ABSTRACT
Water scarcity and increasing urbanization are forcing municipalities to consider alternative water sources, such as stormwater, to fill in water supply gaps or address hydromodification of receiving urban streams. Mounting evidence suggests that stormwater is often contaminated with human feces, even in stormwater drainage systems separate from sanitary sewers. Pinpointing sources of human contamination in drainage networks is challenging given the diverse sources of fecal pollution that can impact these systems and the non-specificity of traditional fecal indicator bacteria (FIB) for identifying these host sources. As such, we used a toolbox approach that encompassed microbial source tracking (MST), FIB monitoring, and bacterial pathogen monitoring to investigate microbial contamination of stormwater in an urban municipality. We demonstrate that human sewage frequently contaminated stormwater (in >50% of routine samples), based on the presence of the human fecal marker HF183, and often exceeded microbial water quality criteria. Arcobacter butzleri, a pathogen of emerging concern, was also detected in >50% of routine samples, with 75% of these pathogen-positive samples also being positive for the human fecal marker HF183, suggesting human municipal sewage as the likely source for this pathogen. MST and FIB were used to track human fecal pollution in the drainage network to the most likely point source of contamination, for which a sewage cross-connection was identified and confirmed using tracer dyes. These results point to the ubiquitous presence of human sewage in stormwater and also provide municipalities with the tools to identify sources of anthropogenic contamination in storm drainage networks.
IMPORTANCE
Water scarcity, increased urbanization, and population growth are driving municipalities worldwide to consider stormwater as an alternative water source in urban environments. However, many studies suggest that stormwater is relatively poor in terms of microbial water quality, is frequently contaminated with human sewage, and therefore could represent a potential health risk depending on the type of exposure (e.g., irrigation of community gardens). Traditional monitoring of water quality based on fecal bacteria does not provide any information about the sources of fecal pollution contaminating stormwater (i.e., animals/human feces). Herein, we present a case study that uses fecal bacterial monitoring, microbial source tracking, and bacterial pathogen analysis to identify a cross-connection that contributed to human fecal intrusion into an urban stormwater network. This microbial toolbox approach can be useful for municipalities in identifying infrastructure problems in stormwater drainage networks to reduce risks associated with water reuse.
KEYWORDS: stormwater, microbial source tracking, water pollution, fecal indicators, enteric pathogens, water quality, Arcobacter
INTRODUCTION
It is currently estimated that by 2050, over half of the urbanized global population will be facing severe challenges posed by water scarcity due to increased urbanization, climate change, and exponential population growth (1). In addition, urbanization has led to hydromodification impacts on urban streams, which necessitates reducing the rate and volume of runoff discharged. In an effort to mitigate the strain on global water supplies and receiving water bodies, the use of alternative water sources, water recycling, and water reuse have been increasingly assessed and utilized, including the use of stormwater (2–5). However, current evidence suggests that stormwater is often of poor microbial water quality based on the levels of fecal indicator bacteria (FIB) such as Enterococcus, Escherichia coli, and total coliforms (6–18). Studies suggest that the use of traditional markers of water quality such as Enterococcus and E. coli only correlate well with gastrointestinal disease when there is an apparent point source of human sewage contamination (19–21) and not when there is no apparent point source of contamination (22–25). This can be partially attributed to the fact that FIB are non-specific to any particular animal host, being found in humans, ruminants, rodents, domestic pets, and waterfowl (26–29). Of particular concern is the public health risk posed by human sewage contamination in environmental waters, considering that the risk from this source has been generally estimated to be higher than that from other animal sources (30, 31).
Consequently, a dominant risk for stormwater use comes from enteric bacterial pathogens sourced from human and non-human feces, such as Campylobacter spp., Salmonella spp., Shiga toxin-producing E. coli (STEC), and Arcobacter (particularly Arcobacter butzleri), with the former three often cited as the respective top three zoonotic causes of bacterial gastrointestinal disease in humans (32, 33). At the same time, Arcobacter spp. (especially Arcobacter cryaerophilus and A. butzleri) have been found to be one of the most dominant pathogenic genera in human sewage (34–37).
Differentiating anthropogenic and non-anthropogenic sources of fecal pollution in stormwater becomes particularly important in characterizing the risks of human exposure to contaminated water (38, 39), and several microbial source tracking (MST) tools have been developed to investigate and help identify sources of pollution in the aquatic environment. While a number of MST technologies have been developed in recent years relying on a diversity of methods (38, 40, 41), popularity has been gaining in methods based on the use of quantitative PCR (9, 39). This method is used to detect and quantify genes or gene fragments specific to microbial populations found in particular hosts, such as the human sewage-specific HF183 marker that has been developed to effectively detect the 16S rRNA of Bacteroides spp. found in human feces (38, 39, 42).
Recent studies across multiple continents using MST quantitative polymerase chain reaction (qPCR) markers suggest that stormwater appears ubiquitously contaminated with human sewage, including where stormwater and sanitary sewer infrastructure are built separately (6, 9–18, 43–46). It is estimated that between 0.1% and 10% of stormwater flows may be comprised of raw human sewage based on concentrations of MST markers in stormwater and raw human sewage (13, 16, 47, 48). Moreover, a number of case studies have recently been successful in investigating and pinpointing sources of human sewage in stormwater by tracking MST markers of human sewage upstream into the drainage network until reaching a terminal point, such as at sanitary sewer infrastructure failures or illicit domestic cross-connections (6, 9, 16, 49).
Due to the limited information that can be gleaned from the enumeration of FIB alone, as well as the increasing capability of MST technologies, several jurisdictions including the province of Alberta, Canada (50), have recently implemented guidelines for stormwater use and established treatment criteria based on quantitative microbial risk assessment (QMRA) (51–53). QMRA is a bi-directional approach that can be used to estimate human health risk based on a number of factors that include an estimation of the concentration of microbial hazards often found in stormwater and can be used to set a benchmark of risk to estimate acceptable levels of microbial contamination (54). One of the most important knowledge gaps for water sources is often in the very first step of QMRA frameworks—the “hazard identification” step (54, 55). Through gathering qPCR-based estimates of human fecal sources of pollution (i.e., HF183) and enteric pathogens, the theory behind these guidelines can be more fully validated and put into practice. Recent QMRA studies have shown, for example, that even relatively low concentrations of the human fecal marker HF183 in environmental waters may increase illness risk appreciably and be detrimental to human health (56–58). In any case, few studies focusing on hazard identification through the lens of QMRA have been performed on stormwater use systems, and more work must be done to understand the sources of pollution and enteric pathogens present (2–5).
In the present study, we set out to identify fecal pollution hazards to public health in a stormwater-impacted creek (Nose Creek) in Airdrie, Alberta, Canada, and, by extension, from the use of that stormwater, by (i) assessing the presence and concentrations of common markers of human sewage contamination (i.e., HF183 and HumM2); (ii) characterizing microbial water quality in terms of FIB and comparing them to traditional recreational water quality criteria standards (59); (iii) identifying the prevalence and concentrations of select enteric bacterial pathogens in stormwater; and (iv) tracking sources of human sewage contamination in storm drainage networks to pinpoint the most likely source(s) of human fecal contamination in the drainage network.
