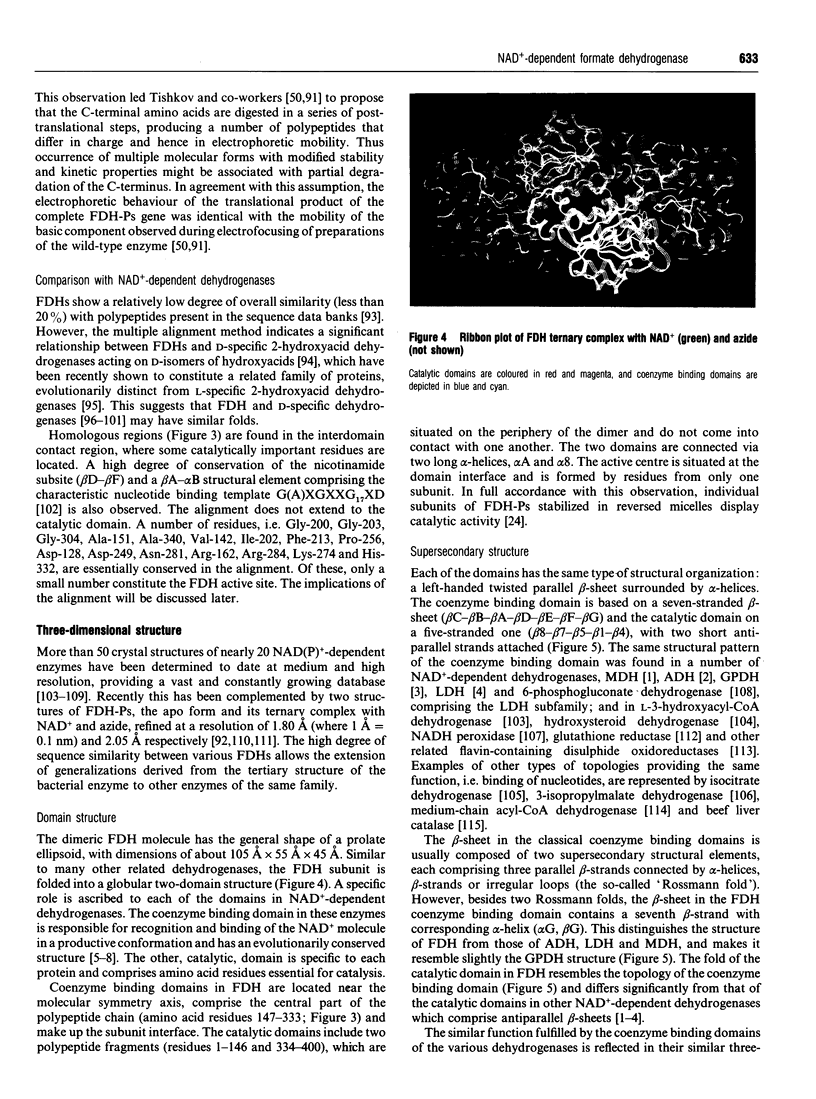
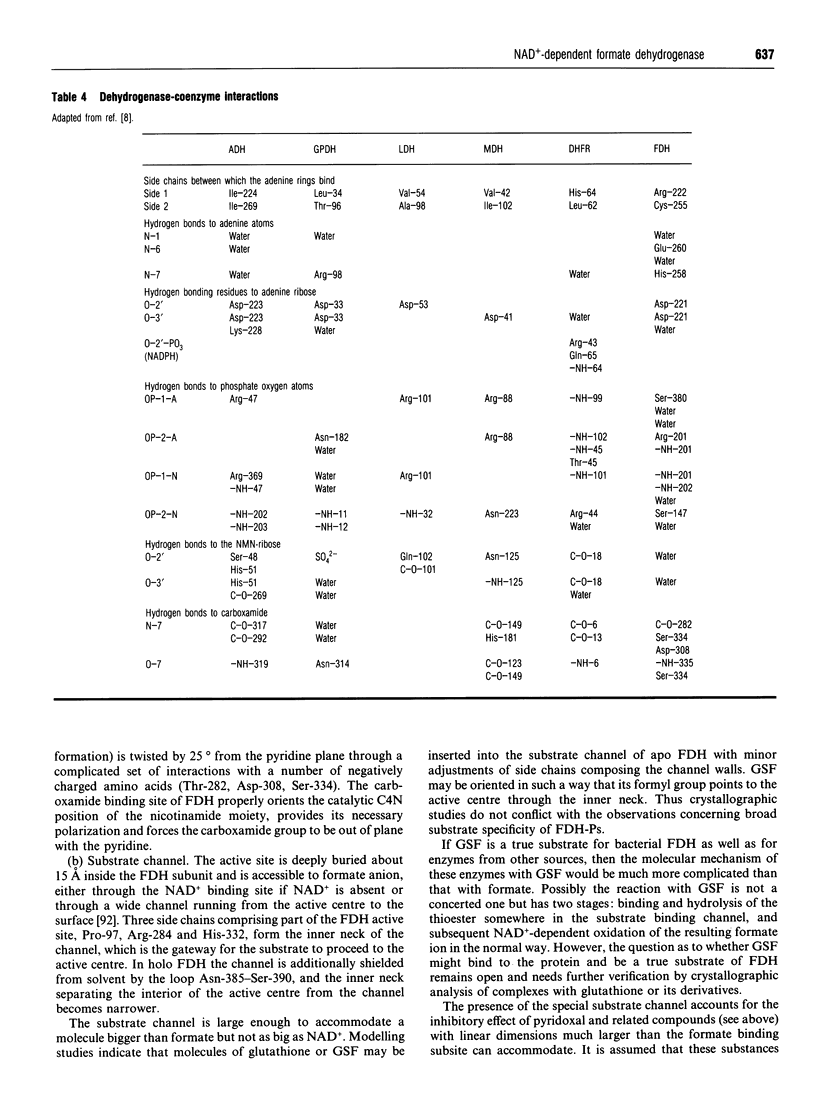
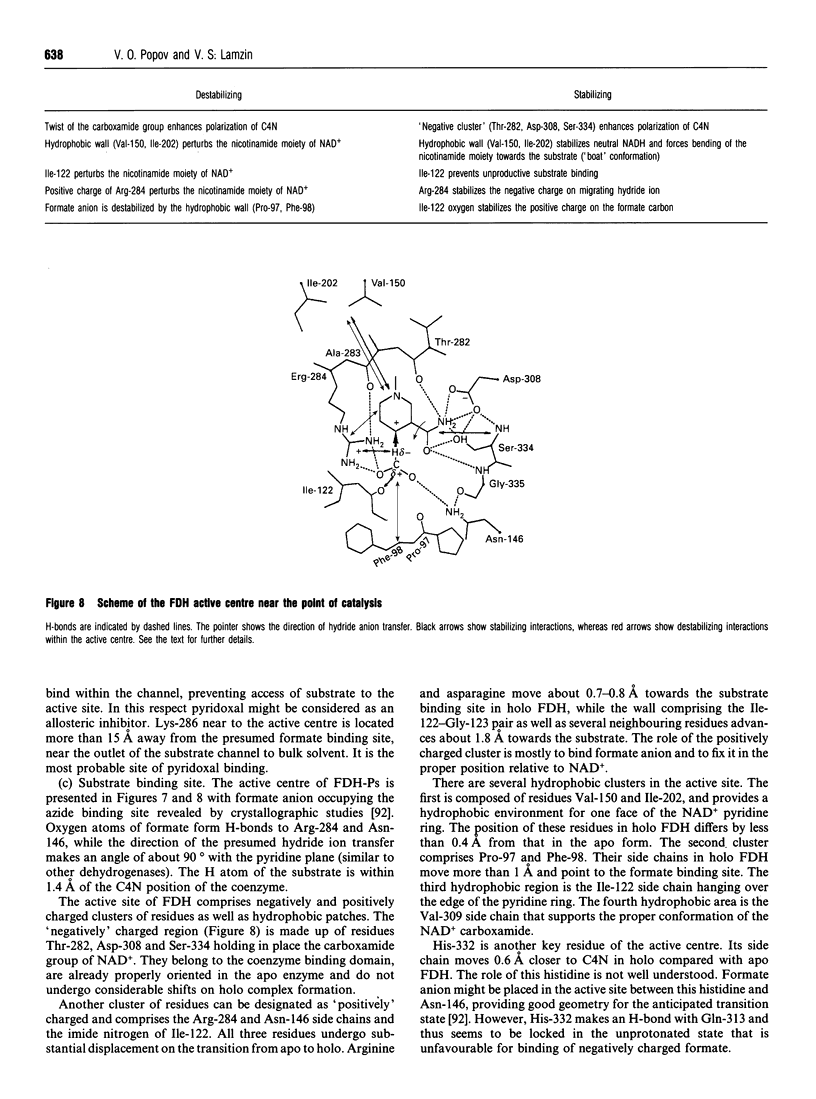
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. J., Archibald I. G., Bugg C. E., Carne A., Gover S., Helliwell J. R., Pickersgill R. W., White S. W. The three dimensional structure of sheep liver 6-phosphogluconate dehydrogenase at 2.6 A resolution. EMBO J. 1983;2(6):1009–1014. doi: 10.1002/j.1460-2075.1983.tb01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreesen J. R., Ljungdahl L. G. Nicotinamide adenine dinucleotide phosphate-dependent formate dehydrogenase from Clostridium thermoaceticum: purification and properties. J Bacteriol. 1974 Oct;120(1):6–14. doi: 10.1128/jb.120.1.6-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. The biochemistry of methylotrophic micro-organisms. Sci Prog. 1975 Summer;62(246):167–206. [PubMed] [Google Scholar]
- Arthur M., Molinas C., Dutka-Malen S., Courvalin P. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene. 1991 Jul 15;103(1):133–134. doi: 10.1016/0378-1119(91)90405-z. [DOI] [PubMed] [Google Scholar]
- Avilova T. V., Egorova O. A., Ioanesyan L. S., Egorov A. M. Biosynthesis, isolation and properties of NAD-dependent formate dehydrogenase from the yeast Candida methylica. Eur J Biochem. 1985 Nov 4;152(3):657–662. doi: 10.1111/j.1432-1033.1985.tb09245.x. [DOI] [PubMed] [Google Scholar]
- Babel W., Mothes G. Rolle der Formiat-Dehydrogenase in "Serinweg-Bakterien". Z Allg Mikrobiol. 1980;20(3):167–175. doi: 10.1002/jobm.3630200303. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Banaszak L. J. The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J Biol Chem. 1983 Jan 10;258(1):472–482. doi: 10.2210/pdb2mdh/pdb. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Holden H. M., Hamlin R., Xuong N. H., Banaszak L. J. Structure of L-3-hydroxyacyl-coenzyme A dehydrogenase: preliminary chain tracing at 2.8-A resolution. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8262–8266. doi: 10.1073/pnas.84.23.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J. S., Cleland W. W. Kinetic and chemical mechanisms of yeast formate dehydrogenase. Biochemistry. 1980 Jul 22;19(15):3543–3550. doi: 10.1021/bi00556a020. [DOI] [PubMed] [Google Scholar]
- Bokranz M., Gutmann M., Körtner C., Kojro E., Fahrenholz F., Lauterbach F., Kröger A. Cloning and nucleotide sequence of the structural genes encoding the formate dehydrogenase of Wolinella succinogenes. Arch Microbiol. 1991;156(2):119–128. doi: 10.1007/BF00290984. [DOI] [PubMed] [Google Scholar]
