Abstract
The NADH-dependent morphinone reductase from Pseudomonas putida M10 catalyses the reduction of morphinone and codeinone to hydromorphone and hydrocodone respectively. Morphinone reductase was purified from crude cell extracts to apparent homogeneity in a single affinity-chromatography step using Mimetic Yellow 2. The purified enzyme was a dimeric flavoprotein with two identical subunits of M(r) 41,100, binding non-covalently one molecule of FMN per subunit. The N-terminal sequence was PDTSFSNPGLFTPLQ. Morphinone reductase was active against morphinone, codeinone, neopinone and 2-cyclohexen-1-one, but not against morphine, codeine or isocodeine. The apparent Km values for codeinone and 2-cyclohexen-1-one were 0.26 mM and 5.5 mM respectively. The steroids progesterone and cortisone were potent competitive inhibitors; the apparent K1 for cortisone was 35 microM. The pH optimum for codeinone reduction was 8.0 in phosphate buffer. No reverse reaction could be detected, and NADPH could not be used as a reducing substrate in place of NADH. Morphinone reductase activity was strongly inhibited by 0.01 mM CuSO4 and p-hydroxymercuribenzoate, suggesting the presence of a vital thiol group. Steady-state kinetic studies suggested a Ping Pong (substituted enzyme) kinetic mechanism; however, product-inhibition patterns were inconsistent with a classical Ping Pong mechanism. Morphinone reductase may, like several other flavoprotein dehydrogenases, operate by a hybrid two-site Ping Pong mechanism.
Full text
PDF


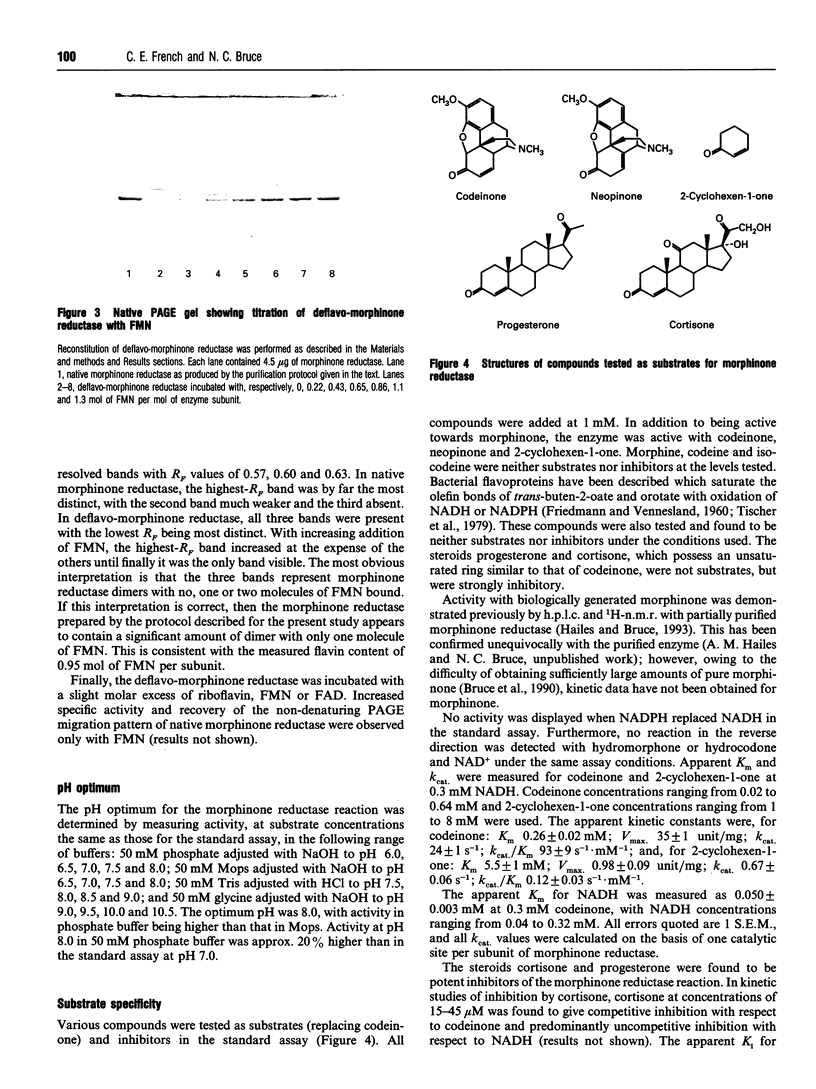
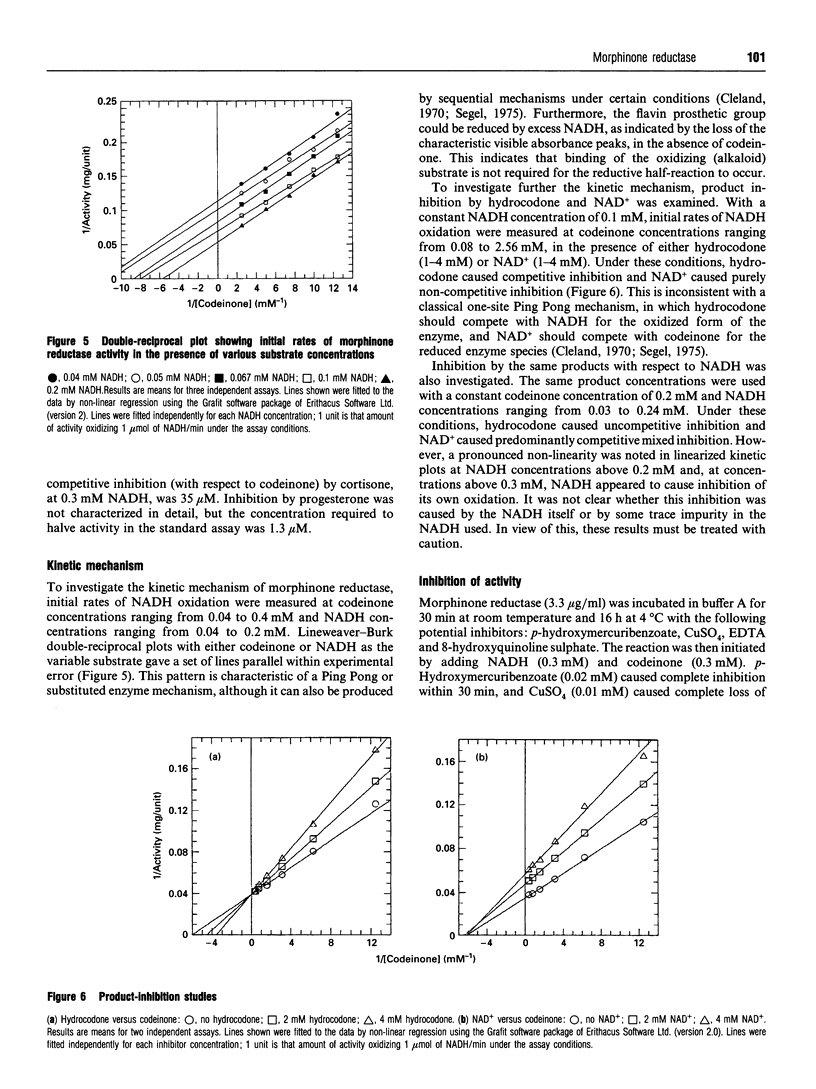


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bruce N. C., Wilmot C. J., Jordan K. N., Trebilcock A. E., Gray Stephens L. D., Lowe C. R. Microbial degradation of the morphine alkaloids: identification of morphine as an intermediate in the metabolism of morphine by Pseudomonas putida M10. Arch Microbiol. 1990;154(5):465–470. doi: 10.1007/BF00245229. [DOI] [PubMed] [Google Scholar]
- Coughlan M. P., Rajagopalan K. V. The kinetic mechanism of xanthine dehydrogenase and related enzymes. Eur J Biochem. 1980 Mar;105(1):81–84. doi: 10.1111/j.1432-1033.1980.tb04476.x. [DOI] [PubMed] [Google Scholar]
