Abstract
Targeted amplicon sequencing is a powerful and efficient tool to interrogate the P. falciparum genome and generate actionable data from infections to complement traditional malaria epidemiology. For maximum impact, genomic tools should be multi-purpose, robust, sensitive and reproducible. We developed, characterized, and implemented MAD4HatTeR, an amplicon sequencing panel based on Multiplex Amplicons for Drug, Diagnostic, Diversity, and Differentiation Haplotypes using Targeted Resequencing, along with a bioinformatic pipeline for data analysis. MAD4HatTeR targets 165 highly diverse loci, focusing on multiallelic microhaplotypes; key markers for drug and diagnostic resistance, including duplications and deletions; and csp and potential vaccine targets. In addition, it can detect non-falciparum Plasmodium species. We used laboratory control and field sample data to demonstrate the high sensitivity and robustness of the panel. The successful implementation of this method in five laboratories, including three in malaria-endemic African countries, showcases its feasibility in generating reproducible data across laboratories. Finally, we introduce an analytical approach to detect gene duplications and deletions from amplicon sequencing data. MAD4HatTeR is thus a powerful research tool and a robust resource for malaria public health surveillance and control.
Introduction
Effective control and eventual elimination of Plasmodium falciparum malaria hinge on the availability and integration of data to inform research and public health strategies. Genomics can augment traditional epidemiological surveillance by providing detailed genetic information about infections1. Molecular markers of drug and diagnostic resistance can guide the selection of antimalarials and diagnostics, respectively2–5. Vaccine target sequences may shed light on vaccine efficacy and identify evidence of selective pressure6. Measures of genetic variation can provide insights into transmission intensity, rate and origin(s) of importation, and granular details of local transmission7–14. Differentiation of infections as either recrudescent or reinfections is critical for measuring outcomes of therapeutic efficacy studies that are used to guide antimalarial use worldwide15–18. Furthermore, the contribution of non-falciparum species to malaria burden is poorly characterized, and could complicate control and elimination efforts19.
To maximize public health and research utility, genomic methods should be robust and provide rich information from field samples, which may be low-density and are often polyclonal in malaria-endemic areas of sub-Saharan Africa13,20–22. While traditional genotyping methods of length polymorphisms and microsatellites can characterize malarial infections, they suffer from low sensitivity and specificity, and difficulties in protocol standardization23–25. Single nucleotide polymorphism (SNP) barcoding approaches have improved throughput, sensitivity and standardization26,27. However, the biallelic nature of most targeted SNPs limits their discriminatory power to compare polyclonal infections. Sequencing of short, highly variable regions within the genome containing multiple SNPs (microhaplotypes) provides multiallelic information that overcomes many of those limitations28. Microhaplotypes can be reconstructed from whole-genome sequencing (WGS) data or amplified by PCR and sequenced. Low abundance variants, especially in low-density samples, may be missed by WGS due to low depth of coverage. Amplicon sequencing offers much higher sensitivity and can target the most informative regions of the genome, increasing throughput and decreasing cost. Several Illumina-based multiplexed amplicon sequencing panels have been developed to genotype P. falciparum infections. SpotMalaria is a panel that genotypes 100 SNPs, most of which are biallelic, for drug resistance and diversity26. Pf AmpliSeq genotypes SNPs, currently focused on Peruvian genetic diversity, and also targets drug and diagnostic resistance markers27. Panels that target multiallelic microhaplotypes, including AMPLseq, provide greater resolution for evaluating polyclonal infections and also include drug resistance markers29,30. Nanopore-based amplicon panels enable the utilization of mobile sequencing platforms31–33. Thus, targeted amplicon sequencing is a flexible approach that has the potential to address multiple use cases. To fully realize this potential, a panel for research and public health would ideally include all necessary targets to answer a wide range of questions, while remaining modular to allow flexible allocation of sequencing resources.
Here, we developed MAD4HatTeR, an Illumina-compatible, multipurpose, modular tool based on Multiplex Amplicons for Drug, Diagnostic, Diversity, and Differentiation Haplotypes using Targeted Resequencing. MAD4HatTeR has 276 targets divided into two modules: A diversity module with 165 targets to assess genetic diversity and relatedness; and a resistance module consisting of 118 targets that cover 15 drug resistance-associated genes and assesses hrp2/3 deletions, along with current and potential vaccine targets. The modules also include targets for non-falciparum Plasmodium species identification. We developed a bioinformatic pipeline to report allelic data, and implemented laboratory and bioinformatic methods in several sites, including countries in malaria-endemic sub-Saharan Africa. We then evaluated the panel’s performance on various sample types, including mosquito midguts, and showed that high quality data can be consistently reproduced across laboratories, including from polyclonal samples with low parasite density.
Results
MAD4HatTeR is a multi-purpose tool that exploits P. falciparum genetic diversity
We designed primers to amplify 276 targets (Figure 1, Supplementary Tables 1–4) and separated them into two modules: (1) Diversity module, a primer pool (D1) targeting 165 high diversity targets and the ldh gene in P. falciparum and in 4 non-falciparum Plasmodium species (P. vivax, P. malariae, P. ovale, and P. knowlesi); and (2) Resistance module, comprised of two complementary and incompatible primer pools (R1 and R2) targeting 118 loci that genotype 15 drug resistance-associated genes (Table 1) along with csp and potential vaccine targets (Table 2), assess for hrp2/3 deletion, and identify non-falciparum species. The protocol involves two initial multiplex PCR reactions, one with D1 and R1 primers, and another with R2 primers (Figure 1C, Supplementary Figure 1). After multiplexed PCR, subsequent reactions continue in a single tube.
Figure 1. MAD4HatTer is a multi-purpose malaria amplicon sequencing panel.
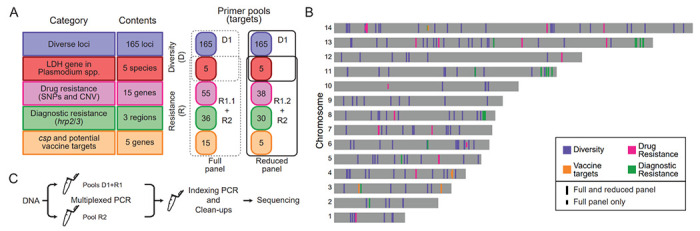
A. Primer pools to amplify targets in 5 categories are grouped into two modules (Diversity and Resistance). R1 refers to two primer pools: R1.1, the original pool, and R1.2, a reduced version of primer pool R1.1 designed to increase sensitivity. The recommended configuration to maximize information retrieval and sensitivity for low parasitemia samples are two mPCR reactions, one with D1 and R1.2 primers, and one with R2 primers (solid lines). Supplementary Tables 1–5 contain complete details on primer pools and targets.
B. Chromosomal locations of all targets in the P. falciparum genome (not including non-falciparum targets). Note that the Diagnostic Resistance category includes targets in and around hrp2 and hrp3 as well as targets in chromosome 11 that are often duplicated when hrp3 is deleted35 and length controls in other chromosomes.
