Abstract
Citrullination plays an essential role in various physiological or pathological processes, however, whether citrullination is involved in regulating tumour progression and the potential therapeutic significance have not been well explored. Here, we find that peptidyl arginine deiminase 4 (PADI4) directly interacts with and citrullinates hypoxia-inducible factor 1α (HIF-1α) at R698, promoting HIF-1α stabilization. Mechanistically, PADI4-mediated HIF-1αR698 citrullination blocks von Hippel-Lindau (VHL) binding, thereby antagonizing HIF-1α ubiquitination and subsequent proteasome degradation. We also show that citrullinated HIF-1αR698, HIF-1α and PADI4 are highly expressed in hepatocellular carcinoma (HCC) tumour tissues, suggesting a potential correlation between PADI4-mediated HIF-1αR698 citrullination and cancer development. Furthermore, we identify that dihydroergotamine mesylate (DHE) acts as an antagonist of PADI4, which ultimately suppresses tumour progression. Collectively, our results reveal citrullination as a posttranslational modification related to HIF-1α stability, and suggest that targeting PADI4-mediated HIF-1α citrullination is a promising therapeutic strategy for cancers with aberrant HIF-1α expression.
Subject terms: Post-translational modifications, Cancer microenvironment
Post-translational modifications on HIF-1α can regulate its stability and activity in tumoral processes. Here, the authors show that PADI4-mediated citrullination avoids HIF-1α degradation to promote hepatocellular carcinoma progression and this can be prevented by a PADI4 antagonist
Introduction
Hypoxia is a hallmark of tumour microenvironment (TME) and is a result of increased cancer cell proliferation and abnormal vasculature development1–3. To overcome severe hypoxic stress in solid tumours, internal tumour cells promote downstream gene transcription by inducing hypoxia-inducible factor 1 (HIF-1) expression4–6, which facilitates various tumour biology processes, including unrestricted cell proliferation7, the epithelial-mesenchymal transition8, and immunosuppression9. However, in addition to its pathological role, HIF-1α also plays a key role under physiological conditions10, and as a consequence, targeting HIF-1α alone will cause unnecessary damage to normal tissues. Therefore, it is of great importance to elucidate the regulatory mechanism of HIF-1α activity and protein stability under pathological conditions so as to target the HIF-1α-mediated network that is activated during tumour progression11.
Under normoxic conditions, HIF-1α is hydroxylated by prolyl hydroxylase domain-containing proteins 1-3 (PHD 1-3), for which oxygen is a cofactor12,13. Hydroxylated HIF-1α is then recognised by the von Hippel-Lindau (VHL) protein, an E3 ubiquitin ligase, and is further degraded through the proteasome pathway14,15. Under hypoxic conditions, with decreased oxygen levels, PHD enzyme activity is downregulated, leading to the stabilisation of HIF-1α protein. Posttranslational modifications (PTMs) have been increasingly recognised for their prominent roles in protein structure, activity, and function16–18. In addition to those on hydroxylation, recent studies revealed that HIF-1α is extensively regulated by other modifications such as acetylation19,20, phosphorylation21,22, methylation23,24, O-GlcNAcylation25 and SUMOylation26. For example, Cheng et al. found that SUMOylation of HIF-1α promotes the binding of HIF-1α to VHL and acts as a signalling molecule for hydroxylation-independent and ubiquitin-dependent degradation under hypoxic conditions. However, the detailed network involving the posttranslational regulation of HIF-1α in disease has not been fully elucidated.
Citrullination, also known as deimination, is a Ca2+-dependent PTM that converts a positively charged and protein-embedded arginine into the electrically neutral and uncoded amino acid citrulline, resulting in a 1 Da loss in molecular mass and a reduction in hydrogen-bond formation27,28. This reaction is catalysed by the family of peptidylarginine deiminases (PADs), including PADI1-4 and PADI629,30. Citrullination is involved in multiple biological processes, including early embryogenesis31, stem cell pluripotency32, neutrophil extracellular trap (NET) formation33,34, and terminal epidermal differentiation by regulating protein folding and protein-protein interactions (PPIs). Citrullinated proteins are the hallmark of rheumatoid arthritis (RA)35, and overactive citrullination has been indicated to be associated with pathological conditions. Yu et al. found that hypoxia upregulates PADI2 and citrullinated proteins in a HIF-1α-dependent manner in human fibroblast-like synoviocytes, proposing hypoxia involved in the mechanism of RA36. However, whether citrullination is involved in remodelling the tumour's hypoxic microenvironment is unclear, and if so, the underlying molecular mechanisms and significance remain unknown.
Here, we reveal a citrullination modification of HIF-1α facilitated by PADI4 expression during tumour progression. Specifically, PADI4 directly binds to HIF-1α and citrullinates it at R698. Thus, PADI4-dependent HIF-1αR698 citrullination enhances HIF-1α stability and transcriptional activity by competitively inhibiting VHL-HIF-1α binding, which promotes tumour progression. Notably, we demonstrate that dihydroergotamine mesylate (DHE), which disrupts the interaction of PADI4 and HIF-1α, promotes HIF-1α degradation and suppresses tumour progression by occupying the enzyme pocket of PADI4 responsible for HIF-1α citrullination, providing a rationale for developing therapeutic strategies against cancers with aberrant HIF-1α expression.
Results
PADI4 directly interacts with and citrullinates HIF-1α at R698
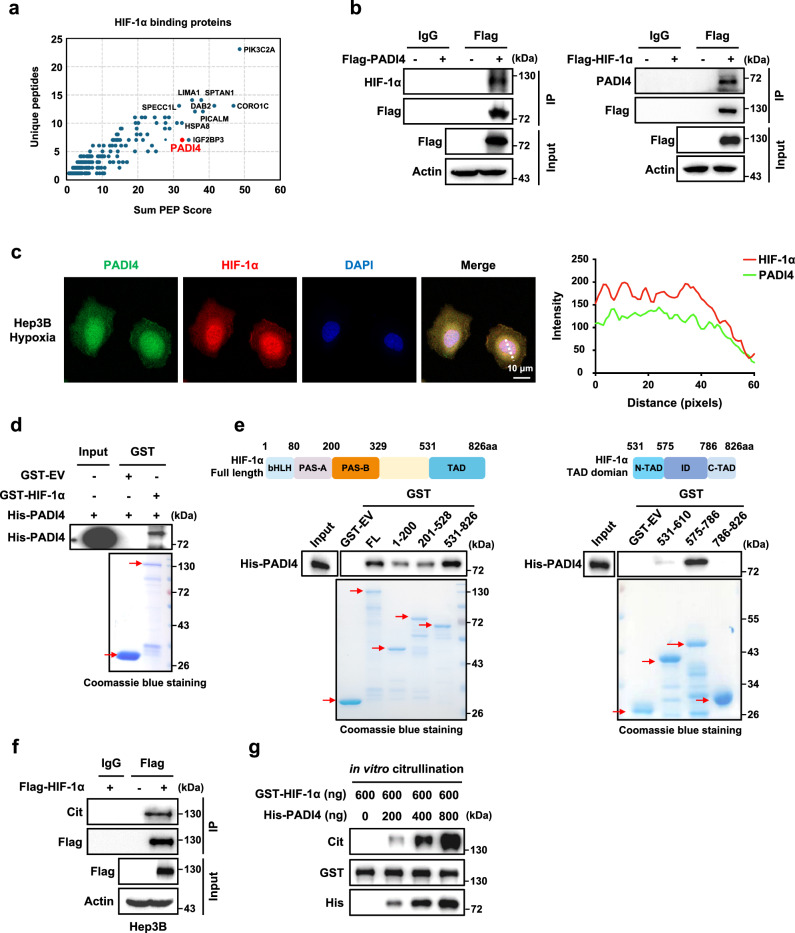
Previous studies demonstrated that acetylation19, methylation22, and SUMOylation26 modifications regulate hypoxia-inducible factor 1 (HIF-1) stability, but few suggested approaches to interfere with these modifications in disease models. To search for novel PTMs of HIF-1α that have been undiscovered to date, we performed immunoprecipitation-mass spectrometry (IP-MS) in Hep3B cells overexpressing Flag-tagged HIF-1α to identify potential HIF-1α-binding proteins known to mediate PTMs. Through a functional cluster analysis of the IP-MS results, we found that PADI4, the protein responsible for citrullination, interacts with HIF-1α (Fig. 1a and Supplementary Fig. 1a). Next, a co-immunoprecipitation (co-IP) assay was performed and confirmed the interaction of HIF-1α and PADI4 in HEK293T cells ectopically coexpressing Flag-tagged HIF-1α and HA-tagged PADI4 (Supplementary Fig. 1b). In addition, overexpression of Flag-PADI4 and Flag-HIF-1α separately in Hep3B cells alone enabled the immunoprecipitation of endogenous HIF-1α and PADI4, respectively (Fig. 1b). Consistent results were also observed in mouse Hepa1-6 liver cancer cells (Supplementary Fig. 1c), indicating that the interaction between HIF-1α and PADI4 is a universal phenomenon in human and mouse liver cancer cells. This phenomenon was also confirmed by immunofluorescence (IF) assay, in which obvious colocalization of HIF-1α and PADI4 was observed in Hep3B cells treated under hypoxic conditions (Fig. 1c).
Fig. 1. PADI4 directly interacts with and citrullinates HIF-1α.
a Cellular extracts from Hep3B cells stably expressing empty vector (EV) or Flag-HIF-1α were immunoprecipitated with an anti-Flag antibody and then eluted. The eluted proteins were separated by SDS-PAGE and the protein bands were retrieved and analyzed by mass spectrometry (MS). b Hep3B cells stably expressing Flag-PADI4 (left panel) or Flag-HIF-1α (right panel) were cultured under hypoxic conditions for 6 h, followed by immunoprecipitation analysis. c Immunofluorescence (IF) analysis of the colocalization of endogenous HIF-1α (red) and PADI4 (green) in Hep3B cells cultured under hypoxic conditions for 6 h. Scale bars: 10 μm (left panel). Intensity profiles of each line were quantified with ImageJ software and drawn with GraphPad Prism 7.0 (right panel). d GST pull-down of His-PADI4 by GST-EV or GST-HIF-1α using proteins purified in Escherichia coli. e GST pull-down of His-PADI4 by GST-EV or GST fusion protein containing the full-length (FL) or indicated truncation mutants of HIF-1α. f Hep3B cells expressing Flag-EV or Flag-HIF-1α were immunoprecipitated with either IgG or anti-Flag antibody, followed by western blotting analysis with antibody against pan citrulline or Flag. g An in vitro citrullination assay was performed by incubating purified His-PADI4 and GST-HIF-1α proteins at 37 °C for 1.5 h in catalytic buffer and probing with an anti-pan citrulline antibody. The samples were derived from the same experiment, but different gels for Cit, His, and another for GST were processed in parallel. Immunoblots and immunofluorescence are representative of three independent experiments (b–g). Source data are provided as a Source Data file.
