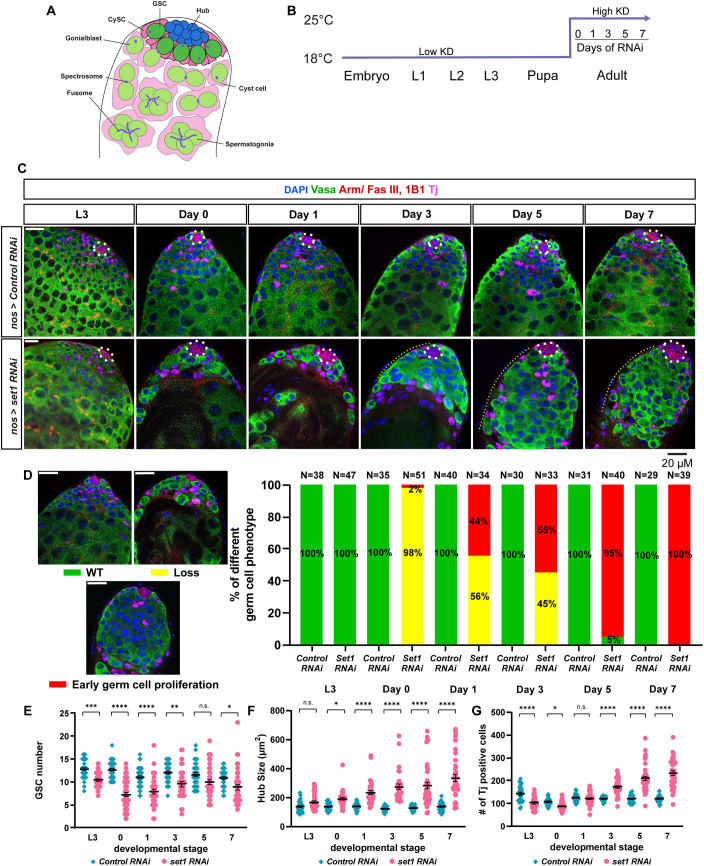
Fig. 1.
Drosophila Set1 is required cell-autonomously for germline maintenance and differentiation. (A) Schematic of the tip of the Drosophila testis. CySC, cyst stem cell; GSC, germline stem cell. (B) nos>Control RNAi (Ctrl KD) and nos>set1 RNAi (Set1 KD) flies were grown at 18°C until eclosion, and then shifted to 25°C for 0, 1, 3, 5 or 7 days. (C) Representative Ctrl KD and Set1 KD testes at the third instar larval stage (L3), day 0, 1, 3, 5 and 7 post-eclosion stained with the GSC lineage marker Vasa (green), Arm (red) or Fas III (red) for the hub area (white dashed outline), 1B1 for the spectrosome and fusome, and Tj (magenta) for the CySC lineage cells. Yellow dotted lines indicate overpopulated early germ cells. (D) Quantification of the percentage of testes with the representative germline phenotypes of germ cell loss (yellow) and early-germline overpopulation (red), present in Ctrl KD and Set1 KD testes at L3, as well as 0, 1, 3, 5 and 7 days post-eclosion. (E) Quantification of GSC number for Ctrl KD testes (L3: n=38; day 0: n=35; day 1: n=40; day 3: n=30; day 5: n=31; day 7: n=29) and Set1 KD testes (L3: n=47; day 0: n=51; day 1: n=34; day 3: n=33; day 5: n=40; day 7: n=39). See also Table S1. (F) Quantification of hub size for Ctrl KD and Set1 KD testes. See also Table S2. (G) Quantification of early cyst cell number for Ctrl KD and Set1 KD testes. See also Table S3. For E-G, individual data points and mean values are shown. Error bars represent s.e.m. ****P<10−4, ***P<10−3, **P<0.01, *P<0.05; n.s., not significant. Two-way ANOVA with interaction and Šidák multiple comparison test was used to compare two individual datasets with each other that have two independent variables. Scale bars: 20 μm.
Two important signaling pathways involved in the maintenance of the GSC niche are the JAnus Kinase Signal Transducer and Activator of Transcription (JAK-STAT) and Bone Morphogenetic Protein (BMP) signaling pathways (Amoyel et al., 2014; Herrera and Bach, 2019; Inaba et al., 2015; Kiger et al., 2001; Leatherman and Dinardo, 2008, 2010; Shivdasani and Ingham, 2003; Tulina and Matunis, 2001). Furthermore, epigenetic mechanisms, such as chromatin remodeling and histone modifications, have been shown to play important roles in the male germline lineage (Cherry and Matunis, 2010; Eun et al., 2017; Feng et al., 2018; Tarayrah and Chen, 2013; Tarayrah et al., 2015). In particular, multiple examples have shown the interplay between extrinsic signaling pathways and intrinsic epigenetic mechanisms in the Drosophila adult testes (Feng et al., 2017; Gleason and Chen, 2023; Vidaurre and Chen, 2021). For example, previous studies have shown that the H3K27me3 histone demethylase, Ubiquitously transcribed Tetratricopeptiderepeat gene on the X chromosome (UTX), acts as a negative regulator of JAK-STAT signaling by maintaining the transcription of Socs36E, proper expression and function of which maintain the balance between GSCs and CySCs (Amoyel et al., 2016; Issigonis et al., 2009; Singh et al., 2010; Tarayrah et al., 2013). Moreover, genes of the Epidermal Growth Factor (EGF) signaling pathway may be directly regulated by the H3K27me3 methyltransferase Enhancer of zestes [E(z)] in cyst cells, to ensure proper germ cell identity (Eun et al., 2014). Additionally, the Drosophila demethylase for H3K4me3, Little imaginal disc (Lid; Lysine demethylase 5, Kdm5), regulates JAK-STAT signaling in the male germline to maintain GSC activity and proliferation (Tarayrah et al., 2015). However, unlike the methyltransferase and demethylase for H3K27me3, the methyltransferases for H3K4me3 have not been studied in the Drosophila male germline.
SET domain containing 1 (Set1) is the main H3K4me3 methyltransferase in Drosophila, and its function is conserved from yeast to mammals (Lee et al., 2007a; Simonet et al., 2007). The post-translational modification of H3K4me3 has previously been shown to correlate with active transcription and is enriched at the promoter and the 5′ coding regions of genes (Bernstein et al., 2002; Heintzman et al., 2007; Pokholok et al., 2005; Santos-Rosa et al., 2002). The role of H3K4me3 in activating transcription is not fully understood, although it is thought that H3K4me3 acts as a docking scaffold for the transcription pre-initiation complex and chromatin remodeling complexes (Vermeulen et al., 2007; Wysocka et al., 2006). In yeast, Set1 catalyzes all methylation forms of H3K4 (i.e. H3K4me1/2/3) and its catalytic activity is modulated via a multi-subunit protein complex known as the Complex of proteins associated with Set1 (COMPASS) (Dehe et al., 2005; Miller et al., 2001; Nagy et al., 2002; Roguev et al., 2001). The Drosophila Set1 also interacts with the COMPASS complex to catalyze H3K4 di- and tri-methylation (H3K4me2/3) at the promoter proximal regions (Ardehali et al., 2011; Hallson et al., 2012; Mohan et al., 2011). In the Drosophila female germline, it has been shown that Set1 regulates GSC maintenance and differentiation; however, the mechanism has not been fully determined (Xuan et al., 2013; Yan et al., 2014).
Here, we use a series of molecular genetics and cell biology tools to study the roles of Set1 in the Drosophila testis. We found that Set1 is required for the normal function of early-stage male germ cells by regulating several key signaling pathways in a methyl-transferase-dependent manner. This study provides an example of how the intrinsic epigenetic mechanisms cooperate with the extrinsic signaling pathways to determine and maintain stem cell fate.
RESULTS
Drosophila Set1 regulates germline survival and proper germ cell differentiation
To explore the function of Set1 in the Drosophila male germline, a short hairpin RNA (shRNA or RNAi) specifically targeting the coding sequence of the Set1 gene driven by the early-stage germline driver nanos-Gal4 (nos-Gal4) (Van Doren et al., 1998) was employed for cell type-specific knockdown (KD) experiments. The Set1 null allele is lethal at the pupal stage and conventional mosaic analysis cannot be performed owing to the genomic location of the Set1 gene, which is very close to the centromere of chromosome 3. Therefore, the KD strategy is required to interrogate its roles in vivo (Ardehali et al., 2011; Hallson et al., 2012). In nos>set1 RNAi (Set1 KD) testes, H3K4me3 signal was greatly reduced in the early-stage germ cells, from GSCs to spermatogonial cells (Fig. S1B), compared with germ cells at the comparable stages in the nos>mCherry RNAi testes (nos>Control RNAi or Ctrl KD; Fig. S1A). In addition, in both Set1 KD and Ctrl KD testes, H3K4me3 signals were present in the somatic gonadal cells, indicating germline-specific and efficient inactivation of the methyltransferase function of Set1 (Fig. S1).
To investigate potential defects in the testis, a time course experiment was performed whereby Set1 KD and Ctrl KD flies were grown at 18°C until eclosion and then shifted to 25°C for 0, 1, 3, 5 or 7 days (Fig. 1B). We also examined an earlier development stage at the third instar larvae (L3), when male GSCs and their niche have already been established (Le Bras and Van Doren, 2006). At L3, 100% of Set1 KD and Ctrl KD testes displayed normal morphology (Fig. 1C,D). At day 0 and day 1 post-eclosion, 98% and 56%, respectively, of the Set1 KD testes, showed a germ cell loss phenotype compared with none of the Ctrl KD testes (Fig. 1C,D). At day 3 post-eclosion, although 45% of Set1 KD testes had germline loss, 55% had an early germ cell overpopulation phenotype, whereby the germ cells surrounding the hub formed a large, disorganized cluster with very few intercalating cyst cells and were often devoid of or had very few late-stage spermatocytes (Fig. 1C,D). By days 5 and 7, 95% and 100%, respectively, of Set1 KD testes exhibited this early-stage germ cell overpopulation phenotype (Fig. 1C,D). Here, we used a membrane marker (anti-Armadillo) to identify cyst cells that encapsulate germ cells, as shown previously (Feng et al., 2017). Any cyst with more than 16 germ cells was considered overpopulated. To test further whether these overpopulated germ cells resulted from over-proliferation, immunostaining using an antibody against a mitosis-enriched marker, H3S10P (phosphorylation at Serine 10 of H3) (Ranjan et al., 2022; Xie et al., 2015), revealed increased H3S10P-positive early-stage germ cells in the Set1 KD testes at days 3, 5 and 7, compared with earlier time points (days 1 and 3) as well as the Ctrl KD testes throughout this time course (Fig. S2).
In addition to the germline phenotypes observed in the Set1 KD testes, two other phenotypes were detected in the somatic cell lineages of the testis. First, the hub area was significantly increased compared with the Ctrl KD testes throughout this time-course experiment, with a higher degree of difference toward the later time points (Fig. 1F). Second, the number of cyst cells, positively stained with both a cyst cell marker, Traffic jam (Tj) (Li et al., 2003), and an early-stage cyst cell marker, Zinc-finger homeodomain protein 1 (Zfh1) (Eun et al., 2014; Issigonis et al., 2009; Leatherman and Dinardo, 2008), was significantly increased in the Set1 KD testes compared with the Ctrl KD testes at the later time points in the time-course experiments (Fig. 1G, Fig. S3). These changes in cyst cell number coincided with the changes in the germline phenotypes over the duration of the time course in the germline Set1 KD testes, indicating that Set1 acts in the germ cells to regulate somatic gonadal cells in a non-cell-autonomous manner.
