Abstract
The olfactory sensory neurons of vinegar flies and mice tend to express a single ligand-specific receptor. While this ‘one neuron-one receptor’ motif has long been expected to apply broadly across insects, recent evidence suggests it may not extend to mosquitoes. We sequenced and analyzed the transcriptomes of 46,000 neurons from antennae of the dengue mosquito Aedes aegypti to resolve all olfactory, thermosensory, and hygrosensory neuron subtypes and identify the receptors expressed therein. We find that half of all olfactory subtypes coexpress multiple receptors. However, coexpression occurs almost exclusively among genes from the same family—among odorant receptors (ORs) or among ionotropic receptors (IRs). Coexpression of ORs with IRs is exceedingly rare. Many coexpressed receptors are recent duplicates. In other cases, the recruitment or co-option of single receptors by multiple neuron subtypes has placed these genes together in the same cells with distant paralogs. Close examination of data from Drosophila reveal rare cases of both phenomena, indicating that the olfactory systems of these two species are not fundamentally different, but instead fall at different locations along a continuum likely to encompass diverse insects.
INTRODUCTION
The olfactory systems of vinegar flies and mice share a common molecular and circuit logic despite what are likely to be independent evolutionary origins1,2. Airborne chemicals are detected by large arrays of olfactory sensory neurons (OSNs) scattered across peripheral tissues. Each sensory neuron tends to express a single ligand-specific receptor, and all neurons that express the same receptor converge on the same spatially discrete glomerulus in the brain2,3. The singular expression of just one receptor per olfactory sensory neuron is often highlighted as a way to limit the tuning breadth and overlap of individual neurons/glomeruli, enabling discrimination among odorants via a combinatorial code4,5.
Regardless of its selective advantages, the fact that both mouse and vinegar fly OSNs canonically express just one receptor suggests that this molecular motif is functionally important and should apply broadly across vertebrates and insects alike. Yet it has been clear for over a decade that the mosquito Aedes aegypti does not conform. Ae. aegypti is a tropical mosquito that specializes in biting humans and serves as the primary vector of dengue, Zika, chikungunya and yellow fever viruses6. Biting females rely heavily on their sense of smell to identify humans7 and express about twice as many receptors in olfactory tissues (n~130)8,9 as they have glomeruli in their antennal lobes (n~60-80)10–13. Recent work suggests that this mismatch is at least partly explained by coexpression of multiple receptors within individual OSNs12. Insect olfactory receptors come from two large families, the odorant receptors (ORs) and ionotropic receptors (IRs). There is evidence that coexpression may occur among both receptors from the same family and receptors from different families (Fig. 1A).
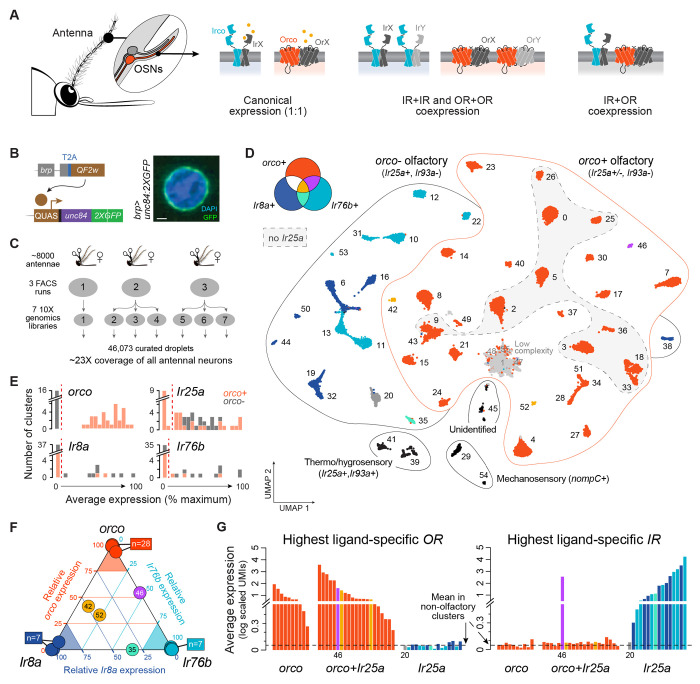
Figure 1. Segregation of ligand-specific ORs and IRs across female Ae. aegypti antennae.
(A) Three models of receptor expression in mosquito olfactory sensory neurons (OSNs): canonical expression of one receptor per neuron (left), within-family coexpression (middle), between-family coexpression (right). Orco and ‘Irco’ stand for conserved OR and IR family coreceptors. (B) Transgenic strategy used to express nuclear membrane-bound GFP in neurons. Confocal image of labeled nucleus. Scale bar, 1 μm. (C) Single nucleus RNAseq data collection schematic. (D) Annotated UMAP of 46,073 nuclei from female antennal neurons (n=52 major clusters). OSN cluster colors reflect on/off expression of orco, Ir8a, and Ir76b as shown. (E) Average expression of olfactory co-receptors in OSN clusters, computed as percent gene-specific maximum. Red dashed lines show cutoffs used to call expression in (D). (F) Triangle plot illustrating segregated expression of orco, Ir8a, and Ir76b. Position of each OSN cluster (circle) reflects relative expression of the 3 genes (where expression is first computed as percent max as in (E)). (G) Expression level of the most highly expressed ligand-specific OR (left) or IR (right) in each OSN cluster. Cluster 46 (purple) is the only cluster that expresses substantial levels of both an OR and an IR. Cluster 20 (dark grey) expresses neither ORs nor IRs (but instead an ammonium transporter, see Fig. 3). Dashed black line shows mean for mechanosensory neurons. Colors in (F–G) as in (D).
Coexpression of ORs with IRs is particularly surprising. The neurons that express these two types of receptors were originally thought to be segregated in developmentally and anatomically distinct sensory hairs and to make up two independent olfactory subsystems14,15. However, a conserved coreceptor for the IR family was recently shown to be expressed in a large fraction of OR neurons in both Drosophila16 and Ae. aegypti12, generating excitement about the possibility that ligand-specific receptors from the two families also enjoy broad coexpression. Indeed, there are hints that both within- and between-family coexpression are widespread in Ae. aegypti, but the best data come from a small subset of OSN subtypes on the maxillary palp12. The full extent and nature of coexpression in this important insect remain fuzzy.
Here we conduct single-nucleus RNA sequencing of 46K neurons from female antennae to generate a comprehensive map of neuronal diversity and expression across the primary olfactory organ of Ae. aegypti. We find that coexpression is common among ligand-specific receptors from the same family but that coexpression between ORs and IRs is extremely rare, contrary to recent expectations. While many examples of within-family coexpression involve tandem duplicates, we also identify an unusual subset of ORs that have been co-opted by multiple OSN subtypes and are now expressed side-by-side with distant paralogs. Our work provides a new perspective on olfactory organization in insects and specific information about a mosquito that uses its sense of smell to transmit dangerous arboviruses to hundreds of millions of people each year.
RESULTS
Segregated expression of ligand-specific ORs and IRs across antennal neurons
We used single-nucleus RNA sequencing to construct a comprehensive molecular atlas of neurons from adult female antennae. Antennae harbor nearly all of the 60+ adult OSN subtypes (excluding only 3 housed on the maxillary palp) and therefore provide a broad picture of OSN diversity and organization. We first developed a transgenic strategy to label the nuclear envelope of all neurons with GFP (Fig. 1B)17,18 so that they could be sorted from other dissociated antennal nuclei and sequenced efficiently (Fig. 1C, Fig. S1). We then optimized in silico mRNA signal detection by updating the AaegL5 genome annotation in two important ways; we added a handful of missing ORs, IRs, and GRs and systematically extended 3’ UTRs where necessary to capture read pileups falling within 750bp of the annotated transcription stop site of any gene (Fig. S2, see Methods). Subsequent preprocessing, ambient RNA decontamination, and doublet removal (Fig. S3) left us with data from a total of 46,073 curated droplets, hereafter referred to as ‘nuclei’ or ‘neurons’. These data represent 23X coverage of the ~2000 neurons present on a single female Ae. aegypti antenna19.
Initial analysis of the full dataset using the UMAP algorithm resulted in 52 well defined clusters (Fig. 1D, Fig. S4–5). We identified two clusters of putative mechanosensory neurons expressing the mechanotransduction channel nompC20, two clusters of putative heat- and humidity-sensing neurons expressing the non-olfactory IR coreceptor Ir93a21, and one cluster of unidentified neurons (Fig. 1D, Fig. S5). The remaining 47 clusters were classified as OSNs due to expression of one or more conserved olfactory coreceptors (see below). While some of these clusters are likely heterogenous, representing multiple related OSN subtypes, they allow an initial look at gross patterns of OR and IR expression.
We identified discrete expression thresholds for each olfactory coreceptor (Fig. 1E) and examined patterns of overlap across OSN clusters. Ligand-specific ORs work in complex with the OR coreceptor orco14,22, which showed a striking on/off pattern of expression leading to a primary classification of OSN clusters as orco− or orco+ (Fig. 1D–E). Ligand-specific IRs work in complex with one or more of three IR coreceptors, Ir25a, Ir8a, and Ir76b14,23,24. We observed minimal expression of Ir8a and Ir76b in orco+ clusters: only 3 of 31 orco+ clusters expressed one or both of these genes (Fig. 1D,1F). In contrast, Ir25a was broadly coexpressed with orco. It was present in 21 of 31 orco+ clusters as well as all orco− OSN clusters (Fig. 1D).