RESULTS
Microbial water quality based on fecal indicators
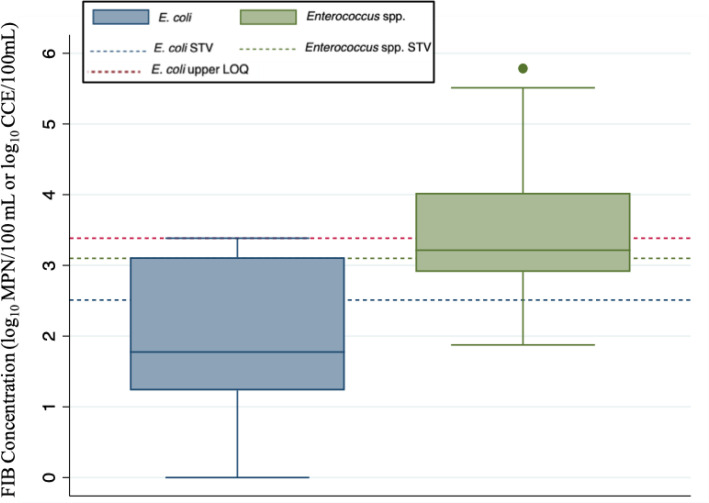
Not surprisingly, all FIB (E. coli, Enterococcus, and total coliforms) were found in 100% of samples and at relatively high concentrations from all storm outfall samples in Nose Creek (though especially Enterococcus—see Fig. 1). For example, total coliforms were frequently found to be ≥3.4 log10 most probable number (MPN)/100 mL (the upper limit of the assay) and in 32 of 38 (84.2%) of routine samples. Enterococcus and E. coli had respective geometric means of 3.5 log10 cell calibrator equivalent (CCE)/100 mL (range: 1.9 log10 CCE/100 mL–5.8 log10 CCE/100 mL) and 2.0 log10 MPN/100 mL (range: <1 log10 MPN/100 mL–>3.4 log10 MPN/100 mL). Despite differences in magnitude, these FIB were found to be highly correlated to each other in routine outfall samples based on the Spearman rank test (see Table 1). Nose Creek sites frequently exceeded traditional water quality guidelines, such as those set by the U.S. EPA for recreational water quality (59). The majority of samples (21 of 38 samples, 55.3%) exceeded the Enterococcus statistical threshold value (STV) of 1,280 CCE/100 mL, while the site-specific Enterococcus geometric mean (GM) of 300 CCE/100 mL was exceeded at every single site (see Table 2). Escherichia coli criteria exceedance was less frequent than for Enterococcus at the routine Nose Creek sites, though every site except NP#1 had at least one sample exceed the E. coli STV of 320 MPN/100 mL, and six of nine (66.6%) outfalls studied also exceeded the site-specific E. coli GM of 100 MPN/100 mL. Importantly, FIB criteria exceedance did not always converge with human sewage detection, with E. coli concentrations exceeding STV criteria in only 7 of 22 samples (31.8%) that were also positive for HF183, while Enterococcus concentrations were above their respective STV in 11 of 22 (50.0%) samples positive for HF183.
Fig 1.
Box and whisker plots of total FIB distributions in all sampled routine stormwater outfalls combined from Nose Creek in Airdrie, Alberta, Canada (sampled in 2021). The E. coli distribution is represented in blue and the Enterococcus distribution in green. The solid line within each box is representative of the median FIB concentration, and the upper and lower horizontal edges of each box represent the 25th and 75th percentile values of concentration, while whiskers represent ±1.5*interquartile range. Outliers are represented by colored dots outside the range of the upper whisker. Note the dotted lines (blue for E. coli, green for Enterococcus) representing acceptable recreational water quality criteria for E. coli (320 MPN/100 mL) and Enterococcus (1,280 CCE/100 mL) (59), as well as the red dotted line representing the upper limit of quantification of the Colilert assay (2,419.60 MPN/100 mL).
TABLE 1.
Spearman correlation coefficients for FIB routinely sampled from Nose Creek stormwater outfalls in Airdrie, Alberta (n = 38)
P < 0.0001.
P < 0.001.
TABLE 2.
Occurrences of human sewage marker HF183 and A. butzleri marker (hsp60) alongside U.S. EPA (59) recreational water quality criteria exceedances (boldface representing criteria that were violated) for routine stormwater outfall samples taken at Nose Creek outfalls (n = 38) in Airdrie, Alberta, Canada
| Nose Creek site | n | A. butzleri marker (hsp60) frequency (%) | HF183 marker frequency (%) | A. butzleri and HF183 marker co-detection frequency (%) | Enterococcus site GM (log10)a | E. coli site GM (log10)a | Enterococcus site STV exceedance (%)b | E. coli site STV exceedance (%)b |
|---|---|---|---|---|---|---|---|---|
| N#1 | 5 | 1/5 (20.0) | 5/5 (100.0) | 1/5 (20.0) | 3.29 | 2.03 | 2/5 (40.0) | 2/5 (40.0) |
| N#2 | 4 | 4/4 (100.0) | 3/4 (75.0) | 3/4 (75.0) | 3.59 | 2.18 | 3/4 (75.0) | 1/4 (25.0) |
| N#3 | 4 | 2/4 (50.0) | 1/4 (25.0) | 1/4 (25.0) | 3.44 | 2.18 | 2/4 (50.0) | 1/4 (25.0) |
| N#4 | 4 | 3/4 (75.0) | 3/4 (75.0) | 3/4 (75.0) | 4.19 | 2.81 | 3/4 (75.0) | 2/4 (50.0) |
| N#5 | 4 | 2/4 (50.0) | 3/4 (75.0) | 2/4 (50.0) | 3.73 | 2.48 | 2/4 (50.0) | 2/4 (50.0) |
| N#6 | 4 | 3/4 (75.0) | 2/4 (50.0) | 2/4 (50.0) | 3.85 | 2.54 | 4/4 (100.0) | 2/4 (50.0) |
| N#7 | 4 | 2/4 (50.0) | 1/4 (25.0) | 1/4 (25.0) | 2.87 | 1.53 | 1/4 (25.0) | 1/4 (25.0) |
| N#8 | 4 | 3/4 (75.0) | 3/4 (75.0) | 3/4 (75.0) | 3.49 | 1.83 | 2/4 (50.0) | 1/4 (25.0) |
| NP#1 | 5 | 1/5 (20.0) | 1/5 (20.0) | 0 | 3.08 | 1.07 | 2/5 (40.0) | 0 |
| Total | 38 | 21/38 (55.3) | 22/38 (57.9) | 16/38 (42.1) | N/A | N/A | 21/38 (55.3) | 12/38 (31.6) |
Enterococcus and E. coli GM of 300 CCE/100 mL and 100 MPN/100 mL, respectively, were used as water quality criteria, with bold values indicating those that exceeded these criteria (59).
Enterococcus and E. coli STVs were 1,280 CCE/100 mL and 320 MPN/100 mL, respectively, with bold values indicating those sites where >10% of samples exceeded these values (59).
Evidence of human sources of fecal pollution impacting stormwater
Human fecal contamination of stormwater, as determined by the presence of the human fecal marker HF183, was detected at every stormwater outfall at least once and, overall, was detected in the majority of routine samples taken from Nose Creek in 2021 (22 of 38 samples or 57.9%) (Fig. 2; Table 2). However, most samples positive for HF183 in Nose Creek were found to have very low concentrations of the marker [detectable but non-quantifiable (DNQ) in 13 of 22 samples (59.1%)], though this marker ranged from 3.6 log10 copies/100 mL to 4.3 log10 copies/100 mL when detected at quantifiable levels. In contrast, the other human fecal marker used (HumM2) was only detected in 6 of 38 (15.8%) total routine samples from Nose Creek and always at a DNQ concentration. While the majority of samples positive for HumM2 were also positive for HF183 (6 of 8 samples, 75.0%), only 27.3% of total samples positive for HF183 (6 of 22 samples) were also positive for HumM2. The higher concentrations found for HF183 compared to HumM2 suggested that HF183 was potentially a more sensitive indicator of human sewage than HumM2. Regardless, the presence of both markers suggested that human fecal wastes were frequently, albeit sporadically, flowing into Nose Creek from stormwater.
Fig 2.
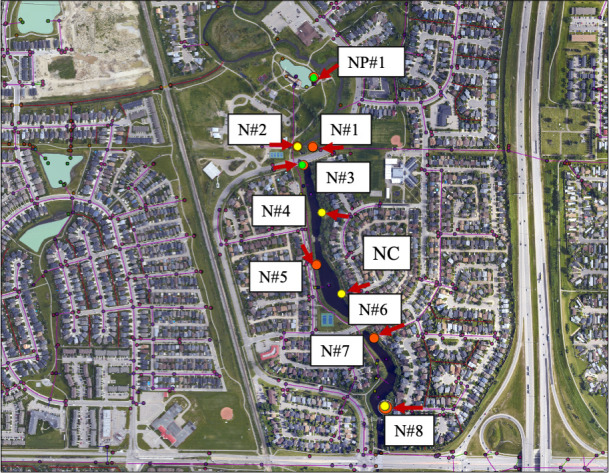
Map of human sewage marker (HF183) detection within stormwater outfalls into Nose Creek (NC) in Airdrie, Alberta, 2021. Note the overlaid colored dots representative of HF183 detection at quantifiable levels (red), detectable but not quantifiable (yellow), and not detected (green) in the Nose Creek Pond (NP#1) and Nose Creek sites (NC#1–8). Red arrows represent the directional flow of stormwater. Note that all maps presented were created and co-owned with the City of Airdrie, and have been used with permission.
