Abstract
Background
Within the family of green fluorescent protein (GFP) homologs, one can mark two main groups, specifically, fluorescent proteins (FPs) and non-fluorescent or chromoproteins (CPs). Structural background of differences between FPs and CPs are poorly understood to date.
Results
Here, we applied site-directed and random mutagenesis in order to to transform CP into FP and vice versa. A purple chromoprotein asCP (asFP595) from Anemonia sulcata and a red fluorescent protein DsRed from Discosoma sp. were selected as representatives of CPs and FPs, respectively. For asCP, some substitutions at positions 148 and 165 (numbering in accordance to GFP) were found to dramatically increase quantum yield of red fluorescence. For DsRed, substitutions at positions 148, 165, 167, and 203 significantly decreased fluorescence intensity, so that the spectral characteristics of these mutants became more close to those of CPs. Finally, a practically non-fluorescent mutant DsRed-NF was generated. This mutant carried four amino acid substitutions, specifically, S148C, I165N, K167M, and S203A. DsRed-NF possessed a high extinction coefficient and an extremely low quantum yield (< 0.001). These spectral characteristics allow one to regard DsRed-NF as a true chromoprotein.
Conclusions
We located a novel point in asCP sequence (position 165) mutations at which can result in red fluorescence appearance. Probably, this finding could be applied onto other CPs to generate red and far-red fluorescent mutants. A possibility to transform an FP into CP was demonstrated. Key role of residues adjacent to chromophore's phenolic ring in fluorescent/non-fluorescent states determination was revealed.
Background
Recently, homologs of the well-known green fluorescent protein (GFP) from jellyfish Aequorea victoria were discovered in Anthozoa species [1-6]. These proteins can be subdivided into two main types. First type, fluorescent proteins (FPs), emit a significant portion (25–80%) of the absorbed photons. Second type, chromoproteins (CPs), effectively absorb but practically do not emit light.
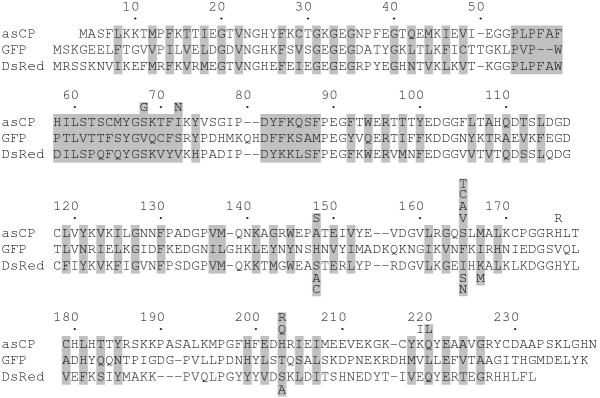
Peculiarities of structure that make each GFP-like protein fluorescent or non-fluorescent are poorly understood to date. Only the importance of position 148 (we will use numbering in accordance to GFP, see Fig. 1) was demonstrated in experiments on appearance of fluorescence in CPs [3,6]. Introduction of Ser148 into several CPs made them clearly fluorescent, although the emission brightness of these mutants was significantly lower in comparison with wild type FPs.
Figure 1.
Sequence alignment of asCP, GFP, and DsRed proteins. The numbering is based on GFP. Introduced gaps are represented by dashes. The residues whose side chains form the interior of the β-can are shaded. Mutations introduced in asCP and DsRed are designated under and below their sequences, respectively.
Due to a great and still growing popularity of GFP and novel FPs in biotechnology, a comprehension of structure-function correlations in GFP-like proteins has both a scientific and a practical significance, showing novel possibilities to achieve desirable protein properties artificially. Here, we applied mutagenesis to a chromoprotein asFP595 (asCP) and a red fluorescent protein drFP583 (DsRed) to study transformation of a chromoprotein into a fluorescent protein and vise verse.
