Abstract
The coordinated action of transcriptional and post-transcriptional machineries shapes gene expression programs at steady state and determines their concerted response to perturbations. We have developed Nanodynamo, an experimental and computational workflow for quantifying the kinetic rates of nuclear and cytoplasmic steps of the RNA life cycle. Nanodynamo is based on mathematical modelling following sequencing of native RNA from cellular fractions and polysomes. We have applied this workflow to triple-negative breast cancer cells, revealing widespread post-transcriptional RNA processing that is mutually exclusive with its co-transcriptional counterpart. We used Nanodynamo to unravel the coupling between transcription, processing, export, decay and translation machineries. We have identified a number of coupling interactions within and between the nucleus and cytoplasm that largely contribute to coordinating how cells respond to perturbations that affect gene expression programs. Nanodynamo will be instrumental in unravelling the determinants and regulatory processes involved in the coordination of gene expression responses.
Subject terms: Computational models, Breast cancer, Gene expression, RNA splicing, Sequencing
The life of RNAs is governed by a series of transcriptional and post-transcriptional steps. Here, authors developed Nanodynamo, an experimental and computational workflow for studying how the coordinated action of these steps shapes breast cancer gene expression programs at the subcellular level.
Introduction
The RNA life cycle is a multi-step program that stems from the birth of novel transcripts till their decay1. The fine regulation of these steps ultimately shapes gene expression programs in physiological and disease conditions. Research in the last decades has revealed that the different steps of the RNA life cycle are extensively coupled, even when the corresponding machineries are in different cellular compartments. This ensures robust and precise coordination of these processes at steady-state conditions, and their concerted regulation when gene expression programs have to be shaped to support cellular responses2,3. The coupling is typically mediated by RNA binding proteins (RBPs) or transcription factors (TFs) that are directly involved in the regulation of multiple machineries4. Several studies focused on characterising the coordination between RNA synthesis with RNA processing5 and with decay6, which were found to be important for the correct execution of transcriptional and post-transcriptional events and for the buffering of RNA levels7,8, respectively. RNA processing was also shown being coordinated with the RNA degradation machinery9. Finally, various studies documented the link between the translation machinery and steps of the RNA life cycle10–15.
The results obtained so far in the field often tackle the coupling between one specific step of the RNA life cycle and another one. Therefore, despite the progress, a comprehensive picture of the intricate crosstalk among the various steps is missing. A key reason is the lack of methods able to determine the efficiency of the various machineries and how this is impacted by perturbation of individual steps. To this end, various approaches were proposed in the last decade to study the dynamics of RNA metabolism, allowing to quantify the kinetic rates of individual RNA life cycle stages16. The ability to profile nascent transcription via RNA metabolic labelling, and the adoption of mathematical modelling were crucial in the field17. Nowadays, available methods allow extracting, for individual cells or populations thereof, precious information on how the abundance of premature and mature RNAs is governed by the kinetic rates of RNA synthesis, processing and degradation18. Other studies tried expanding the covered steps, including the export of RNA into the cytoplasm19 or focusing on the efficiency of translation, through polysomal or ribosomal profiling20. Altogether, each method focuses on specific steps of the RNA life, relying on important assumptions and ultimately falling short in covering all key stages. Few recent studies aim at overcoming these limitations by extending the number of covered RNA life cycle steps21,22.
Following up the development of INSPEcT, a suite of tools for the quantification of the kinetic rates of RNA synthesis, processing and degradation16,23,24 (Supplementary Fig. 1A), we developed Nanodynamo, an experimental and computational framework that markedly expands our ability to quantify RNA dynamics. Nanodynamo relies on RNA metabolic labelling and Nanopore sequencing of native RNA for the profiling of transcripts from cellular fractions and polysomes. Mathematical modelling allows following transcripts from their birth and detachment from chromatin, through their co- or post-transcriptional processing, their export into the cytoplasm, and their translation, up to their final degradation (Supplementary Fig. 1B). We used Nanodynamo to finely characterise transcriptional programs in triple negative breast cancer cells, and how they are modulated in response to drugs blocking the spliceosomal, the export, and the translational machineries. Nanodynamo revealed the prevalence of post-transcriptional RNA processing as an alternative pathway for transcripts maturation and shed light on the extensive crosstalk between steps of the RNA life cycle within and between nucleus and cytoplasm.