Enteric bacterial pathogens
Given that (i) human feces was identified as an important source of microbial pollution flowing into Nose Creek and (ii) pathogens such as Arcobacter spp. are abundant in municipal sewage, we sought to better understand risk by evaluating bacterial pathogen levels in stormwater. Arcobacter butzleri and Campylobacter spp. were detected in 55.3% (21 of 38) and 7.9% (3 of 38) of routine samples collected at Nose Creek outfalls, respectively. Arcobacter butzleri was detected at least once at all Nose Creek outfalls studied. In the 21 samples positive for A. butzleri, six samples had reasonably high quantifiable concentrations, ranging from 3.9 to 4.1 log10 copies/100 mL. In contrast, all three samples positive for Campylobacter spp. were at low levels (i.e., DNQ). Salmonella and STEC were not detected in stormwater flowing into Nose Creek at any of the outfall sites tested.
Of the 21 samples positive for A. butzleri, 16 samples (76%) were also positive for human fecal contamination based on HF183 (see Table 3). An independent analysis of these 16 samples across the 38 (42.1%) total number of stormwater effluent samples flowing into Nose Creek revealed that A. butzleri and HF183 were statistically significantly more likely to be detected together than for either marker to be detected alone [based on Fisher’s exact test (P = 0.013)]. This was in contrast to Campylobacter, where none of the positive samples were also positive for HF183. Enterococcus STV criteria exceedance occurred in 14 of 21 (66.7%) samples positive for A. butzleri, whereas the E. coli STV criteria were only exceeded in 8 of 21 (38.1%) routine outfall samples positive for this pathogen.
TABLE 3.
Two-by-two table of Nose Creek 2021 routine Airdrie stormwater samples positive for A. butzleri (hsp60), human sewage marker (HF183), both, or neither (n = 38)
| HF183 detected | HF183 not detected | |
|---|---|---|
| A. butzleri detected | 16 | 5 |
| A. butzleri not detected | 6 | 11 |
Investigating point sources of human fecal pollution in the stormwater drainage network
Stormwater effluent at the most upstream site on Nose Creek (i.e., Site N#1—see Table 2; Fig. 2) was consistently contaminated with human sewage based on the presence of the human fecal marker HF183. Subsequently, several manholes were systematically sampled upstream of the N#1 stormwater outfall, for which we observed a distinct pattern in both the presence of HF183 and an increasing concentration gradient toward the more northern and distant manholes sampled during the investigation (e.g., Manhole N1-10C41 in the northeast quadrant of the city) (Fig. 3). A manhole (N1-15C62) immediately upstream of the outfall N#1 had approximately a 10-fold increase in HF183 concentration compared to the stormwater effluent flowing into the creek at N#1 itself (i.e., 4.4 log10 copies/100 mL versus 3.6 log10 copies/100 mL, respectively; Fig. 3; Table 4) and which appeared to come from the north trunk of the storm drain feeding into this manhole. Testing water quality in manholes upstream of N1-15C62 revealed an increasing concentration of HF183 in the north trunk sewer, peaking at 5.4 log10 copies/100 mL at Manhole N1-10C49 and 6.1 log10 copies/100 mL at Manhole N1-10C41 (Fig. 3; Table 4). Interestingly, HumM2 (generally a less sensitive human marker of fecal contamination compared to HF183), was also detected at these two upstream manholes at relatively large concentrations (i.e., 4.3 log10 copies/100 mL at Manhole N1-10C49 and 6.1 log10 copies/100 mL at Manhole N1-10C41), which was not detected at downstream manholes nor the outfall into Nose Creek. This “closing-in” on the point of contamination using MST markers was reinforced by FIB concentrations, whereby levels of E. coli at N1-10C49 and N1-10C41 exceeded the upper limits of detection by the Colilert Quanti-Tray method but did not exceed this limit at the downstream Manhole N1-15C62 or the outfall flowing into Nose Creek (Table 4). Likewise, increasing concentrations of Enterococcus were also observed peaking at Manhole N1-10C41 [compare Enterococcus levels at N#1 in Nose Creek (2.2 log10 CCE/100 mL) to Manhole N1-10C41 (5.6 log10 CCE/100 mL)]. Sampling manholes further upstream of Manholes N1-10C49 and N1-1041 revealed no evidence of human fecal pollution and lower FIB counts coming from other directions, suggesting that the actual physical source of contamination was in the vicinity of Manhole N1-10C41. As a result, the City of Airdrie conducted a fluorescein tracer dye test on a nearby multi-user recreational building and confirmed that certain toilets within this facility had been plumbed into the stormwater drains feeding into Manhole N1-10C41. Consequently, this was deemed to be the primary source of human fecal contamination draining into Nose Creek.
Fig 3.
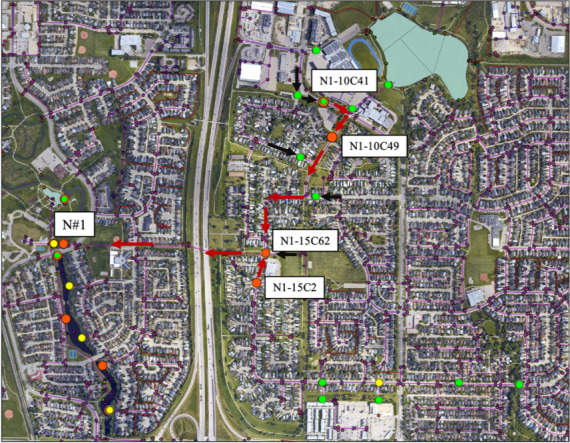
Map of Nose Creek stormwater drainage network summarizing the most relevant manholes tested for the human sewage marker HF183 upstream of the positive outfall site labeled N#1. Note the manholes upstream of N#1 where HF183 was positive and quantifiable (red dots), detectable but not quantifiable (yellow dots), or not detected (green dots). Also note the directional flow of stormwater through the system, represented by drainage networks positive for HF183 (red arrows) as well as stormwater flows with no demonstrable HF183 detection (black arrows). Note that all maps presented were created and co-owned with the City of Airdrie, and have been used with permission.
TABLE 4.
MST and FIB results from select investigative samples highlighting the most direct proposed path of human fecal contamination from a local community center in Airdrie, Alberta, to the N#1 outfall feeding Nose Creek
| Sampling date (date/month/year) | Site | HF183 | HumM2 | Enterococcus | E. coli |
|---|---|---|---|---|---|
| log10 copies/100 mL | log10 copies/100 mL | log10 CCE/100 mL | log10 MPN/100 mL | ||
| 7/9/21 | N#1 | 3.64 | ND | 2.19 | 0.98 |
| 8/9/21 | N1-15C62-N | 4.40 | ND | 2.23 | 1.30 |
| 27/9/21 | N1-10C49-S | 5.36 | 4.31 | 4.36 | >3.38a |
| N1-10C41-N | 6.08 | 6.05 | 5.60 | >3.38a |
Greater than the upper limit of detection (LOD) of assay.