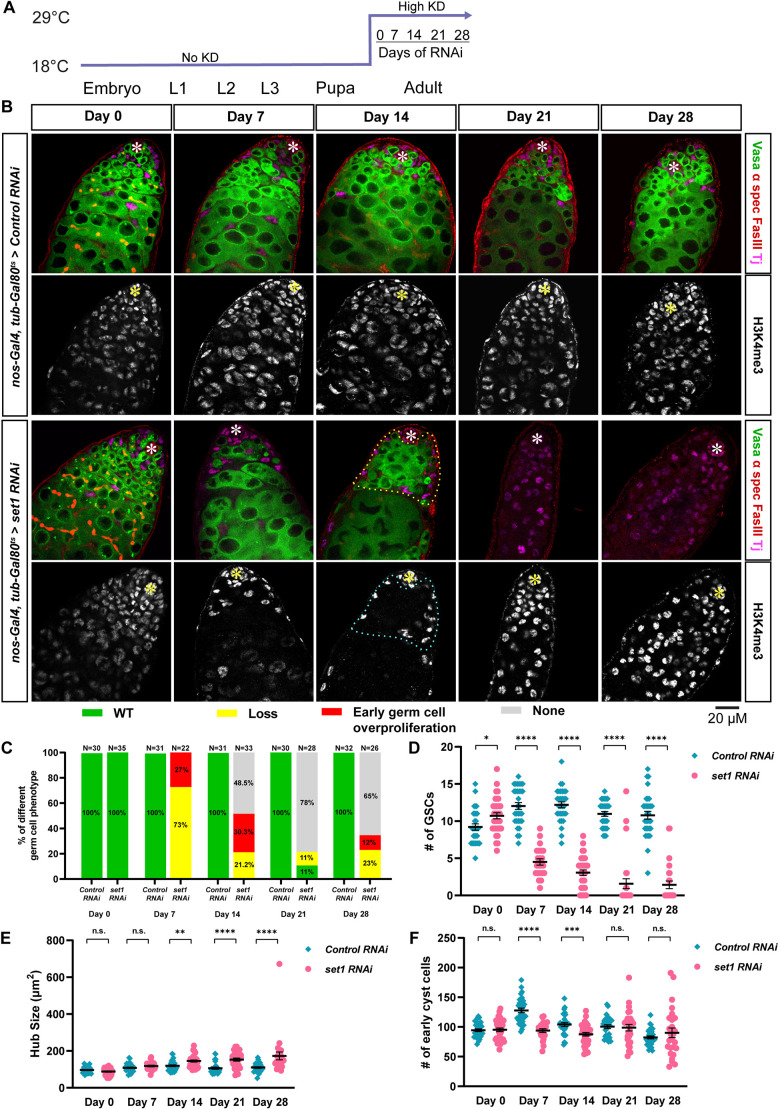
Because the Gal4/UAS system may still be functional at 18°C, knockdown of Set1 at earlier stages of development could contribute to those phenotypes detected in adulthood (Brand and Perrimon, 1993). In Set1 KD and Ctrl KD at L3, the overall early germline morphology in Set1 KD testes resembled that of the Ctrl KD testes with no early-stage germ cell proliferation and less severe germ cell loss compared with adult Set1 KD testes (Fig. 1C,D). Phenotypes such as germline stem cell number and hub size showed much less or no significant difference between the Set1 KD and Ctrl KD testes, respectively (Fig. 1E,F), whereas the cyst cell number continued to increase in the Set1 KD testes compared with the Ctrl KD testes from L3 to day 7 (Fig. 1G). Therefore, to probe further the requirement of Set1 in adulthood, we knocked down Set1 in the early germline using the temperature-sensitive Gal80 controlled by the tubulin promoter (tub-Gal80ts). At the permissive temperature (18°C), functional Gal80 protein inhibits Gal4 from associating with the UAS sequences, thus turning on the RNAi expression, but at the restrictive temperature (29°C) Gal80 is inactivated and Gal4 can associate with UAS and activate RNAi (McGuire et al., 2003). The tub-Gal80ts, nos>set1 RNAi (Set1 ts-KD) and tub-Gal80ts, nos>Control RNAi (Ctrl ts-KD) flies were grown at 18°C until eclosion, when they were shifted to 29°C for 0, 7, 14, 21 and 28 days, respectively, for another time-course experiment (Fig. 2A). At day 0, the Set1 ts-KD and Ctrl ts-KD testes had comparable germline morphology, indicating that active Gal80 at 18°C effectively prevents Set1 knockdown (Fig. 2B,C, day 0). However, 7 days after shifting to 29°C, 73% of the Set1 KD testes showed germline loss, and 27% exhibited the early germline overpopulation phenotype (Fig. 2B,C, day 7). These results indicate that the early germline phenotypes can be induced upon knocking down Set1 in adulthood. In addition to the germline phenotypes detected in the Set1 KD time course (Fig. 1C-E), in Set1 ts-KD testes at later time points, a new phenotype classified as ‘none’ was detected (no Vasa-positive germ cells could be detected at day 21 and day 28; Fig. 2B), whereby the testes are almost completely devoid of germ cells (Fig. 2B,C: 48.5% at day 14, 78% at day 21 and 65% at day 28). Throughout this time course, GSC number was consistently reduced at every time point in Set1 ts-KD testes compared with the Ctrl ts-KD testes (Fig. 2D, days 7, 14, 21 and 28). These results demonstrate that Set1 is required intrinsically for GSC maintenance in adulthood. Furthermore, the hub area was significantly increased at the later time points in the Set1 ts-KD testes (Fig. 2E, days 14, 21 and 28), whereas the cyst cell number was significantly reduced in Set1 ts-KD testes at the earlier time points (Fig. 2F, day 7 and day 14). The variation in somatic gonadal cell phenotypes between Set1 KD and Set1 ts-KD could be due to the non-cell-autonomous effects induced by the strength of germline Set1 KD and/or the different temperature shift regimes, as it has been shown that temperature contributes to germ cell differentiation in Drosophila (Gandara and Drummond-Barbosa, 2022, 2023). Together, both strategies to knock down Set1 in the germline shown in Figs 1 and 2 demonstrate that Set1 is likely required cell-autonomously for adult GSC maintenance and proper differentiation.
Fig. 2.
Knockdown of Set1 exclusively in the adult Drosophila testis leads to germ cell loss and germline differentiation defects. (A) tub-Gal80ts, nos>Control RNAi and tub-Gal80ts, nos>set1 RNAi flies were grown at the permissive temperature (18°C) until eclosion, then shifted to the restrictive temperature (29°C) for 0, 7, 14, 21 or 28 days. (B) Representative images of tub-Gal80ts, nos>Control RNAi and tub-Gal80ts, nos>set1 RNAi testes at day 0, 7, 14, 21 and 28 post-eclosion. vasa-GFP from a knock-in strain label the germline, immunostaining for H3K4me3 (gray), Arm (red) or Fas III (red) labels the hub region, α-spectrin (red) the spectrosome and fusome, and Tj (magenta) the CySC lineage cells. Asterisks indicate the hub. Yellow or cyan dotted outlines indicate overpopulated early-stage germ cells. Notably, for day 21 and 28 Set1 RNAi samples, no Vasa-positive cells could be detected, with all remaining cells being Tj-positive cyst cells. (C,D) Quantification of the percentage of testes with the germline phenotypes (C) and GSC number (D) in tub-Gal80ts, nos>Control RNAi testes (day 0: n=30; day 7: n=31; day 14: n=31; day 21: n=30; day 28: n=32) and tub-Gal80ts, nos>set1 RNAi testes (day 0: n=35; day 7: n=22; day 14: n=33; day 21: n=28; day 28: n=26). See also Table S1. (E) Quantification of hub region size for tub-Gal80ts, nos>Control RNAi and tub-Gal80ts, nos>set1 RNAi testes. See also Table S2. (F) Quantification of early cyst cell number for tub-Gal80ts, nos>Control RNAi and tub-Gal80ts, nos>set1 RNAi testes. See also Table S3. Individual data points and mean values are shown. Error bars represent s.e.m. ****P<10−4, ***P<10−3, **P<10−2, *P<0.05. n.s., not significant. Unpaired t-test was used to compare two individual datasets with each other.
Set1 is not required in somatic gonadal cells or late-stage spermatogonial cells for the GSC loss phenotype
To determine whether the detected GSC loss phenotype (Figs 1E and 2D) is specific to Set1 knockdown in the early germline, we first used cell type-specific strategies to knock down Set1 in the CySC lineage using the tj-Gal4 driver (Li et al., 2003). In tj>set1 RNAi testes, H3K4me3 signal is reduced in the CySCs and cyst cells compared with the signals in other cell types in the adult testis (Fig. S4A). At 7 days post eclosion, no obvious germline phenotype could be detected in the tj>set1 RNAi testes compared with the tj>Control RNAi testes (Fig. S5A). The number of GSCs was actually increased in the tj>set1 RNAi testes (Fig. S5D), contrary to what was detected in the nos>set1 RNAi testes (Figs 1E and 2D). The hub area exhibited no significant difference between the tj>set1 RNAi testes and the tj>Control RNAi testes (Fig. S4A′), but the number of cyst cells was significantly increased in the tj>set1 RNAi testes (Fig. S4A″). In addition, to determine the stage specificity of the GSC loss phenotype in nos>set1 RNAi testes, we used the stage-specific bam-Gal4 (bag of marbles) driver, which turns on target gene expression specifically in 4- to 16-cell spermatogonial cysts (Chen and McKearin, 2003). In bam>set1 RNAi testes, H3K4me3 was diminished in the germline past the 4-cell spermatogonial cyst stage but still present in the GSCs and very early-stage germ cells (Fig. S4B). At 7 days post-eclosion, there were no obvious morphological changes in the germline of bam>set1 RNAi testes compared with bam>Control RNAi testes (Fig. S5B). In addition, the number of GSCs and the hub area in bam>set1 RNAi testes and bam>Control RNAi testes were not significantly different from each other (Fig. S5E), even though the bam>set1 RNAi testes had significantly more cyst cells (Fig. S4B-B″). In summary, these results indicate that knockdown of Set1 in early-stage germ cells is responsible for the detected GSC loss phenotypes shown in Figs 1 and 2.
To confirm these findings, we used nos-Gal4ΔVP16, bam-Gal80 to drive Set1 RNAi only in the GSCs, gonialblasts, and 2-cell spermatogonial cysts as Gal80 protein driven by the bam promoter prevents RNAi expression in 4- to 16-cell spermatogonial cysts (Eliason et al., 2018). In the nos-Gal4ΔVP16, bam-Gal80>set1 RNAi testes, 95% had a reduction in H3K4me3 specifically in GSCs, gonialblasts and early-stage spermatogonial cells (Fig. S4C). At 7 days post-eclosion, the hub size and number of cyst cells in the nos-Gal4ΔVP16, bam-Gal80>set1 RNAi testes were comparable to those in nos-Gal4ΔVP16, bam-Gal80>Control RNAi testes (Fig. S4C′,C″). However, the number of GSCs in the nos-Gal4ΔVP16, bam-Gal80>set1 RNAi testes was significantly reduced compared with the nos-Gal4ΔVP16, bam-Gal80>Control RNAi testes (Fig. S5F). Together, these data demonstrate that knockdown of Set1 in the early-stage germ cells can partially account for the GSC loss phenotype seen in the nos>set1 RNAi testes. The differences in other phenotypes, especially the non-cell-autonomous phenotypes, are likely due to the cell type and stage specificities, as well as the strength of the KD effects, using different drivers. We focused on using nos>set1 RNAi testes for the following experiments to understand the mechanisms underlying the Set1 KD germline phenotypes.