Results
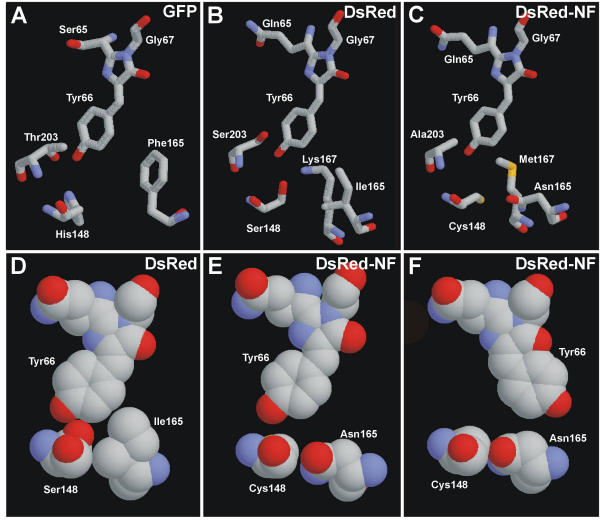
Although sequence comparison of known GFP-like proteins does not reveal absolutely invariable differences between FPs and CPs, one can draw attention to three positions, specifically, 148, 165, and 203, which are occupied by noticeably different residues in the two types of proteins (Fig. 1, Table 1). Since residues at these positions are in a close proximity to chromophore (Fig. 2A,2B) [7-10], it is reasonable to presume that they can participate in the determination of the state (fluorescent or non-fluorescent) of a particular protein.
Table 1.
Amino acids occupying positions 148, 165, and 203 (GFP numbering) in known GFP-like proteins.
| 148 | 165 | 203 | |
| FPs | Ser, His | Ile, Val, Phe | His, Ser, Thr |
| CPs | Cys, Ala, Asn | Asn, Ser | Leu, Ile, His, Arg |
Figure 2.
Schematic outline of the chromophores and selected neighboring residues in GFP (A), DsRed (B, D), and DsRed-NF (C, E, F) in "sticks" and "spacefill" representation. Carbon atoms are gray, nitrogen atoms are blue, and oxygen atoms are red. Images were generated by RasMol 2.6 software. Computer modeling for DsRed-NF was performed using Swiss-PdbViewer and HyperChem 5.01 software.
Random mutagenesis of asCP at position 148
Earlier, we demonstrated for several CPs that Ser-148 containing mutants possess red fluorescence [3,6]. To check other residue we fulfilled mutagenesis using degenerated primers encoding any amino acid at position 148. Visual inspection of about 50 recombinant clones and sequence analysis of the selected clones showed the following. Only Ser148 ensured clear fluorescence. Several intensively colored non-fluorescent clones contained Ala, Cys, Asn, or Gly at position 148 (remarkably, known wild type CPs carry the very Ala, Cys, or Asn at this position). All other substitutions of Ala148 appeared to be intolerable for proper protein folding and chromophore maturation.
Mutagenesis of asCP at position 165
First of all, we tested a substitution S165V because several FPs carry Val at this position. This mutation resulted in the appearance of a clearly visible red fluorescence with a maximum at 620 nm (Fig. 3A, Table 2). Interestingly, in comparison with the wild type asCP, the mutant asCP-S165V showed a strongly modified absorption spectrum which included an additional peak at 390 nm. Absorption at this wavelength produced a very weak (about 10-fold less than the red fluorescence) blue fluorescence at 465 nm.
Figure 3.
Normalized spectra for selected mutants of asCP and DsRed. Absorption (black solid lines), excitation (colored dashed lines), and emission (colored solid lines) spectra are shown for each mutant. Blue, green, or red excitation-emission lines correspond to color of fluorescence. (A) asCP-S165V. Blue fluorescence is about tenfold weaker than red. (B) asCP-S165A. (C) asCP-S165C. (D) asCP-S165T. Green emision is about twofold stronger than red. (E) DsRed-NF. Green emission peak is about threefold lower than red one.
Table 2.
