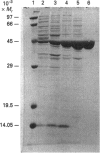
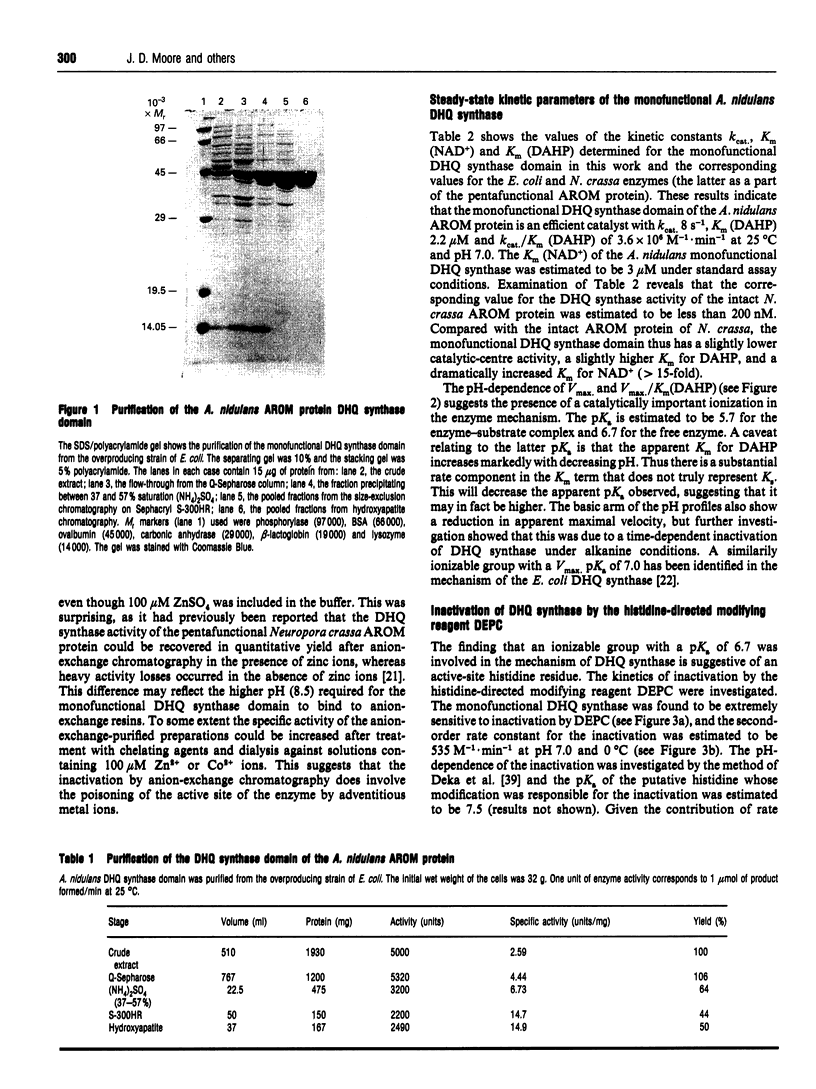
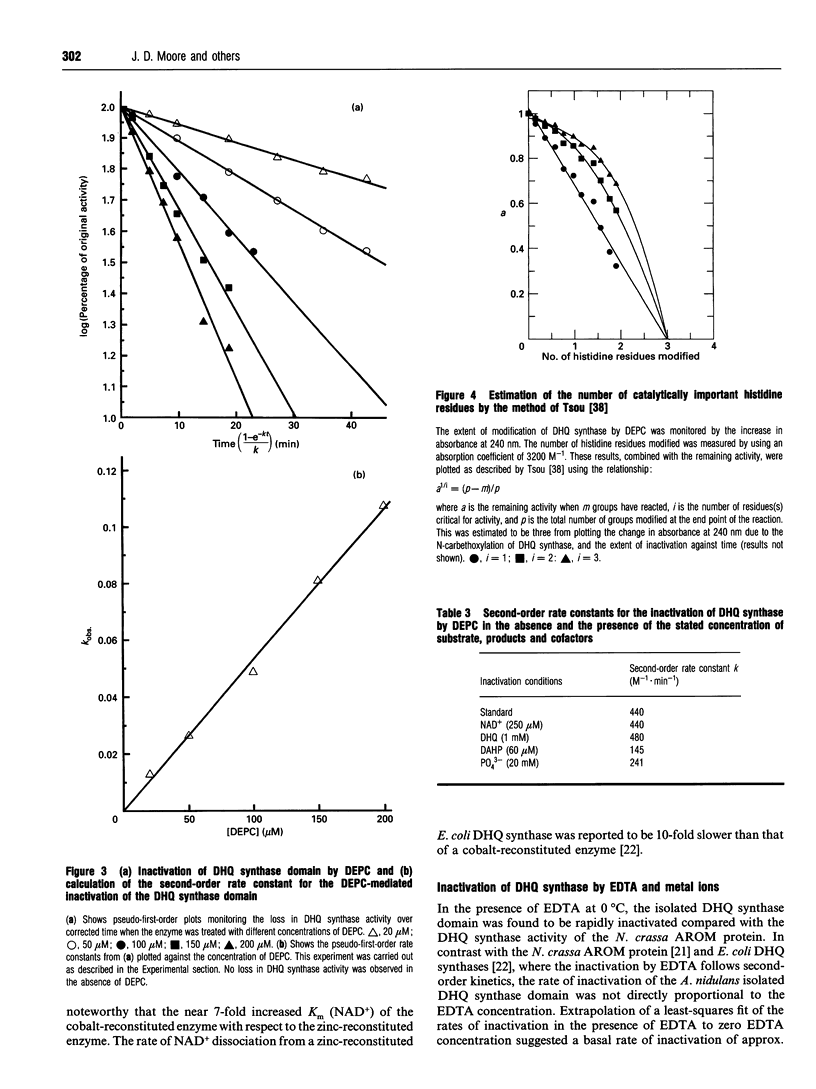
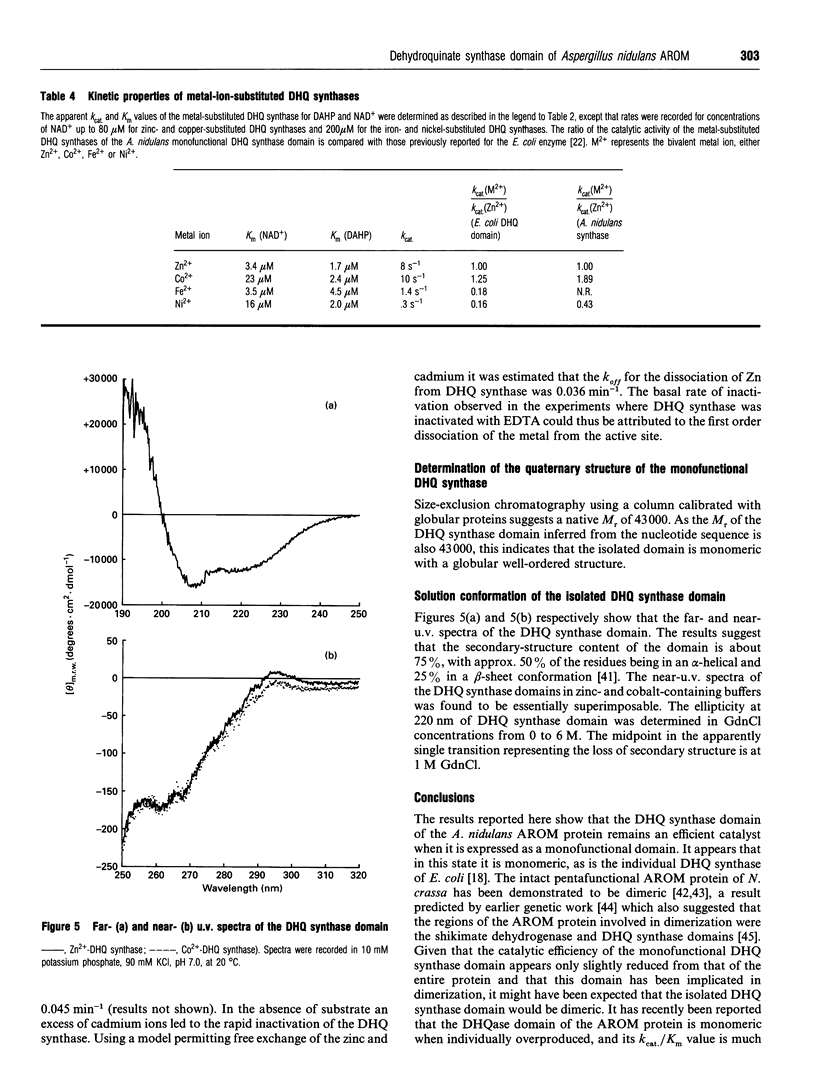
Abstract
The dehydroquinate synthase (DHQ synthase) functional domain from the pentafunctional AROM protein of Aspergillus nidulans has previously been overproduced in Escherichia coli [van den Hombergh, Moore, Charles and Hawkins (1992) Biochem J. 284, 861-867]. We now report the purification of this domain to homogeneity and subsequent characterization. The monofunctional DHQ synthase was found to retain efficient catalytic activity when compared with the intact pentafunctional AROM protein of Neurospora crassa [Lambert, Boocock and Coggins (1985) Biochem J. 226, 817-829]. The apparent kcat. was estimated to be 8 s-1, and the apparent Km values for NAD+ and 3-deoxy-D-arabino-heptulosonate phosphate (DAHP) were 3 microM and 2.2 microM respectively. These values are similar to those reported for the intact N. crassa enzyme, except that the apparent Km for NAD+ reported here is 15-fold higher. The monofunctional DHQ synthase domain is inactivated by treatment with chelating agents in the absence of substrates and is re-activated by the addition of metal ions; among those tested, Zn2+ gave the highest kcat./Km value. The enzyme is inactivated by diethyl pyrocarbonate; both the substrate, DAHP, and the product phosphate protected against inactivation. Size-exclusion chromatography suggested an M(r) of 43,000 for the monofunctional domain, indicating that it is monomeric and compactly folded. The c.d. spectrum confirmed that the domain has a compact globular conformation; the near-u.v. c.d. of zinc- and cobalt-reactivated domains were superimposable.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton I. A., Duncan K., Coggins J. R. A eukaryotic repressor protein, the qa-1S gene product of Neurospora crassa, is homologous to part of the arom multifunctional enzyme. J Mol Biol. 1987 Sep 20;197(2):367–371. doi: 10.1016/0022-2836(87)90130-6. [DOI] [PubMed] [Google Scholar]
- Bender S. L., Mehdi S., Knowles J. R. Dehydroquinate synthase: the role of divalent metal cations and of nicotinamide adenine dinucleotide in catalysis. Biochemistry. 1989 Sep 19;28(19):7555–7560. doi: 10.1021/bi00445a009. [DOI] [PubMed] [Google Scholar]