C. Simplified workflow for library preparation and sequencing, highlighting the need for two multiplexed PCR reactions when using primer pools R1 and R2 which are incompatible due to tiling over some genes of interest. A more detailed scheme can be found in Supplementary Figure 1, and a full protocol, including didactic materials, can be found online49.
Table 1: SNPs associated with antimalarial resistance.
SNPs of interest used to optimize target primer design that are covered by primer pools R1.1, R1.2 or R2. The list excludes copy number variation markers, such as plasmepsins 2 and 3 (piperaquine) or mdr1 (mefloquine). A full list of targets with the amino acid ranges they cover in each gene can be found in the Supplementary File.
| Antimalarial† | Gene | Amino acids covered |
|---|---|---|
| Chloroquine and piperaquine | crt | 72-76, 93, 97, 145, 218, 220, 271*, 326* 343, 350, 353, 356 |
| Chloroquine | aat1 * | 135*, 162*, 185*, 230*, 238*, 380* |
| Piperaquine | exo | 415 |
| Chloroquine and lumefantrine | mdr1 | 86, 184, 186, 371, 1034, 1042, 1246 |
| Sulfadoxine | dhps | 431, 436, 437, 540, 581, 613 |
| Pyrimethamine and proguanil | dhfr | 16, 51, 59, 108, 164 |
| Artemisinin | kelch13 | 441, 446, 449, 458, 469, 476,481, 493, 515, 527, 537, 538, 539, 543, 553, 561, 568, 574, 580, 622, 675 |
| coronin | 50, 100, 107 | |
| fd | 193 | |
| arps10 | 127, 128 | |
| mdr2 | 463, 484, 515 | |
| PF3D7_1322700 | 236 | |
| pib7 | 1484 | |
| ubp-1 | 1525 | |
| pph * | 1157* |
Table 2: SNPs in csp and potential vaccine targets.
SNPs of interest used to optimize target primer design that are covered by primer pools R1.1, R1.2 or R2.1. A full list of targets with the amino acid ranges they cover in each gene can be found in the Supplementary File.
| Gene | Aminoacids covered |
|---|---|
| csp | 280-398 |
| Ripr * | 511, 673, 755, 985, 1039 |
| CyRPA * | 339 |
| Rh5 * | 147, 170, 197, 203, 204, 221, 269, 350, 354, 357, 362 |
| P113 * | 106, 107, 234 |
Included only in R1.1 and not in R1.2.
Based on publicly available WGS data, P. falciparum targets in the diversity module, excluding ldh, had a median of 3 SNPs or indels (IQR 2-5, N=165, Supplementary Table 5). Most (140/165) targets were microhaplotypes (containing > 1 SNP or indel). Global heterozygosity was high, with 35 targets with heterozygosity > 0.75 and 135 with heterozygosity > 0.5. Within African samples, heterozygosity was > 0.75 in 40 targets, > 0.5 in 132 targets, and we observed 2 to 20 unique alleles (median of 5, across a minimum of 3617 samples) in each target. MAD4HatTeR included more high-heterozygosity targets than other published panels (Figure 2A, Supplementary Figures 2 and 3). Additionally, MAD4HatTeR targets better resolved geographical structure globally, within Africa, and even within a country34 (Figure 2B).
Figure 2. In silico analysis demonstrates that MAD4HatTer’s microhaplotypes capture high genetic diversity within African samples.
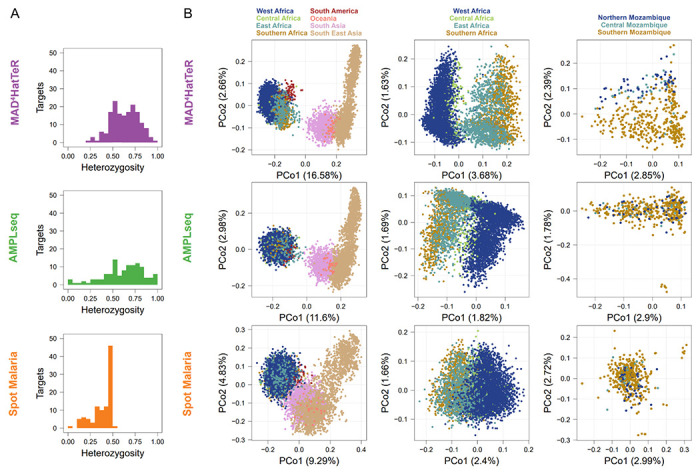
We reconstructed alleles (microhaplotypes) from publicly available WGS data to estimate genetic diversity. For SpotMalaria, SNP barcodes are used instead of microhaplotypes based on intended design and current usage. We note that additional information may be present within the amplified targets if microhaplotype sequences are accurately identifiable with appropriate bioinformatic processing. As such, alternate results for microhaplotypes reconstructed for the targets that contain the SNPs in each of those two panels are shown in Supplementary Figure 2.
A. Diversity module pool D1 includes more highly heterozygous targets than other published highly multiplexed panels. Only targets for diversity in each panel are included and heterozygosity is calculated for samples across Africa.
B. We performed principal coordinate analysis on alleles on global, African or Mozambican data. The percentage of variance explained by each principal component is indicated in parentheses.
We next evaluated the power of the diversity module to detect interhost relatedness between parasites in pairs of simulated infections with complexity of infection (COI) ranging from 1 to 5. We selected one country from each of three continents with the most publicly available WGS data and used reconstructed genotypes for the analysis (Figure 3). MAD4HatTeR identified partially related parasites between polyclonal infections across a range of COI and geographic regions, and generally performed as well or better than the other panels evaluated. For example, in simulated Ghanaian infections sibling parasites (IBD proportion, r=½) were reliably detected with COI of 5 (82% power), half siblings (r=¼) in infections with COI of 3 (73% power), and less related parasites (r=⅛) were still identifiable with COI of 2 (53% power). When using independent SNPs instead of microhaplotypes, the power to identify related parasites between infections was much lower, irrespective of the panel. Constraining the panel to the 50 targets with the highest heterozygosity (mean heterozygosity of 0.8 ± 0.05) reduced the power to infer relatedness by as much as 50%, highlighting the value of highly multiplexed microhaplotype panels for statistical power.
Figure 3. Power to identify relatedness of strains between infections is enhanced by highly multiplexed microhaplotypes.