Spectral characteristics for some mutants of asCP and DsRed.
| Wild type protein | Mutant | Absorption max, nm | Emission max, nm | Extinction coefficient, M-1cm-1 | Quantum yield |
| asCP | wild typea | 568 | 595 | 56,000 | << 0.001 |
| A148S | 572 | 597 | 15,000 | 0.012 | |
| S165V | 583 | 620 | 18,000 | 0.008 | |
| DsRed | wild typeb | 558 | 583 | 75,000 | 0.70 |
| S148A | 568 | 591 | 73,000 | 0.45 | |
| S203A | 562 | 583 | 74,000 | 0.70 | |
| S148A, S203A | 574 | 593 | 73,000 | 0.29 | |
| S148A, K167M | 572 | 595 | 80,000 | 0.22 | |
| S148A, K167M, S203A | 574 | 596 | 104,000 | 0.09 | |
| S148A, I165S, S203A | 574 | 595 | 68,000 | 0.06 | |
| S148A, I165S, K167M, S203A | 552 | 593 | 77,000 | 0.007 | |
| S148C, I165N, S203A | 574 | 591 | 80,000 | 0.009 | |
| S148C, I165N, K167M, S203A | 561 | 600 | 57,000 | << 0.001 | |
To reveal other substitutions at position 165 that could lead to fluorescence appearance we exploited randomization at this position. As a result, several red fluorescent clones of different brightness were selected. The most bright clones carried the already known substitution S165V. All other fluorescent mutants were considerably (5–10 fold) dimmer and contained Ala, Cys, or Thr165 (in decreasing brightness order). Absorption spectra for these mutants have a characteristic peak at about 390 nm, but it produces no detectable blue fluorescence (Fig. 3B,3C,3D). An interesting feature of these low fluorescent mutants is that their excitation spectra for red emission do not coincide with the absorption spectra. This phenomenon implies the existence of different spectral forms within a spectrally heterogeneous population of the mutant protein molecules. Red emitting spectral forms are underrepresented or they possess a very low extinction coefficient. At the same time, the major red light-absorbing spectral forms are non-fluorescent.
Random mutagenesis of asCP
To extend the search of amino acid substitutions that are able to convert asCP into a fluorescent protein, we used random mutagenesis of the whole asCP gene. Visual screening of about 5000 recombinant clones revealed only one brightly fluorescent colony. Sequence analysis showed that this fluorescent mutant contained the already known substitution A148S. After a more thorough visual inspection we found several very weakly fluorescent clones containing the following substitutions: S68G; I72N; H176R/K219I; H203R; H203Q; Q220L (Fig. 1). Importantly, two independent clones carrying different substitutions at position 203 were collected.
Summing up, this experiment has not highlighted novel important sites, because all random mutants were considerably (3–5 fold) dimmer than the mutants at positions 148 and 165 mentioned above. One can conclude that positions 148, 165 are probably the most important sites that influence the state of asCP.
Mutagenesis of DsRed
Finally, we attempted to transform the fluorescent DsRed into a chromoprotein. First of all, mutation S148A was tested. Unexpectedly, this substitution did not exert a strong influence on the fluorescence – quantum yield for DsRed-S148A mutant decreased by a factor of 1.5 only in comparison to the wild type protein (Table 2). Then, on the base of this mutant, a series of mutants carrying substitutions I165S, K167M, and S203A,L in different combinations was generated. Position 167 was added to mutagenesis considering the crystallographic studies that revealed a direct interaction between Lys167 and chromophore's Tyr66 (Fig. 2B) [9,10]. This bond appeared to stabilize the ionized form of the DsRed fluorophore. Mutant proteins containing Leu203 were colorless because of unsatisfactory protein folding in E. coli. Following the spectral properties of other mutants, one can notice a gradient of emission intensity and conclude that all positions mentioned above are important for DsRed fluorescence (Table 2). However, even a quadruple mutant S148A/I165S/K167M/S203A displayed a clearly visible fluorescence comparable to that of some asCP fluorescent mutants (e.g., asCP-S165V). Thus, this DsRed mutant can not be regarded as a true chromoprotein, although it is very close to the CP state because it possesses hundredfold decreased fluorescence in comparison to DsRed.
Then, we tested Cys and Asn that are characteristic for some other known CPs [6] at positions 148 and 165, respectively. Triple mutant DsRed-S148C/I165N/S203A possessed a low quantum yield similarly to the mutant S148A/I165S/K167M/S203A mentioned above. When a substitution K167M was added, the final quadruple mutant S148C/I165N/K167M/S203A became practically non-fluorescent (Table 2). At the same time, this mutant named DsRed-NF intensively absorbed light. Altogether, these properties make DsRed-NF practically indistinguishable from wild type CPs.