To investigate whether HIF-1α directly binds PADI4, an in vitro GST pull-down assay was performed with purified GST-HIF-1α and His-PADI4 proteins, and the results showed that PADI4 directly bound HIF-1α (Fig. 1d). Next, to determine the regions in HIF-1α that bind to PADI4, a panel of GST-HIF-1α truncation mutations was generated and an in vitro GST pull-down assay revealed that His-PADI4 bound strongly to GST-HIF-1α (531-826) and weakly to GST-HIF-1α (1–200) and (201–528) (Fig. 1e, left panel). Further, GST pull-down assay results confirmed that His-PADI4 mainly bound GST-HIF-1α (575-786) (Fig. 1e, right panel).
The above results showed that HIF-1 directly binds PADI4 (Fig. 1d, e), but whether HIF-1α is a substrate of PADI4 was unclear, and whether it was citrullinated by PADI4 was unknown. To test these possibilities, ectopically expressed Flag-tagged HIF-1α was immunoprecipitated and probed with an anti-pan citrulline antibody in Hep3B and HEK293T cells. The IP results showed that HIF-1α was indeed modified by citrullination (Fig. 1f and Supplementary Fig. 1d). More convincingly, we established an in vitro catalytic system by incubating recombinant purified GST-HIF-1α and His-PADI4 proteins in the catalytic buffer. The results showed that PADI4 significantly increased the citrullination levels of HIF-1α in a dose-dependent manner (Fig. 1g). Taken together, these data indicate that HIF-1α directly binds to PADI4 and is an unrecognised substrate that can be citrullinated by PADI4.
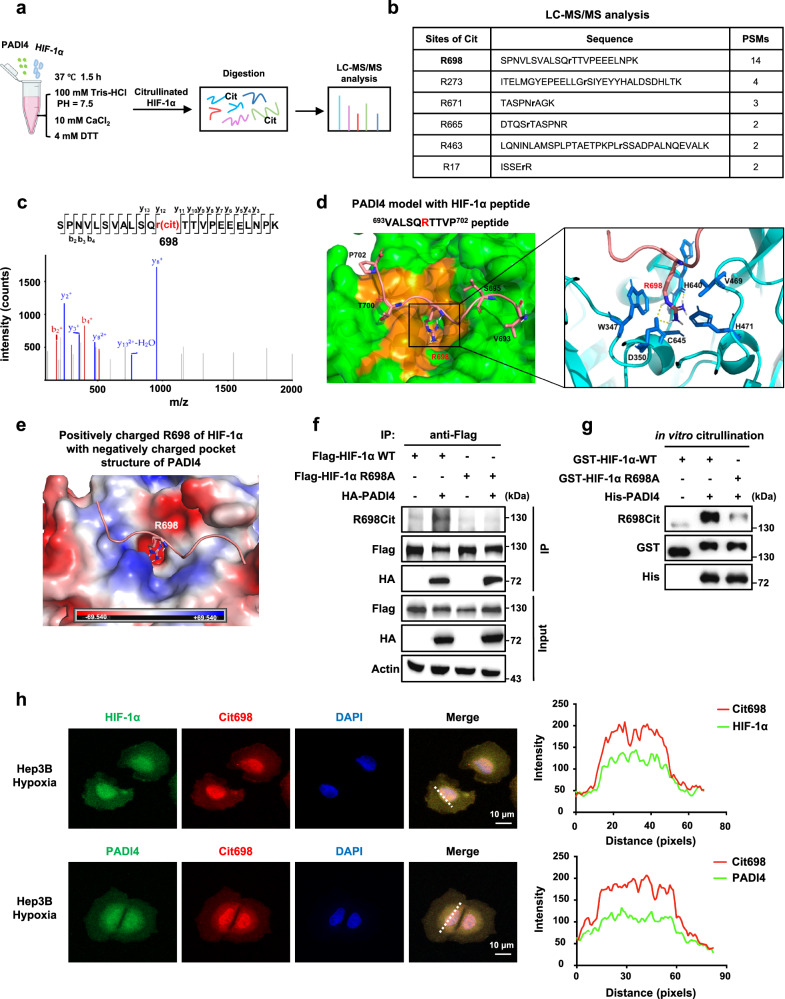
To identify the citrullination site on HIF-1α, an in vitro citrullination assay was performed, and citrullinated HIF-1α proteins were subsequently subjected to mass spectrometry analysis (Fig. 2a). Two independent liquid chromatography-mass spectrometry tandem mass spectrometry (LC-MS/MS) results showed six potential citrullinated arginine (R) residues on HIF-1α protein, including R17, R273, R463, R665, R671 and R698, with the R698 site having the highest peptide-spectrum match (PSM) score (Fig. 2b, c and Supplementary Fig. 2a, b). Next, these six putative R sites were replaced with alanine (A) residues and subjected to IP assay, and the results showed that the citrullination signal intensity of the R698A mutant was significantly reduced (Supplementary Fig. 2c). Furthermore, docking models of the HIF-1α peptide carrying R698 and bound to PADI4 suggested that the positively charged residue R698 was deeply embedded into the negatively charged enzymatic pocket of PADI4 (Fig. 2d, e). These data demonstrate that R698 is the true citrullination site on HIF-1α.
Fig. 2. PADI4 promotes HIF-1α citrullination at R698.
a Diagram showing the in vitro citrullination assay with His-PADI4 protein and GST-HIF-1α proteins in catalytic buffer at 37 °C for 1.5 h. Modified GST-HIF-1α proteins were separated by SDS-PAGE and subsequently subjected to liquid chromatography-mass spectrometry tandem mass spectrometry (LC-MS/MS) analysis. b The citrulline modification sites in HIF-1α were determined by LC-MS/MS. They were R17, R273, R463, R665, R671, and R698. c LC-MS/MS spectrum of the citrullinated peptide containing the R698 site in HIF-1α. d Molecular docking model for the interaction of PADI4 (RCSB PDB: 1WDA) with HIF-1α. A close-up view of the peptide binding site of PADI4 is shown in the cartoon, in which green represents the surface and orange represents the pocket structure. The HIF-1α 693VALSQRTTVP702 peptide is shown as a magenta ribbon and the stick represents side chains (coloured by atom type: oxygen, red; nitrogen, blue; C, grey). The interfacial regions between PADI4 (blue) and HIF-1α (magenta) indicate an interaction between R698 on HIF-1α and the pocket structure of PADI4. W347, D350, V469, H471, H640 and C645 of PADI4 come in close contact. e The surface electrostatic potential of the docking model. Positively charged R698 of the HIF-1α 693VALSQRTTVP702 peptide with the negatively charged pocket structure of PADI4. The electrostatic potential is colour-coded as −69.540 kcal/mol (red) to +69.540 kcal/mol (blue), thus displaying negative or positive charges, respectively. f HEK293T cells expressing WT Flag-HIF-1α or Flag-HIF-1α R698A together with HA-PADI4 were used for immunoprecipitation analysis. g An in vitro citrullination assay was performed by incubating purified WT GST-HIF-1α or GST-HIF-1αR698A with His-PADI4 at 37 °C for 1.5 h, followed by western blotting analysis. h IF analysis of the colocalization of citrullinated HIF-1αR698 with HIF-1α or PADI4 in Hep3B cells under hypoxic conditions. Scale bars: 10 μm (left panel). Intensity profiles of each line were quantified by ImageJ software and drawn with GraphPad Prism 7.0 (right panel). Immunoblots and immunofluorescence are representative of three independent experiments (f–h). Figure 2a, created with BioRender.com, is released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
We then generated a polyclonal antibody that specifically recognised R698-citrullinated HIF-1α. The specificity of the antibody (HIF-1αR698-Cit), obtained from rabbits, was verified by dot blot assay (Supplementary Fig. 2d). Next, an IP assay was performed, and the results showed that the citrullination levels on R698 site are higher in wild-type HIF-1α (HIF-1αWT) but not in the HIF-1αR698A mutant after PADI4 was overexpressed (Fig. 2f). Moreover, in vitro citrullination experiments showed that His-PADI4 catalysed the citrullination of GST-HIF-1αWT, but not the GST-HIF-1αR698A mutant (Fig. 2g). IF assays further showed high colocalization between citrullinated HIF-1αR698 and HIF-1α or PADI4 in Hep3B cells cultured under hypoxic conditions (Fig. 2h). These data indicate that the R698 residue on HIF-1α in Homo sapiens is citrullinated. We then evaluated the homology of this site among different species and found that the arginine residue has been evolutionarily conserved (Supplementary Fig. 2e). The corresponding site of hsa-HIF-1αR698 in mice is mus-HIF-1αR709, and IP assays showed that R709 was the citrullination site in mus-HIF-1α (Supplementary Fig. 2e, f). Taken together, these results demonstrate that PADI4 mainly catalyses HIF-1α citrullination at the R698 residue and that this function has been evolutionarily conserved.
PADI4-mediated HIF-1α citrullination promotes HIF-1α stability and transactivation
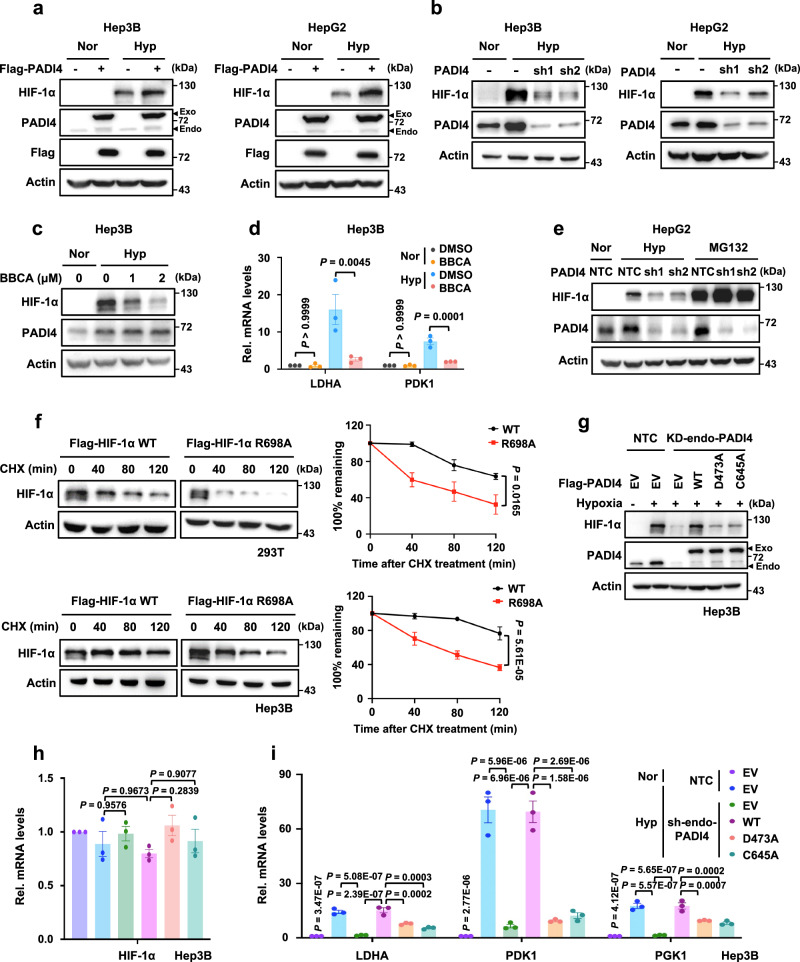
Next, we sought to determine whether and how PADI4 regulates HIF-1α expression. Our western blot results showed that even under hypoxic conditions, ectopic expression of PADI4 led to greater accumulation of HIF-1α proteins, and PADI4 knockdown markedly reduced HIF-1α protein levels, without affecting the HIF-1α mRNA levels (Fig. 3a, b and Supplementary Fig. 3a, b). Moreover, the pan-citrullination inhibitor BB-Cl-amidine (BBCA) clearly inhibited the expression of HIF-1α protein as well as its downstream target genes, including LDHA and PDK1 in Hep3B cells (Fig. 3c, d).