- Bystroff C., Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991 Feb 26;30(8):2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- Chow C. M., RajBhandary U. L. Developmental regulation of the gene for formate dehydrogenase in Neurospora crassa. J Bacteriol. 1993 Jun;175(12):3703–3709. doi: 10.1128/jb.175.12.3703-3709.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C., Ambard-Bretteville F., Small I. D., Rémy R. Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol. 1993 Aug;102(4):1171–1177. doi: 10.1104/pp.102.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. C., Edwards E. S., DeMoss J. A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol. 1981 Mar;145(3):1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorov A. M., Avilova T. V., Dikov M. M., Popov V. O., Rodionov Y. V., Berezin I. V. NAD-dependent formate dehydrogenase from methylotrophic bacterium, strain 1. Purification and characterization. Eur J Biochem. 1979 Sep;99(3):569–576. doi: 10.1111/j.1432-1033.1979.tb13289.x. [DOI] [PubMed] [Google Scholar]
- Egorov A. M., Tishkov V. I., Avilova T. V., Popov V. O. S-Formyl glutathione as a substrate of bacterial formate dehydrogenase. Biochem Biophys Res Commun. 1982 Jan 15;104(1):1–5. doi: 10.1016/0006-291x(82)91932-5. [DOI] [PubMed] [Google Scholar]
- Egorov A. M., Tishkov V. I., Dainichenko V. V., Popov V. O. Chemical modification of lysine residues in bacterial formate dehydrogenase. Biochim Biophys Acta. 1982 Dec 6;709(1):8–12. doi: 10.1016/0167-4838(82)90414-9. [DOI] [PubMed] [Google Scholar]
- Egorov A. M., Tishkov V. I., Popov V. O., Berezin I. V. Study of the role of arginine residues in bacterial formate dehydrogenase. Biochim Biophys Acta. 1981 May 14;659(1):141–149. doi: 10.1016/0005-2744(81)90278-3. [DOI] [PubMed] [Google Scholar]
- Egorova O. A., Avilova T. V., Platonenkova L. S., Egorov A. M. Vydelenie i svoistva NAD-zavisimoi formiatdegidrogenazy iz drozhzhei Candida methylica. Biokhimiia. 1981 Jun;46(6):1119–1126. [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Farinelli M. P., Fry D. W., Richardson K. E. Isolation, purification, and partial characterization of formate dehydrogenase from soybean seed. Plant Physiol. 1983 Nov;73(3):858–859. doi: 10.1104/pp.73.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The NADPH binding site on beef liver catalase. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1604–1608. doi: 10.1073/pnas.82.6.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanukoglu I., Gutfinger T. cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases. Eur J Biochem. 1989 Mar 15;180(2):479–484. doi: 10.1111/j.1432-1033.1989.tb14671.x. [DOI] [PubMed] [Google Scholar]