Results
Nanodynamo: model formulation, parameters identifiability and inference
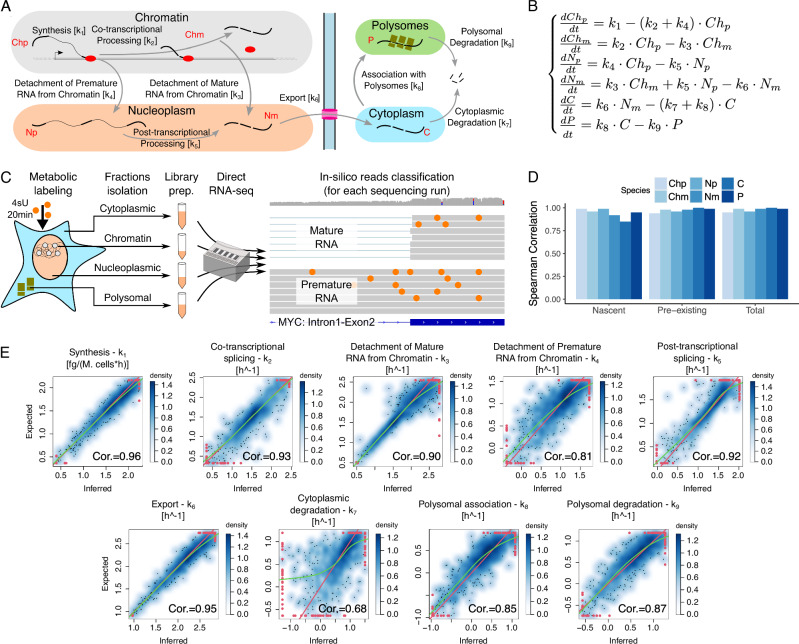
We modelled the RNA life cycle with a set of deterministic Ordinary Differential Equations (ODEs) describing the temporal modulation of RNA species in various RNA pools, including cellular compartments and polysomes. More in detail, premature RNA associated with chromatin is transcribed at rate k1 and is either co-transcriptionally processed into its mature form or detached from chromatin to become premature nucleoplasmic RNA with rates k2 and k4, respectively. Premature nucleoplasmic RNA is then post-transcriptionally spliced into mature nucleoplasmic RNA with rate k5. The latter is also produced from the detachment of mature RNA from chromatin with rate k3. Mature nucleoplasmic RNA is then exported to cytoplasm at rate k6 to become cytoplasmic RNA, which can be finally either directly degraded at rate k7 or associated with actively translating polysomes at rate k8 and subsequently degraded at rate k9 (Fig. 1A, B).
Fig. 1. Nanodynamo: model formulation, parameters identifiability and inference validation.
A The nuclear and cytoplasmic steps of the RNA life cycle considered by Nanodynamo, with the corresponding kinetic rates (squared brackets). Coloured ellipses represent the profiled RNA pools, including cellular and polysomal fractions. B Mathematical formulation in terms of Ordinary Differential Equations of the model presented in A. Ch, N, C, and P denote chromatin associated, nucleoplasmic, cytoplasmic, and polysomal RNA, respectively, while p and m additionally specify premature or mature forms. C Scheme of the experimental workflow, from cell fractionation to reads classification. D Spearman correlation coefficients between modelled expression levels and their experimental counterparts for a simulated dataset composed of 1000 genes generated with CVs retrieved from untreated experimental data, 2 replicates, and 1 labelling time of 20 min; abbreviations of the legend as in B. E Smooth density scatterplots between inferred kinetic rates and their expected counterparts for the same dataset described in D. For each plot, Spearman correlation coefficient (Cor.), identity line (red) and loess (green) lines are reported; red dots represent outliers (i.e., data points in the bottom and top 2.5% of the distribution). For D, E, source data are provided as a Source Data file.