Fig. 3. PADI4-mediated citrullination of HIF-1α promotes HIF-1α stability and transactivation.
a Hep3B (left panel) and HepG2 cells (right panel) expressing Flag-EV or Flag-PADI4 were cultured under normoxic or hypoxic conditions for 6 h, followed by western blotting analysis. The samples were derived from the same experiment, but different gels for HIF-1α, Flag, Actin, and another for PADI4 were processed in parallel. b Hep3B and HepG2 cells expressing NTC or PADI4 shRNAs were cultured under normoxic or hypoxic conditions for 6 h, followed by western blotting analysis. c Hep3B cells were treated with DMSO or 1 or 2 μM BBCA for 24 h, followed by hypoxic treatment for another 6 h before western blotting analysis. d Hep3B cells were treated with DMSO or 2 μM BBCA for 24 h, followed by hypoxic treatment for another 24 h. The mRNA levels of LDHA and PDK1 were analyzed by qPCR. e Western blot analysis of HIF-1α and PADI4 protein levels in HepG2 cells expressing NTC and PADI4 shRNAs cultured under normoxic or hypoxic conditions in the presence or absence of 10 μM MG132 for 6 h. f Western blot analysis (left panel) of HIF-1α protein levels in HEK293T (upper panel) and Hep3B cells (lower panel) expressing Flag-HIF-1αWT or Flag-HIF-1αR698A treated with 20 μg/ml CHX for the indicated times. Quantification of HIF-1α protein levels relative to Actin. g, h Endogenous PADI4-knockdown Hep3B cells were infected with viruses expressing Flag-EV, Flag-PADI4WT, Flag-PADI4D473A, or Flag-PADI4C645A and further cultured under normoxic or hypoxic conditions for 6 h. Cell lysates were harvested, and the protein and mRNA levels of HIF-1α were analyzed by western blotting (g) or qPCR (h), respectively. i The cell lines described in Fig. 3g were cultured under normoxic or hypoxic conditions for 24 h. The mRNA levels of LDHA, PDK1, and PGK1 were analyzed by qPCR. Immunoblots are representative of three independent experiments (a–c, e–g). Data were presented as mean ± SEM of three independent experiments (d, f, h, i). Statistical analyses were performed by ordinary one-way ANOVA (d, h, i) or two-way ANOVA test (f) with Turkey’s multiple comparisons test. Source data are provided as a Source Data file.
PADI4 promoted HIF-1α protein accumulation without regulating its mRNA expression, suggesting that PADI4 may participate in posttranscriptional regulation of HIF-1α. To explore the mechanism by which PADI4 regulates the HIF-1α protein, Hep3B or HepG2 cells with PADI4 knocked down were treated with MG132, a potent 26S proteasome inhibitor, under hypoxic conditions for 6 h. The results showed that MG132 profoundly blocked PADI4 knockdown-induced HIF-1α degradation in a proteasome-dependent pathway (Fig. 3e and Supplementary Fig. 3c). To further demonstrate the involvement of PADI4 in HIF-1α stability, we detected the ubiquitination levels of HIF-1α in 293T-PADI4 cells. Results showed that overexpression of PADI4 could obviously reduce HIF-1α ubiquitination levels (Supplementary Fig. 3d). Furthermore, we investigated the impact of PADI4 knockdown on HIF-1α half-life by cycloheximide (CHX) chase experiments and results showed that the degradation rate of HIF-1α became faster when PADI4 was knocked down (Supplementary Fig. 3e). Collectively, these data demonstrate the effect of PADI4 on the promotion of HIF-1α stability by preventing HIF-1α from ubiquitination-dependent proteasomal degradation pathway. To further investigate whether PADI4-mediated citrullination is essential for HIF-1α stability, the half-lives of HIF-1αWT and the HIF-1αR698A mutant were determined by cycloheximide (CHX) chase experiments, and the results showed that the degradation rate of the HIF-1αR698A mutant was faster than that of HIF-1αWT (Fig. 3f). Taken together, these results suggest that PADI4 resists HIF-1α degradation by promoting HIF-1αR698 citrullination, thereby maintaining HIF-1α stability.
To determine whether enzyme activity is essential for PADI4-enhanced HIF-1α stability, enzymatically inactivated mutants37, namely, PADI4D473A and PADI4C645A were generated and proven that the inactivated PADI4 mutants failed to catalyse the citrullination of HIF-1α at R698 (Supplementary Fig. 3f). As expected, the ectopic expression of PADI4WT, but not that of the inactivated mutants, rescued PADI4 knockdown-induced decrease in HIF-1α protein levels as well as the downregulated expression of HIF-1α target genes, without changing HIF-1α mRNA levels (Fig. 3g–i and Supplementary Fig. 3g–i). These findings show that the enzymatic activity of PADI4 is indispensable for maintaining HIF-1α stability and the transcription of its downstream genes. Taken together, these data indicate that both PADI4 enzyme activity and PADI4-mediated HIF-1αR698 citrullination are required for HIF-1α protein stability and its transcriptional activity under hypoxic conditions.
Citrullination of HIF-1α disrupts the interaction of HIF-1α and VHL
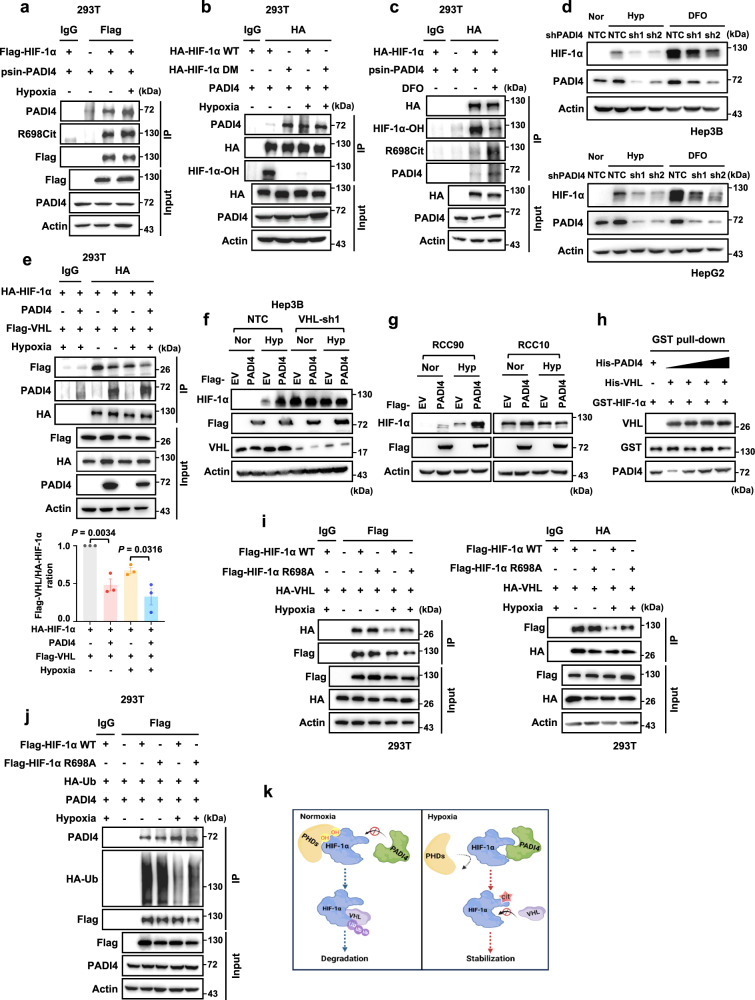
As HIF-1α stability is precisely regulated by oxygen concentration, we examined whether oxygen level affects HIF-1α citrullination. Co-IP results showed that compared with normoxic conditions, the binding between HIF-1α and PADI4 was increased, accompanied by increased citrullinated HIF-1αR698 levels under hypoxic conditions (Fig. 4a), suggesting that oxygen-mediated hydroxylation of HIF-1α may disrupt HIF-1α-PADI4 binding as well as PADI4-mediated HIF-1α citrullination. Therefore, HIF-1αDM (the P402/564A double mutant), which lacked residues that can be hydroxylated, was established, and the binding ability between PADI4 and HIF-1αWT or the HIF-1αDM mutant was measured. Co-IP assays showed that the HIF-1αDM mutant exhibited a higher affinity than HIF-1αWT for PADI4, especially under normoxic conditions (Fig. 4b), suggesting that HIF-1α hydroxylation inhibits HIF-1α-PADI4 binding. Furthermore, we examined the interaction of HIF-1α and PADI4, as well as HIF-1α citrullination under normoxic conditions with PHDs inhibitor (Deferoxamine mesylate, DFO). Results showed that DFO could efficiently repress HIF-1α hydroxylation, accompanied by enhanced interaction of HIF-1α and PADI4 as well as increased HIF-1αR698 citrullination levels, indicating that HIF-1α hydroxylation disrupts HIF-1α-PADI4 interaction as well as PADI4-mediated HIF-1α citrullination (Fig. 4c). Hep3B-shPADI4 and HepG2-shPADI4 cells were treated under hypoxic conditions or DFO for 6 h, respectively. Western blotting results showed that DFO treatments lead to an obvious accumulation of HIF-1α proteins, while PADI4 knockdown could weaken this effect, which is similar to hypoxic treatment, suggesting that PADI4 maintains HIF-1α stability when HIF-1α hydroxylation is inhibited (Fig. 4d).