Spectra for DsRed-NF are shown in Fig. 3D. An extremely weak dual-color fluorescence can be detected at high protein concentration only. Similarly to the low fluorescent mutants of asCP mentioned (see Fig. 3B,C,D), absorption and excitation spectra for DsRed-NF strongly differ from each other. Interestingly, excitation spectrum for green emission displays 2 peaks: a major peak at 410 nm and a minor peak at 490 nm. Such a shape of the excitation curve is similar to that of wild type GFP and has never been detected for DsRed mutants (to date, only EGFP-like single-peak excitation spectra were described for green-emitting mutants of DsRed [11-14]). Probably, the short-wave excitation peak corresponds to a neutral (protonated) form of GFP-like chromophore within DsRed-NF.
Discussion
Great diversity of fluorescent and non-fluorescent colors in the family of GFP-like proteins poses an challenging problem of understanding its structural background. Mutagenetic studies lately demonstrated various transitions of fluorescence color in Anthozoa proteins: from red to green [11-14], from yellow to green, from green to yellow, and from green to red [15]. Also, red and far-red fluorescent mutants of non-fluorescent CPs were generated [3,6].
Basic investigation of relationship between fluorescent and non-fluorescent GFP-like proteins was the main goal of the present work. However, some practical applications of the results obtained can be considered.
The first part of our work, attempts to convert asCP into FP, revealed importance of position 165 for fluorescence appearance. This finding can be applied on other CPs. To date, mutagenesis of natural CPs is the only way to generate a far-red FPs [6] that are in high demand for various applications. Additional far-red fluorescence color broadens abilities of multicolor labeling and assays based on fluorescence resonance energy transfer (FRET). Knowledge about the ways of transforming CPs into FPs could help to generate novel far-red FPs when novel CPs with red-shifted absorption spectra are found.
The second part of our work was to transform DsRed into CP. At first glance, such fluorescence quenching can not be used in practice. However, we found that DsRed-NF mutant can be used to resolve a problem of DsRed tetramerization that is the main disadvantage of this tag [11,14,16,17]. When DsRed is fused with a target protein, especially with oligomeric protein, it often results in improper folding and functioning of the tagged partners as well as intensive aggregation of the fusion protein. To neutralize injurious consequences of DsRed tetramerization we suggest to use a simultaneous co-expression of DsRed-tagged proteins with excess free DsRed-NF. In this case mixed heterotetramers are formed so that DsRed becomes a "monomeric" tag (this approach will be published elsewhere).
It was recently demonstrated that DsRed and asCP carry chemically distinct chromophores [18,19]. Theoretically, spectral differences between DsRed and asCP and generally between FPs and CPs may be explained by the diversity of their chromophores. If so, the appearance of fluorescence in asCP mutants and the disappearance of fluorescence in DsRed mutants should resulted from formation of altered chromophores within these mutants.
Alternatively, one may suggest that each chromophore type in GFP-like proteins can be fluorescent or non-fluorescent depending on the protein environment. Some observations speak in favor of this hypothesis. First, all key residues mentioned above (positions 148, 165, 167, and 203) are grouped in a close proximity to the phenolic ring of Tyr66 (Fig. 2). Thus, they can more likely participate in stabilization and positioning of the chromophore but not in chromophore cyclization events that result in the diversity of chromophores. Second, asCP demonstrates a striking phenomenon of light-induced reversible increasing of fluorescence [3]. This photoconversion clearly shows that an initially non-fluorescent protein molecule can be switched into a fluorescent state due to some conformation changes.
It is well-known that GFP-like chromophores and other chromophores that are capable of cis-trans isomerization are practically non-fluorescent in solution because of fast relaxation of the excited state through chromophore isomerization [20,21]. Probably, chromophore in FPs must be strongly stabilized by the amino acid environment to ensure high quantum yield, while chromophore surrounding within CPs should be more relaxed to allow energy of absorbed light to dissipate into heat.