Our investigations also suggested that the southern trunk of the drainage network flowing into Manhole N1-15C62 (i.e., the manhole immediately upstream of N#1 site draining into Nose Creek) also contributed human sewage into Nose Creek, albeit far less than that coming from the north trunk (see Table S1; Fig. S1). Samples taken the same day at Manhole N1-15C62 and an upstream manhole immediately south (i.e., Manhole N1-15C2) revealed that both these manholes contained HF183, but concentrations were far greater at N1-15C62 (5.3 log10 copies/100 mL) compared to N1-15C2 (4.1 log10 copies/100 mL). This result was also reflected in the higher E. coli concentrations seen at N1-15C62 compared to N1-15C2. Likewise, HumM2 was detected at N1-15C62 at a relatively high concentration (4.32 log10 copies/100 mL), but only at detectable but non-quantifiable levels at N1-15C2 (Table S1). Testing of manholes further to the south resulted in several non-detects, and only one detectable but non-quantifiable sample for HF183 (Table S2). Collectively, the data suggested that the bulk of the human fecal contamination entering Nose Creek through outfall N#1 was due to the cross-connection in the multi-user recreational facility in the north part of the city but that minor contributions of human fecal contamination were coming from the south as well (albeit sources remain unknown).
DISCUSSION
Stormwater is generally considered to be a relatively poor-quality water source, particularly for municipalities that manage stormwater using combined sewer outfalls (CSOs) (60). Nevertheless, even in municipalities where stormwater and sanitary sewers are completely separate, human sewage contamination can be fairly widespread, albeit often at sporadic and frequently low levels. Sources of human fecal pollution within these separated systems can be diverse, ranging from infrastructure failures (e.g., leaky sewer systems or service connections and inadvertent cross-connections), illicit discharges (e.g., dumping of recreational vehicle wastes), or even unhoused populations within urban municipalities.
The City of Airdrie in Alberta, Canada, represented an ideal case study for characterizing sources of urban pollution in a fully separated stormwater drainage system. Herein, we demonstrate that for stormwater outfalls persistently contaminated with human fecal markers, MST, FIB, and pathogen-based monitoring tools can be useful for pinpointing the physical source of pollution within the storm drainage network. In particular, we identified a consistent yet low-level signature of the human fecal marker HF183 (~103 copies/100 mL) flowing from stormwater effluents (both during base flow and storm flows) into an urban creek and tracked these molecular signatures upstream through the drainage network. In some effluent samples, the levels of E. coli and Enterococcus were low and met recreational water quality criteria, yet they still contained a persistent HF183 signature, indicating a potential structural problem in the upstream drainage network that might pose a risk to human health. We subsequently monitored the stormwater quality in the drainage network upstream of this “contaminated” outfall, which revealed an ever-increasing concentration in HF183 as we moved further upstream within the drainage network. HF183 levels peaked at >106 copies/100 mL—a concentration approximately 1,000× greater than that observed in the effluents flowing into the creek, which also correlated with an increase in FIB concentrations (i.e., >250× and >2,500× increase in E. coli and Enterococcus concentrations, respectively). At the most contaminated site in the drainage network, the human fecal marker HumM2 was also detected, even though it was not detected in the effluents flowing into the urban creek. HumM2 has been reported to be less sensitive than HF183, due to the HumM2 marker not always being detected within individual human fecal samples (61), and often typically at lower concentrations (by at least one to two orders of magnitude) in human feces and sewage in comparison to HF183 (62, 63). Collectively, this suggested that the contamination source impacting the storm drainage network was in the near vicinity, and tracer dye testing at a nearby multi-user recreational facility confirmed that several toilets within the facility were cross-connected into the storm drains. This toolbox approach to microbial water quality investigations proved to be extremely valuable in pinpointing infrastructure problems in municipal storm drains that are fully separated from municipal sewage, and offers a novel approach for municipalities to manage cross-connection programs and monitor for infrastructure problems in storm drainage networks.
The prevalence of HF183 in stormwater in Nose Creek is consistent with what is found in other studies around the world, though there is also high variability in marker concentrations (<2 log10–>6 log10 copies/100 mL) (4, 6, 9–11, 13, 16–18, 44, 46). This highlights the variability of this marker in stormwater and reflects the different sources of fecal pollution that range from low-impact cross-connections at single-family residential households to broken sanitary sewer pipes (6, 44, 45). As concentrations of HF183 are typically 7–8 log10 copies/100 mL in raw human municipal sewage (13, 16, 47, 48), our data suggest that approximately 0.1% of the total stormwater effluent (i.e., 1 of every 1,000 L) flowing into the urban creek at this site was made up of raw human sewage.
Recently, multiple similar studies have used the MST toolbox approach to investigate stormwater drainage networks upstream of contaminated outflows, finding similar results [see references (6, 9, 16, 49)]. Three case studies by Gonzalez et al. (49) and one by Hachad et al. (9) were able to pinpoint and identify specific areas of human sewage contamination leakage that included sanitary sewer infrastructure failures, sanitary sewer blockages, and illicit cross-connections. Along with the current case study at Nose Creek, this highlights the effective use of MST technology by municipalities to mitigate fecal pollution and consequent public health risks of stormwater use.
Overall, the microbial water quality of Airdrie stormwater flowing into Nose Creek was found to be generally poor based on traditional (FIB) water quality guidelines/standards, and this is consistent with the literature, which demonstrates the persistent and near-universal exceedance of recreational or ambient water quality FIB criteria in stormwater (6–18). The correlation between Enterococcus and E. coli FIB was significant and relatively high (ρ ≥ 0.8) in Nose Creek outfall samples, being similar to several studies elsewhere (15, 17, 44), though other publications found only weak to moderate correlation between these two (6, 11, 64).
It is notable that high rates of FIB criteria exceedance did not necessarily always co-occur in samples where human sewage marker HF183 was consistently detected, and several studies report relatively low to moderate correlation between the two (9, 13, 14, 18, 45, 46), while Sauer et al. (16) found no significant correlation at all. Differing decay rates between FIB and HF183 (47, 65, 66) as well as animal-specific sources of FIB [see references (26, 27)] may also be contributing to fecal pollution in the drainage network, and indeed, in our study, multiple sources of fecal pollution could be observed (Carson LR, Beaudry M, Valeo C, He J, Banting G, van Duin B, Goodman C, Scott C, and Neumann NF, submitted for publication). These data suggest that FIB monitoring alone is insufficient as a tool for identifying and pinpointing possible infrastructure problems in storm drainage systems and that these methods should be combined with other tools, such as MST (i.e., using human sewage marker HF183 where human sewage contamination is a concern).
While most enteric pathogens were not (or were rarely) observed in the current study (including Campylobacter, Salmonella, and STEC), A. butzleri was observed frequently in Nose Creek samples, found in 21 of 38 samples, and in 16 of these A. butzleri positive samples, HF183 was also detected, reinforcing the finding that human sewage is likely a major source of this pathogen. Arcobacter spp. are abundant (and dominant) in human sewage (35–37) and have been found to correlate well with markers of human sewage in receiving water bodies heavily contaminated by human fecal pollution (34, 67). In some cases, Arcobacter spp. were also found to correlate relatively well with FIB in sewage-contaminated environmental waters (68, 69). This is consistent with the present study, where A. butzleri appeared to be a dominant pathogen in stormwater impacted by human sewage. By contrast, other enteric bacterial pathogens, such as Campylobacter spp., STEC, and Salmonella spp., are often only sporadically detected in stormwater and at relatively low concentrations (43, 70) and are often found to correlate poorly to FIB (8, 64, 70) and human sewage markers (34, 45, 64). These pathogens are often used in QMRA modeling studies as reference pathogens for evaluating risk, but our data, however, suggest that A. butzleri may be far more abundant in stormwater than these commonly used reference pathogens, warranting some consideration for Arcobacter to be used as a potential reference pathogen for QMRA studies on stormwater use. Unfortunately, few studies have looked into the presence of pathogenic Arcobacter spp., such as A. butzleri, in stormwater (67, 71). In the study by Beaudry (71), approximately 75% of stormwater samples analyzed contained culturable Arcobacter butzleri, and based on virulence gene analysis, these isolates appeared to represent pathogenic strains of the bacteria.
