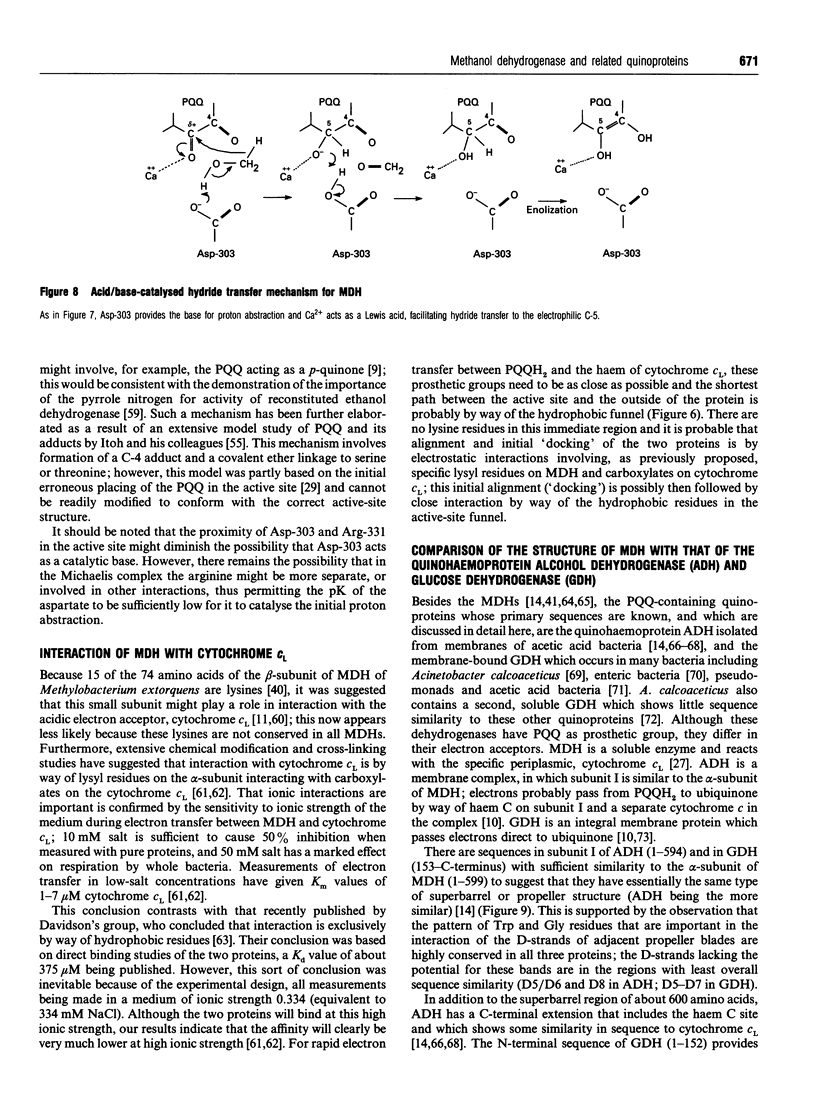
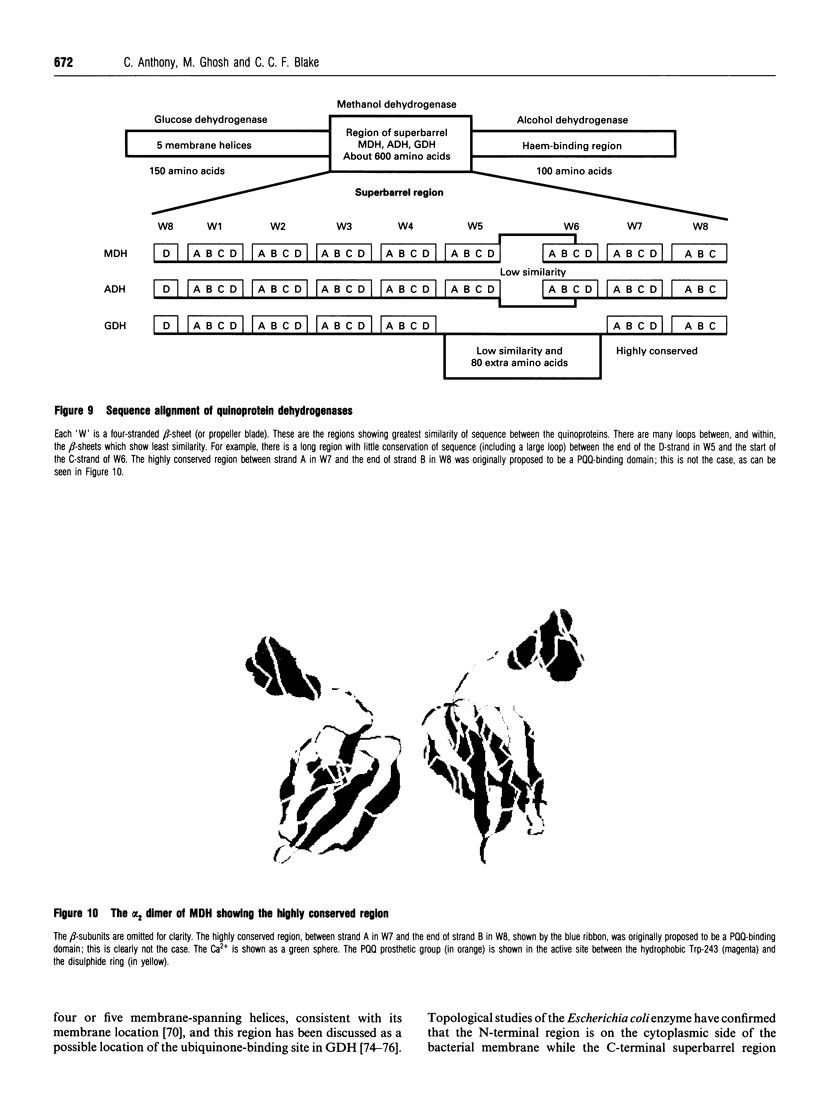
Full text
PDF

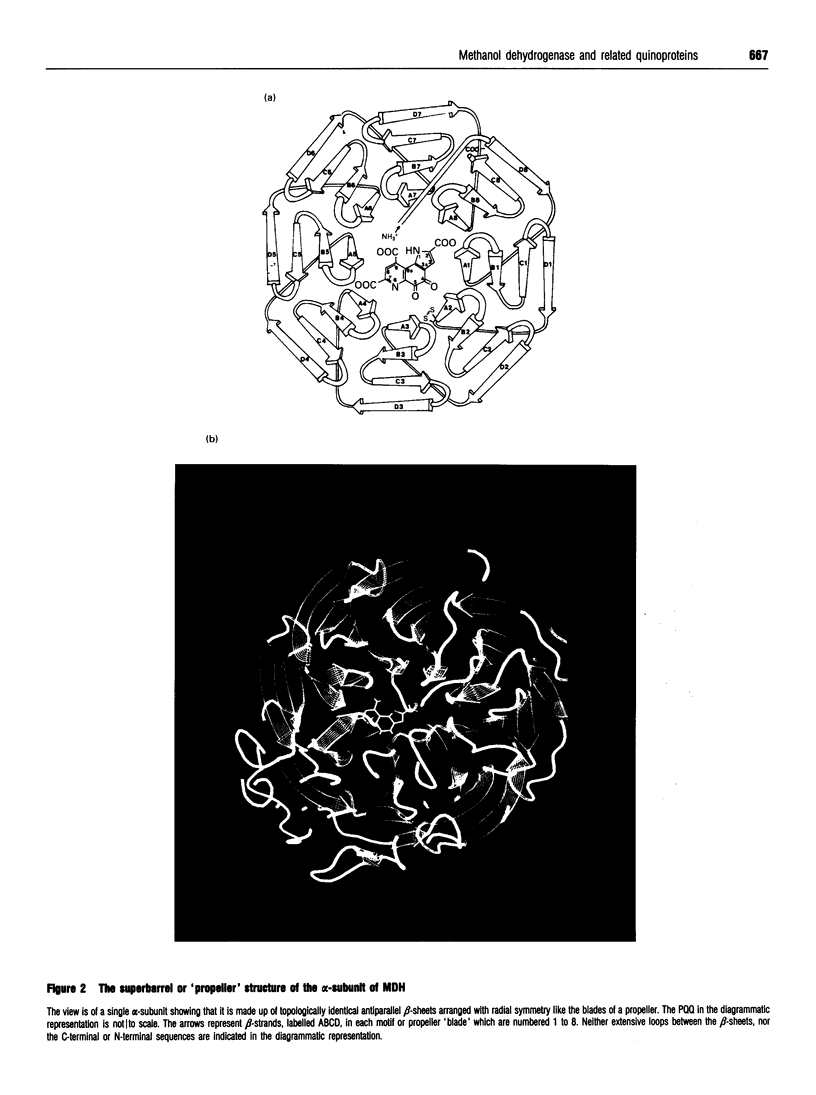
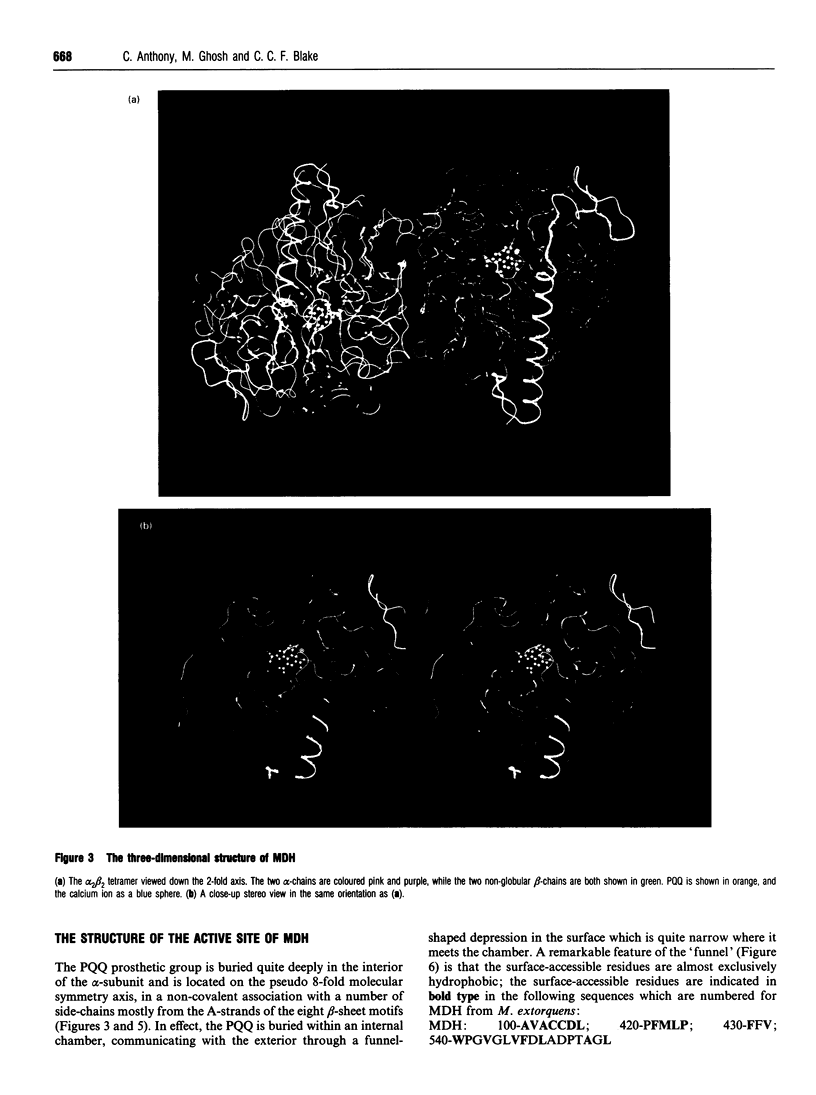
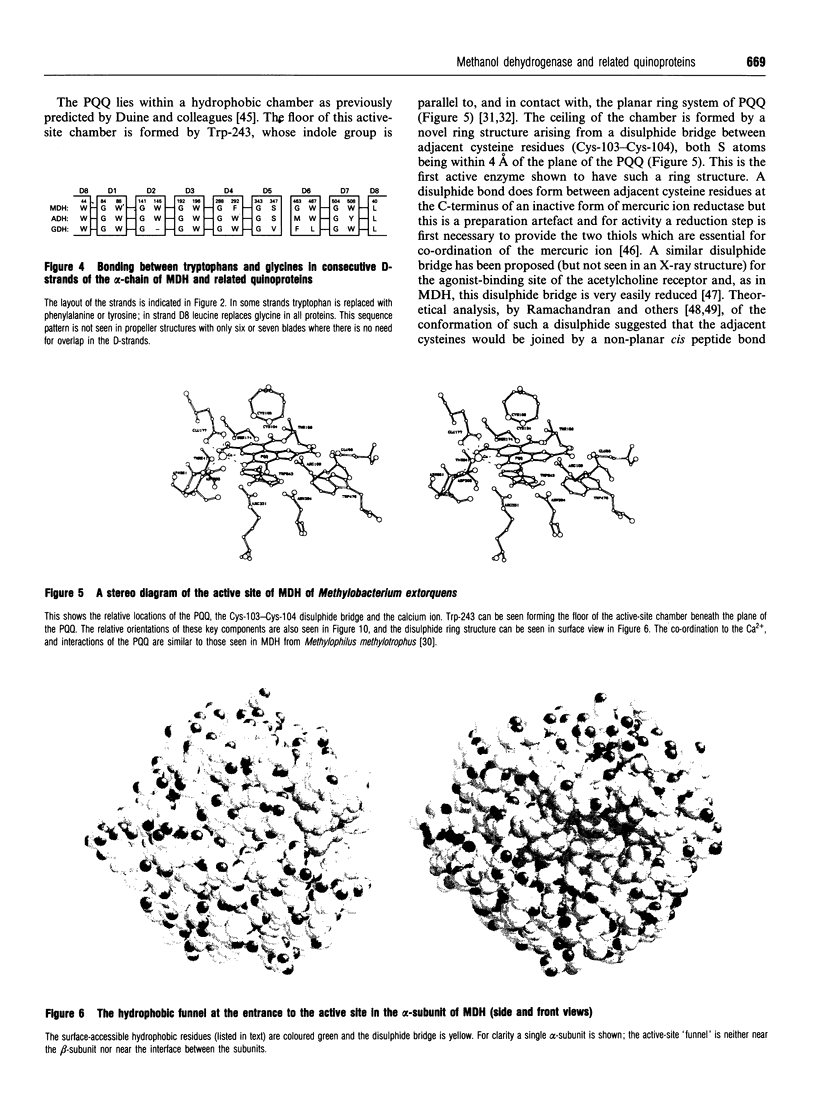
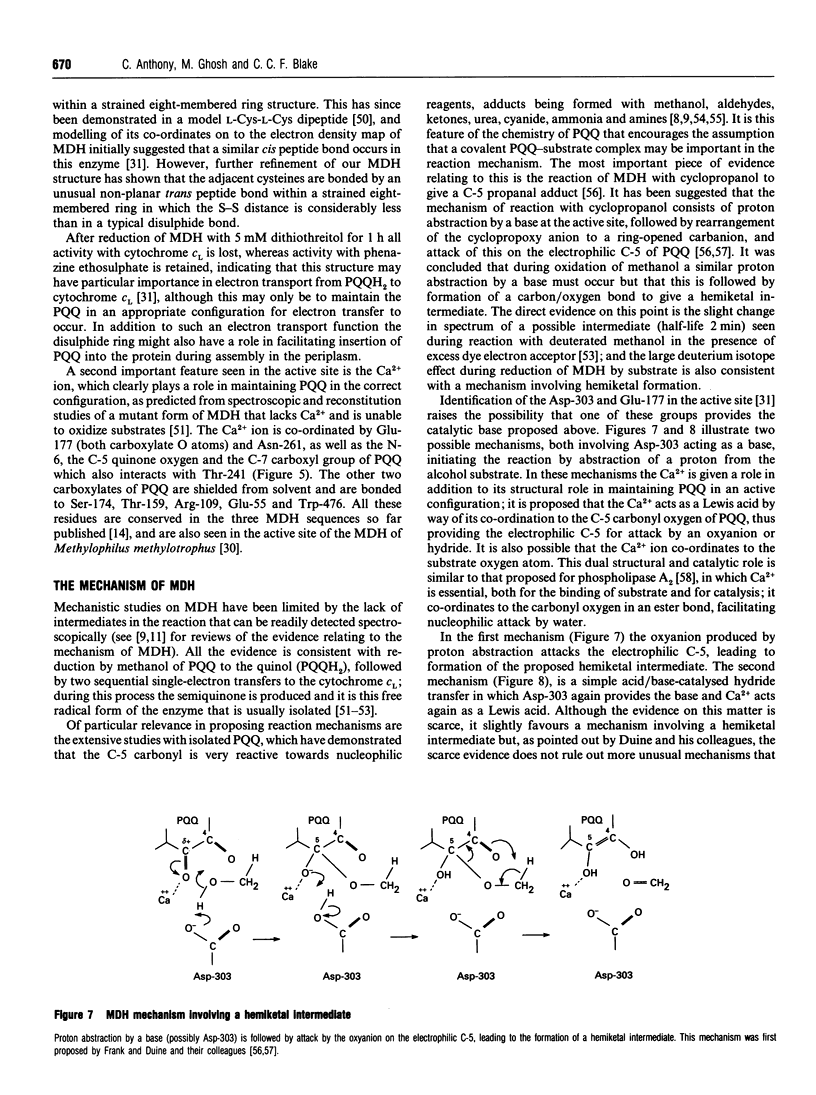


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Morris C. J., Nunn D. N., Anthony C., Lidstrom M. E. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene. 1990 May 31;90(1):173–176. doi: 10.1016/0378-1119(90)90457-3. [DOI] [PubMed] [Google Scholar]
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- Anthony C. The c-type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992 Jan 30;1099(1):1–15. [PubMed] [Google Scholar]
- Anthony C. The oxidation of methanol in gram-negative bacteria. FEMS Microbiol Rev. 1990 Dec;7(3-4):209–214. doi: 10.1111/j.1574-6968.1990.tb04914.x. [DOI] [PubMed] [Google Scholar]
- Anthony C. The structure of bacterial quinoprotein dehydrogenases. Int J Biochem. 1992;24(1):29–39. doi: 10.1016/0020-711x(92)90226-q. [DOI] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J. 1964 Sep;92(3):614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardmore-Gray M., Anthony C. The oxidation of glucose by Acinetobacter calcoaceticus: interaction of the quinoprotein glucose dehydrogenase with the electron transport chain. J Gen Microbiol. 1986 May;132(5):1257–1268. doi: 10.1099/00221287-132-5-1257. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Ghosh M., Harlos K., Avezoux A., Anthony C. The active site of methanol dehydrogenase contains a disulphide bridge between adjacent cysteine residues. Nat Struct Biol. 1994 Feb;1(2):102–105. doi: 10.1038/nsb0294-102. [DOI] [PubMed] [Google Scholar]
- Bork P., Doolittle R. F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994 Mar 11;236(5):1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Bosma G., Braster M., Stouthamer A. H., van Verseveld H. W. Subfractionation and characterization of soluble c-type cytochromes from Paracoccus denitrificans cultured under various limiting conditions in the chemostat. Eur J Biochem. 1987 Jun 15;165(3):665–670. doi: 10.1111/j.1432-1033.1987.tb11492.x. [DOI] [PubMed] [Google Scholar]