The methyltransferase activity of Set1 is required for its proper activity in germ cells
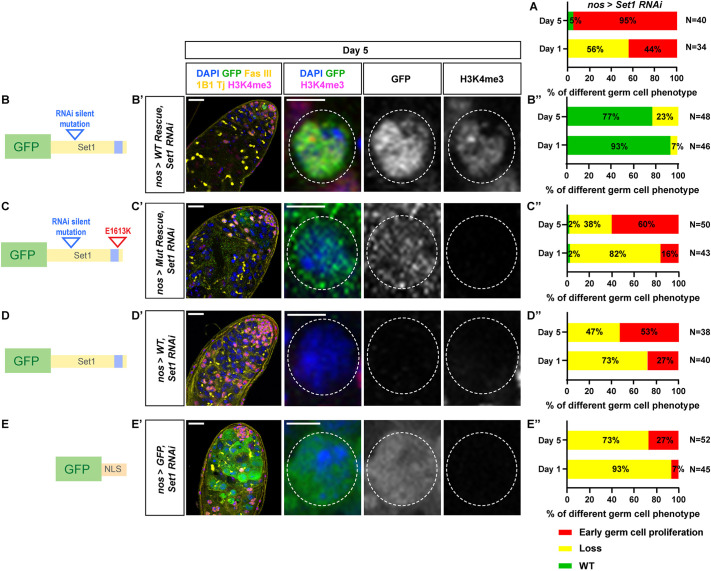
To confirm that the phenotypic effects seen in the nos>set1 RNAi testes (Fig. 3A, adapted from Fig. 1D) are due to the knockdown of the Set1 gene, but not off-targets effects of RNAi, a rescue experiment was performed. In nos>set1 RNAi testes, a GFP-tagged Set1 cDNA transgene was expressed using the same nos-Gal4 driver. Furthermore, because the Set1 RNAi target sequence is within the coding region of Set1, silent mutations were made within the target region to prevent KD of the transgene, namely a GFP-tagged Set1 cDNA transgene with the RNAi silent mutations (WT Rescue; Fig. 3B, Fig. S6A). As controls, three additional transgenes were generated and crossed individually to the same Set1 RNAi background to perform rescuing experiments: the WT Rescue transgene with RNAi silent mutations and an E-to-K mutation in the catalytic SET domain (Mut Rescue; Fig. 3C, Fig. S6B), a GFP-tagged Set1 cDNA transgene without the RNAi silent mutations (WT; Fig. 3D, Fig. S6C), and a GFP transgene with a nuclear localization signal (GFP-NLS; Fig. 3E, Fig. S6D). At day 1 and day 5 post eclosion, 93% and 77% of the nos>WT Rescue, set1 RNAi testes showed full rescue of the Set1 knockdown phenotypes (Fig. 3B′,B″, Fig. S6A′). By contrast, in the nos>Mut Rescue, set1 RNAi testes, only 2% showed full rescue, whereas 82% and 38% exhibited the germline loss phenotype, as well as 16% and 60% displaying the early germ cell proliferation phenotype at day 1 and day 5 post eclosion, respectively (Fig. 3C′,C″, Fig. S6B′), indicating that the catalytic SET domain is indispensable for the normal function of Set1 in the early-stage male germline. Both the nos>WT, set1 RNAi testes and nos>GFP-NLS, set1 RNAi testes show similar results compared with the nos>Mut Rescue, set1 RNAi testes (Fig. 3D′,D″,E′,E″, Fig. S6C′,D′), suggesting that adding back Set1 without changing the RNAi recognition sequences or a non-specific GFP-NLS transgene does not rescue the Set1 knockdown phenotypes in the male germline.
Fig. 3.
The methyltransferase activity of Set1 is required for its function in the male germline. (A) Quantification of the percentage of testes with the representative germline phenotypes, germ cell loss and early-germline overpopulation, present in nos>set1 RNAi testes at 1 day and 5 days post-eclosion (adapted from Fig. 1D). (B-E) Schematics of the Set1 cDNA transgene with the RNAi recognition sequences mutated, named WT rescue (B); the Set1 cDNA transgene with the RNAi recognition sequences mutated and an E→K amino acid change in the SET domain, named Mut rescue (C); the Set1 cDNA transgene without the RNAi recognition sequences mutated, named WT (D); the GFP cDNA with the nuclear localization sequence, named GFP (E). (B′-E′) Representative images of nos>WT Rescue, set1 RNAi testis (B′), nos>Mut Rescue, set1 RNAi testis (C′), nos>WT, set1 RNAi testis (D′) and nos>GFP, set1 RNAi testis (E′). All testes are from males 5 days post-eclosion, immunostained with H3K4me3 (magenta), GFP (green), Fas III (yellow) for the hub region, 1B1 (yellow) for spectrosome and fusome, and Tj (yellow) for the CySC lineage cells. Right-hand panels show higher magnification images of an example germ cell expressing the corresponding transgenes (outlined by white dashed lines). Scale bars: 20 μm (testis); 5 μm (individual germ cell images). (B″-E″) Quantification of the percentage of testes with the representative germline phenotypes, germ cell loss and early-germline overpopulation, present in nos>WT Rescue, set1 RNAi testes (B″), nos>Mut Rescue, set1 RNAi testes (C″), nos>WT, set1 RNAi testes (D″), nos>GFP, set1 RNAi testes (E″), at 1 and 5 days post-eclosion.
Furthermore, the GFP signals from both the nos>WT Rescue and the nos>Mut Rescue GFP-tagged fusion proteins are exclusively detected in the early germline nuclei, indicating that the silent mutations make these transgenes resistant to the RNAi knockdown, allowing for their proper expression and localization (Fig. 3B′,C′, Fig. S6A′,B′). In contrast, no GFP signal could be detected in the nos>WT, set1 RNAi testes without the RNAi silent mutations, indicating effective knockdown. In summary, these results support that the germline phenotypes observed in nos>set1 RNAi testes are due to the methyltransferase activity of the Set1 protein.
Set1 regulates expression of multiple signaling pathway components in the Drosophila testis
To understand the molecular mechanisms underlying the function of Set1 in the germline, RNA sequencing (RNA-seq) was performed with three independent biological replicates to compare the transcriptomes between nos>Control RNAi (Ctrl KD) and nos>set1 RNAi (Set1 KD) testes at 0, 1, 3 and 5 days post-eclosion, respectively (Fig. 4, Fig. S7). To obtain a global picture of the transcriptome changes between Ctrl KD and Set1 KD testes at different time points, principal component analyses were performed for all 24 samples (Fig. S7A). Among the 24 samples, the Ctrl KD samples cluster together and the Set1 KD samples cluster together (Fig. S7A). In addition, the biological replicates for each Set1 KD sample time points cluster more closely with one another than the replicates of the Ctrl KD sample time points (Fig. S7A). This can be explained as all 12 Ctrl KD samples should have similar gene expression patterns to one another given that the age difference between each consecutive time points is only 1-2 days. In contrast, the 12 Set1 KD samples do not, as visible changes could be detected at each time points in the Set1 KD testes. To analyze in more detail the transcriptome changes between the Ctrl KD and Set1 KD samples at every time point, a heatmap of all the differentially expressed genes was created (Fig. S7B). The 16,610 genes included in the heatmap, according to the Drosophila Release 6 reference genome (dos Santos et al., 2015), could be classified into four groups. Group 1 contains genes that are highly expressed at the earlier time points of the Set1 KD samples (days 0 and 1), whereas Group 2 has the genes that are highly expressed at the later time points of the Set1 KD samples (days 3 and 5). Additionally, Group 3 consists of the genes that are downregulated in the Set1 KD samples. Finally, Group 4 comprises the genes that had variable expression profiles across all the time points and thus did not fit the clear expression patterns seen for the genes in groups 1-3 (Fig. S7B).
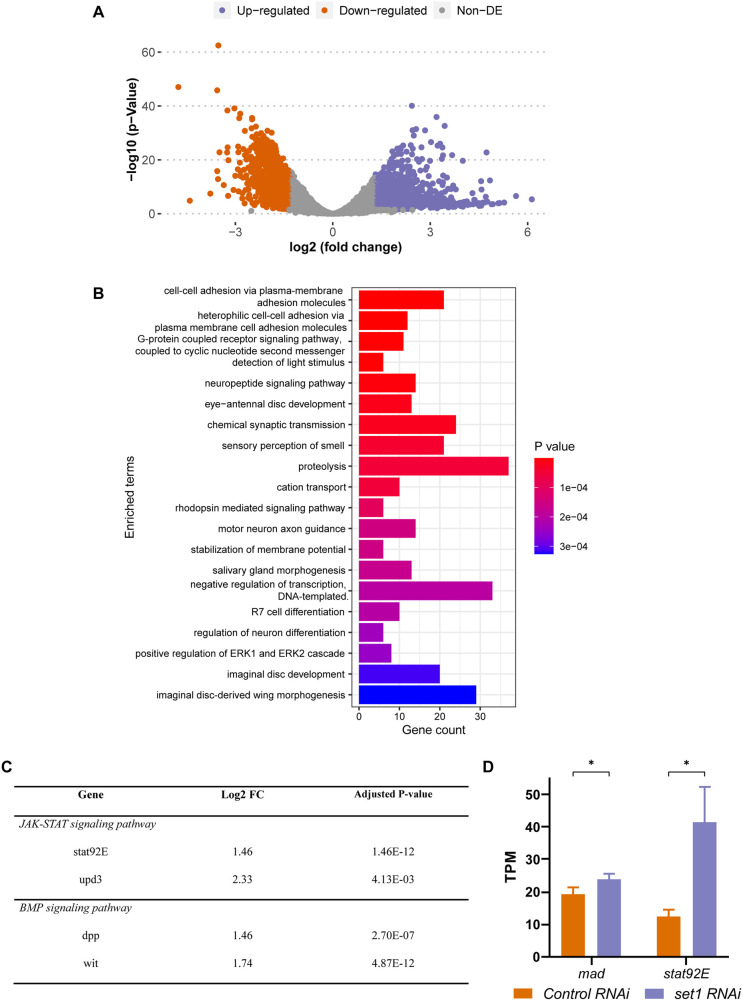
Fig. 4.
JAK-STAT and BMP signaling pathway components have increased expression in Set1 knockdown testis. (A) Volcano plot of differentially expressed genes in Set1 KD testes versus Ctrl KD testes at 3 days post-eclosion (≥log21.3=2.46-fold change, P<0.05). Non-DE, not differentially expressed. (B) GO analysis of the 1216 upregulated genes identified from A. GO analysis of the 764 downregulated genes did not show any significant category. (C) Several JAK-STAT and BMP signaling pathway components that are upregulated in Set1 KD testes compared with Ctrl KD testes 3 days post-eclosion. (D) Expression levels of Mad and Stat92E in Set1 KD testes versus Ctrl KD testes. Error bars represent s.e.m., n=3 biological replicates, *P<0.05 using unpaired t-test. TPM, transcripts per million reads.
