Abstract
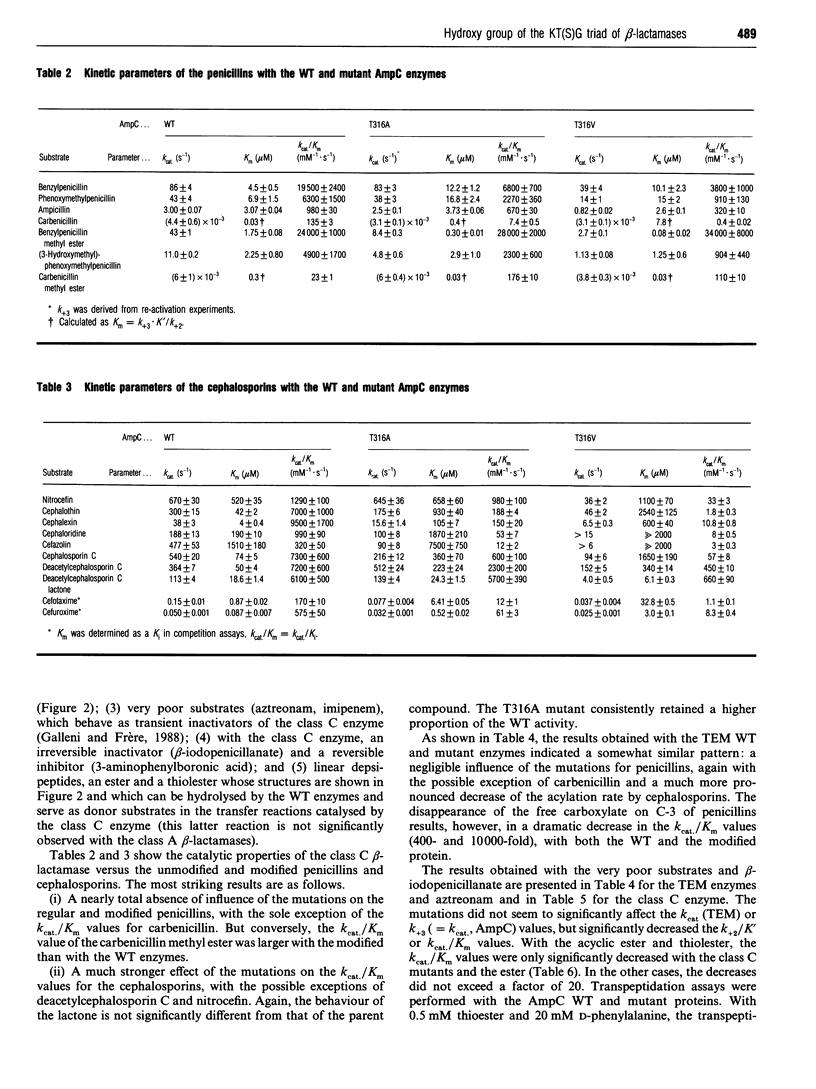
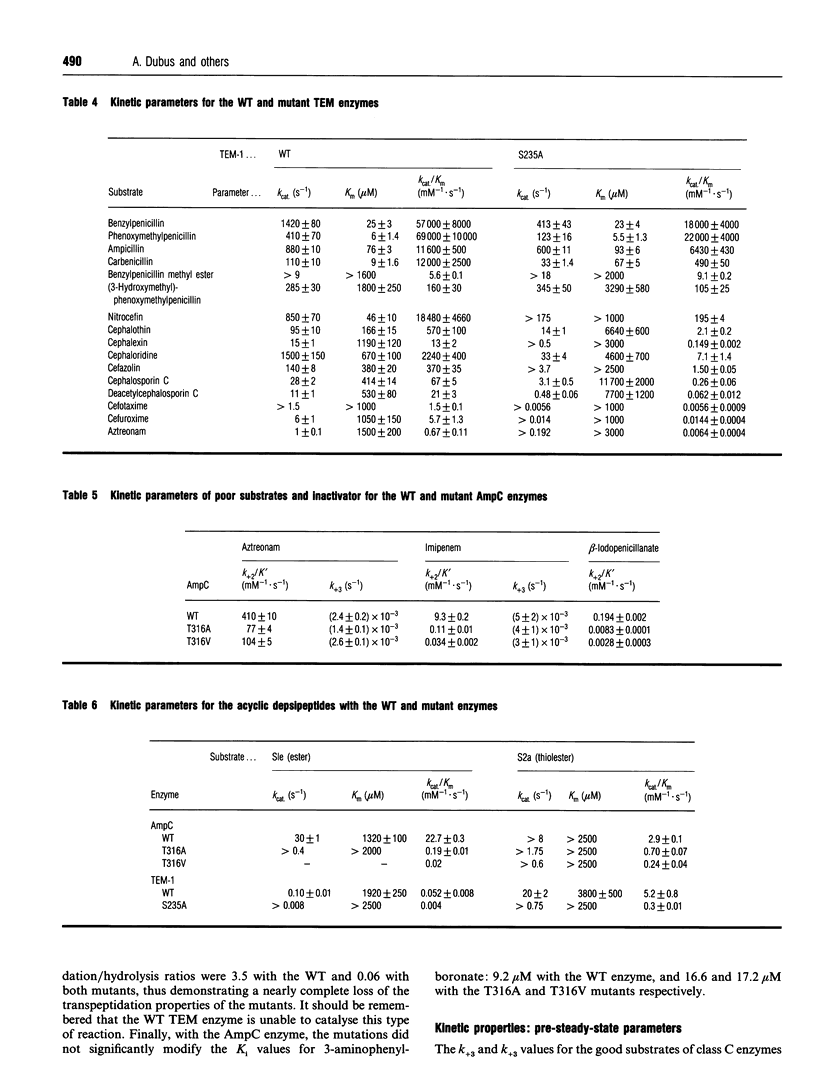
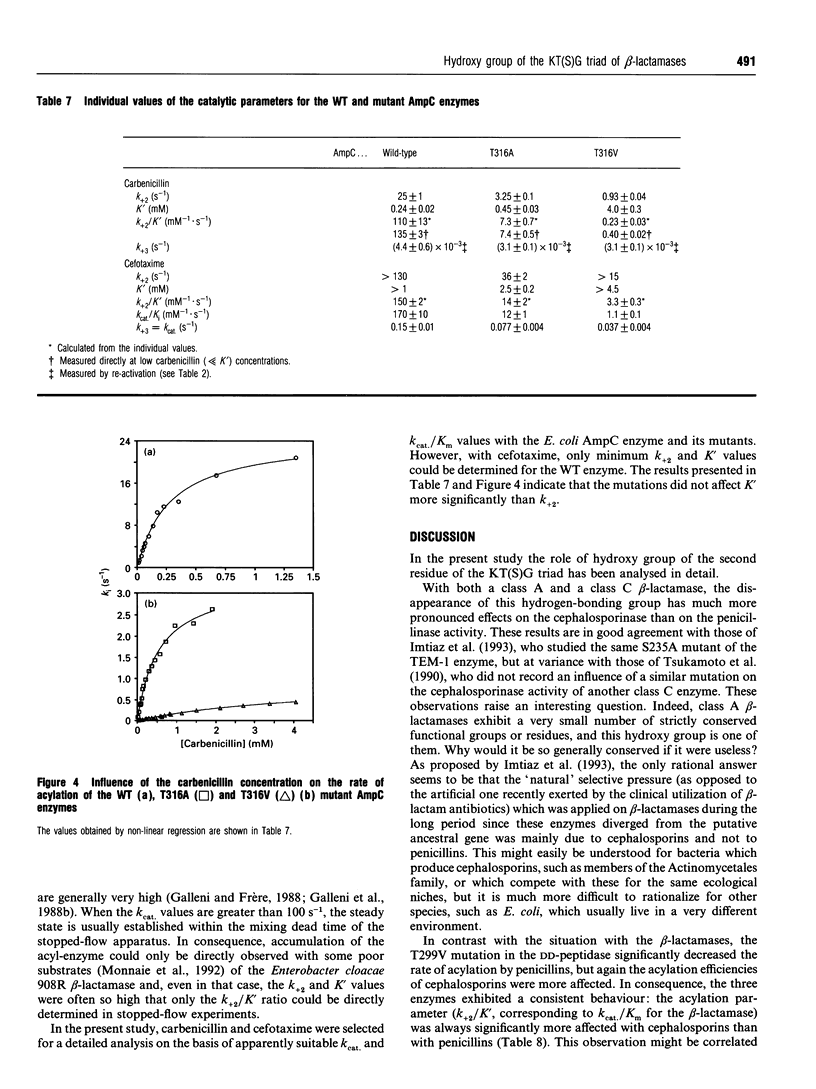
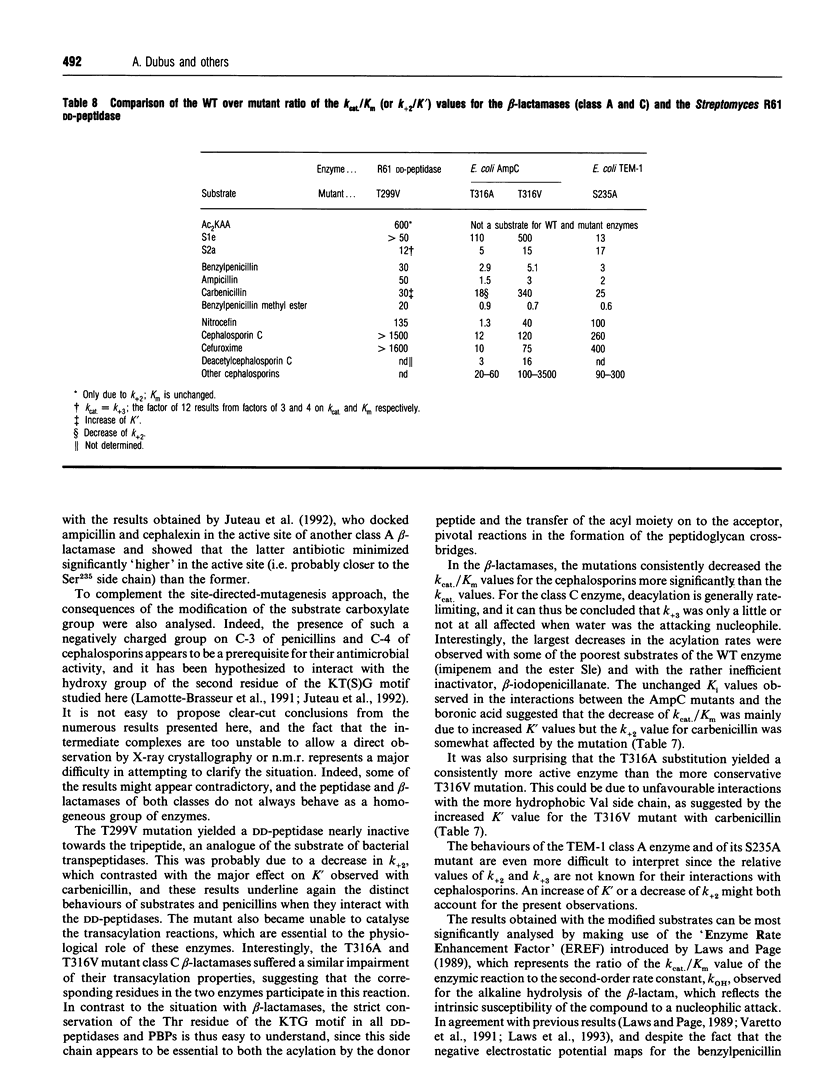
The role of the conserved hydroxy group of the Lys-Thr(Ser)-Gly [KT(S)G] triad has been studied for a class A and a class C beta-lactamase by site-directed mutagenesis. Surprisingly, the disappearance of this functional group had little impact on the penicillinase activity of both enzymes. The cephalosporinase activity was much more affected for the class A S235A (Ser235-->Ala) and the class C T316V (Thr315-->Val) mutants, but the class C T316A mutant was less impaired. Studies were extended to beta-lactams, where the carboxy group on C-3 of penicillins or C-4 of cephalosporins had been modified. The effects of the mutations were the same on these compounds as on the unmodified regular penicillins and cephalosporins. The results are compared with those obtained with a similar mutant (T299V) of the Streptomyces R61 DD-peptidase. With this enzyme the mutation also affected the interactions with penicillins and severely decreased the peptidase activity. The strict conservation of the hydroxy group on the second residue of the KT(S)G triad is thus much more easy to understand for the DD-peptidase and the penicillin-binding proteins than for beta-lactamases, especially those of class C.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam M., Damblon C., Plaitin B., Christiaens L., Frère J. M. Chromogenic depsipeptide substrates for beta-lactamases and penicillin-sensitive DD-peptidases. Biochem J. 1990 Sep 1;270(2):525–529. doi: 