Abstract
The detection of mitochondrial DNA (mtDNA) mutations in single cells holds considerable potential to define clonal relationships coupled with information on cell state in humans. Previous methods focused on higher heteroplasmy mutations that are limited in number and can be influenced by functional selection, introducing biases for lineage tracing. Although more challenging to detect, intermediate to low heteroplasmy mtDNA mutations are valuable due to their high diversity, abundance, and lower propensity to selection. To enhance mtDNA mutation detection and facilitate fine-scale lineage tracing, we developed the single-cell Regulatory multi-omics with Deep Mitochondrial mutation profiling (ReDeeM) approach, an integrated experimental and computational framework. Recently, some concerns have been raised about the analytical workflow in the ReDeeM framework. Specifically, it was noted that the mutations detected in a single molecule per cell are enriched on edges of mtDNA molecules, suggesting they resemble artifacts reported in other sequencing approaches. It was then proposed that all mutations found in one molecule per cell should be removed. We detail our error correction method, demonstrating that the observed edge mutations are distinct from previously reported sequencing artifacts. We further show that the proposed removal leads to massive elimination of bona fide and informative mutations. Indeed, mutations accumulating on edges impact a minority of all mutation calls (for example, in hematopoietic stem cells, the excess mutations on the edge account for only 4.3%−7.6% of the total). Recognizing the value of addressing edge mutations even after applying consensus correction, we provide an additional filtering option in the ReDeeM-R package. This approach effectively eliminates the position biases, leads to a mutational signature indistinguishable from bona fide mitochondrial mutations, and removes excess low molecule high connectedness mutations. Importantly, this option preserves the large majority of unique mutations identified by ReDeeM, maintaining the ability of ReDeeM to provide a more than 10-fold increase in variant detection compared to previous methods. Additionally, the cells remain well-connected. While there is room for further refinement in mutation calling strategies, the significant advances and biological insights provided by the ReDeeM framework are unique and remain intact. We hope that this detailed discussion and analysis enables the community to employ this approach and contribute to its further development.
Lineage tracing using cellular barcoding provides the opportunity to gain insights into the cellular hierarchies and dynamics across tissues in health and disease1. We and others previously demonstrated the potential for mitochondrial DNA (mtDNA) mutations to serve as natural cellular barcodes in humans. Previous methods focus on a limited subset of mtDNA mutations with relatively high heteroplasmy due to the challenge of somatic mutation calling. However, relying solely on this small set of mutations provides incomplete information. Additionally, it has been suggested that some of these higher heteroplasmic mutations might be pre-existing germline variants or subject to bottlenecks or context-dependent functional selection, which introduce biases and hinder their ability to serve as inert lineage tracers as suggested by recent studies2–4. Mitochondrial DNA exhibits a 50-fold higher mutation rate compared to the nuclear genome, providing an opportunity to generate a substantial number of diverse somatic mutations, albeit primarily at lower heteroplasmy, as reported in bulk tissues using ultra-sensitive approaches5. We reason that these intermediate to low heteroplasmy mutations could be particularly valuable because they (1) are more likely to accrue and provide extensive information for clonal and subclonal tracking given their larger number and higher diversity, (2) are overall neutral to selection and thus can serve as an unbiased lineage-tracer, and (3) are less likely to be confounded by preexisting germline variants (Fig. 1a). However, their detection poses a more complex challenge compared to those in the nuclear genome due to the low level of heteroplasmy within individual cells. Consequently, even with high coverage, conventional sequencing cannot easily distinguish these mutations from sequencing errors without appropriate correction (Extended Data Fig. 2 in Ref. 6). We recently presented the single-cell Regulatory multi-omics with Deep Mitochondrial mutation profiling (ReDeeM) method6, with the goal to enhance fine-scale lineage tracing analysis by capturing a rich set of mtDNA somatic mutations (>10-fold more mtDNA somatic mutations) using targeted enrichment and single-molecule consensus error correction, and to link these variants with cell state information. The ability to utilize many high, intermediate, and low heteroplasmy mtDNA somatic mutations with ReDeeM allows us to improve the recording of various clonal and subclonal events, enhancing the informativeness and robustness of lineage tracing.
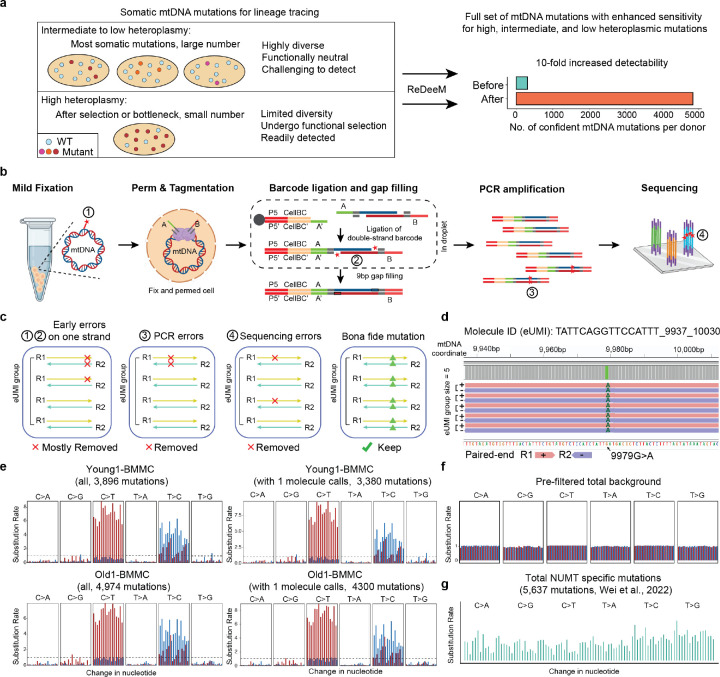
Fig. 1. Enhanced sensitivity of ReDeeM through rigorous single-molecule consensus.
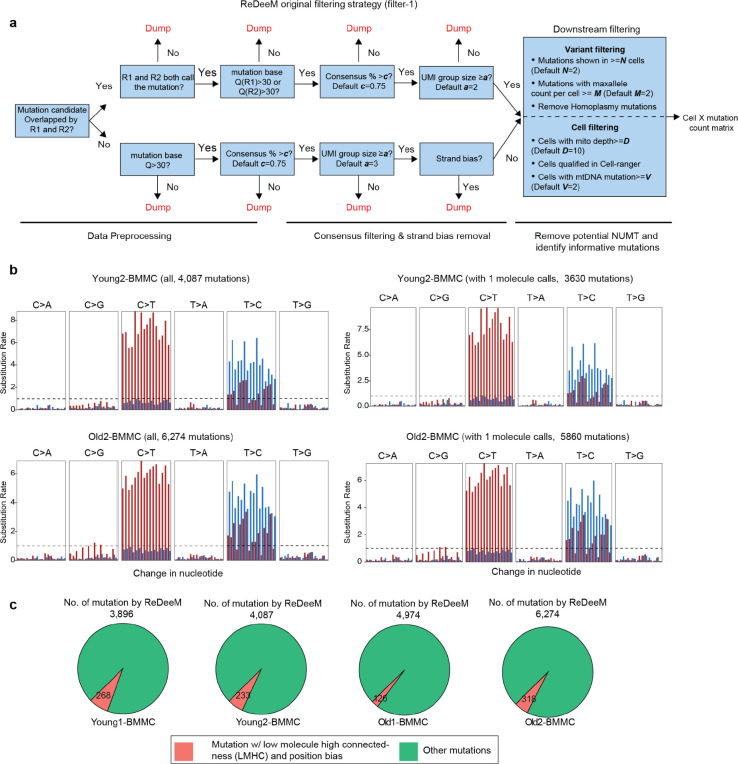
(a) Challenge and motivation of enhancing mtDNA mutation detection. A large majority of somatic mtDNA mutations show intermediate to low heteroplasmy as many are associated with sub-clonal events. The advantages and limitations of different heteroplasmic mutations are discussed. ReDeeM allows detection of a fuller set of mtDNA mutations including high, intermediate, and low heteroplasmic mutations for fine-scale and unbiased lineage tracing. (b) Workflow of the ReDeeM experiment and the potential sources of artifacts within each experimental stage, represented as red stars. See more detailed discussion in Supplementary Notes. (c) Single-molecule consensus error correction strategy that accounts for previously described artifacts. See more detailed discussion in Supplementary Notes. (d) One real data example of the grouped eUMI sequencing read family. The eUMI group size is 5, each is completely overlapping sequenced by both R1 and R2. (e-g) Mutational signatures (frequency unweighted) for all ReDeeM identified confident mtDNA mutations or the collection for 1+-molecule mutations in Young1-BMMC and Old1-BMMC (also see Extended Data Fig. 1b). (e) In comparison, the mutational signatures for prefiltered mutations, and NUMT mutations are also shown (f-g)
A recent commentary from Lareau et al. noted the important advances in ReDeeM implementing experimentally enriched mtDNA and applying consensus mutation calling for higher-quality mtDNA-based lineage tracing, but also brought up concerns about ReDeeM data analysis methods. Specifically, it was noted that the mutations detected in a single molecule per cell are enriched on edges of mtDNA fragments, suggesting they resemble artifacts and proposed to remove all these mutations7. Here, we address these concerns and provide detailed methodologic insights demonstrating that the observed edge mutations are distinct from previously reported artifacts. Of note, for a mutation in one molecule per cell to be included in ReDeeM, it must be supported by at least two molecules (eUMIs) in at least one cell and be detected in multiple cells (for simplicity, hereafter referred to as 1+-molecule mutations). We demonstrate that these 1+-molecule mutations, proposed to be discarded in the commentary, are well supported, mostly not located on fragment edges, show the expected mutational signature, and provide valuable information for downstream lineage inferences. Additional filtering to remove mutations on edges further enhances the true signal rate, while preserves the large majority of unique mutations, and keeps the cells well connected, reinforcing the robustness of the ReDeeM framework. We now provide additional options in the updated ReDeeM-R package for further filtering (see ReDeeM-R for recommended parameters to start with).