- Hermes J. D., Morrical S. W., O'Leary M. H., Cleland W. W. Variation of transition-state structure as a function of the nucleotide in reactions catalyzed by dehydrogenases. 2. Formate dehydrogenase. Biochemistry. 1984 Nov 6;23(23):5479–5488. doi: 10.1021/bi00318a016. [DOI] [PubMed] [Google Scholar]
- Hou C. T., Patel R. N., Laskin A. I., Barnabe N. NAD-linked formate dehydrogenase from methanol-grown Pichia pastoris NRRL-Y-7556. Arch Biochem Biophys. 1982 Jun;216(1):296–305. doi: 10.1016/0003-9861(82)90214-4. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. E., Jr, Stroud R. M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K., Sato M., Tanaka N., Katsube Y., Matsuura Y., Oshima T. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution. J Mol Biol. 1991 Dec 5;222(3):725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- JOHNSON P. A., JONES-MORTIMER M. C., QUAYLE J. R. USE OF A PURIFIED BACTERIAL FORMATE DEHYDROGENASE FOR THE MICRO-ESTIMATION OF FORMATE. Biochim Biophys Acta. 1964 Aug 26;89:351–353. doi: 10.1016/0926-6569(64)90225-1. [DOI] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Karzanov V. V., Correa C. M., Bogatsky Y. G., Netrusov A. I. Alternative NAD(+)-dependent formate dehydrogenases in the facultative methylotroph Mycobacterium vaccae 10. FEMS Microbiol Lett. 1991 Jun 1;65(1):95–99. doi: 10.1016/0378-1097(91)90478-s. [DOI] [PubMed] [Google Scholar]
- Kato N., Sahm H., Wagner F. Steady-state kinetics of formaldehyde dehydrogenase and formate dehydrogenase from a methanol-utilizing yeast, Candida boidinii. Biochim Biophys Acta. 1979 Jan 12;566(1):12–20. doi: 10.1016/0005-2744(79)90243-2. [DOI] [PubMed] [Google Scholar]
- Kato N., Sakazawa C., Nishizawa T., Tani Y., Yamada H. Purificaton and characterization of S-formylglutathione hydrolase from a methanol-utilizing yeast, Kloeckera sp. No. 2201. Biochim Biophys Acta. 1980 Feb 14;611(2):323–332. doi: 10.1016/0005-2744(80)90068-6. [DOI] [PubMed] [Google Scholar]
- Kim J. J., Wu J. Structure of the medium-chain acyl-CoA dehydrogenase from pig liver mitochondria at 3-A resolution. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6677–6681. doi: 10.1073/pnas.85.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar S., Chuard N., Hottinger H. Cloning and overexpression of the Lactobacillus bulgaricus NAD(+)-dependent D-lactate dehydrogenase gene in Escherichia coli:purification and characterization of the recombinant enzyme. Biochem Biophys Res Commun. 1992 Jun 15;185(2):705–712. doi: 10.1016/0006-291x(92)91683-h. [DOI] [PubMed] [Google Scholar]