Each parameter of the model, except for the time (t), is a rate which represents the efficiency of a specific step of the RNA life cycle. In particular, k2-9 are pure rates expressed as h−1, while k1 is the net amount of newly transcribed RNA per hour per million of cells. The rates inference relies as input data on the quantification of RNAs bound to chromatin, present in the nucleoplasm, in the cytoplasm, and associated with actively translating ribosomes. Transcripts associated with different RNA pools are obtained by extracting polyA+ RNAs following the fractionation of the cells into chromatin, nucleoplasm and cytoplasm. Transcripts undergoing translation are obtained by retrieving RNAs associated with more than three ribosomes following polysome fractionation. Direct Nanopore sequencing of native RNA (dRNA-seq) is then performed for each of the four pools. Thus, k7 refers to the rate of degradation of cytoplasmatic transcripts not associated with polysomes, while k9 refers to the rate of degradation of transcripts associated with polysomes. From here on we will refer to k7 as cytoplasmatic degradation, and to k9 as polysomal degradation.
For the quantification of premature and mature RNA species within each pool, we relied on the in silico classification of the reads based on the detection of intronic signal, as widely adopted in the field16,17,23. To validate our ability to classify reads associated with premature transcripts, we reanalysed publicly available dRNA-seq generated by nano-COP on K562 cells (GEO Sample ID GSM4663623)25. Nano-COP relies on the profiling of nascent RNA associated with chromatin, and Nanodynamo confirmed the previously reported enrichment in premature RNA (83% of the reads).
To overcome the undetermined nature of the resulting algebraic system we opted for the inclusion of nascent RNA profiling, similarly to what has been previously proposed by us and others17. For the identification of newly synthetized transcripts, we adopted metabolic labelling with 4-Thiouridine (4sU) for fixed amounts of time. This modified nucleotide can be detected on dRNA-seq data using the nano-ID tool26, which relies on the impact of 4sU on Nanopore ionic current for the supervised classification of the reads (Median gene accuracy ~0.80; Supplementary Fig. 2). This additional piece of data discloses the temporal evolution of the system and guarantees global structural identifiability for all the parameters of the model, i.e., the existence of a unique set of rates for every model output (see the “Parameters identifiability” methods section).
Following the verification of the theoretical feasibility of kinetic rates inference, we developed an R framework to identify the optimal set of rates given a vector of gene expression levels. This framework relies on the minimization of a cost function (sum of absolute logarithmic fold changes) regularised with the L2 norm of the rates and provides the numerical value of the rates at the single gene level.
Evaluation of Nanodynamo through simulated data
The analysis of parameters global identifiability disregards the impact of noise and the numerical issues which may affect the rates inference. Therefore, we relied on simulated data to test the performance of our approach accounting for these additional factors.
Given a set of kinetic rates, the numerical solution of the ODEs system returns pre-existing and nascent RNA expression levels for all the species involved in the model at any time of interest. We bound each rate of the model to the median of the rates of synthesis, processing and degradation previously obtained with INSPEcT for 3T9 mouse fibroblast cells27. In particular, Nanodynamo rates from k2 to k6 were linked to the rate of processing while rates from k7 to k9 to the rate of degradation, in order to maintain a biologically reasonable ranking of the efficiency of the processes (see methods for details). These values were used as means for a set of gaussian distributions from which the rates were independently sampled for 1000 simulated genes. The variation coefficient (CV) of each gaussian distribution was set to 5 in order to explore a broad parameter space covering several orders of magnitude for each rate. The expression level of each RNA species resulting from these rates is exact. To mimic experimental noise, we used this value as the mean of a normal distribution from which we sampled n simulated replicates. Distributions variation coefficients were determined from experimental expression data taking the median CV across all genes for each RNA species (see “Quantification of gene expression levels” and “Data simulation” methods sections).