Principle of error correction in ReDeeM
ReDeeM is designed to rigorously reduce the likelihood for artifacts from diverse origins to achieve high sensitivity and accuracy (see full discussion in Supplementary Notes below). Briefly, we consider 5 potential sources of artifacts spanning the process from cell collection to read alignment that must be considered: (1) formaldehyde (FA)-induced errors; (2) Tn5 9-bp gap filling errors; (3) PCR errors; (4) sequencing errors; and (5) nuclear-embedded mitochondrial DNA (NUMT) misalignments (Fig. 1b). ReDeeM implements both overlapping paired-end (OPE) sequencing and a double-strand single-molecule tagging system (similar to duplex-seq8), which can correct not only downstream PCR errors and sequencing errors (efficiently removing artifacts caused by #3 and #4), but also reduce strand-specific artifacts in the initial molecule, including potential errors during the 9-bp gap-filling reaction or from fixation (#1 and #2) (Fig. 1c, d). ReDeeM implements enzyme-based fragmentation (Tn5 transposase) to avoid artifacts from sonication-induced DNA damage on the edge9, and controls for NUMT misalignments (#5) through multiple steps (Supplementary Notes). Rigorous filtering strategies and artifact removal steps are implemented and have been described in detail in our manuscript. Only high-confidence molecules with multiple supporting reads are considered in downstream analyses (Extended Data. Fig. 1a, Methods)
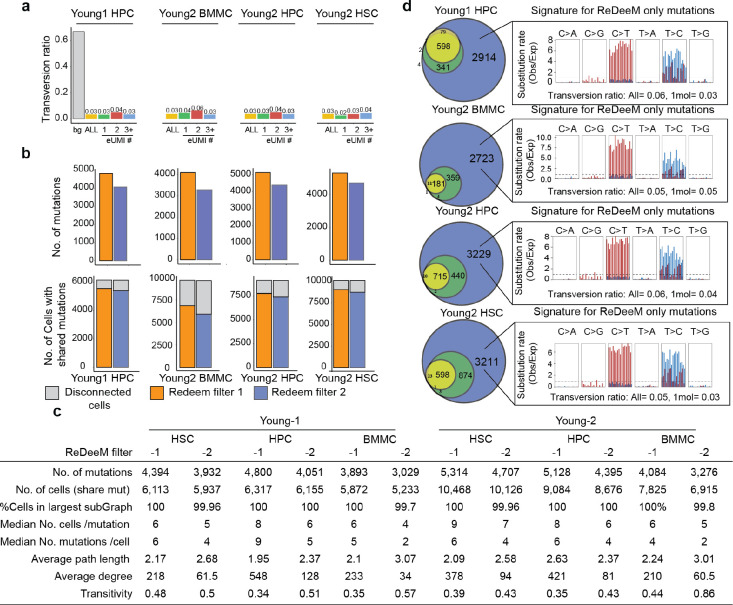
As previously reported, the bona fide mitochondrial mutations have a specific mutational signature enriched in transitions (C:G>T:A and T:A>C:G)10,11, which serves as a critical validation supporting mutation fidelity. Using ReDeeM, we confidently identified several thousand mtDNA somatic mutations (> 10-fold compared to previous methods) for each donor that pass our filtering thresholds (Fig. 1e). This large number of mutations in all donors were further examined for their mutational signatures, where we observed significant enrichment of transitions, as expected for true mitochondrial mutations. In contrast, artifacts such as FA-induced errors (SBS40), or NUMT exhibit distinct mutational signatures12,13, which are not observed in our analysis (Fig. 1f, g). These results suggest that ReDeeM overall maintains high specificity to detect true signals, while achieving substantially increased sensitivity (Fig. 1e, Extended Data Fig. 1b).
Reliability in detecting mtDNA 1+-molecule mutations
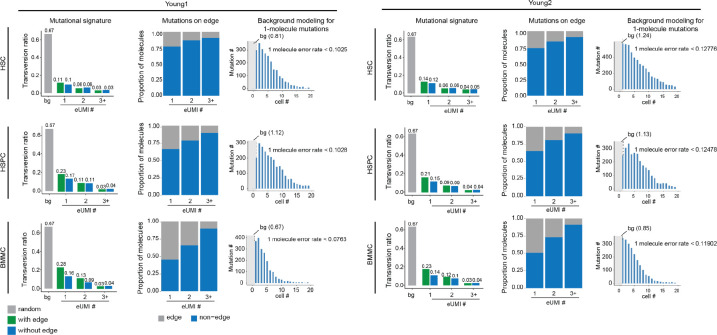
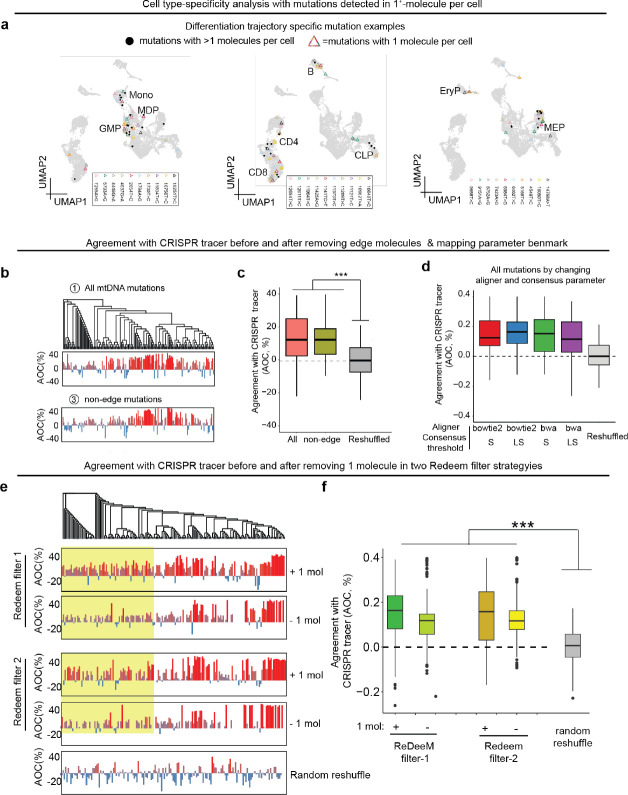
The commentary from Lareau et al. argues that 1+-molecule mutations provide minimal evidence and suggests that all mutations supported by one molecule per cell should be excluded. However, we demonstrate that 1+-molecule mutations are supported by multiple lines of evidence. First, with an average eUMI group size of 4.8, as reported in our manuscript, every variant called by a single molecule is supported by an average of 9.6 reads and has passed multiple filtering steps (Fig. 2a, Extended Data Fig. 1a). Second, the ReDeeM pipeline further strengthens the reliability of these mutations by requiring that any included mutations must be detected by >=2 molecules in at least one cell (Extended Data Fig. 1a). These criteria provide important support for 1+-molecule mutations to be considered in downstream analysis. Third, bona fide mitochondrial mutations typically exhibit a transversion proportion between 0.03 and 0.1, serving as an orthogonal validation that can help estimate the true signal rate of these mutations10,14. We analyzed the mutational signature for mutations identified by ReDeeM with 1, 2, or more molecules per cell and showed a strong enrichment in expected transitions across all samples compared to the random background (Fig. 2b, Extended Data Fig. 4). For example, in hematopoietic stem cells (HSCs), the low transversion proportion (0.11 and 0.14, compared to a random background of 0.67) suggests that approximately 87–88% of 1+-molecule mutations in HSC are true signals (Fig. 2b, see Methods for true signal ratio estimation). This estimated true signal rate for 1+-molecule mutations can be further increased to > 95% across all cell types after additional filtering with minimal edge trimming (please see below). Moreover, we compared the number of cells in which each 1+-molecule mutation was detected to the background distribution. The significant rightward shift in the number of cells suggests that > 90% of the signals are reliable, consistent with estimates based on the transversion proportion. (Fig. 2c, Methods). Finally, we show 1+-molecule mutations provide valuable biological insights, revealing cell type specificity and clonal origins in human hematopoiesis. Moreover, including 1+-molecule mutations improves the concordance with the orthogonal use of CRISPR-based lineage tracing (Fig. 4, Extended Data Fig. 7, more details are discussed below). Taken together, multiple lines of evidence provide support for and illuminate the unique value of mutations detected in one molecule per cell, arguing that the proposed exclusion leads to substantial elimination of bona fide and informative mutations.