- Kochhar S., Hottinger H., Chuard N., Taylor P. G., Atkinson T., Scawen M. D., Nicholls D. J. Cloning and overexpression of Lactobacillus helveticus D-lactate dehydrogenase gene in Escherichia coli. Eur J Biochem. 1992 Sep 15;208(3):799–805. doi: 10.1111/j.1432-1033.1992.tb17250.x. [DOI] [PubMed] [Google Scholar]
- Kochhar S., Hunziker P. E., Leong-Morgenthaler P., Hottinger H. Evolutionary relationship of NAD(+)-dependent D-lactate dehydrogenase: comparison of primary structure of 2-hydroxy acid dehydrogenases. Biochem Biophys Res Commun. 1992 Apr 15;184(1):60–66. doi: 10.1016/0006-291x(92)91157-l. [DOI] [PubMed] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- Kröger A., Winkler E., Innerhofer A., Hackenberg H., Schägger H. The formate dehydrogenase involved in electron transport from formate to fumarate in Vibrio succinogenes. Eur J Biochem. 1979 Mar;94(2):465–475. doi: 10.1111/j.1432-1033.1979.tb12914.x. [DOI] [PubMed] [Google Scholar]
- Kula M. R., Wichmann R., Oden U., Wandrey C. Influence of substrate or product inhibition on the performance of enzyme reactors. Biochimie. 1980;62(8-9):523–536. doi: 10.1016/s0300-9084(80)80097-6. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S., Aleshin A. E., Popov B. O., Arutiunian E. G. NAD-zavisimaia formiatdegidrogenaza metilotrofnykh bakterii Pseudomonas sp. 101. II. Prostranstvennaia struktura fermenta pri razreshenii 3,0 A. Bioorg Khim. 1990 Mar;16(3):336–344. [PubMed] [Google Scholar]
- Lamzin V. S., Aleshin A. E., Shumilin I. A., Ustinnikova T. B., Egorov Ts A., Arutiunian E. G., Popov V. O. NAD-zavisimaia formiatdegidrogenaza metilotrofnykh bakterii Pseudomonas sp. 101. III. Sravnitel'nyi analiz. Bioorg Khim. 1990 Mar;16(3):345–357. [PubMed] [Google Scholar]
- Lamzin V. S., Aleshin A. E., Strokopytov B. V., Yukhnevich M. G., Popov V. O., Harutyunyan E. H., Wilson K. S. Crystal structure of NAD-dependent formate dehydrogenase. Eur J Biochem. 1992 Jun 1;206(2):441–452. doi: 10.1111/j.1432-1033.1992.tb16945.x. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S., Asadchikov V. E., Popov V. O., Egorov A. M., Berezin I. V. Konformatsionnye izmeneniia bakterial'noiformiatdegidrogenazy pri kimpleksoobrazovanii s kofaktorom i ego analogami. Dokl Akad Nauk SSSR. 1986;291(4):1011–1014. [PubMed] [Google Scholar]
- Lamzin V. S., Dauter Z., Popov V. O., Harutyunyan E. H., Wilson K. S. High resolution structures of holo and apo formate dehydrogenase. J Mol Biol. 1994 Feb 25;236(3):759–785. doi: 10.1006/jmbi.1994.1188. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S., Dauter Z., Wilson K. S. Dehydrogenation through the looking-glass. Nat Struct Biol. 1994 May;1(5):281–282. doi: 10.1038/nsb0594-281. [DOI] [PubMed] [Google Scholar]