From this point of view, we can draw the following scheme of DsRed chromophore stabilization. According to the crystal structure of DsRed [9,10] Ser148 and Lys167 hold the chromophore by a direct interaction with phenolate oxygen (Fig. 2B). Bulky Ile165 supports the ring of Tyr66 and prevents its movement required for the chromophore isomerization (Fig. 2D). Although Ser203 has no direct H-bonds with the chromophore in the wild type DsRed, such bonds could be formed in mutants with altered 148, 165 and 167 positions. Possibly, Ser203 in DsRed mutants can turn similarly to GFP Thr203 that forms an H-bond with chromophore's phenolate oxygen [7,8].
Quantitative data on the influence of each substitution on fluorescence intensity speak in favor of this scheme. Comparing in pairs quantum yields for the available DsRed mutants that differ from each other by one substitution (see Table 2), one can note the following. The contribution of each substitution strongly depends on mutation order: the later the substitution is introduced the stronger the impact is. For instance, the mutant S203A demonstrates the same quantum yield as the wild type protein. At the same time, an addition of S203A to the mutant S148A leads to a 1.5-fold decrease in quantum yield. Then, introducing Ala-203 into a double mutant S148A/K167M results in a 2.4-fold decreased fluorescence. Analogously, mutation K167M leads to 2-, 3.2-, or 8.6-fold decrease of quantum yield when Met167 is introduced as second, third or fourth substitution, respectively. Also, 4.8- or 12.9-fold decrease of fluorescence intensity is associated with substitution I165S added to S148A/S203A or S148A/K167M/S203A mutants, respectively. The model of several chromophore-stabilizing interactions mentioned above implies such tendency because the importance of each interaction must progressively increase in absence of one, two or more other bonds.
Computer modeling of the chromophore environment within DsRed-NF showed the following (Fig. 2C,2E,2F). In contrast to Ser148 and Lys167 in DsRed, Cys148 and Met167 in DsRed-NF are incapable of stabilizing the chromophore by H-bonds with phenolate oxygen. Moreover, substitution I165N generates a vacant space near the chromophore (compare Fig. 2D and 2E). We believe that this space is sufficient to ensure the chromophore cis-trans isomerization after light absorption (Fig. 2F). Thus, absence of phenolate-stabilizing interactions together with free space around the chromophore can explain an extremely low fluorescence quantum yield of DsRed-NF.
Unfortunately, no protein structures for CPs were published to date. Obviously, further crystallographic studies of FPs, CPs, and their mutants are required to make valid conclusions about the structural background of differences between FPs and CPs.
Conclusions
The ability for fluorescence of GFP-like proteins depends to a great extent on the surrounding of the phenolic ring of the chromophore. For asCP chromoprotein, mutations at positions 148 and 165 can lead to red fluorescence appearance. For DsRed red fluorescent protein, fluorescence can be quenched by mutagenesis at positions 148, 165, 167, and 203. This knowledge can be applied to other GFP-like proteins in effort of customizing spectral characteristics of FPs and CPs.
Materials and Methods
Mutagenesis and protein expression
Site-directed mutagenesis was performed by PCR with primers containing target substitution using the overlap extension method [22]. The Diversity PCR Random Mutagenesis kit (Clontech) was used for random mutagenesis of asCP, in conditions optimal for 4–5 mutations per 1000 bp. All mutants were cloned into pQE30 vector (Qiagen), so that recombinant proteins contained 6-histidine tag at their N-termini. To express mutant proteins E. coli XL1 Blue cells were transformed with the plasmids according to standard protocols and spread onto 3–4 Petri dishes with LB agar media supplemented with ampicillin for selection. After overnight growth at 37°C the plates were stored for 2–5 days at room temperature or 4°C to allow proteins to mature completely. Then, the plates were washed with PBS. Cells were disrupted by sonication, and soluble recombinant proteins were purified on the TALON metal-affinity resin (Clontech).
Spectroscopy
Absorption spectra were recorded on a Beckman DU520 UV/VIS Spectrophotometer. A Cary Eclipse Fluorescence Spectrophotometer (Varian) was used for measuring excitation-emission spectra.
