Abstract
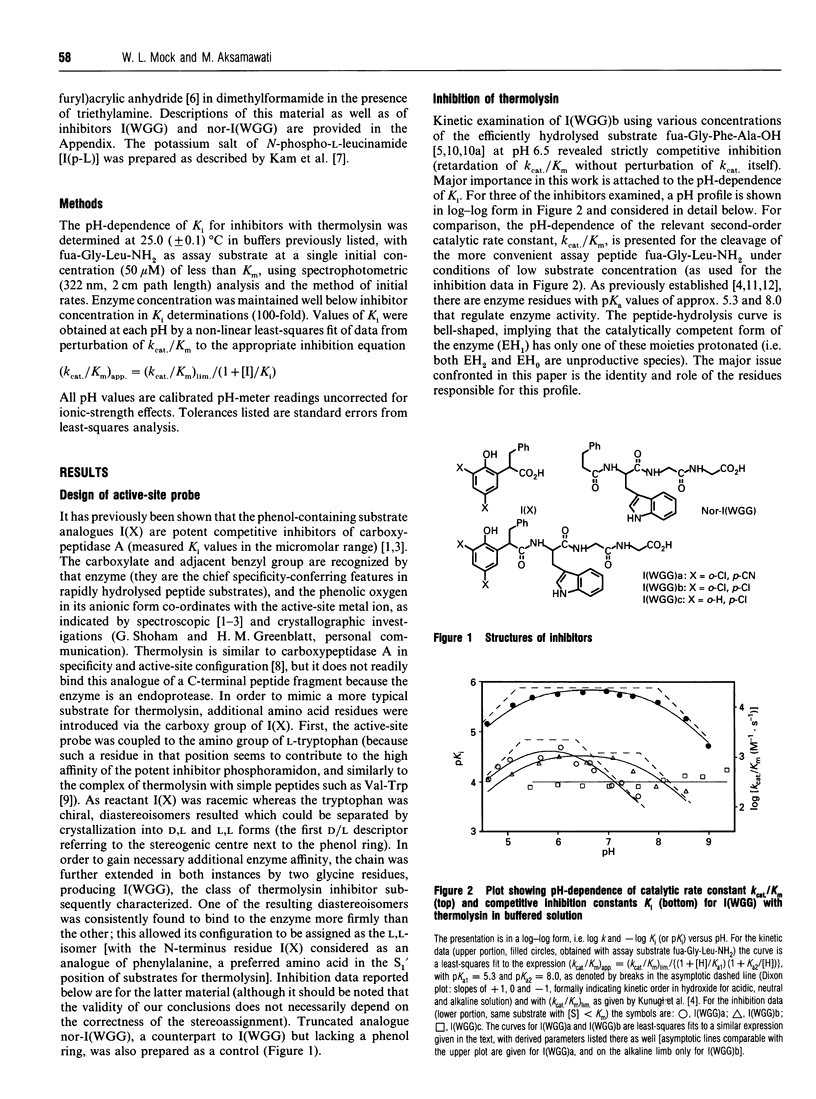
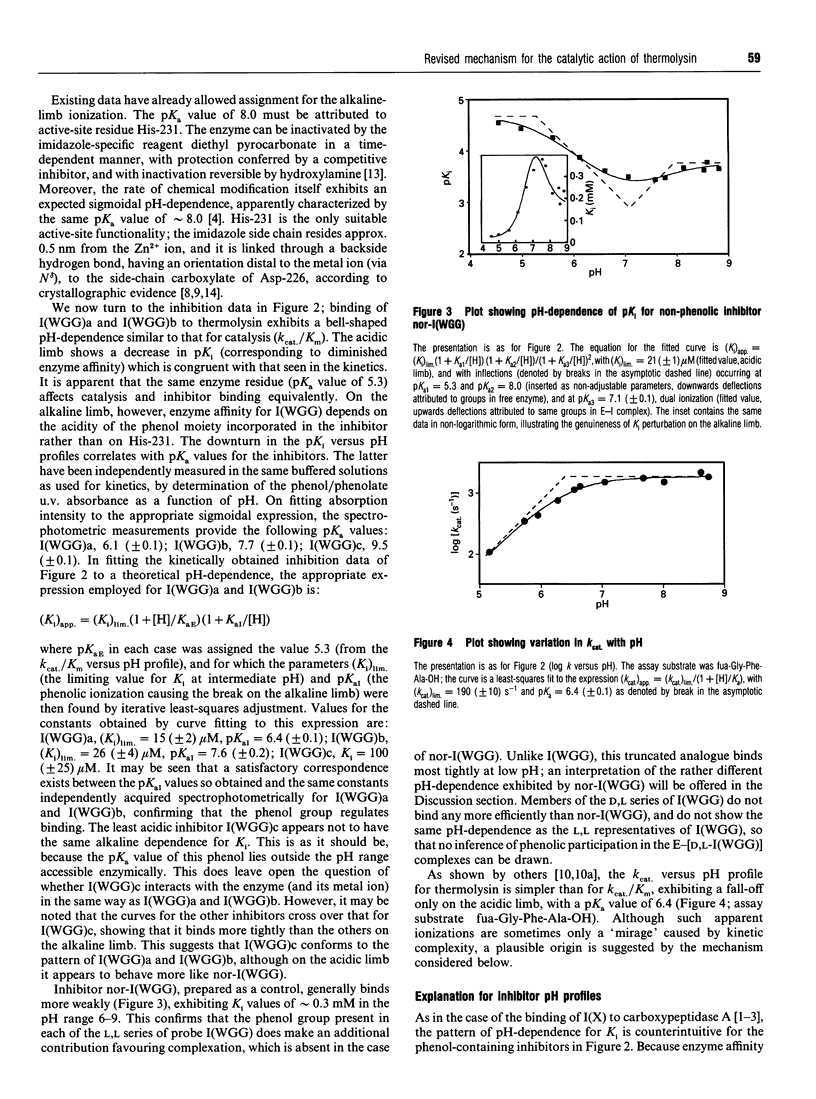
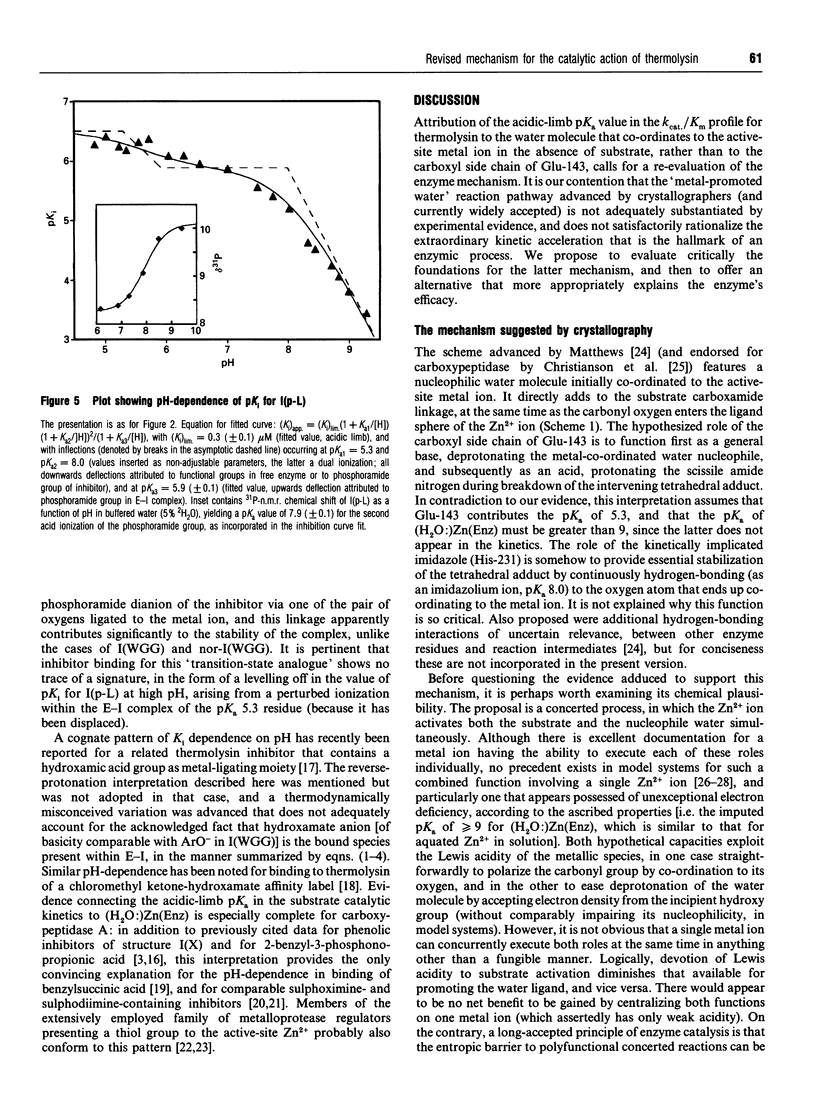
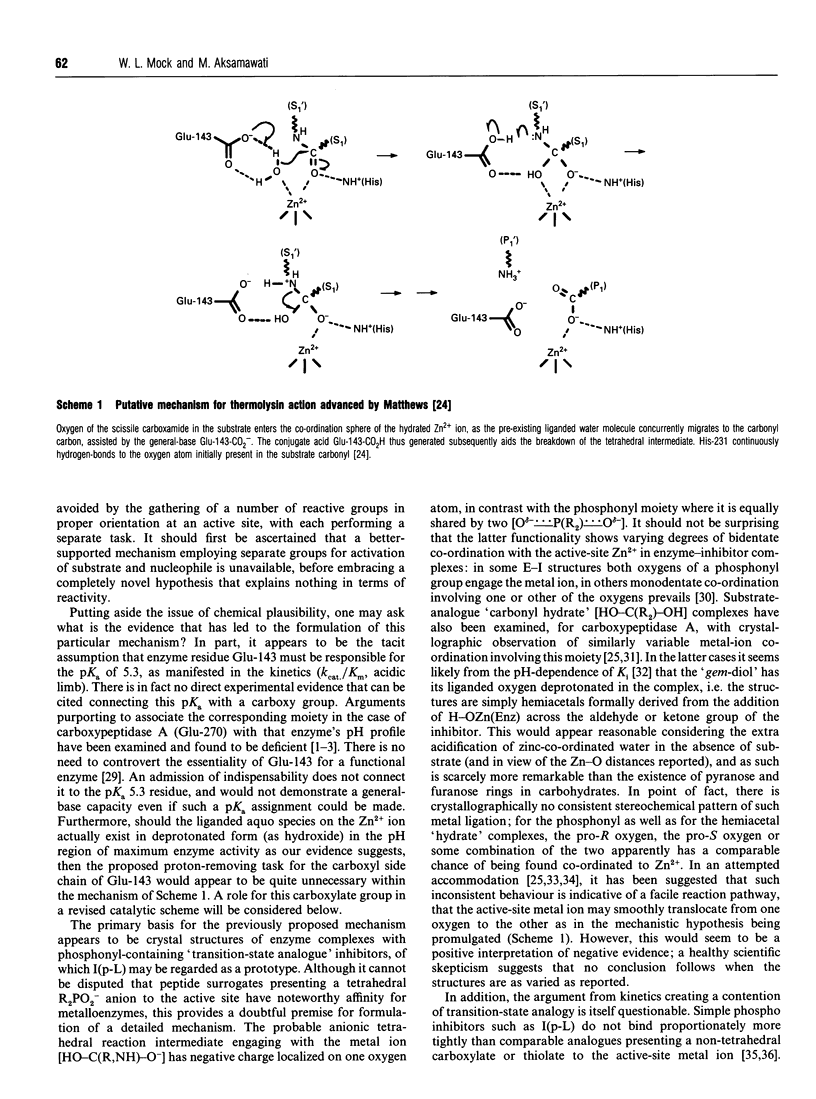
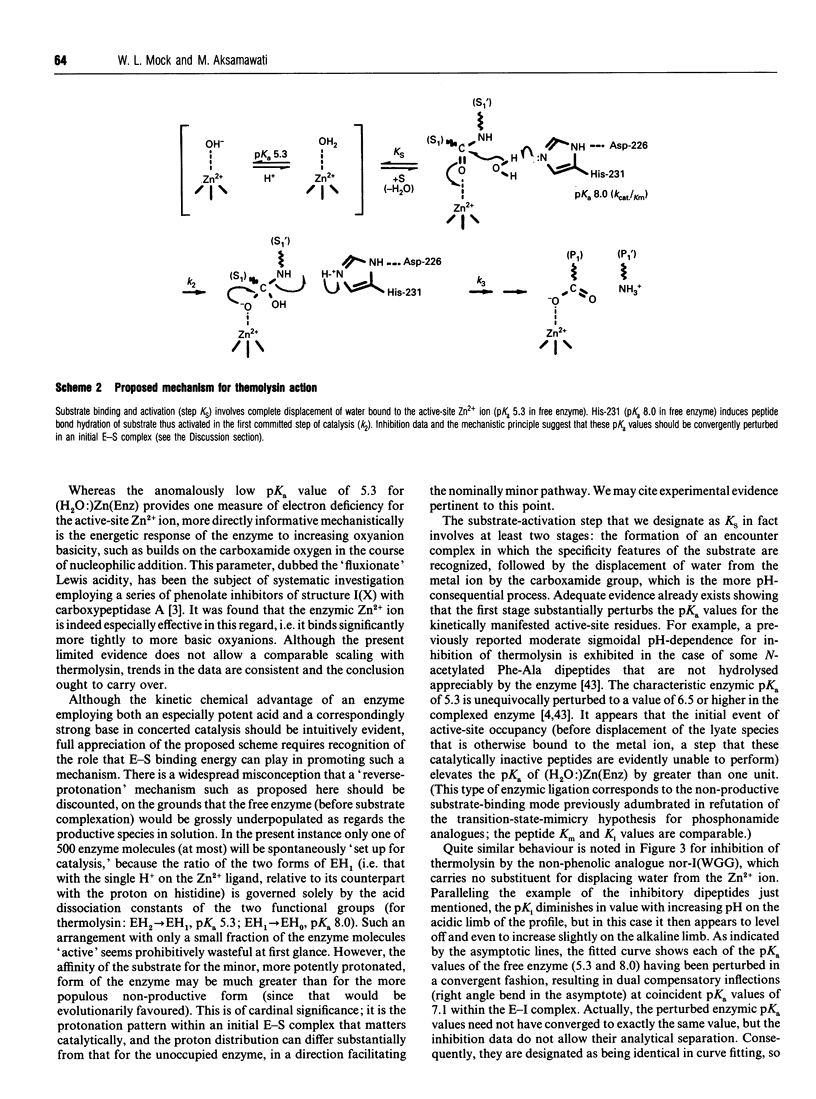
Competitive inhibition as a function of pH for the metalloendoprotease thermolysin by derivatives of L-alpha-(2-hydroxyphenyl)benzenepropanoyl-L- tryptophanylglycylglycine exhibits a diagnostic bell shape. Binding is maximal between two pKa values: on the acidic limb the apparent Ki value is regulated by an unchanging enzymic ionization (pKa 5.3) which is also seen in the substrate-hydrolysis kinetics (kcat/Km), whereas the alkaline limb for inhibition varies and depends specifically on the pKa of the phenolic group in the inhibitor. Although it should be the phenolate form of the inhibitor that co-ordinates more efficiently to the active-site Zn2+, the apparent Ki shifts from pH-independent at pH values immediately below the inhibitor's pKa to progressively weaker binding at higher pH. This is explained by an anomalous acidity for the exchangeable solvent molecule that is attached to enzymic Zn2+ in the absence of substrate or inhibitor. Since OH- cannot be displaced from the enzyme as readily as H2O, a compensating pKa of 5.3 possessed by Zn(2+)-bound water rationalizes the binding characteristics, yielding the level pH profile exhibited at intermediate pH values. Recognition of the implicit heightened Lewis acidity of the metal ion in thermolysin leads to a revision of the mechanism of catalysis. The substrate amide bond becomes activated for hydrolysis by carbonyl-group co-ordination to the especially acidic Zn2+ ion (completely displacing the H2O/OH- species otherwise bound). The imidazole group of enzymic residue His-231, also discerned in the pH profile for kcat/Km from its pKa of 8, provides general-base assistance for hydration of the activated scissile linkage in the first committed step of catalysis. Additional evidence from inhibition patterns shows how substrate-binding energy may be employed in this scheme to promote hydrolysis of peptides by thermolysin.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartlett P. A., Marlowe C. K. Phosphonamidates as transition-state analogue inhibitors of thermolysin. Biochemistry. 1983 Sep 27;22(20):4618–4624. doi: 10.1021/bi00289a002. [DOI] [PubMed] [Google Scholar]
- Bateman R. C., Jr, Rodriguez G., Vijayaraghavan J., Hersh L. B. Effect of electron withdrawing substituents on substrate hydrolysis by and inhibition of rat neutral endopeptidase 24.11 (enkephalinase) and thermolysin. Arch Biochem Biophys. 1990 Jun;279(2):355–362. doi: 10.1016/0003-9861(90)90502-p. [DOI] [PubMed] [Google Scholar]
- Blumberg S., Tauber Z. Inhibition of metalloendopeptidases by 2-mercaptoacetyl-dipeptides. Eur J Biochem. 1983 Oct 17;136(1):151–154. doi: 10.1111/j.1432-1033.1983.tb07719.x. [DOI] [PubMed] [Google Scholar]
- Bode W., Gomis-Rüth F. X., Huber R., Zwilling R., Stöcker W. Structure of astacin and implications for activation of astacins and zinc-ligation of collagenases. Nature. 1992 Jul 9;358(6382):164–167. doi: 10.1038/358164a0. [DOI] [PubMed] [Google Scholar]
- Burstein Y., Walsh K. A., Neurath H. Evidence of an essential histidine residue in thermolysin. Biochemistry. 1974 Jan 1;13(1):205–210. doi: 10.1021/bi00698a030. [DOI] [PubMed] [Google Scholar]