The presence of Ir25a in over half of orco+ clusters corroborates recent findings12,16 and suggests that these unrelated receptor families may collaborate to define the odor tuning of many OSNs. If true, we should see coexpression of not only coreceptors, but also ligand-specific ORs and IRs. Strikingly, however, only 1 of 21 orco+/Ir25a+ clusters expressed ligand-specific receptors from both families at appreciable levels (Fig. 1G, #46). The remaining 20 clusters expressed at least one OR (Fig. 1G, left), but contained IR transcripts at background levels, similar to those observed in orco-only or non-olfactory neurons (Fig. 1G, right). While we cannot rule out the possibility that even very low levels of IR expression mediate the production of functional IRco/IR complexes, we instead propose that Ir25a has a non-odor-tuning function in the vast majority of orco+ OSNs in which it is found and see no reason to abandon the longstanding view of two largely distinct olfactory subsystems in insects.
Twelve subtypes of heat- and humidity-sensing neurons
Before further exploring olfactory neurons, we sought to test the ability of our data to resolve rare cell types by focusing on thermo- and hygrosensory neurons (THSNs). Ae. aegypti antennae harbor at least 4 types of THSNs, termed heating, cooling, dry, and moist cells19,21,25. Most of these cells express the non-olfactory IR coreceptor Ir93a21, and research in both Drosophila and mosquitoes suggests that Ir93a works in concert with modality-specific IR partners, including Ir21a (cooling), Ir40a (dry), and Ir68a (moist)21,26–28. Importantly, THSNs are sparsely distributed across the most basal and distal antennal segments (Fig. 2A), with some subtypes likely comprising just 1-3 neurons per antenna19,21,25.
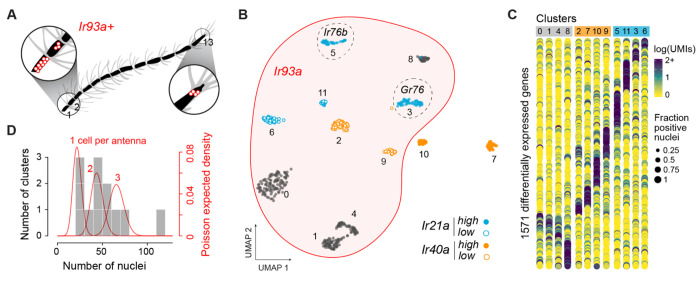
Figure 2. Deep snRNA-sequencing resolves twelve small subpopulations of candidate thermo- and hygrosensors.
(A) Location of Ir93a+ thermo- and hygrosensory neurons on segments 1, 2, and 13 of female Ae. aegypti antennae19,21. (B) Annotated UMAP showing reclustering of 589 nuclei from two Ir93a+ clusters in the original analysis (Fig. 1D). Note that 2 of the 12 new clusters do not expres Ir93a. (C) Dotplot showing expression of 1571 genes differentially expressed across clusters (log2FC>0.3). (D) Histogram showing number of nuclei in each cluster (grey bars) overlaid by the expected densities for cell types comprising 1, 2, or 3 neurons per antenna given 23X coverage (red lines).
In silico subsetting and reclustering of nuclei from the two Ir93a+ clusters in the all-neuron analysis revealed 12 putative subtypes of THSNs (Fig. 2B, Fig. S6–7), each with a strikingly unique transcriptional profile (Fig. 2C).
The cooling receptor Ir21a was expressed in four clusters, one of which was notable for coexpression of the IR coreceptor Ir76b, and another for coexpression of a gustatory receptor (Fig. 2B, Fig. S7), patterns that have not been documented in Drosophila. The dry receptor Ir40a was also expressed in four clusters, two of which did not express Ir93a despite having been lumped with other Ir93a+ cell types in the all-neuron analysis (Fig. 2B, Fig. S7). These Ir40+/Ir93a− nuclei may correspond to elusive heating cells21. As predicted, the number of nuclei in all but one of the 12 THSN clusters ranged from 25-75, consistent with cell types that comprise just 1-3 cells per antenna (Fig. 2D, Fig. S8). Taken together, this analysis reveals exciting cellular diversity among THSNs, along with candidate receptors and a list of marker genes that may be used to gain genetic access for functional studies. It also confirms the ability of our data to resolve rare cell types.
IR coexpression across orco− OSNs
Coexpression of ligand-specific ORs and IRs is rare (Fig. 1G), but coexpression among receptors from the same family may still be widespread. We first set out to explore this possibility among IRs in orco− OSNs. To resolve as many neuron subtypes as possible, we subsetted and iteratively cleaned and reclustered nuclei from the 16 original orco− clusters (Fig. 1D, see Methods). This process identified 4 additional clusters or subclusters (hereafter simply ‘clusters’) for a total of 20 (Fig. 3A, Fig. S6,S9–11). As seen in the original analysis, all clusters expressed Ir25a, and almost all additionally expressed either Ir8a or Ir76b, but rarely both (Fig. 3A–B). The total number of orco−, Ir8a+, and Ir76b+ clusters in our analyses agreed remarkably well with the number of antennal glomeruli identified in each category in recent work (Fig. S9B)12. We therefore conclude that most, if not all, clusters represent single, homogenous OSN subtypes with unique glomerular targets.
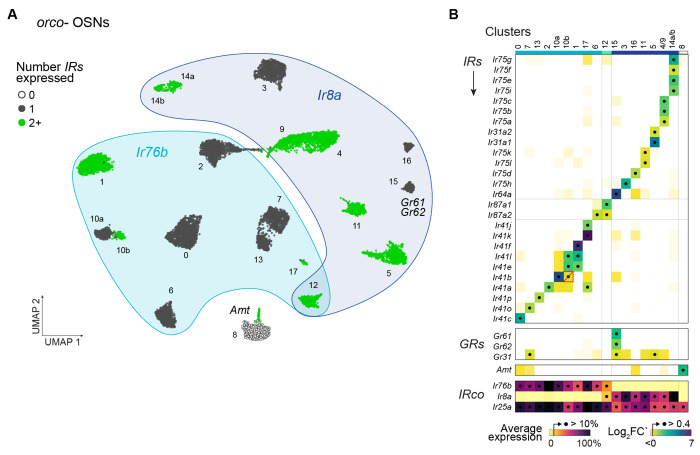
Figure 3. IR coexpression across orco− olfactory neurons.
(A) Annotated UMAP showing reclustering of 12,243 nuclei assigned to orco− OSN clusters in the original analysis (Fig. 1D). (B) Differential expression of ligand-specific receptors across clusters. All IRs, ORs, and GRs, exceeding a log2FC’ cutoff of 0.4 in any cluster are shown, but few GRs and no ORs met this criterion. The red box/slash marks an expression call (Ir41b in cluster 10b) that is likely spurious. Clusters expressing the same receptors were merged (#4/9, 14a/b). See Fig. S12 for log2FC’ and log average expression in all clusters examined separately. IR co-receptor expression shown at bottom as percent gene-specific maximum.
To identify ligand-specific receptors expressed in each orco− subtype, we looked for IRs that were differentially expressed across clusters (Fig. 3B). More specifically, we computed log2FC’ for all IRs in all clusters and chose a cutoff (0.4) that separated a large number of near-zero values from a much smaller number of clearly elevated values (Fig. S12A). Any IR exceeding this threshold was considered expressed in the given cluster. Use of an alternative absolute expression cutoff produced nearly identical expression calls (Fig. S12B–C). We also tested ORs and GRs, but observed no expression of the former and only sporadic expression of the latter (Fig. 3B).
Nine of the 20 putative orco− OSN subtypes expressed a single ligand-specific IR, while 10 expressed two or more IRs (Fig. 3A–B). Importantly, when two IRs were called as expressed in the same cluster, they also showed correlated expression at the neuron level (Fig. S12D–E), corroborating that these represent true cases of coexpression. The only exception involved Ir41b, whose elevated expression in cluster 10b likely results from a clustering artifact (Fig. 3B, red outline/slash; see also Fig. S12E). In two cases, transcriptionally similar clusters appeared to express the same or very similar subsets of IRs (Fig. S10, S12B–C, #4/9 and 14a/b). We conservatively assume these correspond to single OSN subtypes and show merged data in Fig. 3B.
One putative orco− OSN subtype expressed neither Ir8a, Ir76b, nor any ligand-specific IR (Fig. 3A–B, #8). Instead, this subtype expressed the ammonium transporter Amt (Fig. 3A–B; Fig. S11D), which is required for ammonia detection in Drosophila29, though perhaps not in malaria mosquitoes30,31. These neurons should be of great interest in Ae. aegypti as ammonia is a key host odorant, synergizing with lactic acid and carbon dioxide to attract biting females32.
OR coexpression and multi-expression across orco+ OSNs
We next sought to characterize patterns of OR expression among orco+ sensory neurons using a similar approach—by iteratively cleaning and reclustering nuclei from the original orco+ clusters (Fig. 1D) and using differential and absolute expression to identify ligand-specific receptors expressed therein. We identified 40 orco+ clusters in total (Fig. 4A, Fig. S9,S13–14), which conservatively correspond to 37 putative OSN subtypes after merging pairs of clusters that expressed the same or similar receptors (Fig. S15). This number is again consistent with a 1:1 match between clusters and neural subtypes based on recent counts of orco+ antennal lobe glomeruli (Fig. S9B)12.