Fig. 4. Citrullination of HIF-1α disrupts the interaction of HIF-1α and VHL.
a HEK293T cells transfected with Flag-EV or Flag-HIF-1α plasmid together with PADI4 plasmid were cultured under normoxic or hypoxic conditions for 6 h, followed by immunoprecipitation analysis. The IP samples were derived from the same experiment, but different gels for PADI4, R698Cit, another for Flag were processed in parallel. b HEK293T cells transfected with HA-HIF-1αWT or HA-HIF-1αDM (double mutant, P402/564 A) plasmids together with PADI4 plasmids were cultured under normoxic or hypoxic conditions for 6 h, followed by immunoprecipitation analysis. The IP samples were derived from the same experiment, but different gels for PADI4, HA, and another for HIF-1α-OH were processed in parallel. c HEK293T cells were transfected with HA-EV or HA-HIF-1α plasmids together with psin-PADI4 plasmids with or without DFO in the presence of MG132 under normoxic condition for 6 h, followed by immunoprecipitation analysis. The IP samples were derived from the same experiment, but different gels for HA, another for HIF-1α-OH and another for R698Cit, PADI4 were processed in parallel. d Hep3B (upper panel) and HepG2 (lower panel) cells expressing NTC or PADI4 shRNAs were treated by hypoxia or DFO for 6 h, followed by western blotting analysis. e HEK293T cells transfected with EV or PADI4 together with HA-HIF-1α and Flag-VHL plasmids were cultured under normoxic or hypoxic conditions in the presence of 10 μM MG132 for 8 h, followed by immunoprecipitation analysis (upper panel). Quantification of Flag-VHL protein levels relative to HA-HIF-1α protein levels (lower panel). f Hep3B-Flag-EV or Flag-PADI4 cells infected with a virus expressing NTC or VHL shRNA were cultured under normoxic or hypoxic conditions for 6 h, followed by western blotting analysis. g RCC90 (VHL wild type) and RCC10 (VHL-null) cells expressing Flag-EV or Flag-PADI4 were cultured under normoxic or hypoxic conditions for 6 h, followed by western blotting analysis. h In vitro analysis of the interaction between purified GST-HIF-1α and His-VHL with increased amounts of His-PADI4. i HEK293T cells transfected with Flag-HIF-1αWT or Flag-HIF-1αR698A mutant and HA-VHL plasmids were cultured under normoxic or hypoxic conditions for 6 h, followed by immunoprecipitation analysis. j HEK293T cells transfected with Flag-HIF-1αWT or Flag-HIF-1αR698A mutant together with HA-Ub and PADI4 plasmids were cultured under normoxic or hypoxic conditions for 6 h, followed by immunoprecipitation analysis. The IP samples were derived from the same experiment, but different gels for PADI4, Flag, and another for HA were processed in parallel. k Diagram showing the mechanism by which PADI4 promotes the stability of HIF-1α. Immunoblots are representative of three independent experiments (a–j). Error bars denote the mean ± SEM (e). Statistical analyses were performed by ordinary one-way ANOVA with Turkey’s multiple comparisons test (e). Figure 4k created with BioRender.com is released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
Since HIF-1α is degraded mainly through the VHL-mediated ubiquitination and the proteasome pathway, we hypothesised that PADI4-mediated HIF-1α citrullination may antagonise HIF-1α binding to VHL. To test this possibility, we performed IP assays and found that overexpression of PADI4 blocked VHL binding to HIF-1α under both normoxic and hypoxic conditions (Fig. 4e). Next, we knocked down VHL in Hep3B cells with PADI4 overexpressing and found that either PADI4 overexpression or VHL knockdown increased HIF-1α protein levels, but the positive effect of PADI4 on HIF-1α expression was obviously diminished when we knocked down of VHL under both normoxic and hypoxic conditions (Fig. 4f). Furthermore, we overexpressed PADI4 in the RCC90 (with VHL) and RCC10 (VHL-null) cell lines and observed consistent results; that is, PADI4 increased HIF-1α protein levels in the presence of VHL (Fig. 4g, left panel), but exerted no obvious effect on HIF-1α expression in the absence of VHL (Fig. 4g, right panel). These results suggest that PADI4 antagonises the binding of VHL to HIF-1α and stabilises HIF-1α in a VHL-dependent manner.
To investigate whether PADI4 or PADI4-mediated citrullination of HIF-1α blocks the binding of VHL to HIF-1α, an in vitro GST pull-down assay was performed. The results showed that the binding of VHL to non-citrullinated HIF-1α (in vitro) remained unchanged when His-PADI4 protein content was incrementally increased (Fig. 4h), however, the binding activity of HA-HIF-1α and Flag-VHL was greatly reduced in vivo because HIF-1α could be citrullinated (Fig. 4e). Next, in vivo IP assays were performed in which HIF-1α could be citrullinated in 293T cells. Consistent with our previous results, there was little increase in the interaction between VHL and HIF-1αR698A compared with that of VHL and HIF-1αWT, and the ubiquitination levels were largely unchanged under normoxic conditions (Fig. 4i, j). Under hypoxic conditions, the binding of VHL to the HIF-1αR698A mutant was stronger than that to HIF-1αWT, and the ubiquitination level of the HIF-1αR698A mutant was also increased compared to that of HIF-1αWT (Fig. 4i, j). Taken together, these results suggest that it is the action of PADI4-mediated HIF-1α citrullination, but not PADI4 itself, inhibits VHL binding to HIF-1α and plays a crucial role in maintaining HIF-1α stability under hypoxic conditions.
In conclusion, these data demonstrate that under normoxic conditions, PHD-mediated HIF-1α hydroxylation repressed PADI4 binding to HIF-1α, and subsequently led to HIF-1α ubiquitination and degradation. Under hypoxic conditions, the interaction of PADI4 and HIF-1α was obviously enhanced due to reduced HIF-1α hydroxylation. The increase in PADI4-mediated HIF-1αR698 citrullination led to HIF-1α stabilisation by further blocking the interaction between VHL and HIF-1α (Fig. 4k).
Dihydroergotamine mesylate disrupts PADI4-HIF-1α interaction and suppresses HIF-1α expression
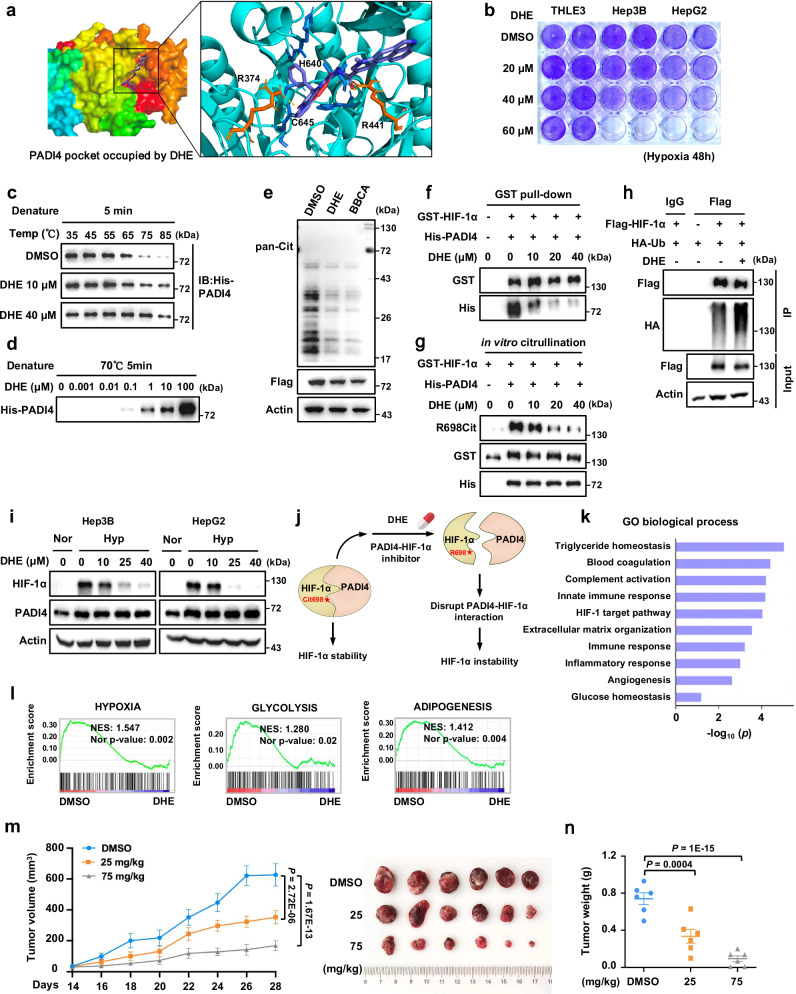
As the accumulation of HIF-1α protein is important for the growth, metastasis, and drug resistance of solid tumours, and is a major cause of cancer-related deaths, HIF-1α has long been considered an attractive target for cancer therapies2. However, similar to other transcription factors, HIF-1α is regarded as “undruggable” by small-molecule agents due to the significant disorder of structure of the HIF-1α protein and lack of a clearly characterised binding pocket38. Based on our findings that the PADI4-HIF-1α interaction is essential for HIF-1α stability, the enzymatic pocket formed by PADI4 might be a drug target that can be leveraged to target HIF-1α indirectly (Fig. 2d and Supplementary Fig. 4a). Therefore, we screened potential antagonists of PADI4 by virtual screening relying on the Autodock-Vina docking. Of the 1379 US Food and Drug Administration (FDA)-approved commercial agents in the ZINC database, we identified 7 candidates through a series of screening (Supplementary Fig. 4b). DHE, an FDA-approved agent for migraine treatment, was selected for further study based on its VINA docking score, interaction partners or types of bonds it forms, and ligand orientation in the PADI4 enzymatic pocket (Fig. 5a). Moreover, the effect of DHE on hepatocellular carcinoma (HCC) cell growth was determined. The results showed that DHE selectively suppressed the growth of Hep3B and HepG2 liver cancer cells without inducing obvious toxicity in the normal THLE3 liver cells (Fig. 5b). Viability assay of DHE-treated Hep3B cells showed that the IC50 of DHE was 30.81 μM under normoxic conditions and 16.46 μM under hypoxic conditions. These results indicate that DHE was more likely to inhibit hypoxic liver cancer cell growth (Supplementary Fig. 4c).