- Byers L. D., Wolfenden R. Binding of the by-product analog benzylsuccinic acid by carboxypeptidase A. Biochemistry. 1973 May 22;12(11):2070–2078. doi: 10.1021/bi00735a008. [DOI] [PubMed] [Google Scholar]
- Christianson D. W., David P. R., Lipscomb W. N. Mechanism of carboxypeptidase A: hydration of a ketonic substrate analogue. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1512–1515. doi: 10.1073/pnas.84.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W. The use of pH studies to determine chemical mechanisms of enzyme-catalyzed reactions. Methods Enzymol. 1982;87:390–405. doi: 10.1016/s0076-6879(82)87024-9. [DOI] [PubMed] [Google Scholar]
- Feder J., Schuck J. M. Studies on the Bacillus subtilis neutral-protease- and Bacillus thermoproteolyticus thermolysin-catalyzed hydrolysis of dipeptide substrates. Biochemistry. 1970 Jul 7;9(14):2784–2791. doi: 10.1021/bi00816a005. [DOI] [PubMed] [Google Scholar]
- Goli U. B., Grobelny D., Galardy R. E. Ionization states of the complex formed between 2-benzyl-3-phosphonopropionic acid and carboxypeptidase A. Biochem J. 1988 Sep 15;254(3):847–853. doi: 10.1042/bj2540847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobelny D., Goli U. B., Galardy R. E. 3-Phosphonopropionic acids inhibit carboxypeptidase A as multisubstrate analogues or transition-state analogues. Biochem J. 1985 Nov 15;232(1):15–19. doi: 10.1042/bj2320015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer D. G., Monzingo A. F., Matthews B. W. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984 Nov 20;23(24):5730–5741. doi: 10.1021/bi00319a011. [DOI] [PubMed] [Google Scholar]