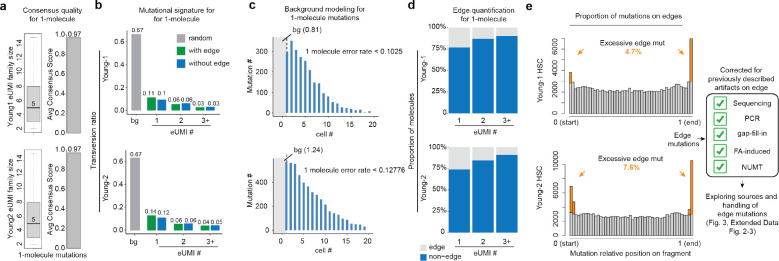
Fig. 2. Limited impact of edge mutations and validation for 1-molecule mutations.
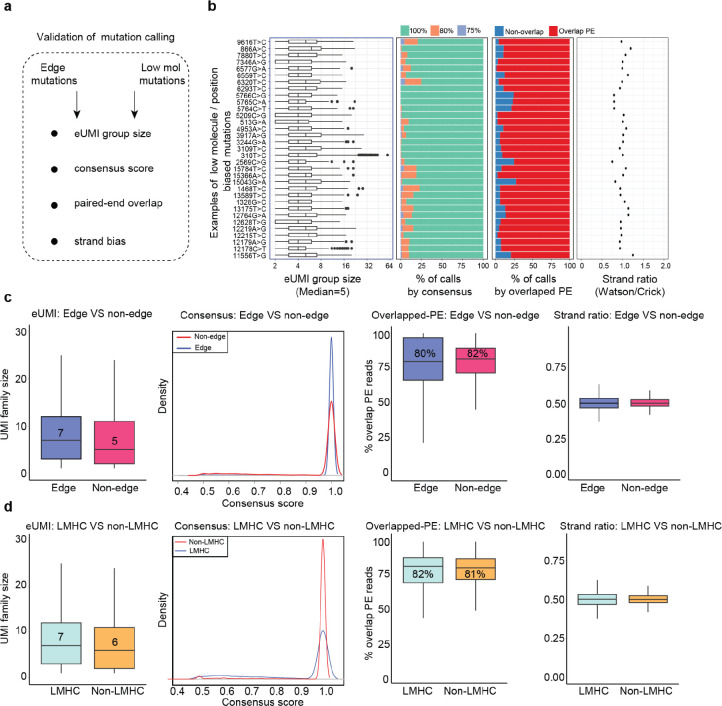
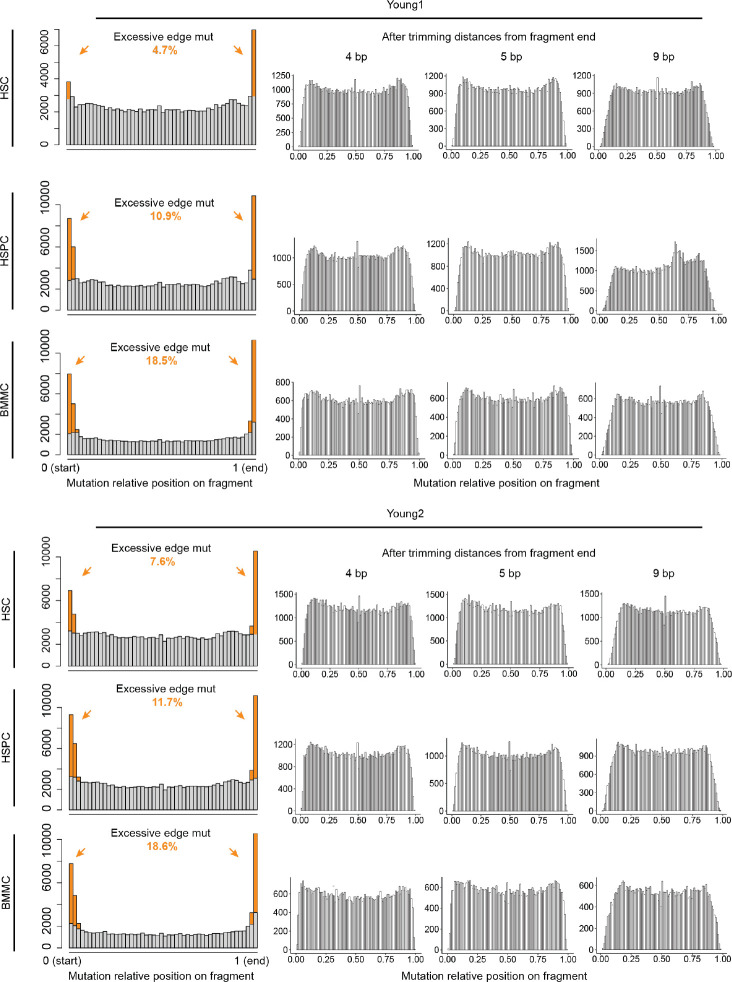
(a) Single molecule consensus metrics for 1+-molecule mutations. eUMI family size: the number of supporting paired-end reads. Average consensus score is the fraction of reads that support the mutation calling. (b) Transversion proportion (mutation frequency weighted) for mtDNA mutations called by ReDeeM with low and high heteroplasmy levels in Young1-HSC and Young2-HSC, including 1+-molecule mutations (1 eUMI). True mtDNA mutations are expected to be enriched in transitions (C>T/T>C), i.e, the lower the transversion proportion, the lower the noise level. The transversion proportion is defined as the fraction of transversion molecule numbers out of all (transversion + transition). eg, HSC shows a transversion proportion of 0.1, suggesting 88% of 1+-molecule mutations in HSCs are likely true signals (see Methods). (c) The observed number of cells that carry a given 1+-molecule mutation, compared to the estimated error background shown as the gray area (Methods). bg: background. (d) Proportion of mutations positioned on edges (defined as <5 bp) across 1, 2, or more molecules (eUMI) per cell. (e) Left: Aggregated distribution of relative position on fragments for Young1-HSC and Young2-HSC (also see Extended Data Fig. 5) Right: enumeration of the post-hoc validation for edge mutations.