- Lerch H. P., Blöcker H., Kallwass H., Hoppe J., Tsai H., Collins J. Cloning, sequencing and expression in Escherichia coli of the D-2-hydroxyisocaproate dehydrogenase gene of Lactobacillus casei. Gene. 1989 May 15;78(1):47–57. doi: 10.1016/0378-1119(89)90313-2. [DOI] [PubMed] [Google Scholar]
- Li H., Goldstein B. M. Carboxamide group conformation in the nicotinamide and thiazole-4-carboxamide rings: implications for enzyme binding. J Med Chem. 1992 Sep 18;35(19):3560–3567. doi: 10.1021/jm00097a013. [DOI] [PubMed] [Google Scholar]
- Liu C. L., Mortenson L. E. Formate dehydrogenase of Clostridium pasteurianum. J Bacteriol. 1984 Jul;159(1):375–380. doi: 10.1128/jb.159.1.375-380.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Willnow P., Ruschig U., Höpner T. Formate dehydrogenase from Pseudomonas oxalaticus. Eur J Biochem. 1978 Feb;83(2):485–498. doi: 10.1111/j.1432-1033.1978.tb12115.x. [DOI] [PubMed] [Google Scholar]
- Nath P. K., Izumi Y., Yamada H. NADH production from NAD+ with a formate dehydrogenase system involving immobilized cells of a methylotrophic Arthrobacter strain. Enzyme Microb Technol. 1990 Jan;12(1):28–32. doi: 10.1016/0141-0229(90)90176-q. [DOI] [PubMed] [Google Scholar]
- Neben I., Sahm H., Kula M. R. Studies on an enzyme, S-formylglutathione hydrolase, of the dissimilatory pathway of methanol in Candida boidinii. Biochim Biophys Acta. 1980 Jul 10;614(1):81–91. doi: 10.1016/0005-2744(80)90169-2. [DOI] [PubMed] [Google Scholar]
- Ohyama T., Yamazaki I. Formate dehydrogenase. Subunit and mechanism of inhibition by cyanide and azide. J Biochem. 1975 Apr;77(4):845–852. doi: 10.1093/oxfordjournals.jbchem.a130792. [DOI] [PubMed] [Google Scholar]
- Oyama T., Yamazaki I. Purification and some properties of formate dehydrogenase. J Biochem. 1974 Jun;75(6):1257–1263. doi: 10.1093/oxfordjournals.jbchem.a130509. [DOI] [PubMed] [Google Scholar]
- Peacock D., Boulter D. Kinetic studies of formate dehydrogenase. Biochem J. 1970 Dec;120(4):763–769. doi: 10.1042/bj1200763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov V. O., Egorov A. M. Issledovanie sushchestvennykh SH-grupp bakterial'noi formiatdegidrogena3y. Biokhimiia. 1979 Feb;44(2):207–213. [PubMed] [Google Scholar]
- Popov V. O., Rodionov KuV, Egorov A. M., Berezin I. V. Izuchenie roli ostatkov gistidina formiatdegidrogenazy iz Bacterium sp.1. Biokhimiia. 1978 Jul;43(7):1212–1221. [PubMed] [Google Scholar]