- Hanson J. E., Kaplan A. P., Bartlett P. A. Phosphonate analogues of carboxypeptidase A substrates are potent transition-state analogue inhibitors. Biochemistry. 1989 Jul 25;28(15):6294–6305. doi: 10.1021/bi00441a022. [DOI] [PubMed] [Google Scholar]
- Holden H. M., Matthews B. W. The binding of L-valyl-L-tryptophan to crystalline thermolysin illustrates the mode of interaction of a product of peptide hydrolysis. J Biol Chem. 1988 Mar 5;263(7):3256–3260. doi: 10.2210/pdb3tmn/pdb. [DOI] [PubMed] [Google Scholar]
- Holmquist B., Vallee B. L. Metal-coordinating substrate analogs as inhibitors of metalloenzymes. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6216–6220. doi: 10.1073/pnas.76.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. P. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Kam C. M., Nishino N., Powers J. C. Inhibition of thermolysin and carboxypeptidase A by phosphoramidates. Biochemistry. 1979 Jul 10;18(14):3032–3038. doi: 10.1021/bi00581a019. [DOI] [PubMed] [Google Scholar]
- Kester W. R., Matthews B. W. Comparison of the structures of carboxypeptidase A and thermolysin. J Biol Chem. 1977 Nov 10;252(21):7704–7710. [PubMed] [Google Scholar]
- Kunugi S., Hirohara H., Ise N. pH and temperature dependences of thermolysin catalysis. Catalytic role of zinc-coordinated water. Eur J Biochem. 1982 May;124(1):157–163. doi: 10.1111/j.1432-1033.1982.tb05919.x. [DOI] [PubMed] [Google Scholar]
- Mock W. L., Freeman D. J., Aksamawati M. Fluxionate Lewis acidity of the Zn2+ ion in carboxypeptidase A. Biochem J. 1993 Jan 1;289(Pt 1):185–193. doi: 10.1042/bj2890185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock W. L., Tsay J. T. A probe of the active site acidity of carboxypeptidase A. Biochemistry. 1986 May 20;25(10):2920–2927. doi: 10.1021/bi00358a028. [DOI] [PubMed] [Google Scholar]