10.1042/bj2700525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- De Meester F., Frère J. M., Waley S. G., Cartwright S. J., Virden R., Lindberg F. 6-beta-Iodopenicillanate as a probe for the classification of beta-lactamases. Biochem J. 1986 Nov 1;239(3):575–580. doi: 10.1042/bj2390575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Joris B., Reckinger G., Bellefroid-Bourguignon C., Frère J. M., Waley S. G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem Pharmacol. 1987 Jul 15;36(14):2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- Dubus A., Monnaie D., Jacobs C., Normark S., Frère J. M. A dramatic change in the rate-limiting step of beta-lactam hydrolysis results from the substitution of the active-site serine residue by a cysteine in the class-C beta-lactamase of Enterobacter cloacae 908R. Biochem J. 1993 Jun 1;292(Pt 2):537–543. doi: 10.1042/bj2920537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Amicosante G., Frère J. M. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem J. 1988 Oct 1;255(1):123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Frère J. M. A survey of the kinetic parameters of class C beta-lactamases. Penicillins. Biochem J. 1988 Oct 1;255(1):119–122. doi: 10.1042/bj2550119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Lindberg F., Normark S., Cole S., Honore N., Joris B., Frere J. M. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem J. 1988 Mar 15;250(3):753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Jaurin B., Edlund T., Normark S. Physical mapping and expression of hybrid plasmids carrying chromosomal beta-lactamase genes of Escherichia coli K-12. J Bacteriol. 