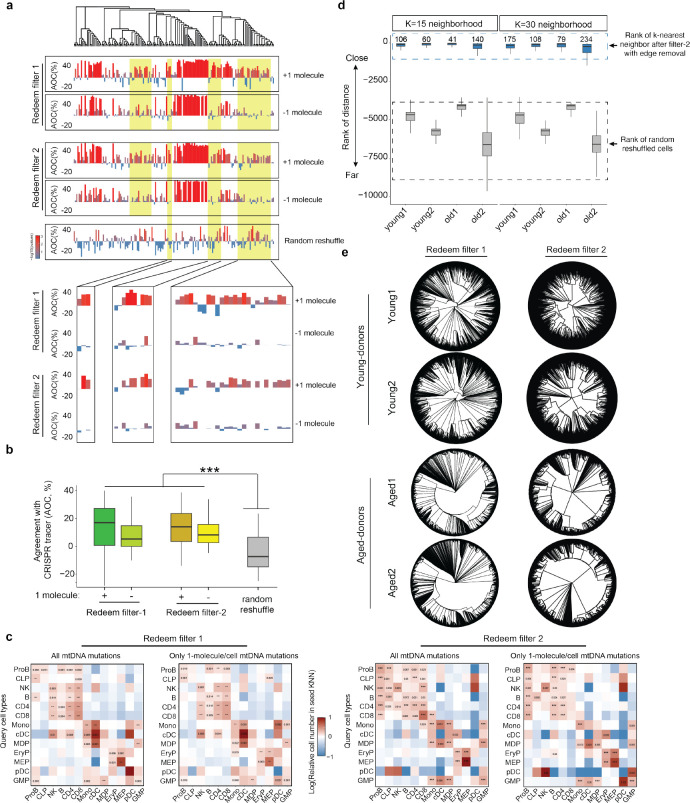
Fig. 4. Robustness of lineage tracing analysis with ReDeeM filter-2.
(a) Reanalysis of the dual lineage-tracer experiment with a Kras;Trp53(KP)-drive lung adenocarcinoma lineage-tracing mouse model. CRISPR-based and ReDeeM-based lineage information were analyzed for the same cells. The agreement of closeness (AOC) between ReDeeM and CRISPR lineage inference is computed across ReDeeM filter-1 (original ReDeeM filtering) and filter-2 (using 5bp trimming and binomial modeling). The phylogenetic trees based on all mtDNA mutations are illustrated and all single cells across 4 panels are in the same order. The AOC is computed for (1) mtDNA mutations using filter-1, with 1+-molecule mutations, (2) mtDNA mutations using filter-1, without 1+-molecule mutations, (3) mtDNA mutations using filter-2, with 1+-molecule mutations, (4) mtDNA mutations using filter-2, without 1+-molecule mutations, The regions with enhanced AOC when including 1+-molecule mutations are highlighted below. (b) AOC distribution across different mtDNA filtering strategies compared to random reshuffled background. P-values (Wilcoxon Rank Sum Test) are 6.7*10−16, 4.8*10−12, 1.2*10−21 and 3.9*10−18 for (1)(2)(3)(4). (c) Reanalysis of cell type origins using lineage informative mtDNA mutations with default setting (Method in original paper) using filter-1 and filter-2 with all mutations or only the 1+-molecule mutations (with max allele>=2). Color intensity indicates the proportion of each target cell type (horizontal axis) within the mtDNA mutation-based k nearest neighborhood (KNN) of the query cell type (vertical axis). Random reshuffling is performed 1000 times to determine enrichment p values. (d) Comparison of k-nearest neighbors between filter-1 and filter-2. k=15, K=30 neighborhoods are defined in distance matrices after different filtering strategies. The distance rank in the original distance matrices before edge removal are shown for these neighborhoods respectively, in comparison to random reshuffle. (e) Phylogenetic tree using mutations after filter-2 across young and old donors. Of note, the phylogenetic tree presented in the commentary from Lareau et al. is derived from removing all 1+-molecule mutations, even though these mutations have >= 2 molecules detected in at least one cell and have multiple technical and biological lines of support. This leads to half the cells not having any shared mutations. These remaining singletons are shown as the area of black on the top part of the circle in the commentary.
Mutation position biases
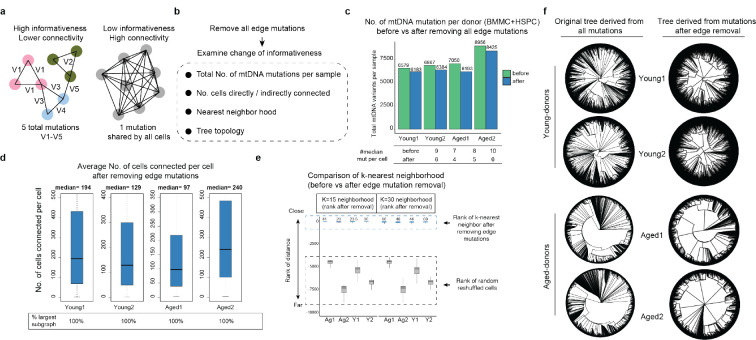
The commentary from Lareau et al. argues that 1+-molecule mutations are enriched at the edges of mtDNA molecules, suggesting artifacts reported in other sequencing approaches. To clarify, the position bias presented in the commentary such as in their Figure 1 and Extended Figure 1–2 represent a small subset of selected mutations (low-mean high-connectedness, or LMHC variants, representing ~5% of total unique mutations, Extended Data Fig. 1c). However, the majority of 1+-molecule mutations are not LMHC. Moreover, we show that overall, the excessive edge mutations impact a limited proportion of molecules when all heteroplasmic mutations are considered, ranging between 4.7% to 18.5% across samples, leaving a substantial majority of mutation calls unaffected (Fig. 2d, Extended Data Fig. 5). Notably, our primary focus, HSCs are the least impacted from edge accumulation, with only 4.3% to 7.6% excessive edge mutations (Fig. 2e, Extended Data Fig. 4). We also investigated the positional biases for 1+-molecule mutations. While a higher proportion of these mutations are located at the edges compared to mutations found in multiple molecules, most are positioned away from the edges (e.g., 75% to 76% of 1+-molecule mutations in HSCs are not on the edges, Fig. 2d, Extended Data Fig. 4).
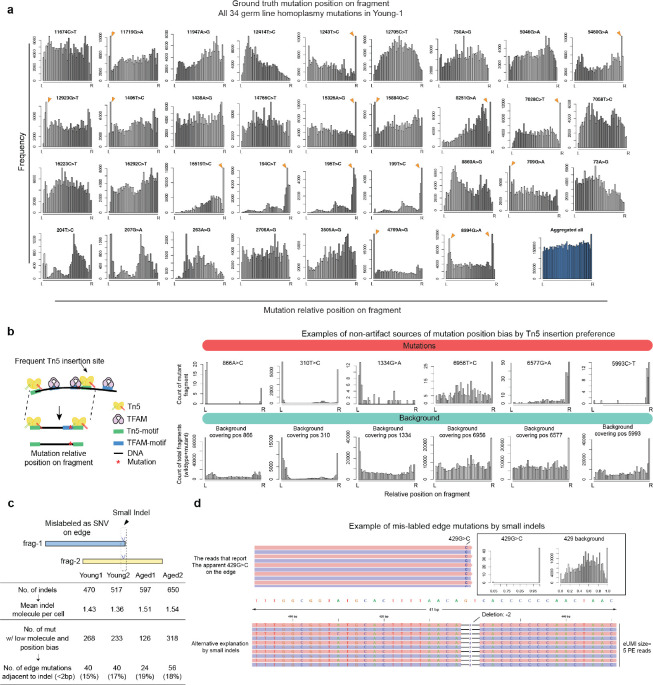
Moreover, it is important to not confuse the observed position bias here with previously reported artifacts9,15,16, given the robust error correction strategy employed (Supplementary notes, Methods, Fig. 2a). We showed comparable supporting reads per molecule between the mutation calls on the edge (within 9-bp to the end) and non-edge regions, suggesting that edge mutations are unlikely to arise from previously reported artifactual sources, such as sequencing and PCR errors (Extended Data Fig. 2). ReDeeM utilizes a double-stranded consensus correction approach and therefore can also reduce single-strand errors such as those arising in Tn5 9-bp gap-filling (Supplementary Notes). While we recognize the possibility of uncorrected artifacts, the observed position biases could also be contributed, at least in part, by error-unrelated sources including Tn5 insertion site preferences and potential small indels17,18 (Extended Data Fig. 3). Of note, many homoplasmic mutations, serving as positive controls for true mutations, also exhibit non-uniform distribution and accumulation on edges, suggesting that observed position bias does not always indicate the presence of artifacts (Extended Data Fig. 3). Taken together, these results suggest that the enriched edge mutations here are distinct from previously reported artifacts and impact an overall limited proportion of molecules.