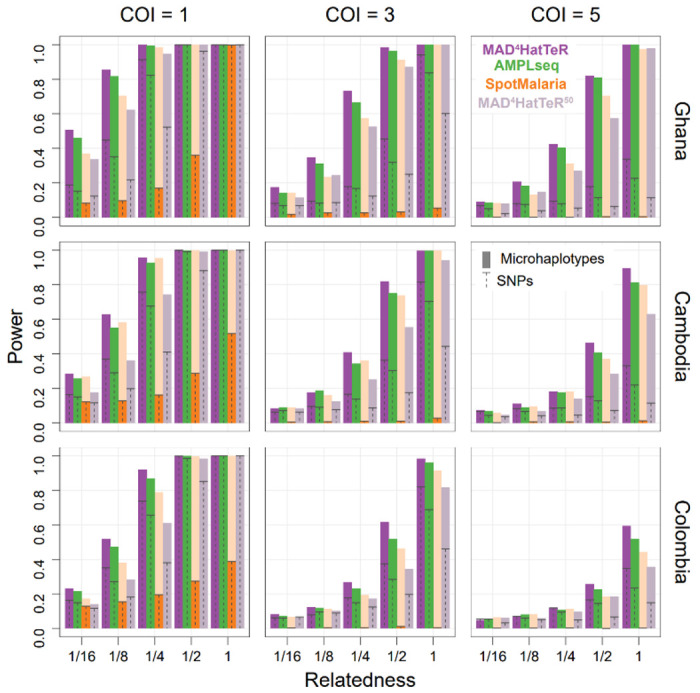
Simulated infections using population allele frequencies from available WGS data were used to estimate the power of testing if a pair of strains between infections is related. Countries in each of three continents with the most available WGS data were selected. Infections were simulated for a range of COI. Only one pair of strains between the infections was related with a given expected IBD proportion (r). The results were compared for reconstructed microhaplotypes and their most highly variable SNP for 3 panels (MAD4HatTeR, SpotMalaria and AMPLseq). Note that SpotMalaria bioinformatics pipeline outputs a 100 SNP barcode, and thus its actual power (dark orange) is not reflective of the potential power afforded by microhaplotypes (light orange). Additionally, the 50 most diverse microhaplotypes and their corresponding SNPs were used to evaluate the effect of down-sizing MAD4HatTeR (MAD4HatTeR50).
MAD4HatTeR allows for genotyping of a variety of sample types and parasite densities
We evaluated MAD4HatTeR’s performance using dried blood spots (DBS) containing up to 7 different cultured laboratory strains each. Sequencing depth was lower for samples amplified with the original resistance R1 primer pool R1.1 than D1 (Supplementary Figure 4A), and primer dimers comprised 58-98% of the reads for R1.1 compared to only 0.1-4% for D1. We thus designed pool R1.2, a subset of targets from R1.1, by selecting the targets with priority public health applications and discarding the primers that accounted for a significant portion of primer dimers in generated data (Figure 1, Supplementary Table 2). Libraries prepared with pools containing R1.2 instead of R1.1 showed higher depth across the range of parasitemia evaluated (Supplementary Figure 4B). With the recommended set of primer pools (D1, R1.2, and R2), sequencing provided > 100 reads for most amplicons from DBS with > 10 parasites/μL, with depth of coverage increasing with higher parasite densities (Figure 4A). Samples with < 10 parasites/μL still yielded data albeit less reliably. Approximately 100,000 total unfiltered reads (the output of sample demultiplexing from a sequencing run) were sufficient to get good coverage across targets; on average, 95% of targets had at least 100 reads, and 98% had at least 10 reads (Supplementary Figure 4 C,D). While results indicate that the protocol provides consistently robust results, different experimental parameters may be optimal for different combinations of primer pools and sample concentration.
Figure 4. MAD4HatTeR produces reproducible and sensitive genetic data from a variety of samples.
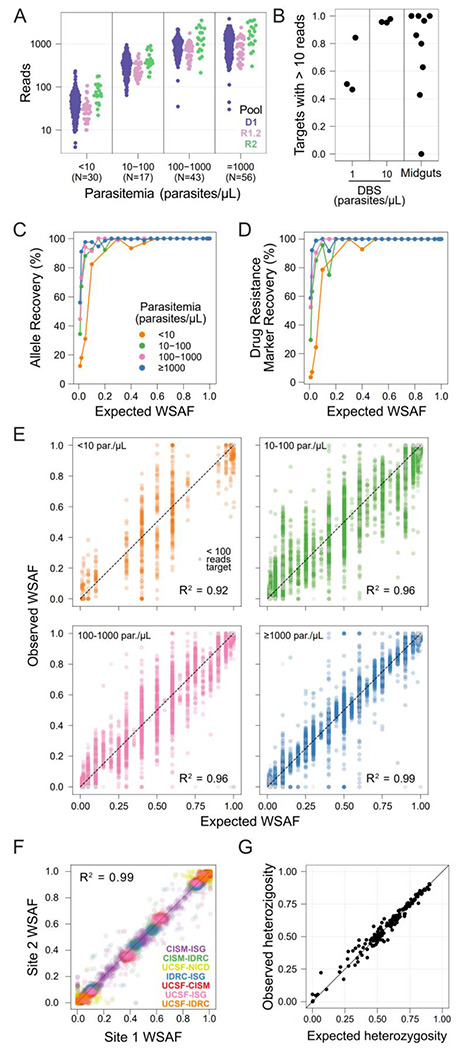
A. Mean read counts for each target in DBS controls (N in parenthesis in x-axis labels for each parasitemia).
B. Proportion of targets with >10 reads in DBS controls with 1 and 10 parasites/μL and 9 midgut samples (median parasite density equivalent to 0.9 parasites/μL in a DBS). 10 targets that generally do not amplify well (>275 bp) were excluded.
C-D. Recovery within-sample allele frequency (WSAF) in the diversity module for 161 loci across 183 samples (C), and biallelic SNPs in drug resistance markers across 20 codons in 165 samples (D).
E. Observed WSAF in laboratory mixed controls of known expected WSAF.
F. WSAF observed in libraries prepared and sequenced in different laboratories from the same DBS mixed control. Participating laboratories are the EPPIcenter at the University of California San Francisco (UCSF); Infectious Diseases Research Collaboration (IDRC), Uganda; Centro de Investigação em Saúde de Manhiça (CISM), Mozambique; National Institutes for Communicable Diseases (NICD), South Africa; and Barcelona Institute for Global Health (ISG), Spain.
G. Observed heterozygosity in field samples from Mozambique22 and the respective expected heterozygosity for each target obtained from available WGS data (which does not include the MAD4HatTeR-sequenced field samples).
False positives are excluded from C-G, as are targets with < 100 reads, except in E.
Depth of coverage per amplicon was highly correlated within technical replicates (Supplementary Figure 5A) with most deviations observed between primer pools. Importantly, coverage was also reproducible when the same samples were tested across five laboratories on 3 continents, with minor quantitative but negligent qualitative differences in coverage (Supplementary Figure 5B). Amplicon coverage was well balanced within a given sample, with differences in depth negatively associated with amplicon length (Supplementary Figure 6). Nine of the 15 worst-performing amplicons were particularly long (>297 bp, Supplementary Table 6). The other worst-performing amplicons covered drug resistance markers in mdr1 and crt (neither covering mdr1 N86Y or crt K76T), 2 high heterozygosity targets, and a target within hrp2. These results indicate that robust coverage of the vast majority of targets can be consistently obtained from different laboratories.