Fig. 5. Dihydroergotamine mesylate disrupts PADI4-HIF-1α interaction and represses HIF-1α expression.
a Molecular docking of dihydroergotamine (DHE) in the enzymatic pocket of PADI4. b THLE3, Hep3B and HepG2 cells were treated with DMSO or DHE as indicated concentration for 48 h under hypoxia, followed by crystal violet staining assay. c His-PADI4 proteins were incubated with DMSO or DHE under the indicated temperature before western blotting analysis. d His-PADI4 proteins were incubated with DHE as indicated concentration before western blotting analysis. e Cell lysates of Hep3B-Flag-PADI4 cells were incubated with DMSO, 50 μM DHE, or 50 μM BBCA at 37 °C for 20 min before western blotting analysis. The samples were derived from the same experiment, but different gels for pan-Cit, another for Flag, Actin were processed in parallel. f Pull-down of His-PADI4 by GST-HIF-1α in the presence of DMSO or DHE as indicated concentration. g An in vitro citrullination assay of GST-HIF-1α was performed in the presence of DMSO or DHE as indicated concentration. The samples were derived from the same experiment, but different gels for R698Cit, His, another for GST were processed in parallel. h HEK293T cells transfected with Flag-EV or Flag-HIF-1αWT together with HA-Ub were treated with DMSO or 25 μM DHE for 24 h and MG132 for 6 h, followed by immunoprecipitation analysis. The IP samples were derived from the same experiment, but different gels for Flag and another for HA were processed in parallel. i Hep3B and HepG2 cells were treated with DMSO or DHE as indicated concentration for 24 h and then cultured under normoxic or hypoxic conditions for another 6 h before western blotting analysis. j Diagram showing the working mechanism of DHE in regulating HIF-1α protein levels. k, l Total mRNA in Hep3B cells treated with DMSO or 20 μM, or 40 μM DHE under hypoxic conditions for 24 h were subjected to RNA-sequencing, followed by GO biological processes (k) and GSEA (l) analysis. m, n Parental Hep3B cells were injected subcutaneously into the flanks of BALB/c nude mice (n = 6 per group). After 12 days, the mice were treated with DMSO or DHE (25 mg/kg or 75 mg/kg) by i.g every 2 days. Tumour volume was determined starting on Day 14 and photographs show xenografts at the end of the experiment (m). Tumour weight was measured (n). Immunoblots are representative of three independent experiments (c–i). Error bars denote the mean ± SEM (m, n). Statistical analyses were performed by ordinary one-way ANOVA (n) or two-way ANOVA (m) with Turkey’s multiple comparisons test. Source data are provided as a Source Data file.
To confirm the direct binding of DHE to PADI4, the thermal stabilisation assay (TSA) was performed based on the biophysical principle of ligand-induced thermal stabilisation of its target protein39,40. The results showed that 10 or 40 μM DHE treatment substantially upregulated the thermal stability of purified His-PADI4 under different temperatures with 5 min denaturation (Fig. 5c). Moreover, the thermal stability of PADI4 was also significantly enhanced by serial increase of DHE concentration (Fig. 5d). These TSA results showed that DHE heat-stabilised PADI4 and indicated that DHE directly interacts with PADI4. To verify whether the binding of DHE to PADI4 regulates PADI4-mediated citrullination, we treated Hep3B-Flag-PADI4 cell lysates with DHE at 37 °C for 20 min and found that DHE treatment resulted in a marked decrease in the overall citrullination levels, similar to the effect of BBCA treatment (Fig. 5e). Next, an in vitro GST pull-down assay showed that DHE treatment effectively blocks the interaction of HIF-1α and PADI4 in a dose-dependent manner (Fig. 5f). A similar dose-dependent reduction in HIF-1αR698 citrullination levels was observed via in vitro citrullination assay (Fig. 5g). Moreover, IP assays showed that the ubiquitination levels of HIF-1α was increased after DHE treatment (Fig. 5h) and western blotting results confirmed that the HIF-1α protein levels were decreased after DHE treatment in a dose-dependent manner, while the HIF-1α mRNA levels and PADI4 protein levels were unaffected (Fig. 5i and Supplementary Fig. 4d), suggesting that DHE disrupts the interaction between PADI4 and HIF-1α, and subsequently promotes HIF-1α degradation through the proteasomal pathway (Fig. 5j). To investigate whether DHE regulates HIF-1 target genes, RNA-sequencing (RNA-seq) was performed and GO biological process and gene set enrichment analysis (GSEA) of the RNA-seq data convincingly demonstrated that the activation of pathways related to HIF-1α, including hypoxia response, glycolysis, and adipogenesis, was significantly downregulated after DHE treatment (Fig. 5k, l). Furthermore, qPCR results confirmed the RNA-seq results and revealed that hypoxia-induced increase of PDK1, LDHA and PGK1 mRNA levels were significantly suppressed by DHE treatment (Supplementary Fig. 4e). These results suggest that DHE, as a PADI4 antagonist, obviously attenuates HIF-1αR698 citrullination, reduces HIF-1α protein levels, and inhibits HIF-1α target gene expression by interfering with the interaction of HIF-1α with PADI4.
Next, we investigated the antitumour efficacy of DHE in the xenograft mouse model by subcutaneous inoculating of Hep3B cells into BALB/c nude mice. The mice were intragastrically administered (i.g.) of 25 mg/kg or 75 mg/kg DHE every 2 days. Compared to the effect on the DMSO control group, DHE inhibited tumour growth in a dose-dependent manner (Fig. 5m, n). In addition, DHE treatment significantly reduced HIF-1α, HIF-1αR698Cit, and Histone H3-Cit protein levels and HIF-1α target genes expression, including LDHA and PDK1 in tumour tissue lysates (Supplementary Fig. 4f). Additionally, to further assess the toxicity of DHE, another xenograft experiment was employed, and no obvious body weight reduction, myelosuppression or hepatorenal toxicity were observed after DHE treatment (Supplementary Fig. 4g–i). Collectively, these data indicate that the PADI4 inhibitor DHE suppresses cancer cell proliferation and tumour growth by disrupting the interaction between HIF-1αR698 and the PADI4 enzymatic pocket and thereby inducing HIF-1α degradation.
PADI4-mediated HIF-1αR698 citrullination promotes HCC tumorigenesis
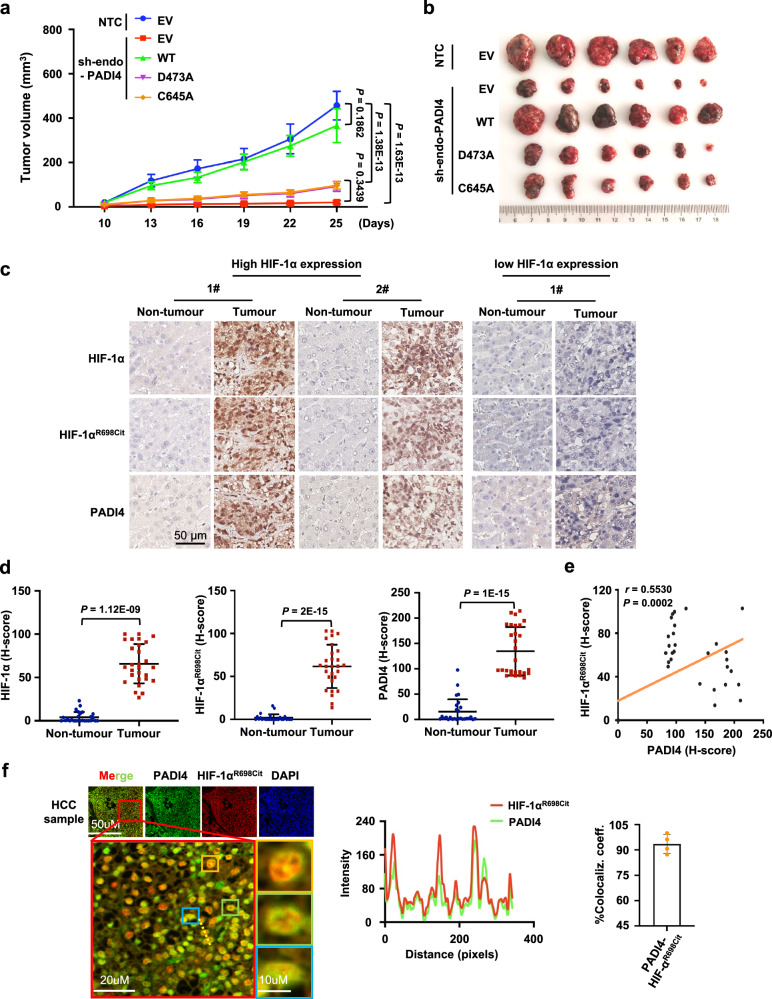
To investigate the role of PADI4-mediated HIF-1α citrullination in cell proliferation and tumour growth, endogenous PADI4-knockdown Hep3B cells were further infected with viruses expressing Flag-EV, Flag-PADI4WT, Flag-PADI4D473A, or Flag-PADI4C645A. Compared with the effect on the NTC group, PADI4 knockdown suppressed cell proliferation in the treatment groups. The reconstituted expression of PADI4WT, but not the expression of the Flag-PADI4D473A or Flag-PADI4C645A mutant, reversed the cell growth inhibition induced by shPADI4 (Supplementary Fig. 5a). Consistent with the assays revealing the cell growth pattern, the mouse xenograft experiments showed that PADI4 knockdown exerted an inhibitory effect on tumour growth in vivo, and reconstituted expression of PADI4WT, but not the expression of the Flag-PADI4D473A or Flag-PADI4C645A mutant reversed PADI4 knockdown-induced suppression of tumour growth (Fig. 6a, b and Supplementary Fig. 5b). Furthermore, western blot and qPCR analysis of tumour tissue lysates confirmed the expression of PADI4, as well as the effect of different PADI4 mutant constructs on the regulation of HIF-1α, LDHA, and PDK1 (Supplementary Fig. 5c, d), and the results were consistent with the trend in tumour growth. These in vivo data revealed that PADI4 maintains HIF-1α stability and promotes tumour growth in an enzyme-dependent manner.
Fig. 6. PADI4-mediated HIF-1αR698 citrullination promotes HCC tumorigenesis.
a, b The cell lines described in Fig. 3g were injected subcutaneously into male BALB/c nude mice (n = 6 per group). Tumour volume was measured starting on Day 10 and determined based on calliper measurements every 3 days (a). Photographs show xenografts at the end of the experiment (Day 25) (b). c Representative results of the IHC analysis of HIF-1α, HIF-1αR698Cit and PADI4 in normal liver tissues (Non-tumour, n = 40) and HCC specimens (Tumour, n = 41) with low or high HIF-1α expression. Scale bars, 50 μm. d Statistical analysis of the IHC results shown in Fig. 6c. HIF-1α, HIF-1αR698Cit and PADI4 protein levels in high HIF-1α expression samples were quantified using HALO software (Non-tumour, n = 27; Tumour, n = 28). e HIF-1αR698Cit protein levels are positively correlated with PADI4 protein levels in clinical HCC lesions (n = 41). Pearson correlation analyses were performed. f Representative multiplex IHC (mIHC) image of HIF-1αR698Cit and PADI4 in HCC samples. Scale bars, 50 µm (upper panel), 20 µm (left layer panel) and 10 µm (right layer panel). The intensity profiles of each line were quantified with ImageJ software and drawn with GraphPad Prism 7.0. Experiments were performed in four HCC tumour specimens. The results are shown as the means of the colocalization coefficient of HIF-1αR698Cit and PADI4. Error bars denote the mean ± SEM (a) and mean ± SD (d). Statistical analyses were performed by ordinary two-way ANOVA with Turkey’s multiple comparisons test (a), two-tailed paired Student’s t-test (d), or Pearson correlation analyses (e). Source data are provided as a Source Data file.