ReDeeM with additional filtering
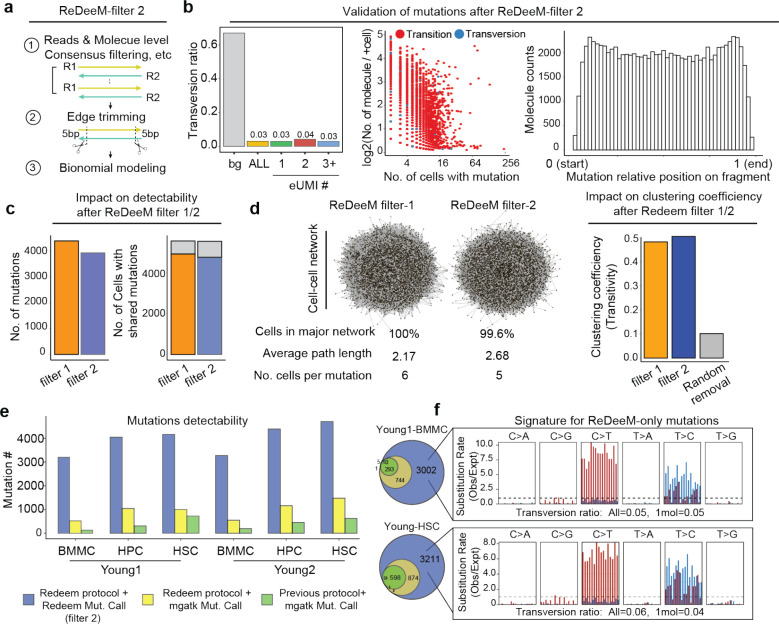
We have shown that edge-biased mutations overall impact a small proportion of molecules. However, recognizing the value of addressing edge mutations even after applying consensus correction, here we offer an additional filtering option to manage the excessive edge mutations. First, we examined various distances to trim from the end of fragment up to 9-bp and found that the excessive edge mutations are primarily restricted to the very end of molecules and trimming 4-bp can effectively remove most excessive edge mutations (Extended Data Fig. 5). We also refined the filtering threshold of max allele (at least 1 cell with two molecules) with binomial modeling to account for different mutation frequencies (chi squared test, Extended Data Fig. 1a, see Methods). We have tested the robustness of various parameters and here we provide a default using 5-bp trimming with binomial modeling (FDR <0.05, termed as filter-2, Fig. 3a) and we encourage further user fine-tuning. We demonstrate that filter-2 effectively removes excessive edge mutations, further reduces the transversion proportion (including 1+-molecule mutations to a level indistinguishable from ground truth), and eliminates excessive LMHC variants (Fig. 3b, Extended Data Fig. 6a). As expected, we demonstrate a limited decrease of the total number of unique mutations identified by filter-2 in comparison to using the original ReDeeM parameters (without trimming, max allele >=2, referred to as filter-1 hereafter), for example in Young1-HSC dataset, the number of variants is reduced from 4,394 (filter-1) to 3,932 (filter-2) (Fig. 3c, Extended Data Fig. 6b). The number of cells with shared mutations are also largely unchanged (median 3.9% decrease, Fig. 3c). ReDeeM filter-2 detects over 10-fold more variants compared to the previous method (Fig. 3e), with significant overlap, further validating our approach. The additional mutations detected exclusively by ReDeeM filter-2 show strong mutational signatures for bona fide mtDNA mutations, including 1+-molecule mutations (estimated accuracy of ≥95% based on transversion proportion, Fig. 3f, Extended Data Fig. 6d). Finally, we demonstrate that after ReDeeM filter-2, the cells remain well connected, with 99.96–100% of the cells being part of an interconnected network through mtDNA mutations (Fig. 3d). While the average degree decreases, several key connectivity metrics, including average path length and transitivity, remain stable, indicating that important substructure within the connectivity graph is maintained (Fig. 3d, Extended Data Fig. 6c, Supplementary Notes). Taken together, we show additional filtering with minimal edge trimming is sufficient to eliminate mutation edge biases, further reduce error rate, and maintain the advanced mutation detection made possible by ReDeeM.
Fig. 3. Robustness of ReDeeM mutation calling using filter-2.
(a) Design of ReDeeM filter-2, including 5bp edge trimming and binomial modeling. (b) Left: Transversion proportion (mutation frequency weighted) for mtDNA mutations called by ReDeeM filter-2 with 1, 2, and more molecules per cell in Young1-HSC. The transversion proportion is defined as the fraction of transversion molecule numbers out of all (transversion + transition). eg, 1+-molecule mutations (or 1 eUMI) shows a transversion proportion of 0.03, indistinguishable from bona fide mitochondrial mutations. Middle: Scatter plot showing the number of cells with the mutation and the average count per cell. Each dot is a mutation. These mutations are colored by transition or transversion. Right: The mutation position on fragments shows flat distribution without edge bias after the filtering. (c) Impact of ReDeeM filter-2 compared to original filter (filter-1). Left: total number of mutations detected in Young1-HSC dataset; Right: the number of cells with connections (or at least share one mutation with other cells). (d) cell-cell network metrics using filter-1 and filter-2. The metric of cells in major network is the percentage of the largest subgraph (see more in Extended Data Fig. 6). (e) Comparison of total number of mutations identified by different combination of experimental protocol and mutation calling algorithm. (f) Overlap analysis among different mutation calling strategies. The ReDeeM-only mutations (those identified by filter-2 algorithm on ReDeeM capture protocol) are further evaluated by mutational signature analysis. The transversion proportion of all or 1+-molecule mutations are both computed.
Robustness of lineage analysis using ReDeeM with additional filtering
The robustness of 1+-molecule mutations and the effects of edge removal have been validated through various orthogonal validations, including significantly enriched mutational signatures, as discussed above. Here, we further assess the impact on lineage tracing performance by implementing different filtering strategies and by including or excluding 1+-molecule (Fig. 4, Extended Data Fig. 7–8).
We reanalyzed the dual-lineage-tracer mouse model data where we detect both engineered CRISPR-based evolving barcodes on the nuclear genome and the mtDNA mutations by ReDeeM in the same single cells. With this system, we compare the CRISPR lineage tracing inferences and the mtDNA mutations, where different filtering strategies are applied, including full ReDeeM mutations with the original filtering strategy (filter-1) or with our additional filtering options that involves edge removal and statistical modeling (filter-2). We also compared mutations with inclusion or exclusion of 1+-molecule mutations in both filtering strategies. As expected, using all mtDNA mutations in filter-1 or filter-2, or only excluding edges all demonstrate significant concordance with CRISPR-based lineage inference (p-values 6.2*10−16, 4.9*10−12, 1.2*10−21, and 3.9*10−18 , Wilcoxon Rank Sum Test, Fig. 4a, Extended Data Fig. 7b–d, Methods). Indeed, including 1+-molecule mutations consistently improves concordance with CRISPR-based lineage inference, underscoring the unique value of these mutations in enhancing lineage tracing (Fig. 4a–b, Extended Data Fig. 7e–f). Notably, this benchmarking offers a conservative estimate of the accuracy of lineage tracing, given the shorter mutation accrual period in the mouse model (6 months) and lower diversity of mtDNA mutations present.
We also reanalyzed the human hematopoiesis dataset we generated and evaluated the impact of mutation subsets on downstream analysis. We found that cell-type-specific mtDNA mutations consistently arise from both single-molecule and multi-molecule mutations, suggesting that single-molecule mutations offer valuable biological insights (Extended Data Fig. 7a). Next, we repeated the cell-type origin analysis using mtDNA mutation-based nearest neighbor analysis, an important demonstration that ReDeeM allows for fine-scale lineage tracing and resolves clonal and subclonal relationships. We evaluated the impact on this analysis by applying filter-1 or filter-2 with only 1+-molecule mutations or the full mutation set. Applying different filtering strategies show reproducible cell-type clonal origin inferences, demonstrating the robustness of ReDeeM in extracting essential biological information (Fig. 4c).
Lastly, we reanalyzed the combined BMMC and HSPC dataset using both filter-1 and filter-2. As expected, trimming edges only slightly decreases the total number of mutations (between 5.9% - 12% across donors, Extended Data Fig. 8c). The nearest neighborhood resolved by ReDeeM remains largely unchanged before and after removal of edge mutations with filter-2 or only excluding edges (Fig. 4d, Extended Data Fig. 8e). This results in robust downstream analysis, including assessment of cell-type origins. Finally, we reanalyzed the phylogenetic trees after implementing filter-2 or only excluding edges. The topology of the tree remains informative. We observe consistent polyclonal structures in young donors and significantly altered clonal structures in aged donors after applying the filtering strategies, which argues for the robustness of the biological findings described in our original study (Fig. 4e, Extended data Fig. 8f). In summary, we present multiple lines of evidence, both technical and biological, that demonstrate the robustness of lineage analysis by ReDeeM with additional filtering and support the value of 1+-molecule mutations.