For molar extinction coefficient determination, we relied on measuring mature chromophore concentration rather than total protein concentration. DsRed and its mutants were alkali-denatured with equal volume of 2 M NaOH. asCP and its mutants were acid-denatured with equal volume of 2 M HCl. Under these conditions, DsRed and asCP chromophores absorb at 452 and 430 nm, respectively [18,19]. The amounts of chromophore (that correspond to amounts of matured protein) were equalized among samples, absorption spectra for the native proteins were collected. Absorbance intensities were compared to that of DsRed (extinction coefficient is 75,000 M-1cm-1[11]) or asCP (extinction coefficient is 56,000 M-1cm-1[3]), and molar extinction coefficient for each mutant was estimated.
For quantum yield determination, the fluorescence of the mutants was compared to equally absorbing DsRed (quantum yield for DsRed was measured to be 0.70 [11]).
Acknowledgments
Acknowledgments
This work was supported by Clontech Laboratories Inc., and by Russian Foundation for Fundamental Research (grant 01-04-49037).
Contributor Information
Maria E Bulina, Email: biomasha@mail.ru.
Dmitry M Chudakov, Email: mitrophan@mail.ru.
Nikolay N Mudrik, Email: mudrikn@rambler.ru.
Konstantin A Lukyanov, Email: kluk@ibch.ru.
References
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Fradkov AF, Chen Y, Ding L, Barsova EV, Matz MV, Lukyanov SA. Novel fluorescent protein from Discosoma coral and its mutants possesses a unique far-red fluorescence. FEBS Lett. 2000;479:127–130. doi: 10.1016/S0014-5793(00)01895-0. [DOI] [PubMed] [Google Scholar]
- Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, Savitsky AP, Markelov ML, Zaraisky AG, Zhao X, Fang Y, Tan W, Lukyanov SA. Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J Biol Chem. 2000;275:25879–25882. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Elke C, Spindler KD, Funke W. Cracks in the beta-can: fluorescent proteins from Anemonia sulcata (Anthozoa, Actinaria). Proc Natl Acad Sci USA. 2000;97:14091–14096. doi: 10.1073/pnas.97.26.14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SG, Hoegh-Guldberg O, Ranganathan S. Major colour patterns of reef-building corals are due to a family of GFP-like proteins. Coral Reefs. 2001;19:197–204. [Google Scholar]
- Gurskaya NG, Fradkov AF, Terskikh A, Matz MV, Labas YA, Martynov VI, Yanushevich YG, Lukyanov KA, Lukyanov SA. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 2001;507:16–20. doi: 10.1016/S0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0 Å resolution. Proc Natl Acad Sci. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. "Fluorescent Timer": protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- Wiehler J, von Hummel J, Steipe B. Mutants of Discosoma red fluorescent protein with a GFP-like chromophore. FEBS Lett. 2001;487:384–389. doi: 10.1016/S0014-5793(00)02365-6. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Savitsky AP, Yanushevich YG, Lukyanov SA, Lukyanov KA. Color transitions in coral's fluorescent proteins by site-directed mutagenesis. BMC Biochem. 2001;2:6. doi: 10.1186/1472-2091-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrzheshch PV, Akovbian NA, Varfolomeyev SD, Verkhusha VV. Denaturation and partial renaturation of a tightly tetramerized DsRed protein under mildly acidic conditions. FEBS Lett. 2000;487:203–208. doi: 10.1016/S0014-5793(00)02344-9. [DOI] [PubMed] [Google Scholar]
- Lauf U, Lopez P, Falk MM. Expression of fluorescently tagged connexins: a novel approach to rescue function of oligomeric DsRed-tagged proteins. FEBS Lett. 2001;498:11–15. doi: 10.1016/S0014-5793(01)02462-0. [DOI] [PubMed] [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynov VI, Savitsky AP, Martynova NY, Savitsky PA, Lukyanov KA, Lukyanov SA. Alternative cyclization in GFP-like proteins family. The formation and structure of the chromophore of a purple chromoprotein from Anemonia sulcata. J Biol Chem. 2001;276:21012–21016. doi: 10.1074/jbc.M100500200. [DOI] [PubMed] [Google Scholar]
- Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji FI. Chemical nature of the light emitter of the Aequorea green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Helms V, McCammon JA, Langhoff PW. Shedding light on the dark and weakly fluorescent states of green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:6177–6182. doi: 10.1073/pnas.96.11.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]