Given the high sensitivity of the method, we evaluated the ability of MAD4HatTeR to generate data from sample types where it is traditionally challenging to obtain high quality parasite sequence data. We amplified DNA extracted from nine infected mosquito midguts with a median P. falciparum DNA concentration equivalent to 0.9 parasites/μL from a DBS. On average, 58% of amplicons had ≥100 reads, 84% had ≥10 reads, and only one sample did not amplify (Figure 4B). These results are comparable to libraries from DBS controls with 1-10 parasites/μL from the same sequencing run, where 45-77% of amplicons with ≥100 reads. Within sample allele frequencies (WSAF) indicated that some of the mosquito midguts contained several genetically distinct P. falciparum clones. These data show the potential for applying MAD4HatTeR to study a variety of sample types containing P. falciparum.
MAD4HatTeR reproducibly detects genetic diversity, including for minority alleles in low density, polyclonal samples
We used DBS controls containing 2 to 7 laboratory P. falciparum strains with minor WSAF ranging from 1 to 50% to evaluate sensitivity of detection and accuracy of WSAF estimation in the diversity pool D1. We optimized and benchmarked the bioinformatic pipeline to maximize sensitivity and precision, which included masking regions of low complexity (tandem repeats and homopolymers) to avoid capturing PCR and sequencing errors in allele calls. Sensitivity to detect minority alleles given that the locus amplified was very high, with alleles present at ≥ 2% reliably detected in samples with > 1,000 parasites/μL and at ≥ 5% in samples with > 10 parasites/μL (Figure 4C). For very low parasitemia samples (< 10 parasites/μL), sensitivity was still 82% for alleles expected at 10% or higher. Similar results were obtained for drug resistance markers targeted by pools R1.2 and R2 (Figure 4D). Overall precision (reflecting the absence of spurious alleles) was also high and could be increased by using a filtering threshold for minimum WSAF. Each sample had a median of 3 false positive alleles (mean = 4.4, N = 161 targets) above 0.75% WSAF, a median of 1 (mean = 2.5) false positives over 2%, and a median of 0 (mean = 0.7) over 5% (Supplementary Figure 7). A strong correlation between expected and observed WSAF was observed in the diversity module targets at all parasite densities and was stronger at higher parasite densities (R2=0.99 for > 1,000 parasites/μL Figure 4E).
Reproducibility is an important feature in generating useful data, particularly given differences in equipment and technique that often exists between laboratories. To evaluate this potential source of variation, we generated data for the same mixed-strain controls in five different laboratories on three continents. Reassuringly, the alleles obtained, along with their WSAF, were highly correlated (Figure 4F). Missed alleles in one or more laboratories were mostly present at < 2% within a sample. Finally, we tested MAD4HatTeR’s ability to recover expected diversity in field samples. Observed genetic heterozygosity in samples from Mozambique22 was correlated with expected heterozygosity based on available WGS data (Figure 4G, Supplementary Figure 8). These results highlight the reliability of MAD4HatTeR as a method to generate high quality genetic diversity data across laboratories.
MAD4HatTeR provides data on copy number variations and detection of non-P. falciparum species
In addition to detecting sequence variation in P. falciparum, amplicon sequencing data can be used to detect gene deletions and duplications, as well as the presence of other Plasmodium species. We tested the ability of MAD4HatTeR to detect hrp2 and hrp3 deletions, and mdr1 and hrp3 duplications (laboratory strain FCR3 has a duplication in hrp335) in DBS controls consisting of one or two laboratory strains, and field samples with previously known genotypes. We applied a generalized additive model to normalize read depth and estimate fold change across several targets per gene, accounting for amplicon length bias and pool imbalances, after using laboratory controls to account for batch effects, e.g. running the assay in different laboratories (Figure 5A, Supplementary Figure 9). The resulting depth fold changes for all loci assayed correlated with the expected sample composition (Figure 5B). At 95% specificity, sensitivity was 100% for all controls composed of > 95% strains with duplications or deletions (Figure 5C). Sensitivity was lower for samples with lower relative abundance of strains carrying duplications or deletions, although this could be increased with a tradeoff in specificity (e.g. if used as a screening test). Fold change data correlated well with quantification by qPCR, indicating that the data obtained from MAD4HatTeR are at a minimum semi-quantitative (Figure 5D). We could also correctly detect deletions in field samples from Ethiopia previously shown to be hrp2- or hrp3-deleted3, and correctly classify the genomic breakpoint profiles within the resolution offered by the targets included (Supplementary Figure 10). Finally, we observed reads in the ldh target for the four non-falciparum species in samples from Uganda known to contain the corresponding species, as previously determined by microscopy or nested PCR. We could distinguish Plasmodium ovale wallikeri from Plasmodium ovale curtisi based on the alleles in the target sequence. These data highlight the potential of MAD4HatTeR to capture non-SNP genetic variation and to characterize mixed species infections.
Figure 5. MAD4HatTeR can be used to screen for deletions and duplications.
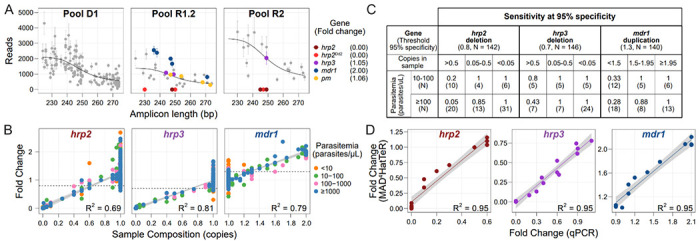
A. Technical replicates of Dd2 (a strain with hrp2 deletion and mdr1 duplication) with similar total reads were used to estimate fold changes in targets in and around hrp2, hrp3, mdr1 and plasmepsin2/3 (pm). A generalized additive model (black line) was applied to raw reads (Supplementary Figure 9) after correction by a control known not to have deletions or duplications in the genes of interest (3D7) to estimate fold changes in each of the genes. Note that there are two groups of hrp2 targets, those that are deleted in field samples (hrp2) and those also deleted in Dd2 (hrp2Dd2). Mean reads and fold changes are shown (N = 3); error bars denote standard deviation.
B. Estimated fold change for hrp2, hrp3, and mdr1 loci in laboratory controls containing 1 or more strains at known proportions, or in field samples from Ethiopia3 with known hrp2 and hrp3 deletions. Sample composition is estimated as the effective number of copies present in the sample based on the relative proportion of the strain carrying a deletion or duplication. Fold changes are obtained using the targets highlighted in A. Fold changes for Dd2-specific targets are shown in Supplementary Figure 10. Linear regression and R2 values were calculated with data with parasitemia > 10 parasites/μL. The thresholds used to flag a sample as containing a duplication or deletion are shown in dashed black lines.
C. Sensitivity in detecting hrp2 and hrp3 deletions and mdr1 duplications in controls, and field samples from Ethiopia with known hrp2 and hrp3 deletions. Effective sample composition (copies in sample) is estimated as in B. Sensitivity was calculated using a threshold to classify samples with 95% specificity. Note that the small number of samples in the 0.05-0.5 copies range may be responsible for the paradoxical lower sensitivity for higher parasitemia samples.