- Chan H. T., Anthony C. The interaction of methanol dehydrogenase and cytochrome cL in the acidophilic methylotroph Acetobacter methanolicus. Biochem J. 1991 Nov 15;280(Pt 1):139–146. doi: 10.1042/bj2800139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran R., Balasubramanian R. Stereochemical studies of cyclic peptides. VI. Energy calculations of the cyclic disulphide cysteinyl-cysteine. Biochim Biophys Acta. 1969 Aug 12;188(1):1–9. doi: 10.1016/0005-2795(69)90039-7. [DOI] [PubMed] [Google Scholar]
- Chen L. Y., Mathews F. S., Davidson V. L., Huizinga E. G., Vellieux F. M., Duine J. A., Hol W. G. Crystallographic investigations of the tryptophan-derived cofactor in the quinoprotein methylamine dehydrogenase. FEBS Lett. 1991 Aug 5;287(1-2):163–166. doi: 10.1016/0014-5793(91)80041-z. [DOI] [PubMed] [Google Scholar]
- Chen L., Durley R., Poliks B. J., Hamada K., Chen Z., Mathews F. S., Davidson V. L., Satow Y., Huizinga E., Vellieux F. M. Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin. Biochemistry. 1992 Jun 2;31(21):4959–4964. doi: 10.1021/bi00136a006. [DOI] [PubMed] [Google Scholar]
- Chen L., Mathews F. S., Davidson V. L., Huizinga E. G., Vellieux F. M., Hol W. G. Three-dimensional structure of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans determined by molecular replacement at 2.8 A resolution. Proteins. 1992 Oct;14(2):288–299. doi: 10.1002/prot.340140214. [DOI] [PubMed] [Google Scholar]
- Chen L., Mathews F. S., Davidson V. L., Tegoni M., Rivetti C., Rossi G. L. Preliminary crystal structure studies of a ternary electron transfer complex between a quinoprotein, a blue copper protein, and a c-type cytochrome. Protein Sci. 1993 Feb;2(2):147–154. doi: 10.1002/pro.5560020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Dekker S., van de Putte P., Goosen N. A single amino acid substitution changes the substrate specificity of quinoprotein glucose dehydrogenase in Gluconobacter oxydans. Mol Gen Genet. 