- Bentley R. The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25(5):307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- Berlyn M. B., Ahmed S. I., Giles N. H. Organization of polyaromatic biosynthetic enzymes in a variety of photosynthetic organisms. J Bacteriol. 1970 Nov;104(2):768–774. doi: 10.1128/jb.104.2.768-774.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E., Burgoyne L., Giles N. H. In vivo and in vitro complementation between DHQ synthetase mutants in the arom gene cluster of Neurospora crassa. Genetics. 1969 Nov;63(3):581–588. doi: 10.1093/genetics/63.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Lambert J. M., McColl L. A., Coggins J. R. Purification and characterization of 3-dehydroquinase from Escherichia coli. Biochem J. 1986 Nov 1;239(3):699–704. doi: 10.1042/bj2390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Campbell M. S., Chaudhuri S., Lambert J. M., Lewendon A., Mousdale D. M., Smith D. D. Functional domains involved in aromatic amino acid biosynthesis. Biochem Soc Trans. 1985 Apr;13(2):299–303. doi: 10.1042/bst0130299. [DOI] [PubMed] [Google Scholar]
- Deka R. K., Kleanthous C., Coggins J. R. Identification of the essential histidine residue at the active site of Escherichia coli dehydroquinase. J Biol Chem. 1992 Nov 5;267(31):22237–22242. [PubMed] [Google Scholar]
- Duncan K., Edwards R. M., Coggins J. R. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J. 1987 Sep 1;246(2):375–386. doi: 10.1042/bj2460375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. R., Bein K., Guy H. I., Liu X., Molina J. A., Zimmermann B. H. CAD gene sequence and the domain structure of the mammalian multifunctional protein CAD. Biochem Soc Trans. 1993 Feb;21(1):186–191. doi: 10.1042/bst0210186. [DOI] [PubMed] [Google Scholar]
- Frost J. W., Bender J. L., Kadonaga J. T., Knowles J. R. Dehydroquinate synthase from Escherichia coli: purification, cloning, and construction of overproducers of the enzyme. Biochemistry. 1984 Sep 11;23(19):4470–4475. doi: 10.1021/bi00314a036. [DOI] [PubMed] [Google Scholar]