- Mock W. L., Tsay J. T. pK values for active site residues of carboxypeptidase A. J Biol Chem. 1988 Jun 25;263(18):8635–8641. [PubMed] [Google Scholar]
- Mock W. L., Zhang J. Z. Mechanistically significant diastereoselection in the sulfoximine inhibition of carboxypeptidase A. J Biol Chem. 1991 Apr 5;266(10):6393–6400. [PubMed] [Google Scholar]
- Mookhtiar K. A., Grobelny D., Galardy R. E., Van Wart H. E. Ketone-substrate analogues of Clostridium histolyticum collagenases: tight-binding transition-state analogue inhibitors. Biochemistry. 1988 Jun 14;27(12):4299–4304. doi: 10.1021/bi00412a016. [DOI] [PubMed] [Google Scholar]
- Morihara K., Tsuzuki H. Thermolysin: kinetic study with oligopeptides. Eur J Biochem. 1970 Aug;15(2):374–380. doi: 10.1111/j.1432-1033.1970.tb01018.x. [DOI] [PubMed] [Google Scholar]
- Nishino N., Powers J. C. Design of potent reversible inhibitors for thermolysin. Peptides containing zinc coordinating ligands and their use in affinity chromatography. Biochemistry. 1979 Oct 2;18(20):4340–4347. doi: 10.1021/bi00587a012. [DOI] [PubMed] [Google Scholar]
- Pank M., Kirret O., Paberit N., Aaviksaar A. Hydrophobic interaction in thermolysin specificity. FEBS Lett. 1982 Jun 7;142(2):297–300. doi: 10.1016/0014-5793(82)80156-7. [DOI] [PubMed] [Google Scholar]