D. Estimated fold change for each gene correlates with qPCR quantification for the same samples.
Discussion
In this study, we developed, characterized and deployed a robust and versatile method to generate sequence data for P. falciparum malaria genomic epidemiology, prioritizing information for public health decision-making. The modular MAD4HatTeR amplicon sequencing panel produces high-resolution data on genetic diversity, key markers for drug and diagnostic resistance, the C-terminal domain of the csp vaccine target, and presence of other Plasmodium species. MAD4HatTeR is highly sensitive, providing data for low parasite density DBS samples and detecting minor alleles at WSAF as low as 1% with good specificity in high parasite density samples; challenging sample types such as infected mosquitos were also successfully amplified. MAD4HatTeR has successfully generated data from field samples from Mozambique and Ethiopia, with particularly good recovery rates for samples with > 10 parasites/μL (~90%)22,36. Deletions and duplications were reliably detected in mono- and polyclonal controls. The data generated by MAD4HatTeR are highly reproducible and have been reliably produced in multiple laboratories, including several in malaria-endemic countries. Thus, MAD4HatTeR is a valuable tool for malaria surveillance and research, offering policymakers and researchers an efficient means of generating useful data.
The 165 diversity and differentiation targets in MAD4HatTeR, of which the majority are microhaplotypes, can be used to accurately estimate within-host and population genetic diversity, and relatedness between infections. These data have promising applications: evaluating transmission patterns, e.g. to investigate outbreaks3; characterizing transmission intensity, e.g. to evaluate interventions10,13,37 or surveillance strategies22; classifying infections in low transmission areas as imported or local11,38; or classifying recurrent infections in antimalarial therapeutic efficacy studies as recrudescence or reinfections18. The high diversity captured by the current microhaplotypes could be further improved with updated WGS data to replace targets with relatively low diversity and amplification efficiency. Fully leveraging the information content of these diverse loci, which are particularly useful for evaluating polyclonal infections, requires bioinformatic pipelines able to accurately call microhaplotype alleles and downstream analysis methods able to incorporate these multi-allelic data. While some targeted sequencing methods and pipelines similarly produce microhaplotype data30,32,39–41, others only report individual SNPs, resulting in the loss of potentially informative data26,27 encoded in phased amplicon sequences. Many downstream analysis tools are similarly limited to evaluating data from binary SNPs42–44. Fortunately, methods to utilize these data are beginning to be developed, providing statistically grounded estimates of fundamental quantities such as population allele frequencies, complexity of infection45, and identity-by-descent46, and highlighting gains in accuracy and power provided by analysis of numerous highly diverse loci.
Multiple targeted sequencing tools designed with different use cases and geographies in mind are being used, raising questions about data compatibility. Comparing diversity metrics from data generated using different target sets is feasible, provided that the panels have equivalent performance characteristics and that the analysis methods appropriately account for differences such as allelic diversity45. Comparing genetic relatedness between infections evaluated with different panels, however, is limited to common loci. Over 25% of SNPs targeted by AMPLseq or SpotMalaria diversity targets were intentionally included in MAD4HatTeR. Other panels have less or no overlap27,39,41 (Supplementary Tables 9–10). Efforts to increase overlap between future versions of amplicon panels would facilitate more direct comparison of relatedness between infections genotyped by different panels.
Depth of coverage and amplification biases were reproducible across samples, with most deviations likely due pipetting volume differences and systematic differences in laboratory equipment and reagent batches. Detection of hrp2/3 deletions and mdr1 duplications was achieved by applying a model that accounts for these factors. MAD4HatTeR detected deletions and duplications in mono- and polyclonal samples, even at low parasitemia. Additional data and analytical developments could improve MAD4HatTeR’s performance in deletion and duplication analysis. The current approach does not make use of COI estimates for inference and relies on controls known not to have duplications or deletions in the target genes within each library preparation batch. While target retrieval was generally uniform, some samples showed target drop-off, indicating the need for multiple targets to avoid falsely calling a deletion. Nonetheless, in its current form, MAD4HatTeR serves as an efficient screening tool for identifying putative duplications and deletions, which can then be validated with gold-standard methodologies.
Continuous improvement of the allele-calling bioinformatic pipeline is planned to increase accuracy and usability. Masking of error-prone regions (e.g. homopolymers and tandem repeats) is useful in reducing common PCR and sequencing errors, but it also removes biological variation. This can be optimized by tailored masking of error hotspots, rather than uniformly masking all low-diversity sequences. To improve the detection of low-abundance alleles, we currently conduct a second inference round using alleles observed within a run as priors, but this approach may also increase the risk of incorporating low-level contaminant reads. Improvements in experimental strategies to detect and prevent cross-contamination47, along with post-processing filtering, could mitigate this. Additionally, curating an evolving allele database from ongoing empiric data generation could replace the run-dependent priors, thereby improving the accuracy and consistency of allele inference.
Integrating genomics into routine surveillance and developing genomic capacity in research and public health institutions in malaria-endemic countries is facilitated by efficient, cost-effective, reliable and accessible tools. MAD4HatTeR is based on a commercially available method for multiplexed amplicon sequencing48. As such, while primer sequences are publicly available (Supplementary Table 2), reagents are proprietary. However, procuring bundled, quality controlled reagents to generate libraries is straightforward, including for laboratories in malaria endemic settings. Procurement costs for laboratory supplies often vary significantly, making direct comparisons with other methods challenging, but we have found the method to be cost-effective compared with other methods. At the time of writing, the list price for all library preparation reagents, excluding plastics, consumables used for other steps (e.g. DNA extraction), sequencing costs, taxes, or handling, was $12-25 per reaction, depending on order volume. Sequencing costs can vary considerably based on the scale of sequencer used. For optimal throughput, we recommend multiplexing up to 96 samples using a MiSeq v2 kit to achieve results comparable to those shown here; much greater efficiency can be obtained with higher throughput sequencers.
This study includes data from five laboratories, three of which are located in sub-Saharan Africa. Beyond this study, MAD4HatTeR is also being used by four other African laboratories for applications ranging from estimating the prevalence of resistance-mediating mutations to characterizing transmission networks. Expertise and computational infrastructure for advanced bioinformatics and data analysis remains a challenge, with fewer users demonstrating autonomy in these areas compared to wet lab procedures. The robustness of the method, along with detailed training activities and materials (available online49), has facilitated easier implementation. Future developments could also expand accessibility, including adaptations for other sequencing platforms and panels targeting a smaller set of key loci for public health decision-making.
In summary, MAD4HatTeR is a powerful and fit-for-purpose addition to the malaria genomic epidemiology toolbox, well-suited for a wide range of surveillance and research applications.
Methods
Participating laboratories
We generated data in five sites: the EPPIcenter at the University of California San Francisco (UCSF), in collaboration with the Chan Zuckerberg Biohub San Francisco, California; Infectious Diseases Research Collaboration (IDRC) at Central Public Health Laboratories (CPHL), Kampala, Uganda; Centro de Investigação em Saúde de Manhiça (CISM), Manhiça, Mozambique; National Institutes for Communicable Diseases (NICD), Johannesburg, South Africa; and Barcelona Institute for Global Health (ISGlobal), Barcelona, Spain. The procedures are described according to the workflows in San Francisco. Minor variations, depending on equipment availability, were implemented at other institutions.