1980 Sep;143(3):1127–1134. doi: 10.1128/jb.143.3.1127-1134.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima Y., Sugimura T., Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991 May 25;19(10):2785–2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz U., Manavathu E. K., Lerner S. A., Mobashery S. Critical hydrogen bonding by serine 235 for cephalosporinase activity of TEM-1 beta-lactamase. Antimicrob Agents Chemother. 1993 Nov;37(11):2438–2442. doi: 10.1128/aac.37.11.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., Lamotte-Brasseur J., Dideberg O., Joris B., Frère J. M. Arginine 220 is a critical residue for the catalytic mechanism of the Streptomyces albus G beta-lactamase. Protein Eng. 1991 Oct;4(7):811–819. doi: 10.1093/protein/4.7.811. [DOI] [PubMed] [Google Scholar]
- Jamin M., Adam M., Damblon C., Christiaens L., Frère J. M. Accumulation of acyl-enzyme in DD-peptidase-catalysed reactions with analogues of peptide substrates. Biochem J. 1991 Dec 1;280(Pt 2):499–506. doi: 10.1042/bj2800499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Grundström T., Edlund T., Normark S. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981 Mar 19;290(5803):221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juteau J. M., Billings E., Knox J. R., Levesque R. C. Site-saturation mutagenesis and three-dimensional modelling of ROB-1 define a substrate binding role of Ser130 in class A beta-lactamases. Protein Eng. 1992 Oct;5(7):693–701. doi: 10.1093/protein/5.7.693. [DOI] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Normark S. Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J Bacteriol. 