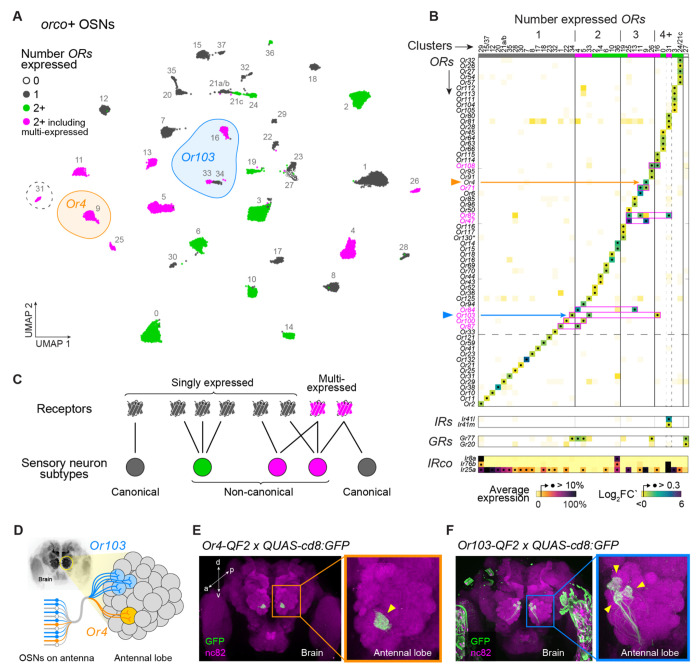
Figure 4. OR coexpression and multi-expression across orco+ olfactory neurons.
(A) Annotated UMAP showing reclustering of 28,807 nuclei assigned to orco+ OSN clusters in the original analysis (Fig. 1D). Dashed circle marks the only cluster (#31) that expressed ligand-specific IRs. (B) Differential expression of ligand-specific receptors across 37 major clusters. All ORs, IRs, and GRs with log2FC’ > 0.3 in any cluster are shown (see Fig. S15 for lower cutoff). Clusters expressing the same receptors were merged (#15/37, 21a/b, 24/21c). See Fig. S15 for log2FC’ and log average expression in all clusters examined separately. IR co-receptor expression shown at bottom as percent gene-specific maximum. Pink boxes highlight expression calls for multi-expressed ORs. (C) Schematic illustrating classification of receptors and OSN subtypes based on patterns of expression. (D) OSNs (bottom left) project axons to antennal lobe glomeruli (right) according to subtype. Data in (B) suggest that Or4 and Or103 are expressed in 1 and 3 OSN subtypes, respectively. (E–F) Antibody stainings show the GFP-expressing axons of Or4 (E) and Or103 (F) neurons in the adult female antennal lobe. Yellow arrowheads mark target glomeruli. See Methods for details.
As seen in the original analysis, approximately two thirds of orco+ subtypes expressed the IR coreceptor Ir25a, but only 1 expressed ligand-specific IRs (Fig. 4B, #31). In contrast, almost all orco+ subtypes expressed at least one OR, and approximately half coexpressed multiple ORs (Fig. 4B, Fig. S15A–B). Without exception, coexpression calls were again supported by elevated correlations at the neuron level (Fig. S15D–E). OR+OR coexpression in orco+ OSNs thus appears to be just as common as IR+IR coexpression in orco− OSNs.
We were intrigued to find not only coexpression, but also a phenomenon we call ‘multi-expression’. This refers to cases where the same receptor is present in multiple neuron subtypes (Fig. 4C). For example, Or103 was expressed in three putative subtypes (Fig. 4A). It was alone in cluster 34, coexpressed with Or125 in cluster 33, and coexpressed with Or108, Or114, and Or115 in cluster 16 (Fig. 4B). Likewise, while Or4 was only expressed in a single cluster (Fig. 4A, cluster 9), it was coexpressed in that cluster with two receptors (Or47 and Or71) that were each present alongside different partners in second clusters (Fig. 4B). In total, our data indicate that 8 of 63 antennal ORs are multi-expressed (Fig. 4B) and that 13 of 37 orco+ subtypes express at least one of these genes.
To provide independent support for multi-expression, we used CRISPR/Cas9 to generate driver lines for both Or4 and Or103, crossed these drivers to an existing GFP effector33, and counted the number of labeled glomeruli.
Importantly, our driver lines leverage the cis-regulatory elements of the target receptors in situ (Fig. S16), which is expected to more faithfully replicate endogenous patterns of expression than alternative approaches. If, as our snRNAseq data suggest, Or4 is expressed in one OSN subtype and Or103 is expressed in three OSN subtypes, then the Or4 and Or103 lines should drive expression of a GFP effector in neurons that target 1 and 3 glomeruli, respectively (Fig. 4D). This is exactly what we observed (Fig. 4E–F).
Two drivers of olfactory receptor coexpression
Receptor coexpression has multiple potential sources. The most likely is recent tandem gene duplication, wherein errors in DNA replication or recombination generate two adjacent copies of an ancestral gene. New duplicates are typically coexpressed because they share or inherit the same regulatory elements34. Importantly, this type of gene duplication is expected to drive coexpression among genes that are both physically and phylogenetically close. A second potential source of receptor coexpression is the recruitment of preexisting receptors to novel OSN subtypes. For example, the multi-expression observed for several ORs may arise when a singly expressed gene is co-opted by a second neural subtype following cis-regulatory evolution, trans-regulatory evolution, or cell type birth. This process would lead to coexpression between the newly recruited receptor and other receptors in the second cell type, which we would not necessarily expect to be closely related or physically adjacent. We can think of these two phenomena as coexpression ‘by descent’ and coexpression ‘by co-option’.
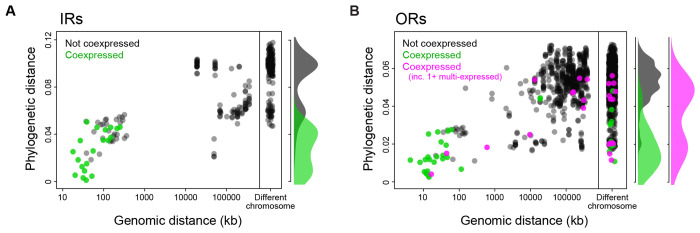
To identify the source of coexpression among Ae. aegypti IRs and ORs, we examined pairwise phylogenetic and chromosomal distances for receptors that were or were not coexpressed. Strikingly, all 20 pairs of coexpressed IRs were closely related and located in the same genomic cluster, consistent with a model of coexpression by descent (Fig. 5A). Many coexpressed ORs were also closely related and physically near—especially those that were singly expressed (green dots/density in Fig. 5B). However, multi-expressed ORs were rarely located in the same genomic cluster as their coexpressed partners and could be either closely or distantly related, consistent with a model of coexpression by co-option (Fig. 5B). Taken together, these patterns strongly suggest that IR+IR coexpression is driven predominantly by the coinheritance of ancestral regulatory elements during gene duplication, while OR+OR coexpression is driven not only by gene duplication, but also but the co-option of a subset of receptors to new neural subtypes.
Figure 5. OR coexpression is driven by both receptor duplication and receptor co-option.
Plots show the genomic and phylogenetic distance for pairs of ligand-specific IRs (A) or ORs (B). Color indicates whether the receptors are coexpressed in the same OSN subtype. Coexpressed ORs are further divided into cases where both receptors are found only in the same, single subtype (green) or at least one receptor is also found in additional subtypes (pink). Marginal densities for phylogenetic distance shown at right of each scatterplot. Phylogenetic distance in units of amino acid changes/site.
DISCUSSION
Recent work in Ae. aegypti mosquitoes challenges the longstanding view that the olfactory systems of most insects will resemble those of vinegar flies and mice, with each sensory cell type expressing a single ligand-specific receptor12. Yet exactly how and to what extent this ‘rule’ is broken has remained uncertain. Here we use deep single-nucleus RNA sequencing data to generate a surprisingly clear picture of neuronal diversity and receptor expression across female Ae. aegypti antenna. We show that half of all olfactory neurons coexpress multiple receptors, but that coexpression occurs almost exclusively among genes from the same receptor family—among IRs or among ORs. Coexpression of genes from different families is rare. We also identify an unexpected evolutionary driver of coexpression—the co-option of single receptors by multiple neural subtypes.
The segregation of ORs and IRs in all but one of ~60 antennal olfactory cell types adds nuance to the recent discovery of coexpression among coreceptors12,16. Ir25a is broadly coexpressed with orco, but it is unlikely to support odorant detection without a ligand-specific partner (let alone the help of a second IR coreceptor23,24). We instead hypothesize that Ir25a has an alternative, non-odor tuning function in most orco+ cells, perhaps acting during development or regulating other aspects of sensory transduction16. At the same time, the rarity of OR+IR coexpression makes the few instances where it does occur interesting from a behavioral perspective. For example, OR+IR neurons are uniquely poised to drive innate behavioral responses to structurally unrelated compounds with the same ecological meaning12. It remains to be seen whether this is true in Ae. aegypti, but it appears likely in a recent example from female hawkmoths35. In addition to the one OR+IR cell we identify on antennae, ligand-specific IRs have been documented in two orco+ cells on the maxillary palp, albeit at relatively low levels12.