- Popov V. O., Shumilin I. A., Ustinnikova T. B., Lamzin V. S., Egorov Ts A. NAD-zavisimaia formiatdegidrogenaza metilotrofnykh bakterii Pseudomonas sp. 101. I. Aminokislotnaia posledovatel'nost'. Bioorg Khim. 1990 Mar;16(3):324–335. [PubMed] [Google Scholar]
- Popov V. O., Tishkov V. I., Dainichenko V. V., Egorov A. M. Khimicheskaia modifikatsiia ostatkov lizina bakterial'noi formiatdegidrogenazy. Biokhimiia. 1983 May;48(5):747–755. [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rodionov Iu B., Avilova T. V., Popov V. O. NAD-zavisimaia formiatdegidrogenaza iz metilotrofnykh bakterii. Izuchenie svoistv i stabil'nosti rastvorimogo i immobilizovannogo fermenta. Biokhimiia. 1977 Nov;42(11):2020–2026. [PubMed] [Google Scholar]
- Rodionov Iu V., Avilova T. V., Zakharova E. V., Platonenkova L. S., Egorov A. M., Berezin I. V. NAD-zavisimaia formiatdegidrogenaza iz metilotrofnykh bakterii. Ochistka i osnovnye svoistva. Biokhimiia. 1977 Oct;42(10):1896–1904. [PubMed] [Google Scholar]
- Rotberg N. S., Cleland W. W. Secondary 15N isotope effects on the reactions catalyzed by alcohol and formate dehydrogenases. Biochemistry. 1991 Apr 23;30(16):4068–4071. doi: 10.1021/bi00230a035. [DOI] [PubMed] [Google Scholar]
- Saleeba J. A., Cobbett C. S., Hynes M. J. Characterization of the amdA-regulated aciA gene of Aspergillus nidulans. Mol Gen Genet. 1992 Nov;235(2-3):349–358. doi: 10.1007/BF00279380. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Properties of formate dehydrogenase in Methanobacterium formicicum. J Bacteriol. 1982 Apr;150(1):1–7. doi: 10.1128/jb.150.1.1-7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüte H., Flossdorf J., Sahm H., Kula M. R. Purification and properties of formaldehyde dehydrogenase and formate dehydrogenase from Candida boidinii. Eur J Biochem. 1976 Feb 2;62(1):151–160. doi: 10.1111/j.1432-1033.1976.tb10108.x. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Stehle T., Claiborne A., Schulz G. E. NADH binding site and catalysis of NADH peroxidase. Eur J Biochem. 1993 Jan 15;211(1-2):221–226. doi: 10.1111/j.1432-1033.1993.tb19889.x. [DOI] [PubMed] [Google Scholar]
- Sukhno A. A., Petukhova R. I., Kiseleva Z. L., Dmitirieva E. B., Rodionov Iu V. Bioélektrokhimicheskoe okislenie formiata. Biokhimiia. 1978 Oct;43(10):1900–1904. [PubMed] [Google Scholar]
- Taguchi H., Ohta T. D-lactate dehydrogenase is a member of the D-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the D-lactate dehydrogenase gene of Lactobacillus plantarum. J Biol Chem. 1991 Jul 5;266(19):12588–12594. [PubMed] [Google Scholar]