- Garbe T., Servos S., Hawkins A., Dimitriadis G., Young D., Dougan G., Charles I. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol Gen Genet. 1991 Sep;228(3):385–392. doi: 10.1007/BF00260631. [DOI] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Partridge C. W., Ahmed S. I. A gene cluster in Nuerospora crassa coding for an aggregate of five aromatic synthetic enzymes. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1453–1460. doi: 10.1073/pnas.58.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Moore J. D., Roberts C. F. Genesis of eukaryotic transcriptional activator and repressor proteins by splitting a multidomain anabolic enzyme. Gene. 1993 Dec 22;136(1-2):49–54. doi: 10.1016/0378-1119(93)90446-a. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Roberts C. F. Structure of the Aspergillus nidulans qut repressor-encoding gene: implications for the regulation of transcription initiation. Gene. 1992 Jan 2;110(1):109–114. doi: 10.1016/0378-1119(92)90452-u. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Smith M., Keyte J. W., Roberts C. F. Molecular organisation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. Mol Gen Genet. 1988 Oct;214(2):224–231. doi: 10.1007/BF00337715. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R. The complex Arom locus of Aspergillus nidulans. Evidence for multiple gene fusions and convergent evolution. Curr Genet. 1987;11(6-7):491–498. doi: 10.1007/BF00384611. [DOI] [PubMed] [Google Scholar]
- Kishore G. M., Shah D. M. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- Koerber S. C., Fink A. L. The analysis of enzyme progress curves by numerical differentiation, including competitive product inhibition and enzyme reactivation. Anal Biochem. 1987 Aug 15;165(1):75–87. doi: 10.1016/0003-2697(87)90203-x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lambert J. M., Boocock M. R., Coggins J. R. The 3-dehydroquinate synthase activity of the pentafunctional arom enzyme complex of Neurospora crassa is Zn2+-dependent. Biochem J. 1985 Mar 15;226(3):817–829. doi: 10.1042/bj2260817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. Evidence from peptide 'maps' for the identity of the subunits. Biochem J. 1978 Feb 1;169(2):441–444. doi: 10.1042/bj1690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes H. W., Zenke W. M., Grundström T., Staub A., Wintzerith M., Chambon P. Simultaneous rapid chemical synthesis of