While OR+IR coexpression is rare, we confirm widespread coexpression of receptors from the same family. Inferences of coexpression based on snRNA sequencing have some inherent limitations. Of greatest concern is the possibility that some of the transcripts we detect in nuclei are not exported and/or translated. For example, recent work in the clonal raider ant revealed cotranscription of dozens of tandem ORs within single nuclei, but only transcripts from the most upstream locus made their way into the cytoplasm36. This interesting phenomenon is likely specific to ants, which have the largest OR repertoires among insects. However, other, more limited forms of post-transcriptional repression have been detected among Drosophila IRs37 and could easily be present in Ae. aegypti. Conversely, the three-prime sequencing used in our study and most other snRNAseq studies will miss cases of coexpression among tandem genes found on a single polycistronic transcript, as has been observed in Anopheles mosquitoes38. We do not expect these phenomena to change our overall conclusions, but further work will be needed to confirm the precise rate at which Ae. aegypti OSNs employ multiple ligand-specific receptors for odor detection. Moving forward it will also be critical to understand how multiple ORs or multiple IRs interact, either directly or indirectly, to define the odor tuning of an OSN.
One of our most surprising findings is that OR+OR coexpression stems not only from gene duplication, but also from the co-option of a subset of receptors by multiple olfactory cell types. We do not know exactly how such co-option occurs, but one intriguing possibility is that it sometimes coincides with the birth of new cell types. Work in Drosophila suggests that new olfactory neurons can evolve via the repression of programmed cell death in precursor cells that would otherwise be eliminated during development, and that these ‘undead’ neurons initially express one or more receptors already present in other olfactory cells39. Multi-expressed genes in Ae. aegypti could represent the vestiges of such a process. Regardless of exactly how, when, or why co-option occurs, multi-expressed ORs are clearly unique in that they are frequently coexpressed with distantly related receptors (Fig. 5B).
Our work and that of others raises the question of why and when we might expect any animal to express only one vs. multiple receptors per OSN. Singular expression is often highlighted as a way to limit the tuning breadth/overlap of individual neurons in support of combinatorial coding4,5. This may explain why OSNs should express few receptors, but it is not a satisfying explanation for why OSNs must express just one receptor. Strict singular expression in vertebrates likely reflects developmental constraints more than any particular coding strategy. Vertebrate olfactory systems have hundreds to thousands of OSN subtypes—perhaps too many to be efficiently specified by conventional transcription factor codes. Instead, each mouse OSN stochastically expresses a single receptor, which then defines the tuning of the neuron and helps direct axon development to ensure proper targeting in the brain40. Interestingly, there are hints that ants, which also have complex olfactory systems, have independently converged on a receptor-dependent developmental strategy that enforces singular expression36,41. It is harder to explain why insects with deterministic olfactory development, like D. melanogaster42,43, would so closely follow the one neuron-one receptor rule. It is possible that singular expression helps to optimize combinatorial coding, even if not strictly required, and is therefore advantageous to species that rely heavily on learning. Coexpression of small numbers of receptors, in contrast, may allow evolution to more precisely tune neurons to important resources or threats in animals for which learning is less important. Or perhaps the number of receptors expressed per neuron reflects non-ecological factors like genome size and dynamics, with selection for compact genomes favoring smaller receptor repertoires and the proliferation of repetitive elements in larger genomes facilitating receptor family expansion.
Regardless of their differences, we would argue that Ae. aegypti and D. melanogaster are fundamentally similar. Drosophila OSNs are more likely to express just one receptor, but exceptions exist3,44,45, and close examination of those exceptions reveals the same signatures of coexpression by descent and coexpression by co-option that we see in mosquitoes (Fig. 6). Patterns of receptor expression are thus qualitatively (if not quantitatively) similar. We propose that the dengue fever mosquito and vinegar fly simply lie at different positions along a continuum that is likely to encompass most insects.
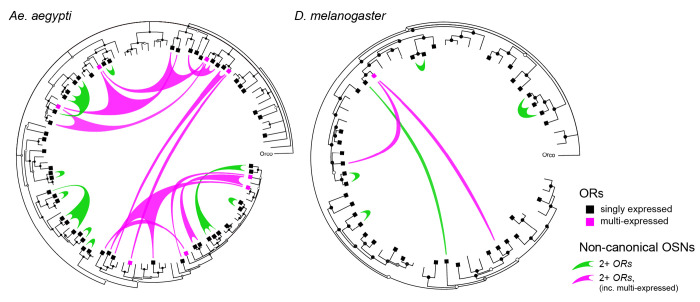
Figure 6. Patterns of OR coexpression in Ae. aegypti and D. melanogaster antennae.
Inward circle phylogenies show the evolutionary relationships among ORs found in the genomes of the dengue mosquito (left) and vinegar flies (right). Squares mark genes expressed in antennae. Green and pink ‘sails’ represent non-canonical OSN subtypes (i.e. those that express multiple receptors) and extend fingers to each receptor expressed therein, with colors distinguishing subtypes that do or do not express at least one multi-expressed gene (as in Fig. 4A). Taken together, coexpression is much less common in vinegar flies than in the dengue mosquito (fewer sails in tree on right), but shows the same signatures of coexpression by descent among singly expressed receptors (green sails tend to connect closely related ORs) and coexpression by co-option involving multi-expressed receptors (pink sails crisscross the center to connect distant ORs). Trees inferred from protein sequences using the program BAli-Phy (see Methods). Circles at tree nodes indicate bootstrap support: open >0.5, grey >0.7, black >0.9. Receptor expression data for Drosophila antennae taken from3,44,45.
MATERIALS and METHODS
Ethics and regulatory information.
Human-blood feeding conducted for mosquito colony maintenance did not meet the definition of human subjects research, as determined by the Princeton University IRB (Non Human-Subjects Research Determination #6870).
Mosquito rearing and colony maintenance.
All female mosquitoes dissected in this study were reared at 26°C, 75% RH on a 14:10 light/dark cycle. Eggs were hatched in a broth containing deoxygenated water and Tetramin Tropical Tablets (Pet Mountain, 16110M) and larvae were fed additional tablets ad libitum through pupation. Male and female pupae were transferred to plastic buckets or bugdorm cages. Adults were allowed unlimited access to 10% sucrose solution. Where applicable, females were allowed to bloodfeed on a human arm and lay eggs on wet filter paper (Whatman, 09-805B). All transgenic strains used in this study were derived from the Orlando (ORL) laboratory strain except for 15XQUAS-mCD8:GFP strain33, which has a Liverpool (LVP) genetic background.
Generation of 15XQUAS-unc84:2XeGFP transgenic strain.
pBac-mediated transposition was used according to previously published methods46. Briefly, the coding sequence of the chimeric protein unc84:2xGFP17 was isolated from the original pMUH_unc84_2XGFP plasmid (Addgene #46023) via PCR using the following primers (5’-AACAGATCTGCGGCGGCAAAATGGCTCCCGCAACGGAAG-3’ and 5’-CGGCCCCTAGGGCGGTCACACCACAGAAGTAAGG-3’). The purified insert was then cloned into the AsiSI (NEBioLabs calatolog number R0630) restriction site of the pBac vector (a gift from Leslie Vosshall) which contains the 15XQUAS promoter sequence from the QF2 expression system47. Transgene plasmid (500 ng/uL; PBac-15XQUAS-unc84:2XeGFP-3XP3-ECFP, AddGene #217614) was sent to the Insect Transformation Facility, Rockville, Maryland, for microinjection into Orlando (ORL) strain embryos alongside pBac transposase mRNA (300 ng/μL). Out of 256 injected G0 eggs, we recovered over twenty G1 families with 3XP3-ECFP expression in the larval eye, and most remained positive at G2. We outcrossed G2 females to ORL males for several more generations and mapped the number and locations of insertions in each line via the TagMap method48. We chose one family (P2) for further use with a single insertion on chromosome 1 far from any annotated coding sequence (Chr1: 61,691,142).
Generation of Or4-T2A-QF2 and Or103-T2A-QF2 strains.
We used CRISPR-mediated homologous recombination46 to insert the QF2 transcription factor47 into the endogenous Or4 (AAEL015147) and Or103 (AAEL017505) loci of the Orlando (ORL) strain of Ae. aegypti (Fig. S16). These inserts disrupt the native coding sequences and are thus putative knock-out alleles. We designed sgRNAs targeting the second exon of each locus (Or4: GGTGGAGATGATCTACGGTCGGG, Or103: GTGCACCTCACGCGCTAGCGG, PAM sequences underlined) and generated dsDNA template for transcription of the sgRNA via template-free PCR with partially overlapping PAGE-purified primers using the NEBNext High-Fidelity polymerase (NEB, M0541S). We then transcribed sgRNA in vitro using the HiScribe T7 Kit (NEB, E2040S) with an 8-9 hr incubation at 37°C. We purified the transcription products using RNAse-free SPRI beads (Agencourt RNAclean XP, Beckman-Coulter A63987) and eluted in nuclease-free water.