Amplicon panel design
We used available WGS data as of June 20213,30,50–58 to identify regions with multiple SNPs within windows of 150-300 bp that lay between tandem repeats, using a local haplotype reconstruction tool (Pathweaver59). We compiled a list of drug resistance-associated and immunity-related SNPs (Tables 1 and 2) and identified regions of 150-300 bp between tandem repeats in and around hrp2 and hrp3 to assess diagnostic resistance-related deletions, as well as a region in chromosome 11 that is often duplicated in hrp3-deleted samples35. Paragon Genomics, Inc. designed amplification primers in multiplexed PCR using the Pf3D7 genome (version=2020-09-01) as a reference and used related species (PvP01 (version=2018-02-28) for P. vivax, PmUG01 (version=2016-09-19) for P. malariae, PocGH01 (version=2017-03-06) for P. ovale, and PKNH (version=2015-06-18) for P. knowlesi) and the human genome to design primers specific for P. falciparum. In addition to the P. falciparum targets, we selected a target in the ldh gene (PF3D7_1325200) and its homologs in the other 4 Plasmodium species listed above for identification of concurrent infections with these species. To minimize PCR bias against longer amplicons, we restricted the design to amplicons of 225-275 bp, which can be covered with a significant overlap in paired-end sequencing in Illumina platforms with 300-cycle kits, except for targets around hrp3 that needed to be 295-300 bp long to design primers successfully. We excluded or redesigned primers that contained more than 1 SNP (including non-biallelic SNPs) or indels in available WGS data or aligned to tandem repeats. To increase coverage of SNPs close to each other, we allowed for overlap in amplicons that targeted drug resistance and immunity-related markers. Primers were grouped in modules, as outlined in the results section (Figure 1 and Supplementary Table 1).
In silico panel performance calculations
Alleles were extracted from available WGS data as of July 20243,30,50–57,60. SNPs, and microhaplotypes were reconstructed using Pathweaver59 for targets in MAD4HatTeR, SpotMalaria26, AMPLseq30, and AmpliSeq27. In silico heterozygosity was calculated using all allele calls in available WGS data. Principal coordinate analysis was performed on the binary distance matrix from presence/absence of alleles using alleles within loci present in both samples for each pair.
To assess statistical power of testing if two (potentially polyclonal) infections are related, we obtained WSAF for the most variable SNP in each diversity target (165, 111 and 100 total SNPs for MAD4HatTeR, AMPLseq and SpotMalaria, respectively) or microhaplotypes (161, 128 and 135, respectively) from WGS data for each of the three panels, and simulated genotypes for mono- and polyclonal samples. In the simulations, COI were fixed and ranged from 1 to 5, and we included genotyping errors with a miss-and-split model46; missing and splitting parameters were 0.05 and 0.01, respectively. Between two samples, only a single pair of parasite strains was related with expected IBD proportion varying from 1/16 to 1/2 (sibling level) to 1 (clones). We then analyzed these simulated datasets to obtain performance measures for combinations of a panel, COI, and a relatedness level: first, we estimated COI and allele frequencies using MOIRE45; we then used these to estimate pairwise interhost relatedness and test the hypothesis that two infections are unrelated at significance level of 0.05 with Dcifer46 and calculated power as the proportion of 1000 simulated pairs where the null hypothesis was correctly rejected.
Samples
We prepared control dried blood spots (DBS) using P. falciparum laboratory strains. We synchronized monocultures in the ring stage. We made polyclonal controls by mixing cultured strains (3D7, Dd2 MRA-156 and MRA-1255, D6, W2, D10, U659, FCR3, V1/S, and HB3), all synchronized and ring-staged at various proportions. We mixed all monocultures and mixtures with uninfected human blood and serially diluted them in blood to obtain a range of parasite densities (0.1-100,000 parasites/μL). We spotted 20 μL of the mixture on filter papers and stored them at −20 °C until processing.
We generated data for 26 field samples from Ethiopia using DNA extracts from a previous study3. Ethical approval for that study was granted by the Ethiopia Public Health Institute (EPHI) Institutional Review Board (IRB; protocol EPHI-IRB-033-2017) and WHO Research Ethics Review Committee (protocol ERC.0003174 001). Processing of de-identified samples and data at the University of North Carolina at Chapel Hill (UNC) was determined to constitute non-human subjects research by the UNC IRB (study 17-0155). The study was determined to be non-research by the Centers for Disease Control (CDC) and Prevention Human Subjects office (0900f3eb81bb60b9). All participants provided informed consent. In addition, we analyzed publicly available data from 436 field samples from Mozambique22. Study protocols were approved by the ethical committees of CISM and Hospital Clinic of Barcelona, and the Mozambican Ministry of Health National Bioethics Committee. All study participants, or guardians/parents in the case of minors, gave written informed consent. The original works detail the sampling schemes and additional sample processing procedures.
Library preparation
We extracted DNA from control DBS using the Chelex-Tween 20 method61, and quantified parasite density by varATS62 or 18S63 qPCR (Supplementary Text).
Libraries were made with a minor adaptation of Paragon Genomics’ CleanPlex Custom NGS Panel Protocol64 (Supplementary Text). A version of the protocol containing any updates can be found at https://eppicenter.ucsf.edu/resources. Library pools were sequenced in Illumina MiSeq, MiniSeq, NextSeq 550, or NextSeq 2000 instruments with 150 paired-end reads. We tested different amplification cycles and primer pool configurations. Based on sensitivity and reproducibility, the following are the experimental conditions we use as a default: primer pools D1+R1.2+R2; 15 multiplexed PCR cycles for moderate to high parasite density samples (equivalent to ≥ 100 parasites/μL in DBS) and 20 cycles for samples with lower parasite density; 0.25X and 0.12X primer pool concentration, respectively.
Bioinformatic pipeline development and benchmarking
We developed a Nextflow-based65 bioinformatic pipeline to filter, demultiplex, and infer alleles from fastq files (Supplementary Text). Briefly, the pipeline uses cutadapt66 and DADA267 to demultiplex reads on a per-amplicon basis and infer alleles, respectively. The pipeline further processes DADA2 outputs to mask low-complexity regions, generate allele read count tables, and extract alleles in SNPs of interest. We developed custom code in Python and R to filter out low-abundance alleles and calculate summary statistics from the data. The current pipeline version, with more information on implementation and usage, can be found at www.github.com/EPPIcenter/mad4hatter.
We processed the data presented in this paper with release 0.1.8 of the pipeline.
We evaluated pipeline performance by estimating sensitivity (ability to identify expected alleles) and precision (ability to identify only expected alleles) from monoclonal and mixed laboratory controls with different proportions of strains (Supplementary Text). We tested the impact of multiple parameters and features on allele calling accuracy, including DADA2’s stringency threshold OMEGA_A and sample pooling treatment for allele recovery, masking homopolymers and tandem repeats, and post-processing filtering of low abundance alleles. Masking removed false positives with the trade-off of masking real biological variation. We obtained the highest precision and sensitivity using sample pseudo-pooling, highly stringent OMEGA_A (10−120), and a moderate postprocessing filtering threshold (minor alleles of > 0.75%). These results indicate that bioinformatic processing of MAD4HatTeR data can be optimized to retrieve accurate sample composition with a detection limit of approximately 0.75% WSAF.