After these initial analyses on all 24 samples, we focused on day 3 of the Set1 KD samples for two reasons. First, it is the time point when the germline mass phenotype became predominant; second, it had the most differentially expressed genes even with a stringent cutoff (≥log21.3=2.46-fold and P<0.05; Fig. 4A, Fig. S7C). At this time point, 1216 genes were significantly upregulated and 764 genes were significantly downregulated in the Set1 KD samples compared with the Ctrl KD samples (Fig. 4A, Fig. S7C). Gene ontology (GO) analysis of the 1216 upregulated genes showed enriched functions, with a significant category being ‘negative regulation of transcription’ (Fig. 4B).
Interestingly, components of multiple signaling pathways, such as JAK-STAT, BMP, EGF, Notch, Hedgehog, Wnt and Hippo signaling pathways, were found to be upregulated in the Set1 KD compared with the Ctrl KD samples at day 3 (Fig. S7D). For the EGF, Hedgehog and Wnt signaling pathways, multiple core components were significantly upregulated, whereas for the Notch and Hippo pathways some components were significantly upregulated and others were significantly downregulated. Among all these, only the JAK-STAT, BMP and EGF signaling pathways had at least two core components that made the significant cut-offs (Fig. 4C, Fig. S7D). For example, the JAK-STAT pathway genes Stat92E and upd3, and the BMP pathway genes dpp and wit, were found to be significantly upregulated (Fig. 4C). Given the importance of both signaling pathways in the testis (Inaba et al., 2015; Kawase et al., 2004; Kiger et al., 2001; Leatherman and Dinardo, 2008, 2010; Schulz et al., 2004; Singh et al., 2010; Tulina and Matunis, 2001), these results indicate that the early-stage germ cell overpopulation phenotype could be due to ectopic expression of both JAK-STAT and BMP signaling pathway genes.
The interactions between Set1 and JAK-STAT or BMP pathway genes are likely responsible for the early-stage germline phenotypes
To investigate further the functional relevance of both JAK-STAT and BMP signaling pathways with the Set1 KD phenotypes, we examined two key downstream genes of JAK-STAT and BMP pathways, Stat92E and Mad, respectively. Both genes were identified to be upregulated in the Set1 KD testes compared with the Ctrl KD testes based on the RNA-seq results (Fig. 4D). However, given that Set1 generates H3K4me3 to activate transcription, the direct target genes of Set1 are likely downregulated in Set1 KD testes. Nevertheless, we reason that the ectopic expression of Stat92E or Mad could contribute to the early-germline overpopulation phenotypes in the Set1 KD testes. Next, we tested potential genetic interactions of them with Set1.
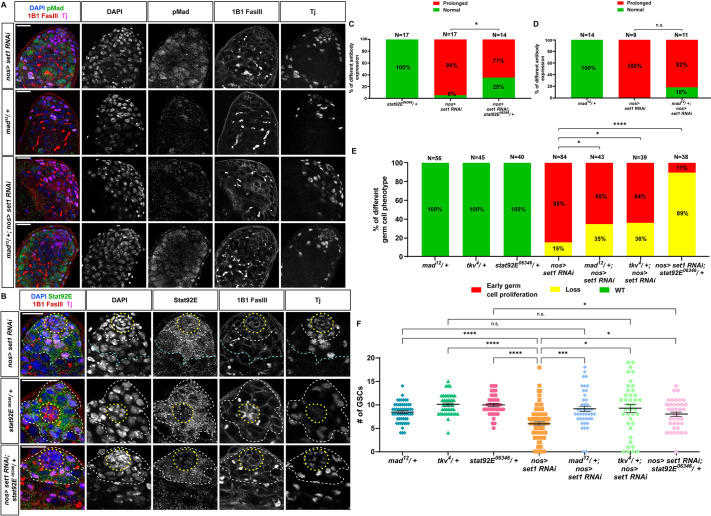
In heterozygous loss-of-function mad12 testes (Raftery et al., 1995) or Ctrl KD testes, pMad was detectable in the GSCs (Fig. 5A,D, mad12/+; Fig. S8A) and 100% showed normal morphology (Fig. 5E). In contrast, in the Set1 KD testes, pMad was detected in GSCs and ectopically in the spermatogonial cells (Fig. 5A,D, nos>set1 RNAi). Overall, 85% of the Set1 KD testes exhibited the early germ cell overpopulation phenotype (Fig. 5E). Even though both pMad patterns were detectable in Set1 KD testes with one copy of the mad12 allele (Fig. 5A,D, mad12/+; nos>set1 RNAi), the addition of one mad12 allele to the Set1 KD background reduced the occurrence of the early germ cell overpopulation phenotype from 85% to 65% (P<0.05; Fig. 5E). Furthermore, we tested thickveins (tkv), which encodes an upstream receptor of the BMP pathway and displays increased expression in the Set1 KD testes compared with the Ctrl KD testes (Fig. S7D). The loss-of-function tkv4 allele (Lecuit and Cohen, 1997; Terracol and Lengyel, 1994) also suppressed the early germ cell overpopulation phenotype from 85% to 64% (P<0.05; Fig. 5E), as well as the GSC loss phenotype (P<0.05; Fig. 5F). Together, these results suggest that Set1 normally represses the BMP signaling pathway. Without this repression, ectopic activity can occur, leading to abnormal germline morphology and composition.
Fig. 5.
Set1 regulates key JAK-STAT and BMP signaling pathway components. (A) Representative images of nos>set1 RNAi, mad12/+ and nos-Gal4/mad12 (on the 2nd chromosome, abbreviated as mad12/+); nos>set1 RNAi testes at 5 days post-eclosion, immunostained for pMad (green), Fas III (red) for the hub region, 1B1 (red) for spectrosome and fusome, and Tj (magenta) for the CySC lineage cells. (B) Representative images of nos>set1 RNAi, stat92E06346/+ and UAS-set1 RNAi/stat92E06346 (on the 3rd chromosome, abbreviated as stat92E06346/+); nos>set1 RNAi testes at 5 days post-eclosion, immunostained for Stat92E (green), Fas III (red) for the hub region, 1B1 (red) for spectrosome and fusome, and Tj (magenta) for the CySC lineage cells. In B, yellow dotted outline delineates the hub, white dashed outline the GSCs and CySCs region, and cyan dashed outline the spermatogonial region. Scale bars: 20 μm. (C) Quantification of the percentage of testes with representative Stat92E antibody expression: normal or ectopic with prolonged expression patterns present in stat92E06346/+ versus nos>set1 RNAi versus nos>set1 RNAi; stat92E06346/+ testes at 5 days post-eclosion. *P<0.05, χ2 test. (D) Quantification of the percentage of testes with representative pMad antibody expression: normal or ectopic with prolonged expression patterns present in mad12/+ versus nos>set1 RNAi versus mad12/+; nos>set1 RNAi testes at 5 days post-eclosion. n.s., not significant, χ2 test. (E) Quantification of the percentage of testes with the representative germline phenotypes, germ cell loss and early-germline overpopulation, using the same criteria as defined in Fig. 1D, present in mad12/+ (n=56), tkv4/+ (n=45), stat92E06346/+ (n=40), nos-Gal4>set1 RNAi (n=84), mad12/+; nos-Gal4>set1 RNAi (n=43), tkv4/+; nos-Gal4>set1 RNAi (n=39) and stat92E06346/+; nos-Gal4>set1 RNAi (n=38) testes at 5 days post-eclosion. *P<0.05, ****P<10−4, χ2 test. (F) Quantification of GSC number for mad12/+ (n=56), tkv4/+ (n=45), stat92E06346/+ (n=40), nos-Gal4>set1 RNAi (n=84), mad12/+; nos-Gal4>set1 RNAi (n=43), tkv4/+; nos-Gal4>set1 RNAi (n=39) and stat92E06346/+; nos-Gal4>set1 RNAi (n=38) testes at 5 days post-eclosion. Individual data points and mean values are shown. See also Table S1. Error bars represent s.e.m. *P<0.05, ***P<10−3, ****P<10−4; n.s., not significant; one-way ANOVA and Dunnett's T3 multiple comparison test was used to compare multiple individual datasets with each other that had one independent variable.
Additionally, in heterozygous testes with the loss-of-function stat92E06346 allele (Hou et al., 1996) or Ctrl KD testes, Stat92E expression was detected in the GSCs (white outline in Fig. 5B,C, stat92E06346/+; Fig. S8B), but in Set1 KD testes Stat92E was not only detectable in GSCs but also in hub cells (yellow outline in Fig. 5B, nos>set1 RNAi) and in spermatogonial cells (cyan outline in Fig. 5B, nos>set1 RNAi). Interestingly, upon compromising Stat92E with stat92E06346 in the Set1 KD background, the expression of Stat92E was no longer found in hub cells (yellow outline in Fig. 5B, nos>set1 RNAi; stat92E06346/+) or in the spermatogonial cells. Furthermore, when the level of Stat was reduced in nos>set1 RNAi; stat92E06346/+ testes, the incidence of the early-stage germ cell overpopulation phenotype decreased from 85% to 11% (P<10−4; Fig. 5E). These results suggest that enhanced Stat92E expression upon Set1 inactivation significantly contributes to the early germ cell overpopulation phenotype in Set1 KD testes. Moreover, compromising the BMP pathway by mad12/+ fully suppresses the GSC loss phenotypes in Set1 KD testes, whereas compromising the BMP pathway by tkv4 can also partially suppress the GSC loss phenotype in Set1 KD testes (P<0.05; Fig. 5F). Halving the level of Stat92E could partially suppress the GSC loss phenotype in Set1 KD testes (P<0.05; Fig. 5F). However, none of the conditions could fully restore the loss of some spermatogonial cells in the Set1 KD testes (Fig. 5E), suggesting that other downstream factors of these two signaling pathways or components from other potentially involved signaling pathways (Fig. S7D) could contribute to the loss of early-stage germ cells at the Set1 KD background. Together, these genetic interaction results support the functional relationships between Set1 and BMP as well as JAK-STAT pathways, with the primary role of BMP signaling relating to the GSC loss phenotype and a predominant contribution of the JAK-STAT pathway to the early-stage germline overpopulation phenotype, consistent with the transcriptome results shown in Fig. 4.
DISCUSSION
This study explores the in vivo roles of Drosophila Set1, a histone methyltransferase responsible for ‘writing’ the H3K4me3 histone modification, in the Drosophila testis. Through a series of cell type- and stage-specific RNAi knockdown via a time course regime, our findings unveil a fascinating progression of germline defects (Fig. 6A-D). These Set1 loss-of-function phenotypes have both cellular and molecular specificities. First, the phenotypes are predominantly linked to the loss of Set1 function in early-stage germ cells, as compromising Set1 in late-stage germ cells or somatic gonadal cells fails to replicate these defects. Moreover, we conducted rescuing experiments utilizing transgenes encoding both the wild-type and catalytic inactive forms of Set1. Only the wild-type Set1 effectively rescues the knockdown phenotypes, underscoring the specificity of these effects to the methyltransferase activity of Set1. To further understand the underlying molecular mechanisms, we performed RNA-seq experiments to identify genes with changed expression upon knocking down Set1 gene. Through this assay, we identified several crucial signaling pathway genes, such as Stat92E and Mad, the downstream components of the JAK-STAT and BMP signaling pathways, respectively, that have increased expression when Set1 is knocked down. Genetic interaction analyses further suggest a functional relationship between Set1 and these signaling pathways, as mutations of the Stat92E and Mad genes suppress the Set1 knockdown phenotypes. Collectively, our investigations shed light on the molecular mechanisms at play, enhancing our comprehension of the pivotal decision-making process governing proliferation versus differentiation within adult stem cell lineages. This insight bears significant implications for both stem cell biology and the broader field of cancer biology.