- Rasnick D., Powers J. C. Active site directed irreversible inhibition of thermolysin. Biochemistry. 1978 Oct 17;17(21):4363–4369. doi: 10.1021/bi00614a002. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Riordan J. F. Inhibition of angiotensin converting enzyme: mechanism and substrate dependence. Biochemistry. 1984 Oct 23;23(22):5225–5233. doi: 10.1021/bi00317a021. [DOI] [PubMed] [Google Scholar]
- Shoham G., Christianson D. W., Oren D. A. Complex between carboxypeptidase A and a hydrated ketomethylene substrate analogue. Proc Natl Acad Sci U S A. 1988 Feb;85(3):684–688. doi: 10.1073/pnas.85.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark W., Pauptit R. A., Wilson K. S., Jansonius J. N. The structure of neutral protease from Bacillus cereus at 0.2-nm resolution. Eur J Biochem. 1992 Jul 15;207(2):781–791. doi: 10.1111/j.1432-1033.1992.tb17109.x. [DOI] [PubMed] [Google Scholar]
- Stoeva S. Modification of a zinc proteinase from Bacillus mesentericus strain 76 by diethylpyrocarbonate. Int J Pept Protein Res. 1991 Apr;37(4):325–330. doi: 10.1111/j.1399-3011.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Toma S., Campagnoli S., De Gregoriis E., Gianna R., Margarit I., Zamai M., Grandi G. Effect of Glu-143 and His-231 substitutions on the catalytic activity and secretion of Bacillus subtilis neutral protease. Protein Eng. 1989 Jan;2(5):359–364. doi: 10.1093/protein/2.5.359. [DOI] [PubMed] [Google Scholar]
- Tronrud D. E., Monzingo A. F., Matthews B. W. Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin. Eur J Biochem. 1986 Jun 2;157(2):261–268. doi: 10.1111/j.1432-1033.1986.tb09664.x. [DOI] [PubMed] [Google Scholar]
- Woolley P. Models for metal ion function in carbonic anhydrase. Nature. 1975 Dec 25;258(5537):677–682. doi: 10.1038/258677a0. [DOI] [PubMed] [Google Scholar]