Conclusions
The commentary from Lareau et al. noted the important advances in ReDeeM implementing targeted capture, deep sequencing, and single-molecule consensus calling for more sensitive mtDNA genotyping. The commentary also suggested that mutations in one molecule per cell (1+-molecule mutations) are likely artifacts, proposing to remove all these mutations. Here we provide various lines of evidence that 1+-molecule mutations are supported by both technical and biological validations, including valid consensus support and strong mutational signatures. Furthermore, these mutations provide critical biological insights for fine-scale lineage analysis, and including them improves the concordance with the orthogonal use of CRISPR-based lineage tracing. We detailed our methodological considerations in mutation calling and error correction, demonstrating that the observed edge mutations are distinct from previously reported artifacts.
The goal of ReDeeM is to provide an integrated experimental and computational framework that enhances mtDNA mutation detection, facilitates finer-scale lineage inference, and enables the integration of lineage structure with multimodal cell state profiling. The bioinformatic methods proposed in ReDeeM aim to provide a flexible platform to implement consensus calling for error correction. We highly encourage fine-tuning of different parameters to fit different applications, including using different consensus thresholds, cell- and variant-level filtering, etc. Here we introduce additional filtering options, including edge trimming and statistical modeling. We show that additional filtering enhances the true signal rate and reproduces the major analyses and conclusions, reinforcing the robustness of the ReDeeM framework.
We appreciate that ReDeeM and mtDNA lineage tracing have limitations. The precise dynamics of mtDNA mutations still requires further investigation as has been demonstrated through a recent study modeling high-frequency mutations and warrants caution against using some of the higher heteroplasmy variants as clonal markers3. There is room to improve the resolution of mtDNA tracing. ReDeeM is an exploration of what is possible with increased mtDNA mutation calling sensitivity. We do not intend to, nor have we made the claim that mtDNA mutations can reconstruct perfect trees for comprehensive resolution of all-encompassing processes. Our goal is to reveal more detailed cell-cell lineage relationships and extract valuable biology, which remains robust and unaffected by the concerns raised by Lareau et al., as we demonstrate by modifying mutation filtering strategies that we present here. We are excited to see further advances as more users apply ReDeeM to a range of biological problems and continue to improve analytical approaches.
Methods
Threshold of ReDeeM-V mutation calling
The commentary from Lareau et al. raises the concern that ReDeeM requires only one mtDNA molecule to call a mutation in a cell (i.e., threshold >0) suggesting it is an unusual choice compared to traditional thresholds (e.g., >10% heteroplasmy) in the previous methods. To clarify, the variants detected at “one molecule per cell” that are mentioned by Lareau et al. have strong consensus support by multiple reads and require both of the following criteria: (1) there are at least 2 cells to have detected 1 or more molecules (eUMIs) and (2) at least 1 cell to have 2+ molecules (eUMIs). The commentary suggests that one molecule per cell is minimal evidence. However, with an average eUMI group size 4.8 as reported in our manuscript, this one molecule has passed multiple filtering steps and is strongly supported by an average of 9.6 reads that are consistently calling mutations from both strands. In addition, these 1+-molecule mutations are supported by multiple orthogonal lines of evidence as discussed.
The commentary suggests that a minimum of 3 eUMIs is a lenient threshold. However, this threshold in a typical dataset with 10,000 single cells represents a global variant allele frequency (VAF) of 10−5 (3/ (10,000*30) with approximately 30 mtDNA copies detected per cell. Indeed, the ReDeeM mtDNA mutation data show the global VAF range between 10−3–10−5 (with a median near 10−4) if all single cells are pooled together as a pseudo bulk population. Aiming to detect mutations with VAFs in this range using a double-stranded eUMI consensus system is not a lenient threshold, as this strategy has been utilized to achieve mutation frequencies down to 10−7 or lower in prior studies19.
ReDeeM’s mutation-calling process benefits from its mtDNA capture and deep profiling protocol, enabling the application of single-molecule consensus correction. This allows for the detection of low heteroplasmy mutations, which cannot be achieved without error correction in previous method. The rationale of comparing the mutation calling thresholds between consensus-based ReDeeM and previous methods (no correction applied) is not clear. To provide a context, the extensive development of consensus error-correction technologies has allowed for the detection of variants at extremely low levels (10−4 to 10−9), far below the conventional sequencing detection threshold, which is typically around 1%.
ReDeeM filter-2
ReDeeM filter-2 applies the same consensus filtering strategies and follows the same downstream filtering procedures except the following two changes. (1) After the consensus error filtering, we further label the distance to the nearest fragment end for every mutation, and remove mutations within the distance d. We have tested the d = 4, 5, 9. We chose d = 5 for main analysis which is sufficient to flat the relative position distribution across all samples (d=4 is sufficient in most samples, Extended Data Fig. 5). (2) In the original downstream filtering, a mutation is only included if it is supported by at least two molecules (eUMIs) in at least one cell and can be detected in multiple cells (The max molecule number per cell all cross cells, or max allele ≥ 2 and detected in ≥ 2 cells, as shown in Extended Data Fig. 1). We further refined this hard cutoff with binomial modeling, which follows the same principle. We assume that the residual noise after consensus filtering follows a binomial distribution. By modeling the observed mutation distribution across cells and testing against the expected binomial distribution (chi-squared test). We filter out mutations if there is insufficient evidence to reject the null hypothesis of a binomial distribution (adjusted p > 0.05). This modeling-based method is largely equivalent to max allele>=2 threshold, but it also effectively removes excessive low molecule high connectedness (LMHC) mutations. In this work, we combined the 5 bp trimming and the binomial modeling with adjusted p-value <0.05 as ReDeeM filter-2. The trimming distances and binomial modeling p-values can be further fine-tuned in the ReDeeM-R package for optimization in different systems.
Comparison with mgatk
We used Cell Ranger pipeline to generate bam files from both the enriched mtDNA library and snATAC library. These bam files were then processed using mgatk v0.6.1 (mgatk tenx mode with default parameters) to call mtDNA mutations. We compared mtDNA mutations from three different datasets: (1) ReDeeM-V variant calling applied to the enriched mtDNA library; (2) mgatk variant calling applied to the enriched mtDNA library; 3) mgatk variant calling applied to the standard (non-enriched) library. Mgatk variant calling parameters keep consistent with previously described (strand_correlation > 0.65 & n_cells_conf_detected >= 3 & (log10(vmr) > −2) & mean < 0.8) Barplots were generated to visualize the number of mtDNA mutations across these datasets. Additionally, we used Venn diagrams (BioVenn v1.1.3) to assess the overlap of mutations across the three datasets. Mutations identified exclusively by Redeem but not by mgatk were subsequently subjected to mutation signature analysis.
Mutational signature analysis
Bona fide mitochondrial mutations typically exhibit a transversion proportion between 0.03 and 0.1, serving as an orthogonal validation that can help estimate the true signal rate of these mutations by comparing to expected background transversion proportion (0.67). We computed both the mutation-frequency weighted and unweighted mutational signature (Indicated in Figure legends). For mutation-frequency weighted mutational signature, each molecule is counted once while for unweighted mutational signature, each unique mutation is counted once regardless of the number of molecules per mutation. The error rate estimated by mutational signature is computed as follows:
Where is the transversion proportion
is the transversion proportion of observed group of mutations
is the the transversion proportion of ground truth mutational signature
is the transversion proportion of random noise. It is set as a constant 0.67 given the expected transversion proportion in background,
For example, if we observe the transversion proportion 0.11 in the HSC 1 eUMI group, and 0.03 in ground truth from the 3+ molecule group. The estimated error rate is (0.11–0.03)/(0.67–0.03)= 0.125. The true signal rate reported in the main text is computed as 1 - error rate.
Maximum background error rate estimation
We use all mutation calls that fail to meet our original cutoff of max allele ≥ 2 after consensus filtering to approximate the “background” error. For each possible mutation that is not located on the edge (i.e., after trimming 5 bp), we counted the number of cells in which the mutation appears and then computed the average number of cells per variant. This value is multiplied by the transversion proportion to estimate the expected number of cells per variant that would arise from background errors. We also counted the observed number of cells for the 1+-molecule mutations that passed the ReDeeM threshold with max allele>=2 and not located on edges (trim 5bp). The estimated error rate for 1+-molecule mutations based on this background modeling is computed as below:
Where is the number total called mutations
is the expected number of cells per variant as background
is the number of 1+-molecule mutation molecules.