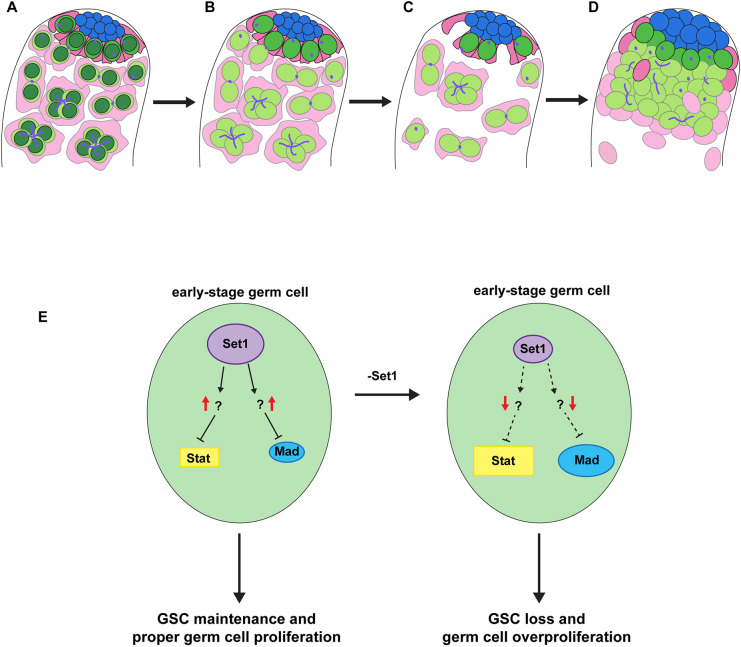
Fig. 6.
Illustration of the progression of Set1 knockdown phenotypes in adult testis. (A) In a wild-type testis, H3K4me3 is present in all cell types of the testis, but for simplicity the presence of H3K4me3 (dark green) is only shown in the germ cells, including GSCs (bright green), gonialblasts and spermatogonia (light green). CySCs are shown in magenta, cyst cells in pink, and hub cells in blue. (B) Knockdown of Set1 in the early germline results in a reduction of H3K4me3 in GSCs, gonialblasts and spermatogonia. (C) The phenotypes at the earlier time points include a reduction of germ cells and cyst cells. (D) The phenotypes at the later time points include overpopulated early-stage germ cells ectopically expressing Stat92E and Mad, as well as increased hub size. (E) Putative pathways of Set1 function in the early-stage germline, including GSCs. Given that both Stat92E and Mad have increased expression in the Set1 KD testes, they are likely indirect targets of Set1. One possible explanation is that Set1 directly regulates inhibitors of both Stat92E and Mad (left). Compromising Set1 could lead to the downregulation of these inhibitors and ectopic expression of Stat92E and Mad, resulting in the GSC loss and early-stage germ cell overproliferation phenotypes (right).
Set1 may regulate JAK-STAT and BMP signaling pathway inhibitors in the Drosophila male germline
Here, our RNA-seq results reveal that upon knockdown of Set1 in the Drosophila male germline, a key downstream factor of the JAK-STAT signaling pathway, Stat92E, is upregulated (Fig. 4D). This is unexpected for the direct targets of Set1, given that Set1 is the methyltransferase for the transcriptional activating mark H3K4me3. In addition, previous ChIP-seq results using progenitor germ cell-enriched bam testes (McKearin and Spradling, 1990) indicate that H3K4me3 is not enriched at the promoter region of the Stat92E gene (Fig. S9A) (Gan et al., 2010). These results suggest that Stat92E is unlikely to be a direct target of Set1. However, given the suppression of the early germ cell overpopulation phenotype when adding a single copy of a Stat92E mutation in the Set1 KD background, they likely interact in vivo. We hypothesize that Set1 acts in the Drosophila male germline by regulating the transcriptional activity of a JAK-STAT inhibitor. When Set1 is compromised, the expression of this inhibitor is possibly downregulated, allowing for the ectopic expression of Stat92E (Fig. 6E). One candidate JAK-STAT inhibitor is protein tyrosine phosphatase 61F (Ptp61F). Ptp61F is a known inhibitor of the JAK-STAT pathway that has been previously identified as a target of Lid, the H3K4 demethylase, in the germline (Baeg et al., 2005; Tarayrah et al., 2015). Additionally, Ptp61F is slightly downregulated in the RNA-seq data (Fig. S9G) and is enriched with the H3K4me3 mark at its promoter regions (Fig. S9C).
In addition to Stat92E, the downstream factor of the BMP signaling pathway Mad is also upregulated in the Set1 KD background (Fig. 4D). Unlike Stat92E, H3K4me3 is enriched at the promoter region of the Mad gene in progenitor germ cells (Fig. S9B) (Gan et al., 2010). However, given that Mad is slightly upregulated (Fig. 4D) and suppression of the early germ cell overpopulation phenotype was detected when a single copy of Mad mutation is combined with the Set1 KD condition (Fig. 5A,E), we hypothesize that BMP inhibitor-encoding genes could be direct targets of Set1, downregulation of which upon inactivation of Set1 could lead to the ultimate upregulation of Mad. When examining the candidate BMP inhibitors Cul-2, MAN1 and Ube3a (Cul-2 is a BMP inhibitor in the Drosophila female germline, whereas both MAN1 and Ub3a are BMP inhibitors in other Drosophila tissues, as shown previously; Ayyub et al., 2015; Duan et al., 2020; Laugks et al., 2017; Li et al., 2016; Wagner et al., 2010), all three BMP inhibitors have downregulated expression to varying degrees in the Set1 KD testes (Fig. S9G) and H3K4me3 enrichment can be detected at their corresponding promoter regions (Fig. S9D-F) (Gan et al., 2010). Additional experiments are needed to determine whether these inhibitors of JAK-STAT and BMP signaling are bona fide direct target genes and have a functional relationship with Set1 in the Drosophila male germline.
Set1 may regulate other signaling pathways in the Drosophila male germline
Notch signaling has been shown to regulate the transit-amplification stage (Ng et al., 2019). Removal of Notch in the cyst cells or Delta in the germline result in spermatogonial cell death. Although Delta is not differentially expressed, Notch is upregulated whereas two other Notch pathway components, Suppressor of Hairless [Su(H)] and Serrate (Ser), have decreased expression in Set1 KD testes (Fig. S7D). Increased expression of Wnt signaling components, such as arm, is detected using RNA-seq (Fig. S7D), which could result in abnormal clusters enriched with undifferentiated germ cell as reported (Pancratov et al., 2013). Finally, the EGF signaling is important for cyst cell differentiation and germline maintenance and differentiation (Feng et al., 2017), which regulates encapsulation of germ cells by the cyst cells (Amoyel et al., 2016; Schulz et al., 2004; Tran et al., 2000). The increased expression of multiple EGF signaling components in Set1 KD testes (Fig. S7D) could act in conjunction with JAK-STAT and BMP signaling pathways. Because our RNA-seq analyses used the entire tissue, further studies are needed to address all these possibilities.
Crosstalk between Set1 and other histone-modifying enzymes in regulating key signaling pathways
In Drosophila, the lid gene encodes the histone demethylase responsible for ‘erasing’ the H3K4 methylation (Eissenberg et al., 2007; Lee et al., 2007b), enzymatic activity of which directly antagonizes the function of Set1. It has been shown that in the male germline Lid is required for the proper levels of Stat92E. In the lid knockdown or mutant cells, Stat92E expression is reduced (Tarayrah et al., 2015). Therefore, the two histone-modifying enzymes with opposite activities, Set1 and Lid, display antagonistic regulation of Stat92E expression. Therefore, this work introduces a new epigenetic regulator within the complex framework of the JAK-STAT signaling pathway in the Drosophila testis niche. Together, these studies highlight the delicate balance maintained by a histone methyltransferase and a histone demethylase in orchestrating a major signaling pathway.
Common and distinct roles of Drosophila Set1 in various stem cell systems
As the primary H3K4 methyltransferase in Drosophila, Set1 has also been demonstrated to play crucial roles in the female germline. For example, it has been shown that Set1 acts with the E3 ubiquitin ligase Bre1 to produce the majority of H3K4me3 modifications in regulating the maintenance and differentiation of female GSCs. Knockdown of Set1 in the early-stage germline results in a significant loss of GSCs in the germaria, due to decreased expression of BMP signaling components, such as Mad or Dad (Bray et al., 2005; Xuan et al., 2013). This phenotype resembles the male GSC loss phenotype observed in this study. However, in the male germline, inactivation of Set1 predominantly affects the JAK-STAT pathway over the BMP pathway in our study. This effect is observed to progress over time, resulting in upregulated Stat92E levels. Consequently, we observe an overpopulation of early-stage germ cells during the late phase of our time-course experiments. Based on an extensive RNAi screen that identified hundreds of candidate genes with potential roles in the female germline, Set1 stands out as a crucial factor required for female GSC maintenance and proper differentiation. When Set1 is knocked down in the female germline, ovaries exhibit a spectrum of phenotypes, including empty ovarioles likely due to female GSC loss as well as so-called ‘pseudoegg’ chambers filled with undifferentiated cells (Yan et al., 2014). The latter phenotype resembles the early-germline overpopulation exhibited in the Set1 male germline knockdown effects shown in this work (Fig. 1C,D). Moreover, for the non-cell-autonomous roles of Set1 in the Drosophila ovary, Set1 acts in somatic gonadal cells to impact the female germline. For example, compromising Set1 in different somatic gonadal cells in the ovary results in either GSC loss or overpopulated GSC-like cells in the germaria (Xuan et al., 2013). By contrast, compromising Set1 in the somatic gonadal cells in the testis does not seem to cause any obvious phenotypes (Fig. S4A). However, in the Drosophila testis Set1 acts in the germline non-cell autonomously to affect niche architecture and cyst cell number (Fig. 1F,G).
In the somatic stem cell lineages, two types of neuroblast (NBs), type I NBs (NB-I) and type II NBs (NB-II), differ in number (e.g. 184 NB-Is and 16 NB-IIs) and lineage progression to ultimately produce the diverse range of neurons and glia in the brain (Bello et al., 2008; Boone and Doe, 2008). Interestingly, Set1 seems to be dispensable for the NB-II differentiation, which is distinct from the profound functions of Set1 in both ovary and testis for proper GSC differentiation (Yan et al., 2014), suggesting different requirements of histone modifications and their modifying enzymes in distinct stem cell systems.
The progression of germline phenotypes resulting from Set1 loss of function underscores the need for caution when considering histone methyltransferase inhibitors in cancer therapy
Many short-lived cell types in the body are produced from adult stem cells, either continuously or in response to physiological signals or trauma. During these processes, genetic lesions or epigenetic misregulations can drive cells to divide continuously instead of undergoing proper differentiation. This shift in cellular properties can initiate diseases such as cancer.