Next, to better illustrate the potential pathological and clinical relevance of PADI4-mediated HIF-1αR698 citrullination, immunohistochemistry (IHC) assays were performed using 41 HCC lesions and 40 adjacent noncancerous tissue samples. As expected, PADI4, HIF-1α, and citrullinated HIF-1αR698 levels were all highly expressed in tumour tissues compared with matched adjacent noncancerous tissues. In addition, in tumour tissues with high HIF-1α expression, the expression of citrullinated HIF-1αR698 and PADI4 was correspondingly increased, and vice versa (Fig. 6c, d). Furthermore, quantification analysis revealed that HIF-1αR698 citrullination expression was positively correlated with both HIF-1α and PADI4 protein levels in liver cancer (Fig. 6e and Supplementary Fig. 5e). Moreover, multiplex IHC (mIHC) assays showed high colocalization between PADI4 and citrullinated HIF-1αR698 (Fig. 6f). Taken together, the xenograft model and clinical data reveal that PADI4-mediated HIF-1α citrullination promotes tumour progression, and that the PADI4-HIF-1αR698-cit axis is a promising therapeutic target for patients with liver cancer.
Discussion
HIF-1α plays a crucial role in the hypoxic responses of cells and gene expression involved in cell survival, angiogenesis, and metabolism. In addition to classical PHD-mediated hydroxylation regulation of HIF-1α protein expression, other modifications can regulate HIF-1α stability41. Herein, we show that PADI4 directly binds to and citrullinates HIF-1α at R698, and that PADI4-dependent HIF-1αR698 citrullination enhances HIF-1α stability by competitively blocking the VHL-HIF-1α interaction. We provide clear evidence to show that PADI4 and citrullinated HIF-1αR698 are highly co-expressed in tumour tissues and suggest that PADI4 is an applicable target for the treatment of liver cancers (Supplementary Fig. 6a).
Protein citrullination is a crucial PTM catalysed by the PAD family, and several studies have shown the physiological importance of PADI4 in biological processes. Genetic variants of the PADI4 gene have been associated with RA, and anti-citrullinated peptide antibodies (ACPAs) are biomarkers of RA used for clinical diagnosis. Since PADI4 plays a critical role in the formation of NETs, it is also an important regulator of the innate immune response42. A few studies have revealed the role of PADI4 in cancer cells30. For example, Yuzhalin et al. found that the extracellular matrix in liver metastases contains a large proportion of citrullinated proteins and the levels of PADI4 were higher in colorectal cancer (CRC) liver metastases than primary CRC, or adjacent colon or liver, demonstrating that PADI4 is essential for liver metastases from CRC43. Positive effects of NETs on tumour proliferation, metastasis and awakening dormant tumour cells also indicate the tumour promoting role of PADI444. In this report, we propose that PADI4 catalyses HIF-1α citrullination via direct interaction, which protects HIF-1α from proteasome-dependent degradation. Knockdown of PADI4 markedly decreased HIF-1α protein expression without affecting its mRNA level, and this outcome was reserved by the treatment with the proteasome inhibitor MG132 under hypoxic conditions (Fig. 3e and Supplementary Fig. 3b, c). Furthermore, we prove that HIF-1αR698 citrullination is essential for HIF-1α protein stability as the uncitrullinated HIF-1αR698A mutant degraded faster than HIF-1αWT (Fig. 3f). Mechanistically, when PHDs bind to and hydroxylate HIF-1α under normoxic conditions, only a few PADI4 proteins could interact with HIF-1α. However, when PHDs lose enzymatic activity under hypoxic conditions, PADI4 significantly binds to HIF-1α and citrullinates the R698 residue on HIF-1α, sequestering HIF-1α away from VHL and preventing its subsequent degradation (Fig. 4b–k). In addition to the long-held view that HIF-1α is stable owing to the decreased activities of PHDs under hypoxic conditions, we provide an extra mechanism based on posttranslational modification findings that PADI4-mediated citrullination of HIF-1α is critical for HIF-1α stabilisation under hypoxic conditions. Further research to address the competitive binding of PADI4 and VHL near HIF-1 will bring this old and fascinating area of research back to life.
Hypoxia is widespread in solid tumours, and its adverse biological effects are closely associated with tumour progression and metastasis, therefore, targeting HIF-1α has been a promising therapeutic strategy for solid tumours2. Interestingly, a recent study showed that hypoxia induced PADI4 expression in a HIF-dependent manner and that PADI4-mediated histone citrullination was required for HIF-1 transcriptional activity45. Combined with our finding that PADI4 catalyses HIF-1α citrullination to maintain HIF-1α stability, these studies collectively indicate that the HIF-1α-PADI4 axis is a positive feedback loop that coordinates tumour development and would be a promising target of tumour therapy. PPIs are important to disease development. With the recent development of high-resolution protein structure studies and virtual screening strategies, PPIs have been considered potential targets for disease treatment46,47. For example, as the interaction of the menin-mix lineage leukaemia gene (MLL) is critical for leukaemia proliferation and survival, Krivtsov et al. used orally administered small-molecule VTP50469 to inhibit the menin-MLL interaction pharmacologically and thus demonstrated curative effects and survival benefits in patient-derived xenograft (PDX) models of MLL-rearranged acute lymphoblastic leukaemia48. Considering this finding and our results showing that the PADI4-HIF-1α interaction is critical for HIF-1α stability and that the HIF-1αR698 residue is deeply embedded into the enzymatic pocket of PADI4, we propose that this interaction can likely be disrupted by pharmacological agents. Through a series of screening and confirmation of cocrystal structures, we found that DHE, an FDA-approved drug for migraine treatment, binds to PADI4 and occupies the enzymatic pocket of PADI4 to block its interaction with HIF-1α and suppresses HIF-1αR698 citrullination (Fig. 5a, c–g). Moreover, a GSEA analysis with RNA-seq data showed that DHE treatment downregulates HIF-1α-related pathways, including hypoxia response, glycolysis and adipogenesis pathways (Fig. 5k, l). An in vivo xenograft mouse model suggested that DHE is a potential antitumour agent that induces HIF-1α degradation by targeting the enzymatic activity of PADI4 (Fig. 5m, n and Supplementary Fig. 4f). In summary, our data proved the binding of DHE to PADI4 and the subsequent inhibitory effects of DHE on PADI4-HIF-1α interaction and HIF-1α stability, making it possible to clinically target HIF-1α for tumour therapy, but more comprehensive studies are required to better elucidate the precise mechanism by which DHE targets PADI4 in the future.
Through an analysis of clinical HCC samples, we found that citrullinated HIF-1αR698 expression positively correlated with HIF-1α and PADI4 expression, and was significantly increased in HCC tumour tissues compared to the adjacent normal liver tissues (Fig. 6c–e and Supplementary Fig. 5e). mIHC experiments confirmed the colocalization of HIF-1αR698 citrullination and PADI4 in clinical HCC tumours (Fig. 6f). These results indicate that PADI4-mediated HIF-1αR698 citrullination is highly correlated with HCC tumorigenesis, and that citrullinated HIF-1αR698 is a potential marker and drug target for HCC. In summary, we show that PADI4-mediated citrullination is a key modification in mediating HIF-1α stability and reveal a mechanism by which HIF-1αR698 citrullination reinforces HIF-1α stability by preventing the VHL-HIF-1α interaction. Our findings also demonstrate the tumour therapeutic potential of targeting the PADI4-HIF-1α complex and identify DHE as a promising agent for liver cancer treatment.
Methods
Our research complies with all relevant ethical regulations of the South China University of Technology and the University of Science and Technology of China. Animal experimental procedures were approved by the Animal Research Ethics Committee of the South China University of Technology (AEC number, 2024061) and were performed following the guidelines for the use of laboratory animals.
Cell culture and reagents
Human cell lines (the HEK293T, Hep3B, HepG2, RCC10 and RCC90 cell lines) and the mouse Hepa1-6 cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, 12800) supplemented with 10% foetal bovine serum (FBS) and 1% penicillin-streptomycin. The cells were cultured at 37 °C with 5% CO2 in humidified incubators. Hypoxia exposure was achieved by placing cells into a modular chamber (Don Whitley Scientific) flushed with a gas mixture consisting of 1% O2, 5% CO2 and 94% N2 at 37 °C. MG132 (MCE, HY-13259), BB-Cl-Amidine (BBCA) (MCE, HY-111347), Cycloheximide (CHX) (MCE, HY-12320) and Dihydroergotamine mesylate (DHE) (MCE, HY-B0670A), Deferoxamine mesylate (DFO) (TargetMol, 138-14-7) were purchased from the indicated vendor.
Plasmid construction and mutagenesis
Polymerase chain reaction (PCR)-amplified hsa-HIF-1α, -PADI4, -VHL and mus-Hif-1α, -Padi4 products were cloned and inserted into the pSin-3×Flag and pSin-HA vectors. PCR-amplified full-length hsa-HIF-1α and different HIF-1α truncation mutants were cloned and inserted into the pGEX-4T1-GST vector. PCR-amplified hsa-PADI4 and hsa-VHL were cloned and inserted into the pET-22b-(N-2Flag-tag)-(C-His) vector. Hsa-HIF-1αR17A, hsa-HIF-1αR273A, hsa-HIF-1αR463A, hsa-HIF-1αR665A, hsa-HIF-1αR671A, hsa-HIF-1αR698A and hsa-PADI4D473A, hsa-PADI4C645A and mus-Hif-1αR709A mutants were generated using QuikChange site-directed mutagenesis methods. shRNAs were constructed via ligation of an oligonucleotide targeting hsa-PADI4 and hsa-VHL into an AgeI/EcoRI-digested pLKO.1 vector. Primers used for plasmid construction and mutagenesis are listed in Supplementary Data 1 and Supplementary Table 1.