We constructed the donor plasmid for the Or4 locus (Or4-T2A-QF2-3XP3-dsRed, AddGene #217647) based on a preexisting plasmid designed for insertion of a different construct at the same locus and cutsite (Or4-T2A-mCD8:GFP-3XP3-dsRed, AddGene #219789). We first amplified the plasmid backbone, including 818bp and 936 bp homology arms and the 3XP3-dsRed marker, but excluding the GSG-T2A-mCD8:GFP element. We then amplified the T2A-QF2-attP construct (1488bp) from different plasmid via PCR, gel extracted and purified both pieces, and cloned them together via InFusion HD cloning (Clontech, 638910). The completed donor plasmid was verified by Sanger sequencing (Genewiz).
We constructed the donor plasmid for the Or103 locus (Or103-T2A-QF2-3XP3-dsRed, AddGene #217629) using the InFusion HD Kit (Clontech, 638910). Homology arms (~1 kb each) flanking the Cas9 cut site were amplified from ORL-strain genomic DNA via PCR and cloned into a T2A-QF2-3XP3-dsRed plasmid backbone (linearized with restriction enzymes NsiI-HF (New England Biolabs #R3127S) and AvrII (New England Biolabs #R0174S). The completed donor plasmid was verified by Sanger sequencing.
For each construct, 1500-2000 embryos were injected with a mixture of donor plasmid (700 ng/μL), sgRNA (110 ng/μL) and Cas9 protein (300 ng/μL; PNA Bio, CP01-200) at the Insect Transformation Facility, Rockville, Maryland. A stable transgenic line was developed from one of 19 g1 families (Or4) and from one of 5 g1 families (Or103) showing 3XP3-dsRed expression in the larval eyes. Proper integration into the genome was verified by PCR and Sanger sequencing. Transformed animals were outcrossed to ORL for 5-7 generations, and then maintained by incrossing with continued selection for 3XP3-dsRed+ individuals. Both strains are homozygous viable and show no obvious fitness deficits.
Or4 primers
sgRNA template: forward 5’-GAAATTAATACGACTCACTATAGGTGGAGATGATCTACGGTCGTTTTAGAGCTAGAAATAGC-3’, reverse5’-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’
Amplification of original homology arms from genomic DNA: Left arm forward 5’-CTGTAACTTTCCAGAACACCACATA-3’, Left arm reverse 5’-CGTAGATCATCTCCACCACC-3’, Right arm forward 5’-GTCGGGTGGTTACCGGAG-3’, Right arm reverse 5’-CGTTTTGTGCGGCAGGTAATAGAG-3’
Amplification of backbone (including homologous arms and 3XP3-dsRed): forward 5’-CGTAGATCATCTCCACCACC-3’, reverse 5’-GATACGCGTACGGCAATTCG-3’
Amplification of T2A-QF2-attP: forward 5’-tggagatgatctacgTGCATGGATCGGGAGAGGG-3’, reverse 5’-tgccgtacgcgtatcGCCGTACGCGTATCTAGAG-3’
Verification of genomic integration: Right arm forward 5’-TGAAGGGCGAGATCCACAAGGC-3’, Right arm reverse 5’-ATGGGCCAAAACTTCCACGCC-3’
Or103 primers
sgRNA template: forward 5’-GAAATTAATACGACTCACTATAGTGCACCTCACGCGCTAGCGGTTTTAGAGCTAGAAATAGC-3’, reverse 5’-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’
Amplification of homology arms: Left arm forward 5’-CAGGCGGCCGCCATATCCCCTTCAAATAGGTAACAATGTATCA-3’, Left arm reverse 5’-CCCTCTCCCGATCCACCCAGGGCGTGCGGGGCCGAGAAAGCTATTTCAGTTGCCTTATTCGGGATT-3’; Right arm forward 5’-TGTATCTTATCCTAGGCGTGGACTAATAAATATGGATCAGCAATT-3’, Right arm reverse 5’-TATTAATAGGCCTAGGGACTTATGAGACTTATATTGATCATGTACTTCTCA-3’.
Verification of genomic integration: Left arm forward 5’-GCCCAAACCCGTACGGTAATAA-3’, Left arm reverse 5’-CGTAGTTGTGGGTCCCAGAC-3’, Right arm forward 5’-CGGCCGCGACTCTAGATCATAATCAG-3’, Right arm reverse 5’-GCAAAGCTATACTAAAATAAAACATCGGGACT-3’
Visualization of Or4 and Or103 target glomeruli via brain immunostaining.
Or4-T2A-QF2 or Or103-T2A-QF2 animals were crossed with 15XQUAS-mCD8:GFP animals (Liverpool background47), and offspring were screened for those inheriting both constructs. Brain immunostaining was carried out as previously described47 on 7–10 day-old mated female mosquitoes. Heads were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 15713-S) for 3 hours at 4°C. Brains were dissected in PBS and blocked in normal goat serum (2%, Fisher Scientific, 005-000-121) for 2 days at 4°C. We then incubated brains in primary antibody solution for 2–3 days, followed by secondary antibody solution for another 2–3 days at 4°C. Brains were mounted in Vectashield (Vector, H-1000) with the anterior side facing the objective. Confocal stacks were taken with a 20X lens with XY resolution of 1024X1024 and Z-step size of 1 mm. Primary antibodies: rabbit anti-GFP (1:10,000 dilution, ThermoFisher, A-11122) and mouse NC82 (1:50 dilution, DHSB, AB_2314866). Secondary antibodies: goat-anti-rabbit Alexa 488 (1:500 dilution, ThermoFisher, A27034SAMPLE), goat-anti-mouse CF680 (1:500 dilution, Biotium, 20065-1).
Female antennal dissection.
We crossed the brp-T2A-QF2w11 driver to our new 15XQUAS-unc84:2XeGFP effector and screened offspring as larvae for those inheriting both constructs (3XP3-dsRed and 3XP3-ECFP positive). Males and females were co-housed with access to 10% sucrose but not blood. Female mosquitoes were dissected at 5–7 days post eclosion. To ensure a sufficient nuclei yield for downstream sorting, we designed a dissection protocol to remove and preserve the integrity of the antennal flagellum (excluding the pedicel). First, mosquitoes were anesthetized at 4°C for 25 minutes and 70 individuals were placed on a prechilled Sylgard® coated 93 mm diameter petri dishes (Living systems instrumentation, DD-90-S). The dissection dish was placed on ice and transferred to a stereoscope at room temperature. Under the stereoscope, we severed the head of each female one at a time with sharp forceps, swirled it gently in a dish of pure ethanol for 3-5 seconds at room temperature, rinsed it twice with vigorous swirls in large amounts of room temperature Dulbecco’s PBS without calcium chloride and magnesium chloride (Sigma-Aldrich, D8537), and then placed it neck down on a second prechilled Sylgard® coated petri dish on ice containing Schneider’s Drosophila Medium (Thermo Fisher, 21720024). When working well, cuticular wax can be seen diffusing away from the head during the dip in ethanol. Once all heads were collected, we placed the second Sylgard coated petri dish on ice under the stereoscope to remove both antennae from each head by (1) holding the head with one forceps, (2) grabbing the base of the flagellum (first flagellomere) with the other forceps, (3) pulling it away from the head, and (4) releasing it in the Schneider’s medium within the petri. With sharp enough forceps, the antennal pedicel, which contains the Johnston’s organ, stays attached to the head, and we confirmed that all dissected antennae were free of pedicel residue. We then used a pipet with wide orifice pipette tips (Thomas Scientific, 1234W) to transfer the flagella from the petri into a 1.5mL non-stick microcentrifuge tube (Neta Scientific, RPI-145530) on ice (Thomas Scientific, 1234W). The antennae naturally sink to the bottom of the tube after 5-7 minutes, after which the flagella can be concentrated by pipetting off the liquid. This procedure was performed simultaneously by two people and both pools of dissected flagella were merged into a single microcentrifuge tube to double the yield of tissue per tube/session. Finally, the flagella were instantly snap frozen in a liquid nitrogen-cooled mini mortar (SP Bel-Art, H37260-0100) and stored at −80°C until nuclear isolation. The entire procedure lasted 45-50 minutes and was repeated approximately 30 times on different days with different batches of mosquitos.
Nuclear isolation.
We followed a previously described method for isolating Drosophila nuclei49 with minor modifications as follows: two microcentrifuge tubes of frozen antennae were transferred to the bench in a −20°C cooling block (one at a time), 100 μl of prechilled HB lysis buffer (250mM Sucrose, 10mM Tris pH 8, 25mM KCl, 5mM MgCl2, 0.1% Triton-x 100, 0.5% RNAse inhibitor Plus (Promega, N2615), 1x Protease inhibitor cocktail dissolved in DMSO (Promega, G6521), 0.1mM DTT) were quickly added to the tube outside of the cooling block and antennae were immediately pulverized with a motor pestle for 30s next supplemented with 400μl HB. Both batches were merged into a single glassTissue Grinder (Wheaton®, 357538) for a total volume of 1mL. The suspension was homogenized with 18 loose Dounce strokes followed by 26 tight Dounce strokes on ice. The lysate was strained through a 35μm meshed 5mL test tube (Corning® Falcon®, 35223535) to remove cuticular aggregates, filtered through a prewet 40μm Flowmi™ cell strainer (Bel-Art, H13680-0040) to remove cellular clumps, and centrifuged at 1000g in swinging buckets (Eppendorf, Rotor S-24-11-AT) at 4°C for 10 minutes.