The COMPASS complex has been shown to be involved in pathogenesis such as cancer (Shilatifard, 2012). Here, our results demonstrate significant phenotypic changes resulting from germline knockdown of Set1 in fly testes, beginning with germ cell loss and progressing to the development of germline tumors within a relatively short timeframe (Fig. 1B-D). This delayed appearance raises an intriguing possibility: a resilient subset of ‘survivor cells’, which evade early cell death, initiates a phase of uncontrolled and rapid over-proliferation of undifferentiated cells, reminiscent of the phenomenon observed for ‘cancer stem cells’ (Beck and Blanpain, 2013). Given that various histone methyltransferases are targeted for therapeutic treatment in cancers, our discovery of initial cell loss followed by over-proliferation in response to knockdown of a key histone methyltransferase raises concerns about the potential use of these enzyme inhibitors in cancer therapy (Morera et al., 2016).
MATERIALS AND METHODS
Fly strains and husbandry
Flies were raised under standard yeast/molasses medium at 25°C unless stated otherwise. The following flies were used: nos-Gal4 (with VP16)/Cyo (Van Doren et al., 1998), nos-Gal4 (without VP16 or ΔVP16) on the second chromosome (from Yukiko Yamashita, University of Michigan, USA), UAS-set1 RNAi (Bloomington Drosophila Stock Center, BL33704), UAS-mCherry RNAi (Bloomington Drosophila Stock Center, BL35785), tj-Gal4/Cyo (Tanentzapf et al., 2007), bam-Gal4/TM6B (Chen and McKearin, 2003), bam-Gal80 on the third chromosome (from Juliette Mathieu and Jean-René Huynh, Collège de France, France), UAS-GFP.nls (Bloomington Drosophila Stock Center, BL4775), UASp-FRT-EGFP-set1 (WT)-PolyA-FRT (this study), UASp-FRT-EGFP-set1 (WT rescue)-PolyA-FRT (this study), UASp-FRT-EGFP-set1 (Mutant rescue)-PolyA-FRT (this study), mad12, FRT40A/Cyo (Bloomington Drosophila Stock Center, BL58785), stat92E06346/TM3, Sb (Bloomington Drosophila Stock Center, BL11681), tkv4 FRT40A/Cyo (Bloomington Drosophila Stock Center, BL58786), vasa-GFP knock-in (from Dr Tatjana Trcek, Johns Hopkins University, Baltimore, MD, USA).
Spatiotemporally controlled experiments
To study the function of Set1 in the Drosophila testis, two RNAi lines, UAS-set1 RNAi and UAS-mCherry RNAi, were crossed with different drivers, nos-Gal4/Cyo, bam-Gal4/TM6B and tj-Gal4/Cyo, at 18°C. Flies at the larval stage L3 were collected and newly eclosed progenies were transferred to new vials at 25°C and aged for 0, 1, 3, 5 or 7 days before dissection.
For Set1 function study in the germline stem cells and gonialblast cells, the two RNAi lines UAS-set1 RNAi and UAS-mCherry RNAi were crossed with nos-Gal4ΔVP16/Cyo; bam-Gal80/MKRS at 25°C. Newly eclosed progeny were transferred to new vials and maintained at 25°C for 7 days before dissection.
To determine the role of Set1 in the Drosophila early germline at adulthood, two RNAi lines, vasa-GFP knock-in/CyO; UAS-set1 RNAi and vasa-GFP knock-in/CyO; UAS-mCherry RNAi, were crossed with nos-Gal4/Cyo; tub-Gal80ts at 18°C. Newly eclosed progenies were transferred to new vials at 29°C and aged for 0, 7, 14, 21 or 28 days before dissection.
To determine whether expression of EGFP-set1 cDNA in the early germline is sufficient to rescue nos>set1 RNAi germ cell phenotypes, flies with the following genotypes were grown at 25°C and dissected at 1 and 5 days post eclosion: nos-Gal4/UASp-FRT-EGFP-set1 (WT rescue)-PolyA-FRT; UAS-set1 RNAi/+, nos-Gal4/UASp-FRT-EGFP-set1 (Mutant rescue)-PolyA-FRT; UAS-set1 RNAi/+, nos-Gal4/UASp-FRT-EGFP-set1 (WT)-PolyA-FRT; UAS-set1 RNAi/+, nos-Gal4/UAS- GFP.nls; UAS-set1 RNAi/+.
To identify whether Set1 genetically interacts with Mad, tkv and Stat92E, the alleles mad12, tkv4 and stat92E06346 were used. Flies with the following genotypes were grown at 25°C and dissected at 5 days post eclosion: nos-Gal4/mad12; UAS-set1 RNAi/+, nos-Gal4/mad12, nos-Gal4/tkv4; UAS-set1 RNAi/+, nos-Gal4/tkv4, nos-Gal4/+; UAS set1 RNAi/stat92E06346, UAS set1 RNAi/stat92E06346; nos-Gal4/+; UAS set1 RNAi/+.
Generation of transgenic fly lines
For the lines UASp-FRT-EGFP-set1 (WT rescue)-PolyA-FRT and UASp-FRT-EGFP-set1 (Mutant rescue)-PolyA-FRT, the RNAi recognition sequence was altered. Because the RNAi recognition sequence is in the coding region of the Set1 gene (AAGGTGCAGAGTATAAGAGTA), the third base for each codon of this sequence was changed to make a silent mutation that would allow for the protein to be translated properly but for the RNAi to not recognize the mRNA. For the line UASp-FRT-EGFP-set1 (Mutant rescue)-PolyA-FRT, a mutation in the SET domain at G4713A to make the E1613K amino acid replacement was made.
Immunofluorescence
Testes were dissected in Schneider's insect media and then fixed in 4% formaldehyde in 1× PBST (1× PBS with 0.1% Triton X-100) for 8 min at room temperature (RT). The testes were rinsed three times in 1×PBST and washed three times for 5 min each time using 1× PBST at RT. Testes were incubated with primary antibodies in 1× PBST+3% bovine serum albumin at 4°C for at least one night. Samples were then rinsed three times in 1× PBST and washed three times for 5 min each time in 1× PBST and then incubated in a 1:1000 dilution of Alexa Fluor-conjugated secondary antibody in PBST+5% normal goat serum for 2 h at RT or rotating for a minimum of 24 h at 4°C. Samples were rinsed three times in 1× PBST and washed three times, 5 min each in 1× PBST and then mounted for microscopy in Vectashield antifade mounting medium (H-1400, Vector Laboratories) with or without DAPI. Samples were imaged using a Leica SP8 or Stellaris 5 confocal microscope with a 63× oil immersion objective. Images were analyzed using ImageJ software. Primary antibodies used were: Vasa (rabbit, 1:5000; from Ruth Lehmann, Skirball Institute of Biomolecular Medicine, NY, USA), Fas III (mouse, 1:50; DSHB, 7G10), Armadillo (mouse, 1:50; DSHB, N2 7A1), α-Spec (mouse, 1:50; DHSB, 3A9), 1B1 (mouse, 1:50; DSHB, 1B1-s), H3S10P (mouse, 1:2000; Abcam, ab14955), Zfh1 (rabbit, 1:5000; from Ruth Lehmann), TJ (guinea pig, 1:1000; from M. Van Doren, Johns Hopkins University, USA), H3K4me3 (rabbit, 1:400, Cell Signaling Technology, 9751S), GFP (chicken, 1:1000; Abcam, ab13970), Stat92E (rabbit, 1:200; gift from Denise Montell, University of Santa Barbara, CA, USA) and pMad (rabbit, 1:100; Abcam, ab52903). All secondary antibodies were Alexa Fluor Conjugated secondaries from Thermo Fisher Scientific used at 1:1000 dilution: goat anti mouse 405 A31553, 488 A23723, 568 A11004, 633 A21050, 647 A21235; goat anti rabbit 488 ab150077, 568 A11011, 633 A21202; goat anti guinea pig 488 A11073, 568 SAB4600080, 633 A21105; goat anti chicken 488 ab150169.
RNA-seq and data analysis
nos-Gal4; UAS-mCherry RNAi and nos-Gal4; UAS-set1 RNAi flies were collected as newly eclosed males and aged for 0, 1, 3 and 5 days at 25°C after shift from 18°C. Approximately 15 pairs of testes for each genotype were dissected in Schneider's media+10% fetal bovine serum as one replicate. Three replicates were generated for each time point and genotype. The testes were then desheathed in 500 μl of lysis buffer (trypsin LE+2 mg/ml collagenase). The samples were incubated in lysis buffer for 10 min in a 37°C water bath with gentle vortex mixing every 2 min. The samples were then filtered through a 40 μm tissue culture filter followed by a 10 min centrifugation at 1200 rpm (106 g). The cells were washed with 200 μl of PBS and pelleted again for 5 min at 1200 rpm (106 g). Total RNA was purified using the Quick-RNA Microprep Kit (R1050, Zymo Research Corporation) following the manufacturer's instructions. The libraries were generated using the reagents provided in NEBNext Ultra II Directional RNA library Prep Kit for Illumina (E7760S, New England Biolabs Inc.) and NEB Next Poly(A) mRNA Magnetic Isolation Module (E7490, New England Biolabs Inc.). The Illumina compatible libraries were sequenced with Illumina Novaseq6000 sequencer at the National Institutes of Health sequencing facility.
The sequencing reads were examined using FastQC quality software (Galaxy Version 0.73+galaxy0) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads that passed the quality filter were then mapped to the Drosophila genome (D. melanogaster August 2014; BDGP Release 6+ISO11 MT/dm6) using Bowtie 2 version 2.5.1 (Langmead and Salzberg, 2012). For gene mapping, the gene model (Drosophila_melanogaster.BDGP6.87.gtf) was utilized, and we followed an RNA-seq data analysis tutorial using Galaxy (https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/ref-based/tutorial.html), as detailed in the work (Batut et al., 2018; Hiltemann et al., 2023). Aligned reads were summarized to genes using featureCounts (Liao et al., 2014) with gene features obtained from NCBI RefSeq dm6 assembly. Raw counts were normalized to transcripts per million reads (TPM) counts. Principal component analysis was performed on 16,610 genes with detectable expression in at least one sample, based on which sample distance matrix was computed. To assess differential gene expression between Set1 knockdown and controls, DESeq2 (Love et al., 2014) was applied on samples from each day, using the Benjamini and Hochberg method for multiple testing correction with default DESeq2 parameters. A threshold of log2 fold change of 1.3 and adjusted P<0.05 was applied to ascertain differentially expressed genes, for which EnrichR (Chen et al., 2013) was applied to assess enrichment of molecular pathways in each set. All statistical analysis was conducted using R software version 4.2.0.
Phenotype quantification
All phenotypic quantification was carried out in Fiji (ImageJ). GSC number was determined by counting every cell that was either Vasa positive or Tj negative and directly next to the hub. Early cyst cell number was quantified by counting every Tj-positive cell in a whole testis z-stack. Hub area quantification was determined by drawing a line around the z-slice with the largest hub size based on Armadillo or Fasciclin III immunostaining and using the area measurement in Fiji. For the early germ cell proliferation phenotype, Armadillo was used as the marker to delineate the germline cysts (Feng et al., 2017). Any cyst with more than 16 cells was considered overpopulated.