Transfection and lentiviral infection
Transfection was performed using PEI (Polysciences, 23966-2) or Lipofectamine (Invitrogen, L3000015) according to the respective manufacturer’s recommendations. For lentiviral infection, 293T cells were transfected with packaging vectors (psPAX2 and pMD2.G) and the indicated plasmids. After 48 h of cultivation, the supernatant containing virus particles was collected for infection. Target cells were infected with lentivirus in the presence of (8 μg/ml) polybrene and selected after (0.5 μg/ml) puromycin treatment.
Western blot analysis
To carry out western blot analyses, total protein was collected and extracted from cells or tissues with RIPA buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% NP-40) containing a protease cocktail, and then was quantified using a Bradford assay kit (Sangon Biotech, C603031). After denaturation, equal amounts of proteins were separated by 8–12% SDS-PAGE and transferred to NC membranes, followed by blocking with 5–10% skim milk in TBST for 1 h. Subsequently, the blots were incubated with the indicated primary antibodies and HRP-conjugated secondary antibodies according to the protocols recommended. Finally, signals were visualised using an ECL kit and a chemiluminescent imaging system (Tanon-5200). The antibodies used are listed in Supplementary Table 2.
Quantitative real-time PCR analysis
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) and 2 μg of purified RNA was used for cDNA synthesis using a HiScript II First Strand cDNA Synthesis Kit (Vazyme, R211-02). Quantitative PCR assays were performed in triplicate with diluted cDNA, the indicated primers and ChamQTM Universal SYBR qPCR Master Mix (Vazyme, Q711-02) according to the manufacturer’s instructions. The qPCR data were analyzed via the 2−ΔΔCt method. Primers used for qPCR are listed in Supplementary Table 3.
Immunoprecipitation assays
To perform IP assays, cells transfected with the indicated plasmids were harvested and lysed in IP lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, and 1% NP-40) supplemented with the protease inhibitor cocktail for 1.5 h on ice. After centrifugation at 12,000×g for 10 min at 4 °C, the supernatants were precleared with protein A/G agarose beads for 1 h, quantified and subsequently incubated with the indicated primary antibodies overnight at 4 °C. Next, 25 μL of beads were incubated for 2 h and washed with IP buffer five times, followed by boiling with SDS loading buffer for denaturation. Finally, the samples were analyzed by western blotting as described above.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 for 1 h, blocked with 2% BSA for 1 h, and incubated with the indicated primary antibodies for 12 h at 4 °C. Next, incubation with fluorescence dye-conjugated secondary antibodies and DAPI was performed according to standard protocols. Images were viewed and obtained with a confocal microscope system (Zeiss LSM 800), and the fluorescence intensity was quantified by Fiji-ImageJ.
GST pull-down
cDNAs encoding PADI4 and VHL were cloned and inserted into a His-tagged expression vector PET-22b (+), and cDNAs encoding GST-fused HIF-1α (full-length or truncations) were cloned and inserted into a pGEX-4T1 vector. Plasmids were transfected into the bacterial strain BL21 (DE3) and the expression of fusion proteins was induced by adding IPTG to the culture at a final concentration of 1 mM at 16 °C for 20–24 h. For purification, His-tagged proteins were selectively bound to a nickel column and washed with elution buffer containing gradually increasing concentrations of imidazole (50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 50 mM, 100 mM or 300 mM imidazole). GST-tagged proteins were purified by GSH-conjugated agarose beads. For pull-down assays, purified recombinant proteins were incubated in pull-down buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.1% NP-40 and 5 mM DTT) at 4 °C for 2 h, washed three times and analyzed by western blotting and Coomassie blue staining.
In vitro citrullination experiments
For the in vitro citrullination experiment, purified GST-tagged HIF-1α proteins were eluted from beads with elution buffer (50 mM Tris-HCl, pH 8.0; 20 mM reduced glutathione) and incubated with His-PADI4 WT or His-PADI4 mutant proteins in reaction buffer (100 mM Tris-HCl, pH 7.4; 10 mM CaCl2 and 4 mM DTT) at 37 °C for 1.5 h. Samples were evaluated by western blotting with an anti-pan citrulline antibody (Invitrogen, MA5-27573).
Thermal shift assay
For the thermal shift assay, purified protein (1 μg) was added to 1.5 mL EP tubes and the volume was adjusted to 120 μL by the addition of buffer or purified proteins and DHE depending on the experimental setup. The samples were heated at a temperature for 5 min. After heating, the samples were immediately cooled down on ice and centrifuged for 10 min at 13000×g at 4 °C. The 80 μL supernatants were transferred to 0.4-μm Ultra-free Centrifugal Filter (Merck) and continually centrifuged for 10 min at 900×g. The 40 μL filtrate were transferred to new EP tubes and analyzed by western blotting.
Protein docking and virtual screening methods
Protein docking and virtual screening were based on the crystal structure of human peptidylarginine deiminase type 4 (PADI4) (RCSB PDB: 1WDA) by Autodock-vina v1.1.2. The protein structure was prepared with AutoDockTools v1.5.7. The protein structure of the HIF-1α peptide was predicted with RoseTTAFold. The library used for the virtual screening of potential PADI4 inhibitors was (FDA)-approved commercial agents library identified by the ZINC15 database. The virtual screening summary is listed in Supplementary Data 2.
RNA-seq methods
Total RNA was extracted from cell lines using TRIzol Reagent (Life Technologies). RNA integrity was assessed by RNA integrity number and determined using an Agilent 2100 Bioanalyzer. A total amount of 3 µg of RNA per sample was used for analysis. Sequencing sampling was performed from one single replicate. Libraries were generated using a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB). RNA-seq was performed on an Illumina NovaSeq 6000 platform by Novogene (Tianjin). Reads were aligned to the human genome hg38. STAR v2.7.10b and RSEM v1.3.1 and used to analyze RNA-seq data. Gene differential expression analysis was carried out with the DEGSeq Rpackage (1.26.0). Gene set enrichment analysis was performed by DAVID Bioinformatics Resources.
MS analysis
To identify the proteins binding to HIF-1α, Hep3B cells were transfected with pSin-3xFlag-EV or HIF-1α plasmid. After culturing under hypoxic conditions for 6 h, the cells were harvested and lysed in IP lysis buffer, and the lysate was subjected to IP assay with an anti-Flag antibody. The immunoprecipitate was digested with trypsin at 37 °C for 12 h. The tryptic peptides were dissolved in 0.1% formic acid and subsequently subjected to Q Exactive plus mass spectrometry (Thermo Fisher Scientific) coupled with an EASY-nLC1200 HPLC system (Thermo Fisher Scientific) via a nano-electrospray ion source in data-dependent mode. The mass spectrometry (MS) raw data were searched against the human UniProt database using Proteomics Discovery Software (version 2.1, Thermo Fisher Scientific). In the Sequest-HT setting part, we selected trypsin as the proteolytic enzyme and peptides length between 6–144, with two missing cleavages sites allowed. The first search mass tolerance and the fragment mass tolerance were 10 p.p.m. and 0.02 Da, respectively. Hereafter, the peptide-spectrum-matches and proteins false discovery rates were set to <1% released and 0.5% strictly. The result was analyzed by Metascape 60 and then, GO enrichment analysis was performed with results visualised via Cytoscape 3.0.
To identify the citrullinated residues on HIF-1α, in vitro citrullination of HIF-1α mediated by PADI4 was carried out. The samples were separated by SDS-PAGE, and bands with a molecular weight of ~130 kDa, as visualised by Coomassie blue staining, were excised and used for further LC-MS/MS analysis (Aimsmass, Shanghai).
Cell proliferation
Cell proliferation was measured by cell counting. The indicated cells were seeded on 12-well plates (40,000 cells per well, n = 3). After 12 h, the medium was replaced with fresh medium containing 10% FBS, and then the cells were placed in a hypoxic incubator. The cells were counted every 24 h.
Xenograft animal model
The experimental procedures were approved by the Animal Research Ethics Committee of the South China University of Technology (AEC number, 2024061). Five-week-old BALB/c male nude mice were obtained from SJA Laboratory Animal Company of China, and were randomly allocated to experimental groups. All animals were housed at a suitable temperature (22–24 °C) and humidity (40–70%) under a 12/12-h light/dark cycle with unrestricted access to food and water for the duration of the experiment. The xenograft experiment follows the humane endpoint. Indicators such as huddled posture, immobility, ruffled fur, failure to eat, hypothermia (colonic temperature of <34 °C), or weight loss (>20%) may be useful objective criteria for early euthanasia. The animals will be euthanized immediately if they are unable to stand or if they display agonal breathing, severe muscular atrophy, severe ulceration, or uncontrolled bleeding. The subcutaneous tumour maximum volume was 2000 mm3 and authorised by the Committees on Animal Research and Ethics, and was not exceeded at any time during the experiments. In xenograft experiments, 5 × 106 Hep3B cells stably expressing EV (with or without endogenous PADI4 knockdown), WT PADI4 or PADI4 mutants (with endogenous PADI4 knocked down) were subcutaneously inoculated into the flanks of the mice. Tumour size was measured every 3 days and converted to volume using the following formula: length (mm) × width (mm) × depth (mm) × 0.52. Tumour volumes, tumour weights, photos and other experimental indicators were obtained at the end of the experimental period.
For DHE treatment, 5 × 106 Hep3B cells were subcutaneously inoculated into 5-week-old male nude mice. Two weeks later, mice bearing tumours were randomly allocated to three groups and given DMSO or DHE (25 mg/kg or 75 mg/kg) via i.g. every 2 days. Tumour volumes, tumour weights, photos and other experimental indicators were obtained at the end of the experimental period.
Clinical HCC samples and immunohistochemistry assays
Formalin-fixed, paraffin-embedded primary HCC specimens were randomly selected from the archives of the First Affiliated Hospital of the University of Science and Technology of China (Hefei, China). Previously obtained patients’ written informed consent and research approval from the Institutional Research Ethics Committee of the First Affiliated Hospital of the University of Science and Technology of China were obtained to use the materials for research purposes. Among the 41 HCC specimens, 40 were paired HCC lesions and adjacent noncancerous tissue samples. Sex and gender were not considered in the study design, nor were sex- and gender-based analyses performed since HCC occurs in both men and women. Of the 41 HCC patients, four were women, and 37 were men, with 32 patients over the age of 50. For the IHC assay, after dewaxing with xylene and rehydrating with graded ethanol, the samples were subjected to antigen retrieval, followed by incubation with 0.3% hydrogen peroxide for 15 min to block endogenous peroxidase activity. Next, 10% goat serum was used to preincubate the samples and thus prevent nonspecific staining, and then, the samples were incubated with the appropriate primary antibody (anti-HIF-1α, anti-HIF-1αR698-cit or anti-PADI4 antibody) at 4 °C for 10 h. Subsequently, secondary antibodies were used, followed by the incubation with a diluted DAB chromogen solution according to the manufacturer’s instructions. Images were obtained with a Digital Pathogen Scanner (Aperio CS2), and the results were analyzed with Halo software (v 3.3.14).