1991 Oct;229(2):206–212. doi: 10.1007/BF00272157. [DOI] [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Goosen N., Fayet O., van de Putte P. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J Bacteriol. 1990 Nov;172(11):6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Goosen N., Odle G., van de Putte P. Nucleotide sequence of the gene coding for quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Nucleic Acids Res. 1988 Jul 11;16(13):6228–6228. doi: 10.1093/nar/16.13.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Goosen N., Vink K., van de Putte P. Cloning, characterization and DNA sequencing of the gene encoding the Mr 50,000 quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Mol Gen Genet. 1989 Jun;217(2-3):430–436. doi: 10.1007/BF02464914. [DOI] [PubMed] [Google Scholar]
- Cox J. M., Day D. J., Anthony C. The interaction of methanol dehydrogenase and its electron acceptor, cytochrome cL in methylotrophic bacteria. Biochim Biophys Acta. 1992 Feb 13;1119(1):97–106. doi: 10.1016/0167-4838(92)90240-e. [DOI] [PubMed] [Google Scholar]
- Crennell S. J., Garman E. F., Laver W. G., Vimr E. R., Taylor G. L. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day D. J., Anthony C. Soluble cytochromes c of methanol-utilizing bacteria. Methods Enzymol. 1990;188:298–303. doi: 10.1016/0076-6879(90)88046-d. [DOI] [PubMed] [Google Scholar]
- Dekker R. H., Duine J. A., Frank J., Verwiel P. E., Westerling J. Covalent addition of H2O, enzyme substrates and activators to pyrrolo-quinoline quinone, the coenzyme of quinoproteins. Eur J Biochem. 1982 Jun 15;125(1):69–73. doi: 10.1111/j.1432-1033.1982.tb06652.x. [DOI] [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jr, Duine J. A. Studies on electron transfer from methanol dehydrogenase to cytochrome cL, both purified from Hyphomicrobium X. Biochem J. 1989 Jan 1;257(1):87–94. doi: 10.1042/bj2570087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duine J. A., Frank J., De Beer R. An electron-nuclear double-resonance study of methanol dehydrogenase and its coenzyme radical. Arch Biochem Biophys. 1984 Sep;233(2):708–711. doi: 10.1016/0003-9861(84)90497-1. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jongejan J. A. Enzymology of quinoproteins. Adv Enzymol Relat Areas Mol Biol. 1987;59:169–212. doi: 10.1002/9780470123058.ch4. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Verwiel P. E. Structure and activity of the prosthetic group of methanol dehydrogenase. Eur J Biochem. 1980;108(1):187–192. doi: 10.1111/j.1432-1033.1980.tb04711.x. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., van Zeeland J. K. Glucose dehydrogenase from Acinetobacter calcoaceticus: a 'quinoprotein'. FEBS Lett. 1979 Dec 15;108(2):443–446. doi: 10.1016/0014-5793(79)80584-0. [DOI] [PubMed] [Google Scholar]
- Duine J. A. Quinoproteins: enzymes containing the quinonoid cofactor pyrroloquinoline quinone, topaquinone or tryptophan-tryptophan quinone. Eur J Biochem. 1991 Sep 1;200(2):271–284. doi: 10.1111/j.1432-1033.1991.tb16183.x. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Microbial oxidation of amines. Spectral and kinetic properties of the primary amine dehydrogenase of Pseudomonas AM1. Biochem J. 1971 Aug;123(5):757–771. doi: 10.1042/bj1230757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Purification and properties of an amine dehydrogenase from Pseudomonas AM1 and its role in growth on methylamine. Biochem J. 1968 Jan;106(1):245–255. doi: 10.1042/bj1060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Jr, Dijkstra M., Duine J. A., Balny C. Kinetic and spectral studies on the redox forms of methanol dehydrogenase from Hyphomicrobium X. Eur J Biochem. 1988 Jun 1;174(2):331–338. doi: 10.1111/j.1432-1033.1988.tb14102.x. [DOI] [PubMed] [Google Scholar]
- Frank J., Jr, van Krimpen S. H., Verwiel P. E., Jongejan J. A., Mulder A. C., Duine J. A. On the mechanism of inhibition of methanol dehydrogenase by cyclopropane-derived inhibitors. Eur J Biochem. 1989 Sep 1;184(1):187–195. doi: 10.1111/j.1432-1033.1989.tb15006.x. [DOI] [PubMed] [Google Scholar]
- Friedrich T., Strohdeicher M., Hofhaus G., Preis D., Sahm H., Weiss H. The same domain motif for ubiquinone reduction in mitochondrial or chloroplast NADH dehydrogenase and bacterial glucose dehydrogenase. FEBS Lett. 1990 Jun 4;265(1-2):37–40. doi: 10.1016/0014-5793(90)80878-m. [DOI] [PubMed] [Google Scholar]