After centrifugation, as much supernatant as possible was removed from the tube and the nuclear pellet (often visible in shades of white to dark grey) was resuspended by pipetting up and down 20 times in 400μL of Washing Buffer (1X Dulbecco’s PBS without calcium chloride and magnesium chloride, 1% MACS BSA stock solution (Miltenyi Biotech,130-091-376), 0.5% RNAse inhibitor Plus). During the centrifugation time, the procedure was repeated on a second round of two batches. During the centrifugation of the second round, the first round was resuspended and then kept on ice until the resuspension of the second round. Once both were resuspended, each isolate was individually filtered through a prewet 40μm Flowmi™ to remove nuclear clumps and collected into the same 1.5mL non-stick microcentrifuge tube (Neta Scientific, RPI-145530) for a total volume of 750-800μL depending on Flowmi™ retention. Hoechst33342 (Thermo Fisher Scientific, 62249) was added 5–10 minutes before sorting at 1μg/ml final concentration. The suspension was kept on ice at all times until FACS loading.
FACS sorting & nuclear imaging.
The full 750-800μL suspension was loaded at 4°C into a FACSAria™ Fusion flow cytometer (BD Biosciences) with a 70 μm nostril running regular 1X PBS at the Flow Cytometry Core Facility of the Molecular Biology Department at Princeton. We empirically refined the optical parameters to isolate healthy single nuclei as follows. We used the first gate [FSC-A(exponential) X DAPI-A(linear x1000)] to select all events in the first of two Hoechst intensity peaks. Most nuclei in the second peak were confirmed by imaging to correspond to doublets or single nuclei with unusually bright Hoechst intensity (possibly in G2/S phase, data not shown). We used a second gate [SSC-W(linear x1000) X SSC-H(exponential)] to discard additional potential doublets or aggregates. We used a third gate [SSC-A(exponential) X GFP FITC-A(exponential)] to select and separate GFP positive neuron nuclei (~35%) from GFP negative nuclei (~65%). We used a fourth gate [DAPI-A(linear) X SSC-A(exponential)] to remove GFP nuclei with heterogeneous granularity which corresponded to damaged single nuclei. This procedure was conducted on three different nuclear suspensions, each processed on a different day and composed by different antennal dissection batches. Together, the three FACS runs yielded 7 samples of approximately 20,000 GFP positive nuclei each (1 sample from the first run and 3 samples from each of the second and third runs). The samples were collected in nonstick 1.5ml microcentrifuge tubes smeared with 12 μl fresh prechilled Washing Buffer.
Single nuclei RNA sequencing.
We measured the volume of each sample, added up to 43μl of nuclease-free water, and loaded it into a 10X Genomics Chromium system using Reagent Kits to generate and amplify cDNAs as recommended by the manufacturer (10X Genomics). We checked the electrophoretic profile of cDNA libraries before and after tagmentation, selected fragments of size 300-700bp using a BluePippin (Sage Science), and generated 100 bp paired-end sequencing reads on an Illumina NovaSeq 6000 SP flowcell following standard Illumina protocols. Only pass-filter reads were retained by Illumina Control Software and were aligned to the Aedes aegypti L5 genome excluding extrachromosomal nuclear contigs50 with CellRanger v6.0.1 with the “include-introns” option on (10X Genomics).
Genome Reannotation.
We made several custom updates to the publicly available AaegL5 genome annotation (VectorBase-55_AaegyptiLVP_AGWG.gff) to facilitate analysis and interpretation of our data. First, we removed the few annotations present for small ‘unplaced’ contigs, leaving only annotations for the three major chromosomal scaffolds and one mitochondrial scaffold. Second, since the annotation was missing several chemoreceptors that might be expressed in female antennae, we revised it to include the manually curated chemoreceptor (OR, GR, IR) annotations provided in the supplement of Matthews et al. 201850. This involved (i) renaming the contigs from the manual annotation file to match those use by VectorBase, (ii) identifying genes in the main gff file that overlapped those in the manual chemoreceptor gff file using sed and bedtools intersect (-wa -u), (iii) removing from the main file all features associated with these genes by feeding grep (-Fvf) a list of gene_ids, (iv) merging the two gff files, and (v) converting to gtf using gffread (-T -o)51. Finally, we used metazoa.ensemble.org52 to infer potential Drosophila orthologs for as many mosquito genes as possible and appended the Drosophila names to the ends of the mosquito names where possible.
We also developed an automated pipeline that leveraged our snRNAseq data to extend 3’ UTRs in the AaegL5 annotation (Fig. S2). Incomplete annotation of 3’ UTRs is a widely recognized problem for the analysis of 3’ sequencing data such as that generated here using the 10X genomics platform53,54. Reads that pile up just downstream of an annotated UTR will not be assigned to the proper gene, causing loss of signal. Briefly, we first extracted the subset of 10X reads that aligned to small windows starting 100bp upstream of the 3’ end of any annotated transcript or transcript-like feature and extending 750bp downstream. We then used StringTie (-p 40 -m 30 --j 1000000000 --fr)55 to assemble these reads into short ‘gene models’, with each ‘gene model’ representing a pile-up of 10X reads just downstream of an annotated gene. Finally, we used a custom python/gffutils script to extend UTRs to the end of any StringTie ‘gene model’ present within its +750bp window. If the extended UTR overlapped with a downstream neighbor on the same strand, we truncated it immediately upstream of that neighboring gene. We used gff3sort.pl56 to sort the updated annotation and gffread (-E -t -o) to again convert to gtf format.
We visually inspected read alignments for ORs and IRs and manually extended them up to 5 kb when the read trace continued further downstream of the 750bp window and/or included one or more extra peaks. For each extra peak, we looked for alternative polyadenylation sequences (AAUAAA) and extended the 3’UTR to either the first A of the alternative sequence (when present) or the last nucleotide of the last mapping reads of the last extra peak.
The final updated gtf file is available at https://doi.org/10.5281/zenodo.12801833.
Data preprocessing, ambient RNA decontamination and doublet removal.
CellRanger-generated UMI count matrices were loaded into singleomics R toolkit Seurat 4.2.057. We merged all 7 libraries and discarded all droplets with low complexity (less than 350 genes detected), high UMI counts (>7000 UMIs detected), and/or high mtDNA content (>0.5% reads). We normalized all droplets together using the Seurat command SCTransform with v2 regularization (vst.flavor=”v2”)58,59, ncells=47388 (equal to the total amount of droplets in the dataset to avoid droplet subsampling), n_genes=10000 (up from 2000 to reduce gene subsampling) and all others at default values. We then explored a variety of clustering parameters in order to identify the most robust. We varied the number of PCs (npcs in the RunPCA) between 10 and 200. We varied the number of dimensions (dims in the RunUMAP) to match the number of PCs in a given run. We varied the number of neighbors (n.neighbors) between 10 to 200. For the FindClusters command, we set the resolution to 1 and chose SLM (algorithm=3) instead of the default Louvain algorithm. Visual comparison of the resulting UMAPs revealed that the number and identity of clusters (i.e. putative cell types) was particularly sensitive to the number of PCs. More specifically, the number of clusters increased non-linearly with the number of PCs up to a saturation point after which the number of clusters remained stable. We therefore chose to use the lowest number of PCs that allowed us to reach the saturation point and then the number of neighbors that provided the best visual segregation pattern given that number of PCs. The final normalization and pre-clustering parameters were as follows: SCTransform(n_cells=47388, n_genes=10000, vst.flavor=“v2”); RunPCA(npcs=60); RunUMAP(reduction=“pca”, dims=1:60, n.neighbors=110); FindNeighbors(reduction=“pca”, dims=1:60); FindClusters(resolution=1, algorithm=3).
We removed ambient RNA contamination using SoupX (autoEstCont maxMarkers=5000)60. We ran SoupX on each library individually to account for library-specific RNA contamination. However, we fed the program cluster identities derived from the unified “pre-clustering” analysis of all 7 libraries described above. After rounding the corrected matrices for each library, we saved them using DropletUtils::write10xCounts61 and removed all predicted doublets with the Python package Solo62 (-p, using recommended model_json parameters). We finally merged all 7 corrected count matrices and obtained a final data matrix with 46073 curated droplets. We further explored the quality of the preprocessing by comparing dispersion, cumulative distribution, and residual variance between pre- and post-processed data.
All neuron normalization and clustering.
We normalized the curated data matrix using the Seurat command SCTransform as described for pre-clustering except we raised the default number of genes to the total number of genes, scaled and centered the residuals, and reduced maximum residual variance to 50, which was empirically defined. We found that reducing the clipping range was essential to avoid artifactual clusters caused by specific genes with unusually high residual variance (e.g. nompC, Fig. S4D). The final normalization parameters were as follows: ncells=total_cells, n_genes=NULL, vst.flavor=“v2”, do.scale=TRUE, do.center=TRUE, clip.range=(−50,50), variable.features.rv.th=1.3. We then clustered the data, again as described for pre-clustering except that we ran UMAP with the uwot method and cosine metric. The final clustering parameters were as follows: RunPCA(npcs=60); RunUMAP(reduction=“pca”, dims=1:60, n.neighbors=110, umap.method=”uwot”, metric-”cosine”); FindNeighbors(reduction=“pca”, dims=1:60); FindClusters(resolution=1, algorithm=3).