Statistical analysis
The data are presented as the mean ± SD or mean ± SEM of three independent experiments. Statistical significance was assessed by Student’s t-test or ANOVA followed by Tukey’s multiple comparisons test. P < 0.05 indicates a significant difference between the indicated groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
This work is supported in part by the National Natural Science Foundation of China (82341013, 82130087, 92357301 to P.G., 82273221 to L.S. and 82103186 to S.S.), the Basic Funding of Guangzhou Municipal Science and Technology Bureau (2024A04J6493 to L.S.) and the Special Programme for Young Scholars of Southern Medical University (G623281133 to L.S.). Illustrations were created with BioRender.com.
Author contributions
P.G. and L.S. conceived and supervised the study. R.C., Z.L., S.S., H.Z., L.S. and P.G. designed the experiments. R.C., Z.L., C.Z., C.S., L.G. and K.F. performed immunoblotting, cloning, cell biology, biochemistry and animal experiments. R.C., Z.L., K.Y., H.W. and R.L. performed and analyzed clinical HCC specimens. S.S. analyzed the RNA-seq and mass spectrometry proteomics data. R.C., Z.L., L.S. and P.G. wrote the paper. All the authors read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Mei-Yee Koh, Yatrik Shah, Arseniy Yuzhalin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the iProX repository with the data set identifier PXD046340 (https://www.iprox.cn//page/project.html?id=IPX0007354000). RNA-seq data supporting the findings of this study have been deposited into GEO and the public with accession no. GSE233772. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rui Chen, Zhiyuan Lin, Shengqi Shen.
Contributor Information
Linchong Sun, Email: sunlc@mail.ustc.edu.cn.
Ping Gao, Email: pgao2@ustc.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51882-w.
References
- 1.Harris, A. L. Hypoxia–a key regulatory factor in tumour growth. Nat. Rev. Cancer2, 38–47 (2002). 10.1038/nrc704 [DOI] [PubMed] [Google Scholar]
- 2.Singleton, D. C., Macann, A. & Wilson, W. R. Therapeutic targeting of the hypoxic tumour microenvironment. Nat. Rev. Clin. Oncol.18, 751–772 (2021). 10.1038/s41571-021-00539-4 [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., Han, F., Du, Y., Shi, H. & Zhou, W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct. Taget. Ther.8, 70 (2023). 10.1038/s41392-023-01332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer3, 721–732 (2003). 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- 5.Semenza, G. L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest.123, 3664–3671 (2013). 10.1172/JCI67230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Missiaen, R., Lesner, N. P. & Simon, M. C. HIF: a master regulator of nutrient availability and metabolic cross-talk in the tumor microenvironment. EMBO J.42, e112067 (2023). 10.15252/embj.2022112067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G. & Thompson, C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab.7, 11–20 (2008). 10.1016/j.cmet.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 8.Rankin, E. B. & Giaccia, A. J. Hypoxic control of metastasis. Science352, 175–180 (2016). 10.1126/science.aaf4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGettrick, A. F. & O’Neill, L. The role of HIF in immunity and inflammation. Cell Metab.32, 524–536 (2020). 10.1016/j.cmet.2020.08.002 [DOI] [PubMed] [Google Scholar]
- 10.Semenza, G. L. Hypoxia-inducible factor 1: control of oxygen homeostasis in health and disease. Pediatr. Res.49, 614–617 (2001). 10.1203/00006450-200105000-00002 [DOI] [PubMed] [Google Scholar]
- 11.Luo, Z. et al. Hypoxia signaling in human health and diseases: implications and prospects for therapeutics. Signal Transduct. Target. Ther.7, 218 (2022). 10.1038/s41392-022-01080-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruick, R. K. & McKnight, S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science294, 1337–1340 (2001). 10.1126/science.1066373 [DOI] [PubMed] [Google Scholar]
- 13.Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell107, 43–54 (2001). 10.1016/S0092-8674(01)00507-4 [DOI] [PubMed] [Google Scholar]
- 14.Ivan, M. et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science292, 464–468 (2001). 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- 15.Kaelin, W. J. & Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell30, 393–402 (2008). 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 16.Pan, C., Li, B. & Simon, M. C. Moonlighting functions of metabolic enzymes and metabolites in cancer. Mol. Cell81, 3760–3774 (2021). 10.1016/j.molcel.2021.08.031 [DOI] [PubMed] [Google Scholar]
- 17.Lee, J. M., Hammaren, H. M., Savitski, M. M. & Baek, S. H. Control of protein stability by post-translational modifications. Nat. Commun.14, 201 (2023). 10.1038/s41467-023-35795-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, H., Yang, L., Liu, M. & Luo, J. Protein post-translational modifications in the regulation of cancer hallmarks. Cancer Gene Ther.30, 529–547 (2023). 10.1038/s41417-022-00464-3 [DOI] [PubMed] [Google Scholar]
- 19.Jeong, J. W. et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell111, 709–720 (2002). 10.1016/S0092-8674(02)01085-1 [DOI] [PubMed] [Google Scholar]
- 20.Kang, J., Chun, Y. S., Huh, J. & Park, J. W. FIH permits NAA10 to catalyze the oxygen-dependent lysyl-acetylation of HIF-1alpha. Redox Biol.19, 364–374 (2018). 10.1016/j.redox.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flugel, D., Gorlach, A., Michiels, C. & Kietzmann, T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol. Cell Biol.27, 3253–3265 (2007). 10.1128/MCB.00015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo, K. S. et al. SIRT2 regulates tumour hypoxia response by promoting HIF-1alpha hydroxylation. Oncogene34, 1354–1362 (2015). 10.1038/onc.2014.76 [DOI] [PubMed] [Google Scholar]
- 23.Bao, L. et al. Methylation of hypoxia-inducible factor (HIF)-1alpha by G9a/GLP inhibits HIF-1 transcriptional activity and cell migration. Nucleic Acids Res.46, 6576–6591 (2018). 10.1093/nar/gky449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, Y. et al. Methylation-dependent regulation of HIF-1alpha stability restricts retinal and tumour angiogenesis. Nat. Commun.7, 10347 (2016). 10.1038/ncomms10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer, C. M. et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell54, 820–831 (2014). 10.1016/j.molcel.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng, J., Kang, X., Zhang, S. & Yeh, E. T. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell131, 584–595 (2007). 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Venrooij, W. J. & Pruijn, G. J. Citrullination: a small change for a protein with great consequences for rheumatoid arthritis. Arthritis Res.2, 249–251 (2000). 10.1186/ar95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuhrmann, J., Clancy, K. W. & Thompson, P. R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev.115, 5413–5461 (2015). 10.1021/acs.chemrev.5b00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darrah, E. & Andrade, F. Rheumatoid arthritis and citrullination. Curr. Opin. Rheumatol. 30, 72–78 (2018). 10.1097/BOR.0000000000000452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuzhalin, A. E. Citrullination in cancer. Cancer Res.79, 1274–1284 (2019). 10.1158/0008-5472.CAN-18-2797 [DOI] [PubMed] [Google Scholar]
- 31.Falcao, A. M. et al. PAD2-mediated citrullination contributes to efficient oligodendrocyte differentiation and myelination. Cell Rep.27, 1090–1102 (2019). 10.1016/j.celrep.2019.03.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christophorou, M. A. et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature507, 104–108 (2014). 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright, H. L., Moots, R. J. & Edwards, S. W. The multifactorial role of neutrophils in rheumatoid arthritis. Nat.Rev.Rheumatol.10, 593–601 (2014). 10.1038/nrrheum.2014.80 [DOI] [PubMed] [Google Scholar]
- 34.Sorensen, O. E. & Borregaard, N. Neutrophil extracellular traps - the dark side of neutrophils. J. Clin. Invest.126, 1612–1620 (2016). 10.1172/JCI84538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curran, A. M., Naik, P., Giles, J. T. & Darrah, E. PAD enzymes in rheumatoid arthritis: pathogenic effectors and autoimmune targets. Nat. Rev. Rheumatol.16, 301–315 (2020). 10.1038/s41584-020-0409-1 [DOI] [PubMed] [Google Scholar]
- 36.Yu, R. et al. Hypoxia induces production of citrullinated proteins in human fibroblast-like synoviocytes through regulating HIF1alpha. Scand. J. Immunol.87, e12654 (2018). 10.1111/sji.12654 [DOI] [PubMed] [Google Scholar]
- 37.Arita, K. et al. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol.11, 777–783 (2004). 10.1038/nsmb799 [DOI] [PubMed] [Google Scholar]
- 38.Henley, M. J. & Koehler, A. N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug. Discov.20, 669–688 (2021). 10.1038/s41573-021-00199-0 [DOI] [PubMed] [Google Scholar]
- 39.Martinez, M. D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science341, 84–87 (2013). 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- 40.Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc.9, 2100–2122 (2014). 10.1038/nprot.2014.138 [DOI] [PubMed] [Google Scholar]
- 41.Albanese, A., Daly, L. A., Mennerich, D., Kietzmann, T. & See, V. The role of hypoxia-inducible factor post-translational modifications in regulating its localisation, stability, and activity. Int. J. Mol. Sci.22, 268 (2020). 10.3390/ijms22010268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, P. et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med.207, 1853–1862 (2010). 10.1084/jem.20100239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuzhalin, A. E. et al. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat. Commun.9, 4783 (2018). [DOI] [PMC free article] [PubMed]
- 44.Adrover, J. M., McDowell, S., He, X. Y., Quail, D. F. & Egeblad, M. NETworking with cancer: the bidirectional interplay between cancer and neutrophil extracellular traps. Cancer Cell41, 505–526 (2023). 10.1016/j.ccell.2023.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y. et al. Histone citrullination by PADI4 is required for HIF-dependent transcriptional responses to hypoxia and tumor vascularization. Sci. Adv.7, eabe3771 (2021). [DOI] [PMC free article] [PubMed]
- 46.Xie, X. et al. Recent advances in targeting the “undruggable” proteins: from drug discovery to clinical trials. Signal. Transduct. Target. Ther.8, 335 (2023). 10.1038/s41392-023-01589-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, D. E., Bayly, A. R., Abell, C. & Skidmore, J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug. Discov.15, 533–550 (2016). 10.1038/nrd.2016.29 [DOI] [PubMed] [Google Scholar]
- 48.Krivtsov, A. V. et al. A menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell36, 660–673 (2019). 10.1016/j.ccell.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the iProX repository with the data set identifier PXD046340 (https://www.iprox.cn//page/project.html?id=IPX0007354000). RNA-seq data supporting the findings of this study have been deposited into GEO and the public with accession no. GSE233772. Source data are provided with this paper.