As stormwater use and water reuse systems have not yet been extensively studied in terms of risk (2–5), understanding the microbial hazards inherent within these systems is of paramount importance. For example, according to several QMRA studies, concentrations of HF183 hovering around 3–4 log10 copies/100 mL in the stormwater ponds themselves (as opposed to in upstream outfalls, where concentrations may be even higher) can be hazardous to human health (56–58), highlighting the increased need to better understand potential human sewage levels in stormwater systems designated for use even when FIB levels do not exceed guideline/regulatory criteria. Schoen et al. (5) also found that risks of gastrointestinal illness could vary for different non-potable uses of stormwater, further suggesting the complex number of factors to consider when exposing people to stormwater as an alternative water source.
In conclusion, the current study improves our understanding of the microbial hazards present in stormwater and promotes the use of a microbial toolbox approach to monitoring FIB, MST, and pathogen occurrence for identifying and mitigating environmental contamination risks in the urban water environment.
Conclusions and recommendations
Stormwater can be a low-quality water source and may not meet ambient or recreational water quality criteria (i.e., Enterococcus and E. coli counts).
Human sewage contamination (as measured by HF183) can be commonly observed in stormwater, even in systems separated from sanitary.
MST, FIB, and pathogen-related methods can be successfully employed to pinpoint the source of human fecal pollution in a storm drainage network.
The bacterial enteric pathogen A. butzleri (but not Campylobacter spp., STEC, or Salmonella spp.) was commonly found in stormwater and significantly associated with human markers of fecal pollution.
MATERIALS AND METHODS
Sampling area and strategy
Two sampling strategies were used when testing stormwater from Nose Creek in the City of Airdrie, Alberta, which consisted of (i) routine outfall sampling (bi-weekly) and (ii) investigative sampling where we interrogated the urban stormwater drainage network (i.e., from downstream outfalls to manholes further upstream) to pinpoint the physical source(s) of sewage contamination when human MST markers of pollution (HF183 and HumM2) were observed. The routine stormwater sampling sites consisted of eight separate outfalls (labeled N#1–N#8) draining into Nose Creek within the city limits, as well as one outfall draining into the Nose Creek Pond (NP#1), which is attached to the creek (see Fig. 2). Each site was sampled on four separate dates (with the exception of N#1 and NP#1, which were sampled on five occasions), for a total of 38 routine samples collected over 4/5 weeks on an approximately bi-weekly basis from 20 July to 7 September 2021.
Investigative sampling of manholes upstream of those outfalls (i.e., N#1) found to be contaminated with human sewage was also performed, with a total of 30 individual samples collected from 15 manholes upstream of this site. Investigative sampling was done between 26 July and 27 September 2021, on an approximately bi-weekly basis. During the investigation, samples were taken when the (base) flow was sufficient and from multiple trunks running in different directions upstream.
Sample collection for both routine and investigative samples consisted of 200 mL grab samples collected in sterile bottles either directly from the stormwater outfall (routine sampling) or directly from pipes flowing into stormwater manholes (investigative sampling). Samples were then shipped overnight on ice from Airdrie to the University of Alberta, in Edmonton, where the samples were fully processed within 24 hours of being collected.
Microbial culture methods
Escherichia coli and total coliforms were enumerated by the defined substrate methods using Colilert in a Quanti-Tray MPN format (Idexx Laboratories, Inc.; Westbrook, ME, USA), as per the manufacturer’s instructions. Briefly, 100 mL of stormwater sample was added to a small vessel alongside one packet of Colilert reagent, then inoculated into a Quanti-Tray/2000 system and incubated at 35°C for 24 hours. Positive and negative controls, respectively, consisted of 100 mL of sterile deionized water that underwent the same process as above but was either spiked with one colony of E. coli ATCC 25922 [incubated previously for 24 hours on trypticase soy agar (BD; Thermo Fisher Scientific, Ottawa, Ontario, Canada) at 37°C for 24 hours] or was not spiked and left as sterile water only. After this 24-hour incubation, results were calculated using a standard MPN table based on a yellow-color change within Quanti-Tray cells (total coliforms) and additional fluorescence under long-wave UV (E. coli).
Molecular-based detection and quantification methods
Quantitative PCR methods based on TaqMan chemistry were used to test for several markers, including for human sewage [HF183 and HumM2—see references (42, 72)], the FIB Enterococcus (73, 74), and enteric pathogens including A. butzleri (75), Campylobacter spp. (76), Salmonella spp. (77), and STEC (78) (also see Table S3).
Sample preparation for qPCR
Stormwater samples were prepared for qPCR testing by first filtering 20 mL of each sample through disposable 0.4-µm pore polycarbonate MicroFunnel filters (Pall Corporation, New York, USA). Sample filters were then extracted and processed according to U.S. EPA Method 1611 (74). Briefly, sample filters [including three filtered calibrators and one filtering blank per day of sampling as described elsewhere (74)] were added to bead tubes (Generite, North Brunswick, NJ, USA) alongside AE buffer (10 mM Tris-Cl, 0.5 mM EDTA, pH 9.0) (Qiagen; Hilden, Germany) and 0.2 µg/mL of Oncorhynchus keta (salmon) sperm as an internal control (74, 79). Tubes were homogenized by a Bead Mill 24 Homogenizer (Thermo Fisher Scientific, Waltham, MA, USA), and the resulting supernatant was transferred to separate tubes and centrifuged twice before the final supernatant was used as the DNA template for qPCR. Templates were frozen at −80°C until qPCR testing.
qPCR reaction conditions
An Applied Biosystems 7500 Real-Time PCR (Applied Biosystems; Thermo Fisher Scientific, Ottawa, Ontario, Canada) was used for the performance of all qPCR assays and under fast cycling conditions. All qPCR runs consisted of two-step reactions with the following cycling conditions: 3 minutes of initial denaturation at 95°C, before denaturation and annealing/extension, respectively, at 95°C for 5 seconds and 60°C for 30 seconds for 40 cycles. Reagents used for all runs included 1× PrimeTime Gene Expression Master Mix (Integrated DNA Technologies, Coralville, IA, USA), 200 µg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MI, USA), and the appropriate primers and probe concentrations dependent on the assay (see Table S3). Primers and probes for all markers are described in Table S3. All reactions consisted of 15 µL of the above reagents and 5 µL of DNA template/control for a total of 20 µL per reaction. All assays were set at a fluorescence threshold of 0.1 with the exception of the VD16S (Campylobacter spp.) marker, which was run at a threshold of 0.05. MicroAmp Fast Optical 96-well plates (Applied Biosystems, Foster City, CA, USA) were used for all qPCR runs; samples and negative controls (no-template controls, filtering blanks) were run in duplicate wells, while positive controls (calibrators or plasmids as appropriate) were run in triplicate wells.
qPCR controls
Positive controls for qPCR reactions consisted either of calibrators or a series of plasmid standard dilutions, depending on whether Enterococcus or qPCR markers for human feces/enteric pathogens were being enumerated, respectively. More specifically, calibrators were used when measuring sample inhibition or determining the concentrations of Enterococcus via the ΔΔCT relative quantification method as specified in U.S. EPA Method 1611 (74). Appropriate plasmid standards were instead used in dilutions of 50K copies/5 µL to 5 copies/5 µL for absolute quantification of other markers, including two human sewage markers (HF183 and HumM2) as well as enteric pathogen markers including those for A. butzleri, Campylobacter spp., Salmonella spp., and STEC (see Table S3). Negative controls consisted of filtering blanks (i.e., filtered sterile phosphate-buffered saline) and no-template controls (i.e., nuclease-free water), while salmon sperm (O. keta) was used as an inhibition control (74, 79). Samples displaying a shift of >3 Cts of salmon sperm concentration in comparison to calibrators were considered inhibited, as specified in U.S. EPA Method 1611 (74), and diluted with water to 1:5 and 1:25 dilutions before being run again for qPCR analysis.