For analyses of allelic data from mixed controls, only samples with ≥ 90% of targets with > 50 reads (183 for diversity, and 165 for drug resistance markers) were included in the analysis. For drug resistance markers, only SNPs with variation between controls were included (20/91 codons from 12/22 targets). Within a sample, targets with less than 100 reads were excluded as alleles with a minor WSAF of 1% are very likely to be missed. The large majority of controls (122/183 and 162/165 for diversity and drug resistance markers, respectively) had very good coverage (at most 2 missing loci).
Heterozygosity was estimated using MOIRE45 version 3.2.0.
Deletions and duplications
We used the following laboratory strains to benchmark deletion and duplication detection using MAD4HatTeR data: pfhrp2 deletions in Dd2 and D10, pfmdr1 duplications in Dd2 and FCR3, pfhrp3 deletion in HB3, and pfhrp3 duplication in FCR335. We also used a set of field samples from Ethiopia previously shown to have deletions in and around pfhrp2 and pfhrp3 at multiple genomic breakpoints3. For sensitivity analysis using field samples, we estimated COI using MOIRE45 and excluded polyclonal samples due to the uncertainty in their true genotypes. Two field samples were excluded from the analysis due to discordance in breakpoint classification, possibly due to sample mislabeling and sequencing depth, respectively.
We applied a generalized additive model (Supplementary Text) to account for target length amplification bias and differences in coverage across primer pools, likely due to pipetting error. We fit the model on controls known not to have deletions or duplications to obtain correction factors for targets of interest within sample batches. We then estimated read depth fold changes from data for each gene of interest (pfhrp2, pfhrp3 and pfmdr1). We did not have sufficient data to validate duplications in plasmepsin 2 and 3.
For a subset of laboratory controls copy numbers were determined by qPCR using previously described methods for pfmdr168, pfhrp2, and pfhrp369.
Supplementary Material
Acknowledgments
We thank Phil Rosenthal and Amy Bei for their input in panel design. We also thank members of the EPPIcenter at UCSF, as well as the Rapid Response Team and the Genomics Platform at the Chan Zuckerberg Biohub for valuable discussions.
Financial support
This work was supported by several grants from the Bill & Melinda Gates Foundation (INV-019032, OPP1132226, INV-037316, INV-024346, INV-031512). This research is also part of the ISGlobal’s Program on the Molecular Mechanisms of Malaria which is partially supported by the Fundación Ramón Areces. We acknowledge support from the grant CEX2023-0001290-S funded by MCIN/AEI/ 10.13039/501100011033, from the Generalitat de Catalunya through the CERCA Program, from the Departament d’Universitats i Recerca de la Generalitat de Catalunya (AGAUR; grant 2017 SGR 664) and from the Ministerio de Ciencia e Innovación (PID2020-118328RB-I00/AEI/10.13039/501100011033). CISM is supported by the Government of Mozambique and the Spanish Agency for International Development (AECID). The parent study from which Ethiopia samples were derived was funded by the Global Fund to Fight AIDS, Tuberculosis, and Malaria through the Ministry of Health - Ethiopia (EPHI5405) and by the Bill & Melinda Gates Foundation through the World Health Organization (OPP1209843). A.A.-D. was supported by the Chan Zuckerberg Biohub Collaborative Postdoctoral fellowship. B.G. was supported by NIH-NIAID K24AI144048. J.B.P. was supported by NIH-NIAD R01 AI77791.
Potential conflicts of interest
J.B.P. reports research support from Gilead Sciences, non-financial Support from Abbott Laboratories, and consulting for Zymeron Corporation, all outside the scope of the current work. All other authors report no potential conflicts of interest.
Footnotes
Disclaimer
The funders had no role in the study design, data collection, data interpretation, or writing of the manuscript.
References
- 1.Dalmat R., Naughton B., Kwan-Gett T. S., Slyker J. & Stuckey E. M. Use cases for genetic epidemiology in malaria elimination. Malar. J. 18, 163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton W. L. et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect. Dis. 19, 943–951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feleke S. M. et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat. Microbiol. 6, 1289–1299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ndwiga L. et al. A review of the frequencies of Plasmodium falciparum Kelch 13 artemisinin resistance mutations in Africa. Int. J. Parasitol. Drugs Drug Resist. 16, 155–161 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenthal P. J. et al. Cooperation in Countering Artemisinin Resistance in Africa: Learning from COVID-19. Am. J. Trop. Med. Hyg. 106, 1568–1570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neafsey Daniel E. et al. Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine. N. Engl. J. Med. 373, 2025–2037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesolowski A. et al. Mapping malaria by combining parasite genomic and epidemiologic data. BMC Med. 16, 190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tessema S. et al. Using parasite genetic and human mobility data to infer local and cross-border malaria connectivity in Southern Africa. eLife 8, e43510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tessema S. K. et al. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar. J. 18, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson O. J. et al. Evaluating the Performance of Malaria Genetics for Inferring Changes in Transmission Intensity Using Transmission Modeling. Mol. Biol. Evol. 38, 274–289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels R. F. et al. Genetic evidence for imported malaria and local transmission in Richard Toll, Senegal. Malar. J. 19, 276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mensah B. A., Akyea-Bobi N. E. & Ghansah A. Genomic approaches for monitoring transmission dynamics of malaria: A case for malaria molecular surveillance in Sub–Saharan Africa. Front. Epidemiol. 2, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaffner S. F. et al. Malaria surveillance reveals parasite relatedness, signatures of selection, and correlates of transmission across Senegal. Nat. Commun. 14, 7268 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fola A. A. et al. Temporal and spatial analysis of Plasmodium falciparum genomics reveals patterns of parasite connectivity in a low-transmission district in Southern Province, Zambia. Malar. J. 22, 208 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeka A. et al. Comparative Efficacy of Artemether-Lumefantrine and Dihydroartemisinin-Piperaquine for the Treatment of Uncomplicated Malaria in Ugandan Children. J. Infect. Dis. 219, 1112–1120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snounou G. & Beck H.-P. The Use of PCR Genotyping in the Assessment of Recrudescence or Reinfection after Antimalarial Drug Treatment. Parasitol. Today 14, 462–467 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Uwimana A. et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. 21, 1120–1128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnoz A. et al. Comparison of different genotyping techniques to distinguish recrudescence from new infection in studies assessing the efficacy of antimalarial drugs against Plasmodium falciparum. 2023.04.24.538072 Preprint at 10.1101/2023.04.24.538072 (2023). [DOI] [Google Scholar]