THSN and OSN renormalization and clustering.
After generating the “all neuron clustering”, we excluded four ‘junk’ clusters (low complexity clusters in the middle of the UMAP; Fig. 1D #1, 47, 48, 55), two nompC+ mechanosensory neuron clusters (#29, 54), and one unidentified cluster (#45) before subsetting the remaining clusters into three categories based on orco and Ir93a expression: thermo/hygrosensory neurons (THSNs; Ir93a+, orco−), orco+ olfactory sensory neurons (orco+ OSNs; orco+, Ir93a−), and orco− olfactory sensory neurons (orco− OSNs; orco−, Ir93a−). We then independently renormalized and reclustered the nuclei belonging to each of the three types of clusters with different parameters based on QC and a clustering sensitivity analysis similar to that described for Data Preprocessing (above). Notably, we used 12, 20, and 60 PCs when running RunPCA on THSNs, orco− OSNs, and orco+ OSNs, respectively, to reflect the differing levels of complexity of these groups of cells. We also reduced the number of neighbors for THSNs and orco− OSNs to 15 and for orco+ OSNs to 50. Inspection and preliminary analysis for each category showed that the smallest orco+ OSN cluster (n=59 droplets) represented a mix of three other orco+ OSN clusters. Importantly, it ‘co-expressed’ groups of ORs that did NOT show correlated expression at the droplet level. We therefore removed these droplets and renormalized/reclustered the remaining orco+ OSN data a second time. We conducted a similar cleaning step for orco− OSN clusters by removing one cluster with putative contamination from orco+ cells (n=248 droplets) and and another very small cluster (n=85 droplets). After renormalization, we again removed the smallest cluster (n=61 droplets), which was a small offshoot of the Amt+ cluster likely caused by a statistical artifact.
Inspection of the UMAPs and receptor expression patterns revealed heterogeneity within a few OSN clusters. In these cases, we looked for evidence of multiple underlying neuron subtypes using the FindSubCluster command. This resulted in the splitting of 1 orco+ OSN cluster (#21) and 2 orco− OSN clusters (#10 and 14). See Fig. S9 for visualization of how the original ‘all neuron’ clusters relate to those in the reanalyzed orco+ OSN and orco− OSN subsets.
Quantification of co-receptor expression in all neuron clustering.
We examined the distribution of average expression values for four olfactory co-receptors (orco, Ir25a, Ir8a, Ir76b) across clusters in the all-neuron analysis (Fig. 1E) and identified a natural break as the on-off cutoff: each co-receptor was considered expressed if its average expression in a given cluster exceeded 10% of the gene-specific maximum across all clusters.
Quantification of receptor expression in THSN, orco+ OSN, and orco− OSN clusters.
We quantified receptor expression within clusters using a median-adjusted log2 fold change (log2FC’). Log2FC compares expression in a focal cluster to expression in all other clusters. The adjustment was made by subtracting the median log2FC value across all clusters from the log2FC in focal clusters. We made this adjustment because low-level background expression led to many low (but positive) values for some receptors. The distribution of log2FC’ values across all receptor-by-cluster combinations (Fig. S12A, S15C) revealed clear breaks or inflection points that we used as on/off thresholds (0.4 for orco− OSNs, 0.3 for orco+ OSNs). Alternative absolute log average expression thresholds produced similar results (0.15 for orco− OSNs, 0.16 for orco+; Fig. S12C, S15B).
More than one ligand-specific receptor was called as expressed in many OSN clusters. To confirm co-expression, we looked for correlated expression at the droplet level and found significantly elevated Pearson correlations in all but one case. Ir41b was called as expressed in orco− OSN cluster 10b, alongside Ir41e and Ir41l, but showed no sign of correlated expression with the latter two receptors at the droplet level.
In a few cases, two OSN clusters expressed the same set of receptors (e.g. orco− OSN clusters 14a/14b, 4/9 and orco+ OSN clusters 15/37, 21a/21b, 21c/24). These may represent (i) distinct OSN subtypes that express the same set of receptors, (ii) the same OSN subtype from individuals with different genotypes (given the presence of genetic variation within the ORL lab strain) or in different biological states, or (iii) clustering artifacts. Conservatively, we decided to merge such clusters for the purposes of downstream analyses.
Receptor gene tree inference.
We inferred phylogenetic trees for the Ae. aegypti IR, Ae. aegypti OR, and D. melanogaster OR families using Bali-phy 3.6.163. We first tested different parameter combinations and settled on the following substitution frequencies, heterogenous rates and indel models for all gene families: -S wag+f+Rates.gamma+inv -I rs07. We then (1) ran ten simultaneous iterations of 50000 generations each, (2) merged all iterations discarding the minimum burnin, (3) calculated posterior probabilities for all nodes, and (4) only retained phylogenetic relationships with probabilities above 50%. Trees were visualized as inward phylogenies using the Rstudio package ggtree64. Phylogenetic distances between pairs of receptors was computed using the cophenetic.phylo function in the Rstudio package phytools65.
Supplementary Material
Acknowledgements:
We thank Richard Benton for comments on the manuscript, Hongjie Li for sharing the antennal nucleus dissociation protocol pre-publication, Tina DeCoste of the Princeton Flow Cytometry Core Facility for help optimizing our FACS protocol, Wei Wang and Jennifer Miller of the Princeton Genomics Core Facility for help with 10X library prep and sequencing, and Vanessa Ruta and members of the McBride laboratory for discussion.
Funding:
This work was funded in part by the following grants and fellowships: National Institutes of Health grant R01AI175490 (CSM), New York Stem Cell Foundation Robertson Neuroscience Investigator Award (CSM), Princeton Neuroscience Institute Innovation Award (CSM), Helen Hay Whitney Postdoctoral Fellowship (NHR).
Footnotes
Competing interests. The authors declare that they have no competing interests.
Materials, data, and code availability. Plasmids generated in this study have been deposited with AddGene (accession #217614, 217647, 217629). Genetically modified mosquito strains are available upon request. Raw snRNAseq reads are available in the NCBI-SRA (BioProject ID PRJNA1138769). Processed snRNAseq data (pre-clustered R objects) and an updated AaegL5 gtf annotation file are available through Zenodo (doi.org/10.5281/zenodo.12797292, doi.org/10.5281/zenodo.12801833). Scripts used for genome reannotation and snRNAseq data analysis are available on github (github.com/mcbridelab/Adavi_2024_snRNAseqAaegAntennae, copy archived at swh:1:rev:ff813a7183ae56cbf3a03a5458450fdf057e674e).