- Tishkov V. I., Egorov A. M., Popov V. O. Bakterial'naia formiatdegidrogenaza. Substratnaia spetsifichnost' i kineticheskii mekhanizm okisleniia S-formilglutationa. Biokhimiia. 1983 Jul;48(7):1172–1180. [PubMed] [Google Scholar]
- Tishkov V. I., Galkin A. G., Egorov A. M. Kinetic isotope effect and the presteady-state kinetics of the reaction catalyzed by the bacterial formate dehydrogenase. Biochimie. 1989 Apr;71(4):551–557. doi: 10.1016/0300-9084(89)90186-7. [DOI] [PubMed] [Google Scholar]
- Tishkov V. I., Galkin A. G., Marchenko G. N., Egorova O. A., Sheluho D. V., Kulakova L. B., Dementieva L. A., Egorov A. M. Catalytic properties and stability of a Pseudomonas sp.101 formate dehydrogenase mutants containing Cys-255-Ser and Cys-255-Met replacements. Biochem Biophys Res Commun. 1993 Apr 30;192(2):976–981. doi: 10.1006/bbrc.1993.1511. [DOI] [PubMed] [Google Scholar]
- Triebig G., Schaller K. H. A simple and reliable enzymatic assay for the determination of formic acid in urine. Clin Chim Acta. 1980 Dec 22;108(3):355–360. doi: 10.1016/0009-8981(80)90341-1. [DOI] [PubMed] [Google Scholar]
- Uotila L., Koivusalo M. Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys. 1979 Aug;196(1):33–45. doi: 10.1016/0003-9861(79)90548-4. [DOI] [PubMed] [Google Scholar]
- Vinals C., Depiereux E., Feytmans E. Prediction of structurally conserved regions of D-specific hydroxy acid dehydrogenases by multiple alignment with formate dehydrogenase. Biochem Biophys Res Commun. 1993 Apr 15;192(1):182–188. doi: 10.1006/bbrc.1993.1398. [DOI] [PubMed] [Google Scholar]
- White J. L., Hackert M. L., Buehner M., Adams M. J., Ford G. C., Lentz P. J., Jr, Smiley I. E., Steindel S. J., Rossmann M. G. A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J Mol Biol. 1976 Apr 25;102(4):759–779. doi: 10.1016/0022-2836(76)90290-4. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Yomano L. P., Scopes R. K., Ingram L. O. Cloning, sequencing, and expression of the Zymomonas mobilis phosphoglycerate mutase gene (pgm) in Escherichia coli. J Bacteriol. 1993 Jul;175(13):3926–3933. doi: 10.1128/jb.175.13.3926-3933.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaks A. M., Avilova T. V., Egorova O. A., Popov V. O., Egorov A. M. Kineticheskii mekhanizm deistviia NAD-zavisimoi formiatdegidrogenazy metilotrofnykh drozhzhei Candida methylica. Biokhimiia. 1982 Apr;47(4):546–551. [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken J. P., Oostra-Demkes G. J., Otto R., Harder W. S-formylgluthathione: the substrate for formate dehydrogenase in methanol-utilizing yeasts. Arch Microbiol. 1976 Dec 1;111(1-2):77–83. doi: 10.1007/BF00446552. [DOI] [PubMed] [Google Scholar]