- 19.Lover A. A., Baird J. K., Gosling R. & Price R. N. Malaria Elimination: Time to Target All Species. Am. J. Trop. Med. Hyg. 99, 17–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mwesigwa A. et al. Plasmodium falciparum genetic diversity and multiplicity of infection based on msp-1, msp-2, glurp and microsatellite genetic markers in sub-Saharan Africa: a systematic review and meta-analysis. Malar. J. 23, 97 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs J. et al. Within-household clustering of genetically related Plasmodium falciparum infections in a moderate transmission area of Uganda. Malar. J. 20, 68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brokhattingen N. et al. Genomic malaria surveillance of antenatal care users detects reduced transmission following elimination interventions in Mozambique. Nat. Commun. 15, 2402 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viriyakosol S. et al. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull. World Health Organ. 73, 85–95 (1995). [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson T. J. C., Su X.-Z., Bockarie M., Lagog M. & Day K. P. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119, 113–125 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Anderson T. J. C. et al. Microsatellite Markers Reveal a Spectrum of Population Structures in the Malaria Parasite Plasmodium falciparum. Mol. Biol. Evol. 17, 1467–1482 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Jacob C. G. et al. Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination. eLife 10, e62997 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kattenberg J. H. et al. Molecular Surveillance of Malaria Using the PF AmpliSeq Custom Assay for Plasmodium falciparum Parasites from Dried Blood Spot DNA Isolates from Peru. Bio-Protoc. 13, e4621 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor A. R., Jacob P. E., Neafsey D. E. & Buckee C. O. Estimating Relatedness Between Malaria Parasites. Genetics 212, 1337–1351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessema S. K. et al. Sensitive, Highly Multiplexed Sequencing of Microhaplotypes From the Plasmodium falciparum Heterozygome. J. Infect. Dis. 225, 1227–1237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaVerriere E. et al. Design and implementation of multiplexed amplicon sequencing panels to serve genomic epidemiology of infectious disease: A malaria case study. Mol. Ecol. Resour. 22, 2285–2303 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesare M. de et al. Flexible and cost-effective genomic surveillance of P. falciparum malaria with targeted nanopore sequencing. 2023.02.06.527333 Preprint at 10.1101/2023.02.06.527333 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holzschuh A. et al. Using a mobile nanopore sequencing lab for end-to-end genomic surveillance of Plasmodium falciparum: A feasibility study. PLOS Glob. Public Health 4, e0002743 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girgis S. T. et al. Drug resistance and vaccine target surveillance of Plasmodium falciparum using nanopore sequencing in Ghana. Nat. Microbiol. 8, 2365–2377 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva C. et al. Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique. Commun. Biol. 6, 1–11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hathaway N. J. et al. Interchromosomal segmental duplication drives translocation and loss of P. falciparum histidine-rich protein 3. eLife 13, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emiru T. et al. Evidence for a role of Anopheles stephensi in the spread of drug- and diagnosis-resistant malaria in Africa. Nat. Med. 29, 3203–3211 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels R. F. et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc. Natl. Acad. Sci. 112, 7067–7072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H.-H. et al. Mapping imported malaria in Bangladesh using parasite genetic and human mobility data. eLife 8, e43481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holzschuh A. et al. Multiplexed ddPCR-amplicon sequencing reveals isolated Plasmodium falciparum populations amenable to local elimination in Zanzibar, Tanzania. Nat. Commun. 14, 3699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hathaway N. J., Parobek C. M., Juliano J. J. & Bailey J. A. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res. 46, e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerch A. et al. Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections. BMC Genomics 18, 864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaffner S. F., Taylor A. R., Wong W., Wirth D. F. & Neafsey D. E. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar. J. 17, 196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henden L., Lee S., Mueller I., Barry A. & Bahlo M. Identity-by-descent analyses for measuring population dynamics and selection in recombining pathogens. PLOS Genet. 14, e1007279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang H.-H. et al. THE REAL McCOIL: A method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLOS Comput. Biol. 13, e1005348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy M. & Greenhouse B. MOIRE: A software package for the estimation of allele frequencies and effective multiplicity of infection from polyallelic data. 2023.10.03.560769 Preprint at 10.1101/2023.10.03.560769 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerlovina I., Gerlovin B., Rodríguez-Barraquer I. & Greenhouse B. Dcifer: an IBD-based method to calculate genetic distance between polyclonal infections. Genetics 222, iyac126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagerborg K. A. et al. Synthetic DNA spike-ins (SDSIs) enable sample tracking and detection of inter-sample contamination in SARS-CoV-2 sequencing workflows. Nat. Microbiol. 7, 108–119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.CleanPlex amplicon sequencing for targeted DNA and RNA Seq. Paragon Genomics; https://www.paragongenomics.com/targeted-sequencing/amplicon-sequencing/cleanplex-ngs-amplicon-sequencing/. [Google Scholar]
- 49.Resources | EPPIcenter. https://eppicenter.ucsf.edu/resources. [Google Scholar]
- 50.Melnikov A. et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12, R73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villena F. E., Lizewski S. E., Joya C. A. & Valdivia H. O. Population genomics and evidence of clonal replacement of Plasmodium falciparum in the Peruvian Amazon. Sci. Rep. 11, 21212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathieu L. C. et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife 9, e51015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerqueira G. C. et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 18, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parobek C. M. et al. Partner-Drug Resistance and Population Substructuring of Artemisinin-Resistant Plasmodium falciparum in Cambodia. Genome Biol. Evol. 9, 1673–1686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelleau S. et al. Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc. Natl. Acad. Sci. 112, 11672–11677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dara A. et al. New var reconstruction algorithm exposes high var sequence diversity in a single geographic location in Mali. Genome Med. 9, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tvedte E. S. et al. Evaluation of a high-throughput, cost-effective Illumina library preparation kit. Sci. Rep. 11, 15925 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.An open dataset of Plasmodium falciparum … | Wellcome Open Research. https://wellcomeopenresearch.org/articles/6-42. [Google Scholar]
- 59.Hathaway N. A suite of computational tools to interrogate sequence data with local haplotype analysis within complex Plasmodium infections and other microbial mixtures. (2018) doi: 10.13028/M2039K. [DOI] [Google Scholar]
- 60.Malaria GEN et al. Pf7: an open dataset of Plasmodium falciparum genome variation in 20,000 worldwide samples. Wellcome Open Res. 8, 22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teyssier N. B. et al. Optimization of whole-genome sequencing of Plasmodium falciparum from low-density dried blood spot samples. Malar. J. 20, 116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofmann N. et al. Ultra-Sensitive Detection of Plasmodium falciparum by Amplification of Multi-Copy Subtelomeric Targets. PLOS Med. 12, e1001788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayor A. et al. Sub-microscopic infections and long-term recrudescence of Plasmodium falciparum in Mozambican pregnant women. Malar. J. 8, 9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paragon Genomics Product Documents. Paragon Genomics; https://www.paragongenomics.com/customer-support/product_documents/. [Google Scholar]
- 65.Di Tommaso P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 67.Callahan B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta H. et al. Drug-Resistant Polymorphisms and Copy Numbers in Plasmodium falciparum, Mozambique, 2015. Emerg. Infect. Dis. 24, 40–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grignard L. et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine 55, 102757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010-2019). https://www.who.int/publications/i/item/9789240012813. [Google Scholar]
- 71.Miotto O. et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tørresen O. K. et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 47, 10994–11006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