- Govindaraj S., Eisenstein E., Jones L. H., Sanders-Loehr J., Chistoserdov A. Y., Davidson V. L., Edwards S. L. Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme. J Bacteriol. 1994 May;176(10):2922–2929. doi: 10.1128/jb.176.10.2922-2929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen B. W., van Kleef M. A., Duine J. A. Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J. 1986 Mar 15;234(3):611–615. doi: 10.1042/bj2340611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAUGE J. G. GLUCOSE DEHYDROGENASE OF BACTERIUM ANITRATUM: AN ENZYME WITH A NOVEL PROSTHETIC GROUP. J Biol Chem. 1964 Nov;239:3630–3639. [PubMed] [Google Scholar]
- Harms N., de Vries G. E., Maurer K., Hoogendijk J., Stouthamer A. H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987 Sep;169(9):3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. K., Davidson V. L. Binding and electron transfer reactions between methanol dehydrogenase and its physiologic electron acceptor cytochrome c-551i: a kinetic and thermodynamic analysis. Biochemistry. 1993 Dec 28;32(51):14145–14150. doi: 10.1021/bi00214a011. [DOI] [PubMed] [Google Scholar]
- Heinrich H., Azevedo J. E., Werner S. Characterization of the 9.5-kDa ubiquinone-binding protein of NADH:ubiquinone oxidoreductase (complex I) from Neurospora crassa. Biochemistry. 1992 Nov 24;31(46):11420–11424. doi: 10.1021/bi00161a021. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Rogozinski J., Toczko M. Lupanine hydroxylase, a quinocytochrome c from an alkaloid-degrading Pseudomonas sp. Biochem J. 1991 Oct 1;279(Pt 1):105–109. doi: 10.1042/bj2790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga E. G., van Zanten B. A., Duine J. A., Jongejan J. A., Huitema F., Wilson K. S., Hol W. G. Active site structure of methylamine dehydrogenase: hydrazines identify C6 as the reactive site of the tryptophan-derived quinone cofactor. Biochemistry. 1992 Oct 13;31(40):9789–9795. doi: 10.1021/bi00155a036. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sunagawa M., Mori A., Imai C., Fukuda M., Takagi M., Yano K. Cloning and sequencing of the gene encoding the 72-kilodalton dehydrogenase subunit of alcohol dehydrogenase from Acetobacter aceti. J Bacteriol. 1989 Jun;171(6):3115–3122. doi: 10.1128/jb.171.6.3115-3122.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Phillips S. E., Stevens C., Ogel Z. B., McPherson M. J., Keen J. N., Yadav K. D., Knowles P. F. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991 Mar 7;350(6313):87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- Ito N., Phillips S. E., Yadav K. D., Knowles P. F. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994 May 20;238(5):794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- Janes S. M., Mu D., Wemmer D., Smith A. J., Kaur S., Maltby D., Burlingame A. L., Klinman J. P. A new redox cofactor in eukaryotic enzymes: 6-hydroxydopa at the active site of bovine serum amine oxidase. Science. 1990 May 25;248(4958):981–987. doi: 10.1126/science.2111581. [DOI] [PubMed] [Google Scholar]
- Janes S. M., Palcic M. M., Scaman C. H., Smith A. J., Brown D. E., Dooley D. M., Mure M., Klinman J. P. Identification of topaquinone and its consensus sequence in copper amine oxidases. Biochemistry. 1992 Dec 8;31(48):12147–12154. doi: 10.1021/bi00163a025. [DOI] [PubMed] [Google Scholar]
- Kao P. N., Karlin A. Acetylcholine receptor binding site contains a disulfide cross-link between adjacent half-cystinyl residues. J Biol Chem. 1986 Jun 25;261(18):8085–8088. [PubMed] [Google Scholar]
- Klinman J. P., Dooley D. M., Duine J. A., Knowles P. F., Mondovi B., Villafranca J. J. Status of the cofactor identity in copper oxidative enzymes. FEBS Lett. 1991 Apr 22;282(1):1–4. doi: 10.1016/0014-5793(91)80431-2. [DOI] [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K., Takahashi K., Adachi O. A novel quinoprotein methanol dehydrogenase containing an additional 32-kilodalton peptide purified from Acetobacter methanolicus: identification of the peptide as a MoxJ product. Biochemistry. 1993 Jun 1;32(21):5576–5582. doi: 10.1021/bi00072a012. [DOI] [PubMed] [Google Scholar]