Network analysis
Following QC of cells and mutations, we binarized the cell~variants count matrix , and then compute the adjacency matrix by multiplying by its transpose , representing the connections between cells based on shared mutations. An undirected graph was subsequently constructed using the function graph_from_adjacency_matrix from the igraph R package. Key network metrics, including “average degree”, “average path length”, and “transitivity”, were calculated using the degree, mean_distance, and transitivity functions in igraph. The largest subgraph was defined as the proportion of cells connected within the largest component of the graph.
CRISPR lineage-tracing validation using subsets of mtDNA mutations
The detailed methods have been described in the original manuscript. Briefly, to test the accuracy of phylogenetic reconstructions generated by ReDeeM, we used a Kras;Trp53-drive lung adenocarcinoma lineage-tracer mouse model for detection of both engineered CRISPR-based evolving barcodes in the nuclear genome and naturally occurring mitochondrial somatic mutations by ReDeeM in the same single cells. The measure of cell–cell relatedness and clonal groupings as determined by ReDeeM was significantly supported by CRISPR-based methods at both the clonal cluster level and the single-cell level as measured by the agreement of closeness (AOC). The AOC is defined as follows. Given a single cell X, we firstly identified k (default is 15) nearest neighbors M1,M2.. Mk in mtDNA mutation derived graph Gmt (Jaccard distance matrix). Then we computed the average distance from M1,M2..Mk to the cell X on CRISPR derived graph Gcr (hamming distance matrix). The ranks of these distances on Gcr (The closest is indicated as rank #1) were compared with that from randomly picked k cells. The random process was repeated 1000 times and the rank % closer toward cell X in observed data than expected was defined as “agreement of closeness” or AOC. (i.e., a positive value of AOC indicates a closer distance to cell X than expected, while a negative score indicates a farther distance). For example, if the real rank of nearest neighbors from Gmt in Gcr is 8th while the random reshuffled rank is 16th (with a total of 32 cells), the AOC is (16–8)/32= 25%. The empirical p value was also generated by i permutations (default is 1000). Here, we applied the same method to compute AOC by inputting different subsets of mtDNA mutations to compute the distance matrix. The subset of mtDNA mutations includes: (1) using mutations from RedeeM filter-1. (2) excluding 1+-molecule mutations from RedeeM filter-1. (3) using mutations from RedeeM filter-2. (4) excluding 1+-molecule mutations from RedeeM filter-2.
Mouse ReDeeM data
We have deposited all raw data of the CRISPR lineage tracing experiments in the GEO (GSE219015) and provided the preprocessed data using the latest pipeline that is the same as was used to process human data. We have described the version of the pipeline used in the GEO records with details.
We clarify some misunderstanding of the experimental design and data processing presented from the Lareau et al. commentary. Specifically: (1) As stated in our manuscript, dedicated mtDNA libraries were generated following the ReDeeM protocol for mouse data. The dedicated mtDNA library and ATAC library are both generated and are then pooled in a ratio (2:1) to be sequenced in one Illumina sequencing lane. The only difference in human experiments is that the mtDNA library and ATAC library are sequenced in two separate Illumina lanes. (2) The reason for the lower saturation rate in mouse data is due to the mouse tumor cells having more unique fragments per cell, which made us choose to not fully saturate the sequencing for cost-effectiveness, not that as stated in the commentary that “no dedicated mtDNA library was isolated.” Therefore, additional sequencing can be beneficial, and this benchmarking provides a conservative estimate of the accuracy of the mtDNA-based lineage tracing we perform.
We also benchmarked the robustness of this validation analysis by comparing the agreement of closeness using mtDNA mutations by the ReDeeM pipeline with different parameters, with different read-mapping strategies (bowtie2 with mt-genome masking vs bwa without mt-genome masking) and different consensus thresholds. All 4 pipelines yield consistent positive AOC, significantly higher than random reshuffle (Extended Data Fig. 7d).
Lineage origin analysis using subsets of mtDNA mutations
To validate if mtDNA mutations can provide valuable fine-scale lineage information for different subsets of mutations, we performed the lineage origin analysis as described in the original manuscript. Briefly, we choose lineage-informative mtDNA mutations to generate matrix Cbin and computed weighted Jaccard distance. We then generated KNN graph G that describes cell-to-cell lineage relationships based on shared mutations. We then integrated cell-type annotations from the multiomics analysis with graph G. For any given cell (query cell), the proportion of each cell type (target cell types) within KNN on graph G was computed. Target cell-type proportions for each query cell type were then aggregated and scaled. Randomly reshuffled cell type information was used to compute the background proportion. Random reshuffling was performed 1000 times to determine enrichment p values. We performed the same analysis by inputting different subsets of mtDNA mutations to compute the KNN graph G: (1) using mutations from RedeeM filter-1. (2) Only using 1+-molecule mutation from RedeeM filter-1. (3) using mutations from RedeeM filter-2. (4) Only using 1+-molecule mutation from RedeeM filter-2.
Indel analysis
We adapted our ReDeeM framework to call indels with consensus error correction. Briefly, after eUMI single-molecule grouping, we extract the indel information from the CIGAR string and perform a more stringent consensus filtering using family size >= 4 and consensus score > 0.9. The average number of molecules per indels are summarized and the number of adjacent edge mutations is counted.
Neighborhood and Tree analysis
Next, we examined the impact on the k-nearest neighbors. We computed the weighted jaccard distance for the cell-variant matrix using filter2 (or only remove edges) and defined the k-nearest neighbors using K=15 and 30. We then examined the closeness (rank of distance to the given cell) for the defined nearest neighbor in the filter-1. Finally, we repeated the phylogenetic tree analysis by using filter-2 and removing hypermutable mutations (defined as mutations with >=0.5% across all donors, and those located in D-loop region), followed by tree reconstruction using the neighbor joining algorithm from weighted Jaccard distance as described previously. We have also tested only removing edge mutations and LMHC, followed by tree reconstruction. Reproducible results showing topology difference between young and old are observed.
Extended Data
Extended Data Fig. 1. Error correction strategy and mutational signature.
(a) Workflow of the ReDeeM variant calling pipeline (b) Related to Fig. 1e. Mutational signatures (frequency unweighted) for all ReDeeM identified confident mtDNA mutations or the collection for 1+-molecule mutations in Young2-BMMC and Old2-BMCM. (c) Number of total ReDeeM mtDNA somatic mutations in BMMC for 4 donors and the number of mutations defined as low molecule high connectedness (LMHC) and position bias (using Kolmogorov–Smirnov test (KS)>0.35 and defined as LMHC as Lareau et al commentary proposed) takes account for small proportion (6.9%, 5.7%, 2.5%, 5%);
Extended Data Fig. 2. Consensus validations for edge and low molecule mutations.
(a) key benchmarks for validating edge and low molecule mutations. (b) Individual mutation examples with edge position bias and/or low molecule of proposed LMHC mutations. For each mutation, the following key benchmarks are conducted: the eUMI group size (number of supporting paired-end reads for each eUMI group), the consensus score distribution of the calls (fraction of reads that supports the mutation calling), the proportion of mutation calls with OPE (overlapping paired-end (OPE) sequencing), and the level of strand bias. (c) Summary statistics that compare each of the key benchmarks between edge biased mutations (less than 9bp from the ends) and non-edge mutations (greater than 9 bp from the ends) for consensus mutation calling quality control, including eUMI group size, the consensus score distribution of the calls, the proportion of mutation calls with OPE, and the level of strand bias. (d) Same summary statistics with c, a comparison between low-molecule mutations (LMHC) versus other mutations.
Extended Data Fig. 3. Alternative sources potentially contributing to position biases.
(a) The relative position distribution on fragments for individual homoplasmic mutations. All 34 homoplasmic mutations of donor Young-1 are shown. These homoplasmic mutations serve as true mutation controls. Notably, the majority of these mutations are not uniformly distributed. Some exhibit strong “position bias” and excessive edge enrichment. L and R on the x axis indicate left and right side of the fragment. (b) Left: Schematics of the apparent position bias associated with frequent Tn5 insertion sites. Right: Examples of position biased mutations where the background fragment (including wildtype and mutant) are consistently skewed. Top row indicates the position-on-molecule distribution for mutants. The bottom row indicates the position-on-molecule distribution for wildtype+mutant. (c) Top: Schematics to illustrate that some indels can be mislabeled as point mutations on edges and show apparent position biased edge mutations. Bottom: summary of the number of indels, the mean molecule number of indels per cell, and the number of edge mutations that are adjacent to an indel. (d) An Example of position biased mutation that can be potentially explained by small indels.