Statistics and reproducibility
Data were subjected to the Shapiro–Wilk test to determine whether the data were normally distributed or skewed. For normally distributed data, an unpaired, two-sample t-test was used to compare two individual datasets with each other. For skewed data, the Wilcoxon signed rank test was used to compare two individual datasets with each other. A χ2 test was used to compare distributions of datasets containing data from different categories. Two-way ANOVA with interaction and Šidák multiple comparison test were used to compare two individual datasets with each other that had two independent variables. One-way ANOVA and Dunnett's T3 multiple comparison test were used when more than two individual datasets were compared with each other that had one independent variable. Data are presented with error bars representing the s.e.m. Significant differences based on these statistical analyses are noted by asterisks (*P<0.05, **P<10−2, ***P<10−3, ****P<10−4).
Supplementary Material
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data
Genetic interaction data
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data
Acknowledgements
We thank Drs S. Myong, D. Drummond-Barbosa, C. Wu and Y. Kim for critical comments. We thank Chen lab members for insightful suggestions. We thank Yingying Li for generating the transgenes. We thank Johns Hopkins Integrated Imaging Center for confocal imaging.
Footnotes
Author contributions
Conceptualization: V.V., X.C.; Methodology: V.V., A.S., T.L.; Software: V.V., T.L., W.L.K.; Validation: V.V.; Formal analysis: V.V., A.S., T.L., W.L.K.; Investigation: V.V., A.S.; Data curation: V.V., T.L., W.L.K.; Writing - original draft: V.V., X.C.; Writing - review and editing: V.V., T.L., X.C.; Visualization: V.V., A.S., T.L.; Supervision: X.C., J.Q., K.Z.; Project administration: X.C.; Funding acquisition: V.V., X.C., K.Z.
Funding
This work was supported by the National Institutes of Health (5T32GM007231 and F31GM134641 to V.V.; R35GM127075 and R01HD102474 to X.C.), the Division of Intramural Research of the National Heart, Lung, and Blood Institute (K.Z.), and the Howard Hughes Medical Institute (X.C.). Open Access funding provided by Howard Hughes Medical Institute. Deposited in PMC for immediate release.
Data availability
RNA-seq data have been deposited in Gene Expression Omnibus under accession number GSE254876.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.202729.reviewer-comments.pdf
References
- Amoyel, M., Anderson, A. M. and Bach, E. A. (2014). JAK/STAT pathway dysregulation in tumors: a Drosophila perspective. Semin. Cell Dev. Biol. 28, 96-103. 10.1016/j.semcdb.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel, M., Anderson, J., Suisse, A., Glasner, J. and Bach, E. A. (2016). Socs36E controls Niche competition by repressing MAPK signaling in the Drosophila testis. PLoS Genet. 12, e1005815. 10.1371/journal.pgen.1005815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali, M. B., Mei, A., Zobeck, K. L., Caron, M., Lis, J. T. and Kusch, T. (2011). Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30, 2817-2828. 10.1038/emboj.2011.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyub, C., Banerjee, K. K. and Joti, P. (2015). Reduction of Cullin-2 in somatic cells disrupts differentiation of germline stem cells in the Drosophila ovary. Dev. Biol. 405, 269-279. 10.1016/j.ydbio.2015.07.019 [DOI] [PubMed] [Google Scholar]
- Baeg, G.-H., Zhou, R. and Perrimon, N. (2005). Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 19, 1861-1870. 10.1101/gad.1320705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut, B., Hiltemann, S., Bagnacani, A., Baker, D., Bhardwaj, V., Blank, C., Bretaudeau, A., Brillet-Guéguen, L., Čech, M., Chilton, J.et al. (2018). Community-driven data analysis training for biology. Cell Syst. 6, 752-758.e751. 10.1016/j.cels.2018.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, B. and Blanpain, C. (2013). Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727-738. 10.1038/nrc3597 [DOI] [PubMed] [Google Scholar]
- Bello, B. C., Izergina, N., Caussinus, E. and Reichert, H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5. 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., Kouzarides, T. and Schreiber, S. L. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99, 8695-8700. 10.1073/pnas.082249499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone, J. Q. and Doe, C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185-1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. 10.1242/dev.118.2.401 [DOI] [PubMed] [Google Scholar]
- Bray, S., Musisi, H. and Bienz, M. (2005). Bre1 is required for Notch signaling and histone modification. Dev. Cell 8, 279-286. 10.1016/j.devcel.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Chen, D. and McKearin, D. M. (2003). A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159-1170. 10.1242/dev.00325 [DOI] [PubMed] [Google Scholar]
- Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., Clark, N. R. and Ma'ayan, A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Tiyaboonchai, A., Yamashita, Y. M. and Hunt, A. J. (2011). Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development 138, 831-837. 10.1242/dev.057901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, C. M. and Matunis, E. L. (2010). Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell 6, 557-567. 10.1016/j.stem.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, M. F. and Fuller, M. (2006). Stem cells and cancer: two faces of eve. Cell 124, 1111-1115. 10.1016/j.cell.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Clevers, H. (2005). Stem cells, asymmetric division and cancer. Nat. Genet. 37, 1027-1028. 10.1038/ng1005-1027 [DOI] [PubMed] [Google Scholar]
- Davies, E. L. and Fuller, M. T. (2008). Regulation of self-renewal and differentiation in adult stem cell lineages: lessons from the Drosophila male germ line. Cold Spring Harb. Symp. Quant. Biol. 73, 137-145. 10.1101/sqb.2008.73.063 [DOI] [PubMed] [Google Scholar]
- de Cuevas, M. and Matunis, E. L. (2011). The stem cell niche: lessons from the Drosophila testis. Development 138, 2861-2869. 10.1242/dev.056242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé, P.-M., Pamblanco, M., Luciano, P., Lebrun, R., Moinier, D., Sendra, R., Verreault, A., Tordera, V. and Géli, V. (2005). Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 353, 477-484. 10.1016/j.jmb.2005.08.059 [DOI] [PubMed] [Google Scholar]
- dos Santos, G., Schroeder, A. J., Goodman, J. L., Strelets, V. B., Crosby, M. A., Thurmond, J., Emmert, D. B., Gelbart, W. M. and FlyBase, C. (2015). FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43, D690-D697. 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, T., Kitzman, S. C. and Geyer, P. K. (2020). Survival of Drosophila germline stem cells requires the chromatin-binding protein Barrier-to-autointegration factor. Development 147, dev186171. 10.1242/dev.186171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., Lee, M. G., Schneider, J., Ilvarsonn, A., Shiekhattar, R. and Shilatifard, A. (2007). The Trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 14, 344-346. 10.1038/nsmb1217 [DOI] [PubMed] [Google Scholar]
- Eliason, J., Afify, A., Potter, C. and Matsumura, I. (2018). A GAL80 collection to inhibit GAL4 transgenes in Drosophila olfactory sensory neurons. G3 (Bethesda) 8, 3661-3668. 10.1534/g3.118.200569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun, S. H., Shi, Z., Cui, K., Zhao, K. and Chen, X. (2014). A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker. Science 343, 1513-1516. 10.1126/science.1246514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun, S. H., Feng, L., Cedeno-Rosario, L., Gan, Q., Wei, G., Cui, K., Zhao, K. and Chen, X. (2017). Polycomb group gene E(z) is required for spermatogonial dedifferentiation in Drosophila adult testis. J. Mol. Biol. 429, 2030-2041. 10.1016/j.jmb.2017.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L., Shi, Z. and Chen, X. (2017). Enhancer of polycomb coordinates multiple signaling pathways to promote both cyst and germline stem cell differentiation in the Drosophila adult testis. PLoS Genet. 13, e1006571. 10.1371/journal.pgen.1006571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L., Shi, Z., Xie, J., Ma, B. and Chen, X. (2018). Enhancer of polycomb maintains germline activity and genome integrity in Drosophila testis. Cell Death Differ. 25, 1486-1502. 10.1038/s41418-017-0056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, M. T. (1993). Spermatogenesis. In The Development of Drosophila melanogaster, Vol. I (ed. Bate M. and Martinez Arias A.), pp. 71-147. Cold Spring Harbor: Cold Spring Harbor Press. [Google Scholar]
- Fuller, M. T. and Spradling, A. C. (2007). Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402-404. 10.1126/science.1140861 [DOI] [PubMed] [Google Scholar]
- Gan, Q., Schones, D. E., Ho Eun, S., Wei, G., Cui, K., Zhao, K. and Chen, X. (2010). Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 11, R42. 10.1186/gb-2010-11-4-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara, A. C. P. and Drummond-Barbosa, D. (2022). Warm and cold temperatures have distinct germline stem cell lineage effects during Drosophila oogenesis. Development 149, dev200149. 10.1242/dev.200149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara, A. C. P. and Drummond-Barbosa, D. (2023). Chronic exposure to warm temperature causes low sperm abundance and quality in Drosophila melanogaster. Sci. Rep. 13, 12331. 10.1038/s41598-023-39360-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, R. J. and Chen, X. (2023). Epigenetic dynamics during germline development: insights from Drosophila and C. elegans. Curr. Opin. Genet. Dev. 78, 102017. 10.1016/j.gde.2022.102017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, R. J., Anand, A., Kai, T. and Chen, X. (2018). Protecting and diversifying the germline. Genetics 208, 435-471. 10.1534/genetics.117.300208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson, G., Hollebakken, R. E., Li, T., Syrzycka, M., Kim, I., Cotsworth, S., Fitzpatrick, K. A., Sinclair, D. A. R. and Honda, B. M. (2012). dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190, 91-100. 10.1534/genetics.111.135863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., Barrera, L. O., Van Calcar, S., Qu, C., Ching, K. A.et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311-318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Herrera, S. C. and Bach, E. A. (2019). JAK/STAT signaling in stem cells and regeneration: from Drosophila to vertebrates. Development 146, dev167643. 10.1242/dev.167643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltemann, S., Rasche, H., Gladman, S., Hotz, H.-R., Larivière, D., Blankenberg, D., Jagtap, P. D., Wollmann, T., Bretaudeau, A., Goué, N.et al. (2023). Galaxy training: a powerful framework for teaching! PLoS Comput. Biol. 19, e1010752. 10.1371/journal.pcbi.1010752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. S., Melnick, M. B. and Perrimon, N. (1996). Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84, 411-419. 10.1016/S0092-8674(00)81286-6 [DOI] [PubMed] [Google Scholar]
- Inaba, M., Buszczak, M. and Yamashita, Y. M. (2015). Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature 523, 329-332. 10.1038/nature14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis, M., Tulina, N., de Cuevas, M., Brawley, C., Sandler, L. and Matunis, E. (2009). JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 326, 153-156. 10.1126/science.1176817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahney, E. W., Snedeker, J. C. and Chen, X. (2019). Regulation of Drosophila germline stem cells. Curr. Opin. Cell Biol. 60, 27-35. 10.1016/j.ceb.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase, E., Wong, M. D., Ding, B. C. and Xie, T. (2004). Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131, 1365-1375. 10.1242/dev.01025 [DOI] [PubMed] [Google Scholar]