- Matsushita K., Toyama H., Adachi O. Respiratory chains and bioenergetics of acetic acid bacteria. Adv Microb Physiol. 1994;36:247–301. doi: 10.1016/s0065-2911(08)60181-2. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Wemmer D. E., Chistoserdov A., Lidstrom M. E. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991 May 10;252(5007):817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- McIntire W. S. Wither PQQ. Essays Biochem. 1992;27:119–134. [PubMed] [Google Scholar]
- Murzin A. G. Structural principles for the propeller assembly of beta-sheets: the preference for seven-fold symmetry. Proteins. 1992 Oct;14(2):191–201. doi: 10.1002/prot.340140206. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Anthony C. The nucleotide sequence and deduced amino acid sequence of the cytochrome cL gene of Methylobacterium extorquens AM1, a novel class of c-type cytochrome. Biochem J. 1988 Dec 1;256(2):673–676. doi: 10.1042/bj2560673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Day D., Anthony C. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem J. 1989 Jun 15;260(3):857–862. doi: 10.1042/bj2600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Richardson I. W., Anthony C. Characterization of mutant forms of the quinoprotein methanol dehydrogenase lacking an essential calcium ion. Biochem J. 1992 Nov 1;287(Pt 3):709–715. doi: 10.1042/bj2870709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury S. A., Forrest H. S., Cruse W. B., Kennard O. A novel coenzyme from bacterial primary alcohol dehydrogenases. Nature. 1979 Aug 30;280(5725):843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- Schiering N., Kabsch W., Moore M. J., Distefano M. D., Walsh C. T., Pai E. F. Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature. 1991 Jul 11;352(6331):168–172. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- Scott D. L., White S. P., Otwinowski Z., Yuan W., Gelb M. H., Sigler P. B. Interfacial catalysis: the mechanism of phospholipase A2. Science. 1990 Dec 14;250(4987):1541–1546. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki T., Fukaya M., Takemura H., Tayama K., Okumura H., Kawamura Y., Nishiyama M., Horinouchi S., Beppu T. Cloning and sequencing of the gene cluster encoding two subunits of membrane-bound alcohol dehydrogenase from Acetobacter polyoxogenes. Biochim Biophys Acta. 1991 Feb 16;1088(2):292–300. doi: 10.1016/0167-4781(91)90066-u. [DOI] [PubMed] [Google Scholar]
- Van Spanning R. J., Wansell C. W., De Boer T., Hazelaar M. J., Anazawa H., Harms N., Oltmann L. F., Stouthamer A. H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991 Nov;173(21):6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J. N., Laver W. G., Colman P. M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983 May 5;303(5912):35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- Vellieux F. M., Huitema F., Groendijk H., Kalk K. H., Jzn J. F., Jongejan J. A., Duine J. A., Petratos K., Drenth J., Hol W. G. Structure of quinoprotein methylamine dehydrogenase at 2.25 A resolution. EMBO J. 1989 Aug;8(8):2171–2178. doi: 10.1002/j.1460-2075.1989.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwiel P. E., Frank J., Verwiel E. J. Characterization of the second prosthetic group in methanol dehydrogenase from hyphomicrobium X. Eur J Biochem. 1981 Aug;118(2):395–399. doi: 10.1111/j.1432-1033.1981.tb06415.x. [DOI] [PubMed] [Google Scholar]
- White S., Boyd G., Mathews F. S., Xia Z. X., Dai W. W., Zhang Y. F., Davidson V. L. The active site structure of the calcium-containing quinoprotein methanol dehydrogenase. Biochemistry. 1993 Dec 7;32(48):12955–12958. doi: 10.1021/bi00211a002. [DOI] [PubMed] [Google Scholar]
- Xia Z. X., Dai W. W., Xiong J. P., Hao Z. P., Davidson V. L., White S., Mathews F. S. The three-dimensional structures of methanol dehydrogenase from two methylotrophic bacteria at 2.6-A resolution. J Biol Chem. 1992 Nov 5;267(31):22289–22297. [PubMed] [Google Scholar]
- Yamada M., Sumi K., Matsushita K., Adachi O., Yamada Y. Topological analysis of quinoprotein glucose dehydrogenase in Escherichia coli and its ubiquinone-binding site. J Biol Chem. 1993 Jun 15;268(17):12812–12817. [PubMed] [Google Scholar]