Data analysis
Prior to data analysis, estimates of qPCR markers (both MST and enteric pathogens), Enterococcus, and E. coli were all reported, respectively, as either copy, CCE, and MPN per 100 mL of stormwater sampled. Data analysis began with first log10 transforming quantifiable estimates of FIB, MST markers, and enteric pathogen markers. As some samples for E. coli were at the lower (<1 MPN/100 mL) or upper (>3.4 log10 MPN/100 mL) detection limits of the Colilert assay, these samples were simply set to these limits for the sake of analysis. In terms of qPCR testing, marker estimates were considered either non-detects (ND) if the marker amplification was absent over 40 cycles, DNQ if amplification occurred but marker estimates were found to be below the 95% percentile of the limit of detection (LOD95), or quantifiable if marker estimates were found to exceed this limit. Testing by the Shapiro–Wilk test found that FIB were not normally distributed. As a result, the non-parametric Spearman rank test was used to determine if there was any significant correlation between Enterococcus, E. coli, and/or total coliforms, while the non-parametric Fisher’s exact test was used to determine whether HF183 and A. butzleri were significantly more likely to be detected within the same sample, rather than independently.
ACKNOWLEDGMENTS
Special thanks are due to the City of Airdrie’s environmental monitoring team (Kevin Kerr, Jennifer Sugden, Terry Parks, and Kelly McKague) who organized and performed on-site sampling for routine and investigative sample collection. Special thanks are due to Candis Scott and Dr. Graham Banting for providing laboratory support for this project.
Funding for this project was provided by grants to N.F.N. from Alberta Innovates (Project Number AI-EES2333), the Natural Sciences and Engineering Research Council (NSERC) (University of Alberta Project Number: NSERC CRDPJ 520869-17), and the City of Calgary (University of Alberta Project Number CC CRD 520869), through in-kind logistic support from the City of Airdrie, Alberta, Canada. Other than the affiliation of some authors to municipal-level funding agencies [B.V.D. (City of Calgary) and C.G. (City of Airdrie)], the funding agencies themselves did not play a role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Please see the following URLs for representative funding agencies: Alberta Innovates: https://albertainnovates.ca/, Natural Sciences and Engineering Research Council of Canada: https://www.nserc-crsng.gc.ca/index_eng.asp, City of Calgary: https://www.calgary.ca/home.html, City of Airdrie: https://www.airdrie.ca/.
Contributor Information
Norman F. Neumann, Email: nfneuman@ualberta.ca.
Blaire Steven, Connecticut Agricultural Experiment Station, New Haven, Connecticut, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.00337-24.
Fig. S1; Tables S1 to S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. He C, Liu Z, Wu J, Pan X, Fang Z, Li J, Bryan BA. 2021. Future global urban water scarcity and potential solutions. Nat Commun 12:4667. doi: 10.1038/s41467-021-25026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy HM, Meng Z, Henry R, Deletic A, McCarthy DT. 2017. Current stormwater harvesting guidelines are inadequate for mitigating risk from Campylobacter during nonpotable reuse activities. Environ Sci Technol 51:12498–12507. doi: 10.1021/acs.est.7b03089 [DOI] [PubMed] [Google Scholar]
- 3. Petterson SR, Mitchell VG, Davies CM, O’Connor J, Kaucner C, Roser D, Ashbolt N. 2016. Evaluation of three full-scale stormwater treatment systems with respect to water yield, pathogen removal efficacy and human health risk from faecal pathogens. Sci Total Environ 543:691–702. doi: 10.1016/j.scitotenv.2015.11.056 [DOI] [PubMed] [Google Scholar]
- 4. Sales-Ortells H, Medema G. 2015. Microbial health risks associated with exposure to stormwater in a water plaza. Water Res 74:34–46. doi: 10.1016/j.watres.2015.01.044 [DOI] [PubMed] [Google Scholar]
- 5. Schoen ME, Ashbolt NJ, Jahne MA, Garland J. 2017. Risk-based enteric pathogen reduction targets for non-potable and direct potable use of roof runoff, stormwater, and greywater. Microb Risk Anal 5:32–43. doi: 10.1016/j.mran.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahmed W, Payyappat S, Cassidy M, Harrison N, Besley C. 2020. Sewage-associated marker genes illustrate the impact of wet weather overflows and dry weather leakage in urban estuarine waters of Sydney, Australia. Sci Total Environ 705:135390. doi: 10.1016/j.scitotenv.2019.135390 [DOI] [PubMed] [Google Scholar]
- 7. Converse RR, Piehler MF, Noble RT. 2011. Contrasts in concentrations and loads of conventional and alternative indicators of fecal contamination in coastal stormwater. Water Res 45:5229–5240. doi: 10.1016/j.watres.2011.07.029 [DOI] [PubMed] [Google Scholar]
- 8. de Man H, van den Berg HHJL, Leenen EJTM, Schijven JF, Schets FM, van der Vliet JC, van Knapen F, de Roda Husman AM. 2014. Quantitative assessment of infection risk from exposure to waterborne pathogens in urban floodwater. Water Res 48:90–99. doi: 10.1016/j.watres.2013.09.022 [DOI] [PubMed] [Google Scholar]
- 9. Hachad M, Lanoue M, Vo Duy S, Villlemur R, Sauvé S, Prévost M, Dorner S. 2022. Locating illicit discharges in storm sewers in urban areas using multi-parameter source tracking: field validation of a toolbox composite index to prioritize high risk areas. Sci Total Environ 811:152060. doi: 10.1016/j.scitotenv.2021.152060 [DOI] [PubMed] [Google Scholar]
- 10. Kinzelman JL, McLellan SL. 2009. Success of science-based best management practices in reducing swimming bans—a case study from Racine, Wisconsin, USA. Aquat Ecosyst Health Manag 12:187–196. doi: 10.1080/14634980902907466 [DOI] [Google Scholar]
- 11. Lee S, Suits M, Wituszynski D, Winston R, Martin J, Lee J. 2020. Residential urban stormwater runoff: a comprehensive profile of microbiome and antibiotic resistance. Sci Total Environ 723:138033. doi: 10.1016/j.scitotenv.2020.138033 [DOI] [PubMed] [Google Scholar]
- 12. Monteiro S, Queiroz G, Ferreira F, Santos R. 2021. Characterization of stormwater runoff based on microbial source tracking methods. Front Microbiol 12:674047. doi: 10.3389/fmicb.2021.674047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nshimyimana JP, Ekklesia E, Shanahan P, Chua LHC, Thompson JR. 2014. Distribution and abundance of human‐specific Bacteroides and relation to traditional indicators in an urban tropical catchment. J Appl Microbiol 116:1369–1383. doi: 10.1111/jam.12455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olds HT, Corsi SR, Dila DK, Halmo KM, Bootsma MJ, McLellan SL. 2018. High levels of sewage contamination released from urban areas after storm events: a quantitative survey with sewage specific bacterial indicators. PLoS Med 15:e1002614. doi: 10.1371/journal.pmed.1002614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parker JK, McIntyre D, Noble RT. 2010. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, USA. Water Res 44:4186–4194. doi: 10.1016/j.watres.2010.05.018 [DOI] [PubMed] [Google Scholar]
- 16. Sauer EP, Vandewalle JL, Bootsma MJ, McLellan SL. 2011. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res 45:4081–4091. doi: 10.1016/j.watres.2011.04.049 [DOI] [PubMed] [Google Scholar]