REFERENCES
- 1.Strausfeld N.J., and Hildebrand J.G. (1999). Olfactory systems: common design, uncommon origins? Curr. Opin. Neurobiol. 9, 634–639. [DOI] [PubMed] [Google Scholar]
- 2.Fulton K.A., Zimmerman D., Samuel A., Vogt K., and Datta S.R. (2024). Common principles for odour coding across vertebrates and invertebrates. Nat. Rev. Neurosci. 25, 453–472. [DOI] [PubMed] [Google Scholar]
- 3.Vosshall L.B., and Stocker R.F. (2007). Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533. [DOI] [PubMed] [Google Scholar]
- 4.Malnic B., Hirono J., Sato T., and Buck L.B. (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 5.Wang J.W., Wong A.M., Flores J., Vosshall L.B., and Axel R. (2003). Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282. [DOI] [PubMed] [Google Scholar]
- 6.Christophers S.R. Aedes aegypti: the yellow fever mosquito (Cambridge University Press; ). [Google Scholar]
- 7.McBride C.S. (2016). Genes and Odors Underlying the Recent Evolution of Mosquito Preference for Humans. Curr. Biol. 26, R41–R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohbot J., Pitts R.J., Kwon H.-W., Rutzler M., Robertson H.M., and Zwiebel L.J. (2007). Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol. 16, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews B.J., McBride C.S., DeGennaro M., Despo O., and Vosshall L.B. (2016). The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ignell R., Dekker T., Ghaninia M., and Hansson B.S. (2005). Neuronal architecture of the mosquito deutocerebrum. Preprint, https://doi.org/10.1002/cne.20800 https://doi.org/10.1002/cne.20800. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z., Zung J.L., Hinze A., Kriete A.L., Iqbal A., Younger M.A., Matthews B.J., Merhof D., Thiberge S., Ignell R., et al. (2022). Mosquito brains encode unique features of human odour to drive host seeking. Nature 605, 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herre M., Goldman O.V., Lu T.-C., Caballero-Vidal G., Qi Y., Gilbert Z.N., Gong Z., Morita T., Rahiel S., Ghaninia M., et al. (2022). Non-canonical odor coding in the mosquito. Cell 185, 3104–3123.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shankar S., and McMeniman C.J. (2020). An updated antennal lobe atlas for the yellow fever mosquito Aedes aegypti. PLoS Negl. Trop. Dis. 14, e0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wicher D., and Miazzi F. (2021). Functional properties of insect olfactory receptors: ionotropic receptors and odorant receptors. Cell Tissue Res. 383, 7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silbering A.F., Rytz R., Grosjean Y., Abuin L., Ramdya P., Jefferis G.S.X.E., and Benton R. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Task D., Lin C.-C., Vulpe A., Afify A., Ballou S., Brbic M., Schlegel P., Raji J., Jefferis G., Li H., et al. (2022). Chemoreceptor co-expression in Drosophila melanogaster olfactory neurons. Elife 11. 10.7554/eLife.72599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry G.L., Davis F.P., Picard S., and Eddy S.R. (2012). Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 40, 9691–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin C.N., Brbić M., Xie Q., Li T., Horns F., Kolluru S.S., Kebschull J.M., Vacek D., Xie A., Li J., et al. (2021). Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. Elife 10. 10.7554/eLife.63856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mclver S.B. (1982). Sensilla of Mosquitoes (Diptera: Culicidae). J. Med. Entomol. 19, 489–535. [DOI] [PubMed] [Google Scholar]
- 20.Walker R.G., Willingham A.T., and Zuker C.S. (2000). A Drosophila mechanosensory transduction channel. Science 287, 2229–2234. [DOI] [PubMed] [Google Scholar]
- 21.Laursen W.J., Budelli G., Tang R., Chang E.C., Busby R., Shankar S., Gerber R., Greppi C., Albuquerque R., and Garrity P.A. (2023). Humidity sensors that alert mosquitoes to nearby hosts and egg-laying sites. Neuron 111, 874–887.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benton R., Sachse S., Michnick S.W., and Vosshall L.B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abuin L., Bargeton B., Ulbrich M.H., Isacoff E.Y., Kellenberger S., and Benton R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vulpe A., and Menuz K. (2021). Ir76b is a Co-receptor for Amine Responses in Drosophila Olfactory Neurons. Front. Cell. Neurosci. 15, 759238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis E.E., and Sokolove P.G. (1975). Temperature responses of antennal receptors of the mosquito, Aedes aegypti. J. Comp. Physiol. A 96, 223–236. [Google Scholar]
- 26.Knecht Z.A., Silbering A.F., Ni L., Klein M., Budelli G., Bell R., Abuin L., Ferrer A.J., Samuel A.D., Benton R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife 5. 10.7554/eLife.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knecht Z.A., Silbering A.F., Cruz J., Yang L., Croset V., Benton R., and Garrity P.A. (2017). Ionotropic Receptor-dependent moist and dry cells control hygrosensation in Drosophila. Elife 6. 10.7554/eLife.26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greppi C., Laursen W.J., Budelli G., Chang E.C., Daniels A.M., van Giesen L., Smidler A.L., Catteruccia F., and Garrity P.A. (2020). Mosquito heat seeking is driven by an ancestral cooling receptor. Science 367, 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vulpe A., Kim H.S., Ballou S., Wu S.-T., Grabe V., Nava Gonzales C., Liang T., Sachse S., Jeanne J.M., Su C.-Y., et al. (2021). An ammonium transporter is a non-canonical olfactory receptor for ammonia. Curr. Biol. 31, 3382–3390.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z., Liu F., Ferguson S.T., Baker A., Pitts R.J., and Zwiebel L.J. (2021). Ammonium transporter AcAmt mutagenesis uncovers reproductive and physiological defects without impacting olfactory responses to ammonia in the malaria vector mosquito Anopheles coluzzii. Insect Biochem. Mol. Biol. 134, 103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitts R.J., Derryberry S.L. Jr, Pulous F.E., and Zwiebel L.J. (2014). Antennal-expressed ammonium transporters in the malaria vector mosquito Anopheles gambiae. PLoS One 9, e111858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geier M., Bosch O.J., and Boeckh J. (1999). Ammonia as an attractive component of host odour for the yellow fever mosquito, Aedes aegypti. Chem. Senses 24, 647–653. [DOI] [PubMed] [Google Scholar]
- 33.Matthews B.J., Younger M.A., and Vosshall L.B. (2019). The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. Elife 8. 10.7554/eLife.43963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno S. (2013). Evolution by Gene Duplication (Springer Science & Business Media; ). [Google Scholar]
- 35.Schuh E., Cassau S., Ismaieel A.R., Stieber R., Krieger J., Hansson B.S., Sachse S., and Bisch-Knaden S. (2024). Females smell differently: characteristics and significance of the most common olfactory sensilla of female silkmoths. Proc. Biol. Sci. 291, 20232578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brahma A., Frank D.D., Pastor P.D.H., Piekarski P.K., Wang W., Luo J.-D., Carroll T.S., and Kronauer D.J.C. (2023). Transcriptional and post-transcriptional control of odorant receptor choice in ants. Curr. Biol. 33, 5456–5466.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mika K., and Benton R. (2021). Olfactory Receptor Gene Regulation in Insects: Multiple Mechanisms for Singular Expression. Front. Neurosci. 15, 738088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karner T., Kellner I., Schultze A., Breer H., and Krieger J. (2015). Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae. Frontiers in Ecology and Evolution 3. 10.3389/fevo.2015.00026. [DOI] [Google Scholar]
- 39.Prieto-Godino L.L., Silbering A.F., Khallaf M.A., Cruchet S., Bojkowska K., Pradervand S., Hansson B.S., Knaden M., and Benton R. (2020). Functional integration of “undead” neurons in the olfactory system. Sci Adv 6, eaaz7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monahan K., and Lomvardas S. (2015). Monoallelic expression of olfactory receptors. Annu. Rev. Cell Dev. Biol. 31, 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryba A.R., McKenzie S., McKenzie S., Olivos-Cisneros L., Clowney E.J., Pires P.M., and Kronauer D. (2020). Comparative development of the ant chemosensory system. Curr. Biol. 30, 3223–3230.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komiyama T., and Luo L. (2006). Development of wiring specificity in the olfactory system. Curr. Opin. Neurobiol. 16, 67–73. [DOI] [PubMed] [Google Scholar]
- 43.Li Q., Barish S., Okuwa S., Maciejewski A., Brandt A.T., Reinhold D., Jones C.D., and Volkan P.C. (2016). A functionally conserved gene regulatory network module governing olfactory neuron diversity. PLoS Genet. 12, e1005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishilevich E., and Vosshall L.B. (2005). Genetic and Functional Subdivision of the Drosophila Antennal Lobe. Curr. Biol. 15, 1548–1553. [DOI] [PubMed] [Google Scholar]
- 45.Couto A., Alenius M., and Dickson B.J. (2005). Molecular, Anatomical, and Functional Organization of the Drosophila Olfactory System. Curr. Biol. 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES for Methods
- 46.Kistler K.E., Vosshall L.B., and Matthews B.J. (2015). Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M.N., Luo L., and Potter C.J. (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12, 219–222, 5 p following 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stern D.L. (2016). Tagmentation-Based Mapping (TagMap) of Mobile DNA Genomic Insertion Sites. bioRxiv, 037762. 10.1101/037762. [DOI] [Google Scholar]
- 49.Li H., Janssens J., De Waegeneer M., Kolluru S.S., Davie K., Gardeux V., Saelens W., David F.P.A., Brbić M., Spanier K., et al. (2022). Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matthews B.J., Dudchenko O., Kingan S.B., Koren S., Antoshechkin I., Crawford J.E., Glassford W.J., Herre M., Redmond S.N., Rose N.H., et al. (2018). Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertea G., and Pertea M. (2020). GFF Utilities: GffRead and GffCompare. F1000Res. 9. 10.12688/f1000research.23297.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates A.D., Allen J., Amode R.M., Azov A.G., Barba M., Becerra A., Bhai J., Campbell L.I., Carbajo Martinez M., Chakiachvili M., et al. (2022). Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50, D996–D1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shields E.J., Sorida M., Sheng L., Sieriebriennikov B., Ding L., and Bonasio R. (2021). Genome annotation with long RNA reads reveals new patterns of gene expression and improves single-cell analyses in an ant brain. BMC Biol. 19, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agarwal V., Lopez-Darwin S., Kelley D.R., and Shendure J. (2021). The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., and Salzberg S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu T., Liang C., Meng Z., Guo S., and Zhang R. (2017). GFF3sort: a novel tool to sort GFF3 files for tabix indexing. BMC Bioinformatics 18, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M. 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafemeister C., and Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhary S., and Satija R. (2022). Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 23, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young M.D., and Behjati S. (2020). SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9. 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., participants in the 1st Human Cell Jamboree Atlas, and Marioni J.C. (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernstein N.J., Fong N.L., Lam I., Roy M.A., Hendrickson D.G., and Kelley D.R. (2020). Solo: Doublet Identification in Single-Cell RNA-Seq via Semi-Supervised Deep Learning. Cell Syst 11, 95–101.e5. [DOI] [PubMed] [Google Scholar]
- 63.Redelings B.D. (2021). BAli-Phy version 3: model-based co-estimation of alignment and phylogeny. Bioinformatics 37, 3032–3034. [DOI] [PubMed] [Google Scholar]
- 64.Yu G., Smith D.K., Zhu H., Guan Y., and Lam T.T.-Y. (2017). Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. [Google Scholar]
- 65.Revell L.J. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. [Google Scholar]
- 66.Diao F., and White B.H. (2012). A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Z., Tian D., and McBride C.S. (2021). Development of a pan-neuronal genetic driver in Aedes aegypti mosquitoes. Cell Rep Methods 1. 10.1016/j.crmeth.2021.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.