- FRIEDMANN H. C., VENNESLAND B. Crystalline dihydroorotic dehydrogenase. J Biol Chem. 1960 May;235:1526–1532. [PubMed] [Google Scholar]
- Hailes A. M., Bruce N. C. Biological synthesis of the analgesic hydromorphone, an intermediate in the metabolism of morphine, by Pseudomonas putida M10. Appl Environ Microbiol. 1993 Jul;59(7):2166–2170. doi: 10.1128/aem.59.7.2166-2170.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines V., Johnston M. Analysis of the kinetic mechanism of the bovine liver mitochondrial dihydroorotate dehydrogenase. Biochemistry. 1989 Feb 7;28(3):1222–1226. doi: 10.1021/bi00429a040. [DOI] [PubMed] [Google Scholar]
- Husain M., Massey V. Reversible resolution of flavoproteins into apoproteins and fee flavins. Methods Enzymol. 1978;53:429–437. doi: 10.1016/s0076-6879(78)53047-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Rendina A. R., Orme-Johnson W. H. Glutamate synthase: on the kinetic mechanism of the enzyme from Escherichia coli W. Biochemistry. 1978 Dec 12;17(25):5388–5393. doi: 10.1021/bi00618a011. [DOI] [PubMed] [Google Scholar]
- Renosto F., Ornitz D. M., Peterson D., Segel I. H. Nitrate reductase from Penicillium chrysogenum. Purification and kinetic mechanism. J Biol Chem. 1981 Aug 25;256(16):8616–8625. [PubMed] [Google Scholar]
- Stokes N. A., Hylemon P. B. Characterization of delta 4-3-ketosteroid-5 beta-reductase and 3 beta-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim Biophys Acta. 1985 Sep 11;836(2):255–261. doi: 10.1016/0005-2760(85)90073-6. [DOI] [PubMed] [Google Scholar]
- Tischer W., Bader J., Simon H. Purification and some properties of a hitherto-unknown enzyme reducing the carbon-carbon double bond of alpha, beta-unsaturated carboxylate anions. Eur J Biochem. 1979 Jun;97(1):103–112. doi: 10.1111/j.1432-1033.1979.tb13090.x. [DOI] [PubMed] [Google Scholar]
- Willey D. L., Caswell D. A., Lowe C. R., Bruce N. C. Nucleotide sequence and over-expression of morphine dehydrogenase, a plasmid-encoded gene from Pseudomonas putida M10. Biochem J. 1993 Mar 1;290(Pt 2):539–544. doi: 10.1042/bj2900539. [DOI] [PMC free article] [PubMed] [Google Scholar]