- 17. Sidhu JPS, Hodgers L, Ahmed W, Chong MN, Toze S. 2012. Prevalence of human pathogens and indicators in stormwater runoff in Brisbane, Australia. Water Res 46:6652–6660. doi: 10.1016/j.watres.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 18. Staley ZR, Boyd RJ, Shum P, Edge TA. 2018. Microbial source tracking using quantitative and digital PCR to identify sources of fecal contamination in stormwater, river water, and beach water in a Great Lakes area of concern. Appl Environ Microbiol 84:e01634-18. doi: 10.1128/AEM.01634-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wade TJ, Pai N, Eisenberg JNS, Colford JM. 2003. Do U.S. environmental protection agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ Health Perspect 111:1102–1109. doi: 10.1289/ehp.6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wade TJ, Calderon RL, Sams E, Beach M, Brenner KP, Williams AH, Dufour AP. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ Health Perspect 114:24–28. doi: 10.1289/ehp.8273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, Wymer L, Rankin CC, Love D, Li Q, Noble R, Dufour AP. 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ Health 9:66. doi: 10.1186/1476-069X-9-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnold BF, Schiff KC, Griffith JF, Gruber JS, Yau V, Wright CC, Wade TJ, Burns S, Hayes JM, McGee C, Gold M, Cao Y, Weisberg SB, Colford JM. 2013. Swimmer illness associated with marine water exposure and water quality indicators: Impact of widely used assumptions. Epidemiology 24:845–853. doi: 10.1097/01.ede.0000434431.06765.4a [DOI] [PubMed] [Google Scholar]
- 23. Colford JM, Wade TJ, Schiff KC, Wright CC, Griffith JF, Sandhu SK, Burns S, Sobsey M, Lovelace G, Weisberg SB. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27–35. doi: 10.1097/01.ede.0000249425.32990.b9 [DOI] [PubMed] [Google Scholar]
- 24. Fleisher JM, Fleming LE, Solo-Gabriele HM, Kish JK, Sinigalliano CD, Plano L, Elmir SM, Wang JD, Withum K, Shibata T, Gidley ML, Abdelzaher A, He G, Ortega C, Zhu X, Wright M, Hollenbeck J, Backer LC. 2010. The BEACHES study: health effects and exposures from non-point source microbial contaminants in subtropical recreational marine waters. Int J Epidemiol 39:1291–1298. doi: 10.1093/ije/dyq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griffith JF, Weisberg SB, Arnold BF, Cao Y, Schiff KC, Colford JM. 2016. Epidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Res 94:371–381. doi: 10.1016/j.watres.2016.02.036 [DOI] [PubMed] [Google Scholar]
- 26. Ahmed W, O’Dea C, Masters N, Kuballa A, Marinoni O, Katouli M. 2019. Marker genes of fecal indicator bacteria and potential pathogens in animal feces in subtropical catchments. Sci Total Environ 656:1427–1435. doi: 10.1016/j.scitotenv.2018.11.439 [DOI] [PubMed] [Google Scholar]
- 27. Ervin JS, Russell TL, Layton BA, Yamahara KM, Wang D, Sassoubre LM, Cao Y, Kelty CA, Sivaganesan M, Boehm AB, Holden PA, Weisberg SB, Shanks OC. 2013. Characterization of fecal concentrations in human and other animal sources by physical, culture-based, and quantitative real-time PCR methods. Water Res 47:6873–6882. doi: 10.1016/j.watres.2013.02.060 [DOI] [PubMed] [Google Scholar]
- 28. Layton BA, Walters SP, Boehm AB. 2009. Distribution and diversity of the enterococcal surface protein (ESP) gene in animal hosts and the Pacific coast environment. J Appl Microbiol 106:1521–1531. doi: 10.1111/j.1365-2672.2008.04113.x [DOI] [PubMed] [Google Scholar]
- 29. Layton BA, Walters SP, Lam LH, Boehm AB. 2010. Enterococcus species distribution among human and animal hosts using multiplex PCR. J Appl Microbiol 109:539–547. doi: 10.1111/j.1365-2672.2010.04675.x [DOI] [PubMed] [Google Scholar]
- 30. Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi: 10.1016/j.watres.2010.06.049 [DOI] [PubMed] [Google Scholar]
- 31. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ Sci Technol 44:2286–2291. doi: 10.1021/es903523q [DOI] [PubMed] [Google Scholar]
- 32. Collado L, Figueras MJ. 2011. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter . Clin Microbiol Rev 24:174–192. doi: 10.1128/CMR.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. European Food Safety Authority, European Centre for Disease Prevention and Control . 2021. The European Union one health 2019 zoonoses report. EFS2 19. doi: 10.2903/j.efsa.2021.6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui Q, Huang Y, Wang H, Fang T. 2019. Diversity and abundance of bacterial pathogens in urban rivers impacted by domestic sewage. Environ Pollut 249:24–35. doi: 10.1016/j.envpol.2019.02.094 [DOI] [PubMed] [Google Scholar]
- 35. Fisher JC, Levican A, Figueras MJ, McLellan SL. 2014. Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 5:525. doi: 10.3389/fmicb.2014.00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu X, Zhang X-X, Wang Z, Huang K, Wang Y, Liang W, Tan Y, Liu B, Tang J. 2015. Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One 10:e0125549. doi: 10.1145/2818302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L, Cheng Y, Qian C, Lu W. 2020. Bacterial community evolution along full-scale municipal wastewater treatment processes. J Water Health 18:665–680. doi: 10.2166/wh.2020.092 [DOI] [PubMed] [Google Scholar]
- 38. Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538. doi: 10.1016/j.watres.2007.06.056 [DOI] [PubMed] [Google Scholar]
- 39. Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031 [DOI] [PubMed] [Google Scholar]
- 40. Sidhu JPS, Ahmed W, Gernjak W, Aryal R, McCarthy D, Palmer A, Kolotelo P, Toze S. 2013. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ 463–464:488–496. doi: 10.1016/j.scitotenv.2013.06.020 [DOI] [PubMed] [Google Scholar]
- 41. Van De Werfhorst LC, Murray JLS, Reynolds S, Reynolds K, Holden PA. 2014. Canine scent detection and microbial source tracking of human waste contamination in storm drains. Water Environ Res 86:550–558. doi: 10.2175/106143013x13807328848496 [DOI] [PubMed] [Google Scholar]
- 42. Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. 2010. Evaluation of genetic markers from the 16s rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst Appl Microbiol 33:348–357. doi: 10.1016/j.syapm.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 43. Ahmed W, Hamilton K, Toze S, Cook S, Page D. 2019. A review on microbial Contaminants in stormwater runoff and outfalls: potential health risks and mitigation strategies. Sci Total Environ 692:1304–1321. doi: 10.1016/j.scitotenv.2019.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hart JD, Blackwood AD, Noble RT. 2020. Examining coastal dynamics and recreational water quality by quantifying multiple sewage specific markers in a North Carolina estuary. Sci Total Environ 747:141124. doi: 10.1016/j.scitotenv.2020.141124 [DOI] [PubMed] [Google Scholar]
- 45. Steele JA, Blackwood AD, Griffith JF, Noble RT, Schiff KC. 2018. Quantification of pathogens and markers of fecal contamination during storm events along popular surfing beaches in San Diego, California. Water Res 136:137–149. doi: 10.1016/j.watres.2018.01.056 [DOI] [PubMed] [Google Scholar]
- 46. Williams NLR, Siboni N, Potts J, Campey M, Johnson C, Rao S, Bramucci A, Scanes P, Seymour JR. 2022. Molecular microbiological approaches reduce ambiguity about the sources of faecal pollution and identify microbial hazards within an urbanised coastal environment. Water Res 218:118534. doi: 10.1016/j.watres.2022.118534 [DOI] [PubMed] [Google Scholar]
- 47. Ahmed W, Toze S, Veal C, Fisher P, Zhang Q, Zhu Z, Staley C, Sadowsky MJ. 2021. Comparative decay of culturable faecal indicator bacteria, microbial source tracking marker genes, and enteric pathogens in laboratory microcosms that mimic a sub-tropical environment. Sci Total Environ 751:141475. doi: 10.1016/j.scitotenv.2020.141475 [DOI] [PubMed] [Google Scholar]
- 48. Mayer RE, Reischer GH, Ixenmaier SK, Derx J, Blaschke AP, Ebdon JE, Linke R, Egle L, Ahmed W, Blanch AR, et al. 2018. Global distribution of human-associated fecal genetic markers in reference samples from six continents. Environ Sci Technol 52:5076–5084. doi: 10.1021/acs.est.7b04438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gonzalez D, Keeling D, Thompson H, Larson A, Denby J, Curtis K, Yetka K, Rondini M, Yeargan E, Egerton T, Barker D, Gonzalez R. 2020. Collection system investigation microbial source tracking (CSI-MST): applying molecular markers to identify sewer infrastructure failures. J Microbiol Methods 178:106068. doi: 10.1016/j.mimet.2020.106068 [DOI] [PubMed] [Google Scholar]
- 50. Alberta Health Services . 2021. Public health guidelines for water reuse and stormwater use. Available from: https://open.alberta.ca/dataset/6a57d29c-d437-4dd9-94e3-d96bedc01bb4/resource/d533afcb-2933-43da-9199-eea030148c00/download/health-public-health-guidelines-water-reuse-stormwater-use-2021. Retrieved 3 Apr 2024.