- Kiger, A. A., Jones, D. L., Schulz, C., Rogers, M. B. and Fuller, M. T. (2001). Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542-2545. 10.1126/science.1066707 [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849-860. 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugks, U., Hieke, M. and Wagner, N. (2017). MAN1 restricts BMP signaling during synaptic growth in Drosophila. Cell. Mol. Neurobiol. 37, 1077-1093. 10.1007/s10571-016-0442-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras, S. and Van Doren, M. (2006). Development of the male germline stem cell niche in Drosophila. Dev. Biol. 294, 92-103. 10.1016/j.ydbio.2006.02.030 [DOI] [PubMed] [Google Scholar]
- Leatherman, J. (2013). Stem cells supporting other stem cells. Front. Genet. 4, 257. 10.3389/fgene.2013.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman, J. L. and Dinardo, S. (2008). Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44-54. 10.1016/j.stem.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman, J. L. and Dinardo, S. (2010). Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell Biol. 12, 806-811. 10.1038/ncb2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit, T. and Cohen, S. M. (1997). Proximal-distal axis formation in the Drosophila leg. Nature 388, 139-145. 10.1038/40563 [DOI] [PubMed] [Google Scholar]
- Lee, J.-H., Tate, C. M., You, J.-S. and Skalnik, D. G. (2007a). Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419-13428. 10.1074/jbc.M609809200 [DOI] [PubMed] [Google Scholar]
- Lee, N., Zhang, J., Klose, R. J., Erdjument-Bromage, H., Tempst, P., Jones, R. S. and Zhang, Y. (2007b). The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 14, 341-343. 10.1038/nsmb1216 [DOI] [PubMed] [Google Scholar]
- Li, M. A., Alls, J. D., Avancini, R. M., Koo, K. and Godt, D. (2003). The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol. 5, 994-1000. 10.1038/ncb1058 [DOI] [PubMed] [Google Scholar]
- Li, W., Yao, A., Zhi, H., Kaur, K., Zhu, Y.-C., Jia, M., Zhao, H., Wang, Q., Jin, S., Zhao, G.et al. (2016). Angelman syndrome protein Ube3a regulates synaptic growth and endocytosis by inhibiting BMP signaling in Drosophila. PLoS Genet. 12, e1006062. 10.1371/journal.pgen.1006062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lim, J. G. Y. and Fuller, M. T. (2012). Somatic cell lineage is required for differentiation and not maintenance of germline stem cells in Drosophila testes. Proc. Natl. Acad. Sci. USA 109, 18477-18481. 10.1073/pnas.1215516109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick, V. P., Morris, L. X., Fox, D. T. and Spradling, A. (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 21, 159-171. 10.1016/j.devcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, E. L., Stine, R. R. and de Cuevas, M. (2012). Recent advances in Drosophila male germline stem cell biology. Spermatogenesis 2, 137-144. 10.4161/spmg.21763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. and Davis, R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765-1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- McKearin, D. M. and Spradling, A. C. (1990). bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4, 2242-2251. 10.1101/gad.4.12b.2242 [DOI] [PubMed] [Google Scholar]
- Miller, T., Krogan, N. J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J. F. and Shilatifard, A. (2001). COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98, 12902-12907. 10.1073/pnas.231473398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, M., Herz, H.-M., Smith, E. R., Zhang, Y., Jackson, J., Washburn, M. P., Florens, L., Eissenberg, J. C. and Shilatifard, A. (2011). The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31, 4310-4318. 10.1128/MCB.06092-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera, L., Lübbert, M. and Jung, M. (2016). Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenet. 8, 57. 10.1186/s13148-016-0223-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S. J. and Kimble, J. (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068-1074. 10.1038/nature04956 [DOI] [PubMed] [Google Scholar]
- Morrison, S. J. and Spradling, A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598-611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Kong, J. and Brat, D. J. (2015). Cancer stem cell division: when the rules of asymmetry are broken. Stem Cells Dev. 24, 405-416. 10.1089/scd.2014.0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P. L., Griesenbeck, J., Kornberg, R. D. and Cleary, M. L. (2002). A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99, 90-94. 10.1073/pnas.221596698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C. L., Qian, Y. and Schulz, C. (2019). Notch and Delta are required for survival of the germline stem cell lineage in testes of Drosophila melanogaster. PLoS ONE 14, e0222471. 10.1371/journal.pone.0222471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancratov, R., Peng, F., Smibert, P., Yang, J.-S., Jr, Olson, E. R., Guha-Gilford, C., Kapoor, A. J., Liang, F.-X., Lai, E. C., Flaherty, M. S.et al. (2013). The miR-310/13 cluster antagonizes β-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development 140, 2904-2916. 10.1242/dev.092817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok, D. K., Harbison, C. T., Levine, S., Cole, M., Hannett, N. M., Lee, T. I., Bell, G. W., Walker, K., Rolfe, P. A., Herbolsheimer, E.et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517-527. 10.1016/j.cell.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Raftery, L. A., Twombly, V., Wharton, K. and Gelbart, W. M. (1995). Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics 139, 241-254. 10.1093/genetics/139.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, R., Snedeker, J., Wooten, M., Chu, C., Bracero, S., Mouton, T. and Chen, X. (2022). Differential condensation of sister chromatids acts with Cdc6 to ensure asynchronous S-phase entry in Drosophila male germline stem cell lineage. Dev. Cell 57, 1102-1118.e1107. 10.1016/j.devcel.2022.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W. W., Wilm, M., Aasland, R. and Stewart, A. F. (2001). The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137-7148. 10.1093/emboj/20.24.7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C. T., Schreiber, S. L., Mellor, J. and Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407-411. 10.1038/nature01080 [DOI] [PubMed] [Google Scholar]
- Schulz, C., Kiger, A. A., Tazuke, S. I., Yamashita, Y. M., Pantalena-Filho, L. C., Jones, D. L., Wood, C. G. and Fuller, M. T. (2004). A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 167, 707-723. 10.1534/genetics.103.023184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell, S. (2010). On the stem cell origin of cancer. Am. J. Pathol. 176, 2584-2594. 10.2353/ajpath.2010.091064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard, A. (2012). The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65-95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani, A. A. and Ingham, P. W. (2003). Regulation of stem cell maintenance and transit amplifying cell proliferation by TGF-beta signaling in Drosophila spermatogenesis. Curr. Biol. 13, 2065-2072. 10.1016/j.cub.2003.10.063 [DOI] [PubMed] [Google Scholar]
- Simonet, T., Dulermo, R., Schott, S. and Palladino, F. (2007). Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 312, 367-383. 10.1016/j.ydbio.2007.09.035 [DOI] [PubMed] [Google Scholar]
- Singh, S. R., Zheng, Z., Wang, H., Oh, S. W., Chen, X. and Hou, S. X. (2010). Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J. Cell. Physiol. 223, 500-510. 10.1002/jcp.22073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A., Fuller, M. T., Braun, R. E. and Yoshida, S. (2011). Germline stem cells. Cold Spring Harb. Perspect. Biol. 3, a002642. 10.1101/cshperspect.a002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf, G., Devenport, D., Godt, D. and Brown, N. H. (2007). Integrin-dependent anchoring of a stem-cell niche. Nat. Cell. Biol. 9, 1413-1418. 10.1038/ncb1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayrah, L. and Chen, X. (2013). Epigenetic regulation in adult stem cells and cancers. Cell. Biosci. 3, 41. 10.1186/2045-3701-3-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayrah, L., Herz, H.-M., Shilatifard, A. and Chen, X. (2013). Histone demethylase dUTX antagonizes JAK-STAT signaling to maintain proper gene expression and architecture of the Drosophila testis niche. Development. 140, 1014-1023. 10.1242/dev.089433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayrah, L., Li, Y., Gan, Q. and Chen, X. (2015). Epigenetic regulator Lid maintains germline stem cells through regulating JAK-STAT signaling pathway activity. Biol. Open 4, 1518-1527. 10.1242/bio.013961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracol, R. and Lengyel, J. A. (1994). The thick veins gene of Drosophila is required for dorsoventral polarity of the embryo. Genetics 138, 165-178. 10.1093/genetics/138.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, J., Brenner, T. J. and DiNardo, S. (2000). Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature 407, 754-757. 10.1038/35037613 [DOI] [PubMed] [Google Scholar]
- Tulina, N. and Matunis, E. (2001). Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546-2549. 10.1126/science.1066700 [DOI] [PubMed] [Google Scholar]
- Van Doren, M., Williamson, A. L. and Lehmann, R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243-246. 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Venkei, Z. G. and Yamashita, Y. M. (2018). Emerging mechanisms of asymmetric stem cell division. J. Cell Biol. 217, 3785-3795. 10.1083/jcb.201807037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W. M. P., van Schaik, F. M. A., Varier, R. A., Baltissen, M. P. A., Stunnenberg, H. G., Mann, M. and Timmers, H. T. M. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58-69. 10.1016/j.cell.2007.08.016 [DOI] [PubMed] [Google Scholar]
- Vidaurre, V. and Chen, X. (2021). Epigenetic regulation of drosophila germline stem cell maintenance and differentiation. Dev. Biol. 473, 105-118. 10.1016/j.ydbio.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, N., Weyhersmüller, A., Blauth, A., Schuhmann, T., Heckmann, M., Krohne, G. and Samakovlis, C. (2010). The Drosophila LEM-domain protein MAN1 antagonizes BMP signaling at the neuromuscular junction and the wing crossveins. Dev. Biol. 339, 1-13. 10.1016/j.ydbio.2009.11.036 [DOI] [PubMed] [Google Scholar]
- Wysocka, J., Swigut, T., Xiao, H., Milne, T. A., Kwon, S. Y., Landry, J., Kauer, M., Tackett, A. J., Chait, B. T., Badenhorst, P.et al. (2006). A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86-90. 10.1038/nature04815 [DOI] [PubMed] [Google Scholar]
- Xie, J., Wooten, M., Tran, V., Chen, B.-C., Pozmanter, C., Simbolon, C., Betzig, E. and Chen, X. (2015). Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the Drosophila male germline. Cell 163, 920-933. 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, T., Xin, T., He, J., Tan, J., Gao, Y., Feng, S., He, L., Zhao, G. and Li, M. (2013). dBre1/dSet1-dependent pathway for histone H3K4 trimethylation has essential roles in controlling germline stem cell maintenance and germ cell differentiation in the Drosophila ovary. Dev. Biol. 379, 167-181. 10.1016/j.ydbio.2013.04.015 [DOI] [PubMed] [Google Scholar]
- Yamashita, Y. M., Jones, D. L. and Fuller, M. T. (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547-1550. 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yan, D., Neumüller, R. A., Buckner, M., Ayers, K., Li, H., Hu, Y., Yang-Zhou, D., Pan, L., Wang, X., Kelley, C.et al. (2014). A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 28, 459-473. 10.1016/j.devcel.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. and Hsu, Y. C. (2017). Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip. Rev. Dev. Biol. 6, e282. 10.1002/wdev.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zion, E. H., Chandrasekhara, C. and Chen, X. (2020). Asymmetric inheritance of epigenetic states in asymmetrically dividing stem cells. Curr. Opin. Cell Biol. 67, 27-36. 10.1016/j.ceb.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data
Genetic interaction data
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data
nanos-Gal4 driven Control RNAi data
nanos-Gal4 driven set1 RNAi data
nos-Gal4, tub-Gal80ts driven Control RNAi data
nos-Gal4, tub-Gal80ts driven set1 RNAi data
tj-Gal4 driven RNAi data
bam-Gal4 driven RNAi data
nos-Gal4, bam-Gal80 driven RNAi data