Extended Data Fig. 4. Validation for 1+-molecule mutations.
Related to Fig. 2. HSC, HSPC, and BMMC for Young-1 and Young-2 are shown. For each sample, the left sample shows transversion proportion for mtDNA mutations called by ReDeeM with low and high heteroplasmy levels including 1, 2, and more molecules (eUMI) per cell. True mtDNA mutations are expected to be enriched in transitions (C>T/T>C), i.e, the lower the transversion proportion, the lower the noise level. The transversion proportion is defined as the fraction of transversion molecule numbers out of all (transversion + transition). The middle panel shows the proportion of mutations positioned on edges (defined as <5 bp) across 1, 2, and more molecules (eUMI) per cell. The right panel shows the observed number of cells that carry a given 1+-molecule mutation, compared to the estimated error background shown as the gray area (Methods).
Extended Data Fig. 5. Evaluation of edge trimming distances to remove edge biases.
Related to Fig. 2. Aggregated distribution of relative position on fragments for HSC, HSPC, and BMMC for Young-1 and Young-2. Both before and after trimming (4 bp, 5 bp, and 9 bp are shown).
Extended Data Fig. 6. Robustness of ReDeeM mutation calling using filter-2 across samples. Related to Fig. 3.
(a) transversion proportion for mtDNA mutations called by ReDeeM filter-2 with 1, 2, and more molecules per cell across samples in Young-1 and Young-2 donors. i.e, the lower the transversion proportion, the lower the noise level. The transversion proportion is defined as the fraction of transversion molecule numbers out of all (transversion + transition). (b) Impact of ReDeeM filter-2 compared to original filter (filter-1). Top: total number of mutations detected across samples; Bottom: the number of cells with connections (at least share one mutation with other cells). Disconnected cells are labeled in grey. (c) comparison between filter-1 and filter-2, including mutation and cell number, and various connectivity metrics. (d) Overlap analysis among different mutation calling strategies. The ReDeeM-only mutations (those identified by filter-2 algorithm on ReDeeM capture protocol) but not by mgatk are further evaluated by mutational signature analysis. The transversion proportion of all and1+-molecule mutations are shown on the bottom.
Extended Data Fig. 7. Extended reproducible lineage tracing analysis with ReDeeM filtering.
(a) Example mtDNA mutations that show lineage specificity. The 1+-molecule mutations are represented in triangles with colors. The mutations with more molecules per cell are represented in black dots. (b-d) Related to Fig. 4a. (b) Reanalysis of the agreement of closeness (AOC) between ReDeeM and CRISPR lineage inference using ReDeeM pipeline by cell-ranger (bwa) as aligner (without mitochondrial genome masking) is computed with or without edge trimming. (c) AOC distribution compared to random reshuffled for panel a with or without edge trimming. (d) AOC distribution using all mtDNA mutations called by 4 varieties of ReDeeM pipelines, where two aligners: bowtie2, and cell-ranger (bwa) and two consensus calling thresholds are tested. (ef) Extended example related to Fig. 4a–b, The agreement of closeness (AOC) between ReDeeM and CRISPR lineage inference is computed across ReDeeM filter-1 and filter-2. 4 panels are in the same order. The AOC is computed for (1) mtDNA mutations using filter-1 with 1+-molecule mutations, (2) mutations with 1 molecule per cell with filter-1 (3) all mtDNA mutations with filter-2 (4) mutations with 1 molecule per cell with filter-2. The regions with enhanced AOC when including 1-molecule mutations are highlighted. AOC distribution across different mtDNA filtering strategies compared to random reshuffled.
Extended Data Fig. 8. Extended reproducible lineage tracing analysis using filter-1 with 4 bp edge trimming.
(a) Comparison between informativeness and connectivity. See more detailed discussion in Supplementary Notes. (b) Enumerate the downstream benchmarks if one were to exclude 4 bp edge mutations. The edge mutations are defined as ones that are <= 4 bp to the end of a fragment throughout this analysis (c) Number of total mutations before and after excluding all edge mutations with filter-1. (d) Number of cells connected for individual cell across donors (sharing >=1 mutation), the % largest subgraph is shown below (includes the most cells that can be connected directly or indirectly.) (e) Comparison of k-nearest neighbors before and after excluding edge mutations. k=15, K=30 neighborhoods are defined in distance matrices after removing edge mutations. The distance rank in the original distance matrices before edge removal are shown for these neighborhoods respectively, in comparison to random reshuffle. (f) Phylogenetic tree using mutations without edge and LMHC across young and old donors.
Supplementary Material
Acknowledgments
We thank W. N. Colgan, S. Zhang, M. Poeschla, T. Gao, J. A. Gudera, P. van Galen, S. Shelton for helpful discussions and suggestions. The laboratory of V.G.S. is supported by National Institutes of Health (NIH) grants R01CA265726, R01DK103794, R01HL146500, R01CA292941 and R33CA278393, the Mathers Foundation, the RUNX1 Research Program, and Alex’s Lemonade Stand Foundation. The laboratory of J.S.W. is supported by the Mathers Foundation, NCI Cancer Target Discovery And Development (CTD2) and the NIH Centers of Excellence in Genomic Science (RM1HG009490), the Howard Hughes Medical Institute, and the Ludwig Center at MIT.
Footnotes
Conflict of interest
J.S.W. serves as an advisor to and/or has equity in 5 AM Ventures, Amgen, Chroma Medicine, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics and Tessera Therapeutics, all unrelated to this work. V.G.S. is an advisor to Ensoma, unrelated to this work.
Code availability
Additional filtering strategies are implemented in redeemR, available at: https://github.com/chenweng1991/redeemR
Reproducibility code for this work is available at: https://github.com/chenweng1991/redeem_robustness_reproducibility
References
- 1.Sankaran V. G., Weissman J. S. & Zon L. I. Cellular barcoding to decipher clonal dynamics in disease. Science 378, eabm5874 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotrys A. V. et al. Single-cell analysis reveals context-dependent, cell-level selection of mtDNA. Nature 629, 458–466 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X. et al. Clonal expansion dictates the efficacy of mitochondrial lineage tracing in single cells. bioRxiv 2024.05.15.594338 (2024) doi: 10.1101/2024.05.15.594338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Árnadóttir E. R. et al. The rate and nature of mitochondrial DNA mutations in human pedigrees. Cell (2024) doi: 10.1016/j.cell.2024.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez-Contreras M. et al. The multi-tissue landscape of somatic mtDNA mutations indicates tissue-specific accumulation and removal in aging. Elife 12, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng C. et al. Deciphering cell states and genealogies of human haematopoiesis. Nature 627, 389–398 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lareau C. A. et al. Artifacts in single-cell mitochondrial DNA mutation analyses misinform phylogenetic inference. bioRxiv 2024.07.28.605517 (2024) doi: 10.1101/2024.07.28.605517. [DOI] [Google Scholar]
- 8.Schmitt M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 109, 14508–14513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abascal F. et al. Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Ju Y. S. et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M. H. et al. DNA mismatch and damage patterns revealed by single-molecule sequencing. Nature 630, 752–761 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thapa M. J., Fabros R. M., Alasmar S. & Chan K. Analyses of mutational patterns induced by formaldehyde and acetaldehyde reveal similarity to a common mutational signature. G3 12, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W. et al. Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Nature 611, 105–114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lareau C. A. et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 39, 451–461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickrell J. K., Gilad Y. & Pritchard J. K. Comment on ‘Widespread RNA and DNA sequence differences in the human transcriptome’. Science 335, 1302; author reply 1302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W., Piskol R., Tan M. H. & Li J. B. Comment on ‘Widespread RNA and DNA sequence differences in the human transcriptome’. Science 335, 1302; author reply 1302 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y. & Greenleaf W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg D. E., Schmandt M. A. & Lowe J. B. Specificity of transposon Tn5 insertion. Genetics 105, 813–828 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salk J. J., Schmitt M. W. & Loeb L. A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 19, 269–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Townsend J. P. Profiling phylogenetic informativeness. Syst. Biol. 56, 222–231 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Isaac R. S. et al. Single-nucleoid architecture reveals heterogeneous packaging of mitochondrial DNA. Nat. Struct. Mol. Biol. 31, 568–577 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.