- 51. National Academies of Sciences, Engineering, and Medicine (NASEM) . 2016. Using graywater and stormwater to enhance local water supplies: an assessment of risks, costs, and benefits. National Academies Press, Washington, D.C [Google Scholar]
- 52. Natural Resource Management Ministerial Council (NRMMC), Environment Protection and Heritage Council (EPHC), National Health and Medical Research Council (NHMRC) . 2009. Australian guidelines for water recycling: Managing health and environmental risks (phase 2) stormwater harvesting and reuse
- 53. San Francisco Public Utilities Commission (SFPUC) . 2020. Onsite water reuse program guidebook. Available from: https://sfpuc.org/sites/default/files/documents/OnsiteWaterReuseGuidebook2020.pdf. Retrieved 3 Apr 2024.
- 54. Haas CN, Rose JB, Gerba CP. 2014. Quantitative microbial risk assessment. 2nd ed. John Wiley & Sons, Inc, New York. [Google Scholar]
- 55. World Health Organization . 2016. Quantitative microbial risk assessment: application for water safety management. World Health Organization, Geneva. Available from: https://www.who.int/publications/i/item/9789241565370. Retrieved 3 Apr 2024. [Google Scholar]
- 56. Ahmed W, Hamilton KA, Lobos A, Hughes B, Staley C, Sadowsky MJ, Harwood VJ. 2018. Quantitative microbial risk assessment of microbial source tracking markers in recreational water contaminated with fresh untreated and secondary treated sewage. Environ Int 117:243–249. doi: 10.1016/j.envint.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 57. Boehm AB, Graham KE, Jennings WC. 2018. Can we swim yet? systematic review, meta-analysis, and risk assessment of aging sewage in surface waters. Environ Sci Technol 52:9634–9645. doi: 10.1021/acs.est.8b01948 [DOI] [PubMed] [Google Scholar]
- 58. Brown KI, Graham KE, Soller JA, Boehm AB. 2017. Estimating the probability of illness due to swimming in recreational water with a mixture of human- and gull-associated microbial source tracking markers. Environ Sci Proc Impacts 19:1528–1541. doi: 10.1039/C7EM00316A [DOI] [PubMed] [Google Scholar]
- 59. United States Environmental Protection Agency (US EPA) . 2012. Recreational water quality criteria (820-F-12-058). Available from: https://www.epa.gov/sites/default/files/2015-10/documents/rwqc2012.pdf. Retrieved 3 Apr 2024.
- 60. Ahn JH, Grant SB, Surbeck CQ, Jiang S, DiGiacomo PM, Nezlin NP.. 2008. Urban runoff impact study phase III: size distribution, sources, and transport of suspended particles along an inland-to-ocean transect. National Water Res Institute, Fountain Valley, California [Google Scholar]
- 61. Ahmed W, Hughes B, Harwood V. 2016. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231. doi: 10.3390/w8060231 [DOI] [Google Scholar]
- 62. Boehm AB, Van De Werfhorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, Wang D, Weisberg SB. 2013. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res. 47:6812–6828. doi: 10.1016/j.watres.2012.12.046 [DOI] [PubMed] [Google Scholar]
- 63. Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG, Sivaganesan M. 2016. Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Appl Environ Microbiol 82:2773–2782. doi: 10.1128/AEM.03661-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schriewer A, Miller WA, Byrne BA, Miller MA, Oates S, Conrad PA, Hardin D, Yang H-H, Chouicha N, Melli A, Jessup D, Dominik C, Wuertz S. 2010. Presence of Bacteroidales as a predictor of pathogens in surface waters of the central California coast. Appl Environ Microbiol 76:5802–5814. doi: 10.1128/AEM.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl Environ Microbiol 76:3255–3262. doi: 10.1128/AEM.02636-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walters SP, Field KG. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ Microbiol 11:1410–1421. doi: 10.1111/j.1462-2920.2009.01868.x [DOI] [PubMed] [Google Scholar]
- 67. Carney RL, Brown MV, Siboni N, Raina J-B, Kahlke T, Mitrovic SM, Seymour JR. 2020. Highly heterogeneous temporal dynamics in the abundance and diversity of the emerging pathogens Arcobacter at an urban beach. Water Res 171:115405. doi: 10.1016/j.watres.2019.115405 [DOI] [PubMed] [Google Scholar]
- 68. Collado L, Inza I, Guarro J, Figueras MJ. 2008. Presence of Arcobacter spp. In environmental waters correlates with high levels of fecal pollution: correlation of Arcobacter with fecal pollution. Environ Microbiol 10:1635–1640. [DOI] [PubMed] [Google Scholar]
- 69. Webb AL, Taboada EN, Selinger LB, Boras VF, Inglis GD. 2017. Prevalence and diversity of waterborne Arcobacter butzleri in southwestern Alberta, Canada. Can J Microbiol 63:330–340. doi: 10.1139/cjm-2016-0745 [DOI] [PubMed] [Google Scholar]
- 70. McGinnis S, Spencer S, Firnstahl A, Stokdyk J, Borchardt M, McCarthy DT, Murphy HM. 2018. Human Bacteroides and total coliforms as indicators of recent combined sewer overflows and rain events in urban creeks. Sci Total Environ 630:967–976. doi: 10.1016/j.scitotenv.2018.02.108 [DOI] [PubMed] [Google Scholar]
- 71. Beaudry M. 2019. MSc Thesis. From nuisance to resource: understanding microbial sources of contamination in urban stormwater-impacted bodies of water intended for water reuse activities. University of Alberta, Edmonton. Available from: https://era.library.ualberta.ca/items/6f834aa3-56d3-4829-8c9e-5b294d95b11c. Retrieved Apr 3 Apr 2024. [Google Scholar]
- 72. Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl Environ Microbiol 75:5507–5513. doi: 10.1128/AEM.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ludwig W, Schleifer K-H. 2000. How quantitative is quantitative PCR with respect to cell counts?. Syst Appl Microbiol 23:556–562. doi: 10.1016/S0723-2020(00)80030-2 [DOI] [PubMed] [Google Scholar]
- 74. United States Environmental Protection Agency (US EPA) . 2012. Method 1611: enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) Assay (EPA-821-R-12-008). Available from: https://www.epa.gov/sites/default/files/2015-08/documents/method_1611_2012.pdf. Retrieved 3 Apr 2024.
- 75. de Boer RF, Ott A, Güren P, van Zanten E, van Belkum A, Kooistra-Smid AMD. 2013. Detection of Campylobacter species and Arcobacter butzleri in stool samples by use of real-time multiplex PCR. J Clin Microbiol 51:253–259. doi: 10.1128/JCM.01716-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Van Dyke MI, Morton VK, McLellan NL, Huck PM. 2010. The occurrence of Campylobacter in river water and waterfowl within a watershed in Southern Ontario, Canada. J Appl Microbiol 109:1053–1066. doi: 10.1111/j.1365-2672.2010.04730.x [DOI] [PubMed] [Google Scholar]
- 77. Daum LT, Barnes WJ, McAvin JC, Neidert MS, Cooper LA, Huff WB, Gaul L, Riggins WS, Morris S, Salmen A, Lohman KL. 2002. Real-time PCR detection of Salmonella in suspect foods from a gastroenteritis outbreak in Kerr County, Texas. J Clin Microbiol 40:3050–3052. doi: 10.1128/JCM.40.8.3050-3052.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chui L, Lee M-C, Allen R, Bryks A, Haines L, Boras V. 2013. Comparison between immunoCard STAT!® and real-time PCR as screening tools for both O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in Southern Alberta, Canada. Diagn Microbiol Infect Dis 77:8–13. doi: 10.1016/j.diagmicrobio.2013.05.015 [DOI] [PubMed] [Google Scholar]
- 79. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 39:559–568. doi: 10.1016/j.watres.2004.11.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1; Tables S1 to S3.