1987 Feb;169(2):758–763. doi: 10.1128/jb.169.2.758-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., Frère J. M. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990 Jan 1;265(1):131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnaie D., Virden R., Frère J. M. A rapid-kinetic study of the class C beta-lactamase of Enterobacter cloacae 908R. FEBS Lett. 1992 Jul 20;306(2-3):108–112. doi: 10.1016/0014-5793(92)80979-q. [DOI] [PubMed] [Google Scholar]
- Normark S., Burman L. G. Resistance of Escherichia coli to penicillins: fine-structure mapping and dominance of chromosomal beta-lactamase mutations. J Bacteriol. 1977 Oct;132(1):1–7. doi: 10.1128/jb.132.1.1-7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto K., Nishida N., Tsuruoka M., Sawai T. Function of the conserved triad residues in the class C beta-lactamase from Citrobacter freundii GN346. FEBS Lett. 1990 Oct 1;271(1-2):243–246. doi: 10.1016/0014-5793(90)80416-g. [DOI] [PubMed] [Google Scholar]
- Varetto L., De Meester F., Monnaie D., Marchand-Brynaert J., Dive G., Jacob F., Frère J. M. The importance of the negative charge of beta-lactam compounds in the interactions with active-site serine DD-peptidases and beta-lactamases. Biochem J. 1991 Sep 15;278(Pt 3):801–807. doi: 10.1042/bj2780801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin J. M., Dubus A., Joris B., Frère J. M. The mechanism of action of DD-peptidases: the role of Threonine-299 and -301 in the Streptomyces R61 DD-peptidase. Biochem J. 1994 Jul 15;301(Pt 2):477–483. doi: 10.1042/bj3010477. [DOI] [PMC free article] [PubMed] [Google Scholar]