over one hundred oligonucleotides on a microscale. EMBO J. 1984 Apr;3(4):801–805. doi: 10.1002/j.1460-2075.1984.tb01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. D., Hawkins A. R. Overproduction of, and interaction within, bifunctional domains from the amino- and carboxy-termini of the pentafunctional AROM protein of Aspergillus nidulans. Mol Gen Genet. 1993 Jul;240(1):92–102. doi: 10.1007/BF00276888. [DOI] [PubMed] [Google Scholar]
- Moore J. D., Lamb H. K., Garbe T., Servos S., Dougan G., Charles I. G., Hawkins A. R. Inducible overproduction of the Aspergillus nidulans pentafunctional AROM protein and the type-I and -II 3-dehydroquinases from Salmonella typhi and Mycobacterium tuberculosis. Biochem J. 1992 Oct 1;287(Pt 1):173–181. doi: 10.1042/bj2870173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966 Apr;91(4):1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher S. W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981 Jan 6;20(1):33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- SRINIVASAN P. R., ROTHSCHILD J., SPRINSON D. B. THE ENZYMIC CONVERSION OF 3-DEOXY-D-ARABINO-HEPTULOSONIC ACID 7-PHOSPHATE TO 5-DEHYDROQUINATE. J Biol Chem. 1963 Oct;238:3176–3182. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproat B. S., Gait M. J. Chemical synthesis of a gene for somatomedin C. Nucleic Acids Res. 1985 Apr 25;13(8):2959–2977. doi: 10.1093/nar/13.8.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L., Jackson T. R., Hawkins P. T. Activation of phosphatidylinositol 4,5-bisphosphate supply by agonists and non-hydrolysable GTP analogues. Biochem J. 1993 Dec 1;296(Pt 2):481–488. doi: 10.1042/bj2960481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSOU C. L. Relation between modification of functional groups of proteins and their biological activity. I.A graphical method for the determination of the number and type of essential groups. Sci Sin. 1962 Nov;11:1535–1558. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van den Hombergh J. P., Moore J. D., Charles I. G., Hawkins A. R. Overproduction in Escherichia coli of the dehydroquinate synthase domain of the Aspergillus nidulans pentafunctional AROM protein. Biochem J. 1992 Jun 15;284(Pt 3):861–867. [PMC free article] [PubMed] [Google Scholar]