The temporal design of metabolic labelling is an important experimental aspect which could strongly impact the inference performance26,28. The optimal labelling time(s) should allow capturing the sharp increase of nascent transcription, while accommodating the slower dynamics of nascent RNA accumulation for steps further downstream the RNA life cycle. The chosen labelling time(s) should also allow producing non-negligible amounts of labelled RNA in the various RNA pools. By means of mathematical simulations (see methods), we identified a labelling time of 20 min as a good trade-off for all the RNA species where the time derivatives of their nascent RNA saturation curves are significantly higher than zero, i.e., far from the steady state, but also distant from the initial exponential transition (Supplementary Fig. 3). Altogether, we simulated three datasets (CVs assigned as previously explained, 1000 genes, 2 replicates) with an increasing number of labelling pulses of 20, 60 and 120 min. We ran the inference pipeline, and we compared the resulting rates against the real counterparts to quantify the goodness of fit. The increase in performance gained with multiple labelling times was minor (maximum increase of 15%; Supplementary Fig. 4A).
We adopted a similar approach to evaluate the optimal number of replicates. We simulated five datasets (CVs assigned as previously explained, 1000 genes, 20 min labelling) with an increasing number of replicates. The gain in correlation between inferred and real rates at an increasing number of experiments suggested that two samples are sufficient to have a performance remarkably close to the optimal one (maximum increase of 20%; Supplementary Fig. 4B).
The abundances of RNA species, inferred with two replicates and 20 min labelling pulse, were highly correlated with their expected expression levels (median Spearman correlations >0.98; Fig. 1D and Supplementary Fig. 5). The inferred kinetic rates were well correlated with the expected counterparts (Spearman coefficients in the 0.68–0.96 range; Fig. 1E). The variability in rates correlations for the last steps of the RNA life cycle likely derived from the increased complexity of the model moving away from the RNA synthesis step. In fact, the determination of k7-9 involved upstream rates in defining cytoplasmic and polysomal RNA species, complicating the inferences based on these data. Instead, k4 and k7 were affected by the presence of branching points in the model. Indeed, the simplification of the equations disregarding certain steps of the RNA life cycle improved the correlation coefficients of the remaining ones (see Supplemental Material). Nevertheless, we considered this performance a reasonable compromise between inference quality, experimental workload, and sequencing cost.
Nanodynamo unravels complex RNA dynamics of SUM159 triple-negative breast cancer cells
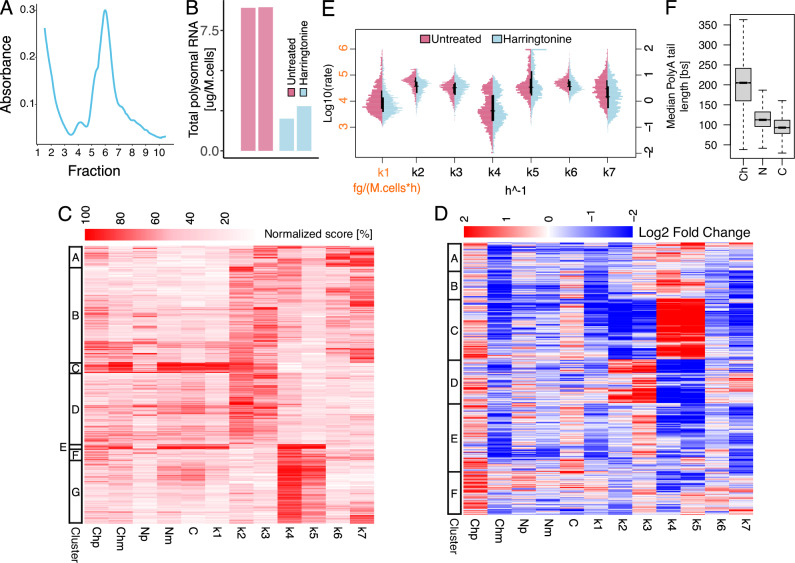
We used Nanodynamo to profile the RNA dynamics of SUM159 triple-negative breast cancer cells. We profiled transcription by dRNA-seq with two replicates for each RNA pool: chromatin-associated RNA, nucleoplasmic RNA, cytoplasmic RNA and transcripts associated with actively translating polysomes (Figs. 1C, 2A, and Supplementary Fig. 6A). See the methods for details on reads number and statistics for the sequencing runs.
Fig. 2. Nanodynamo unravels complex RNA dynamics of SUM159 triple-negative breast cancer cells.
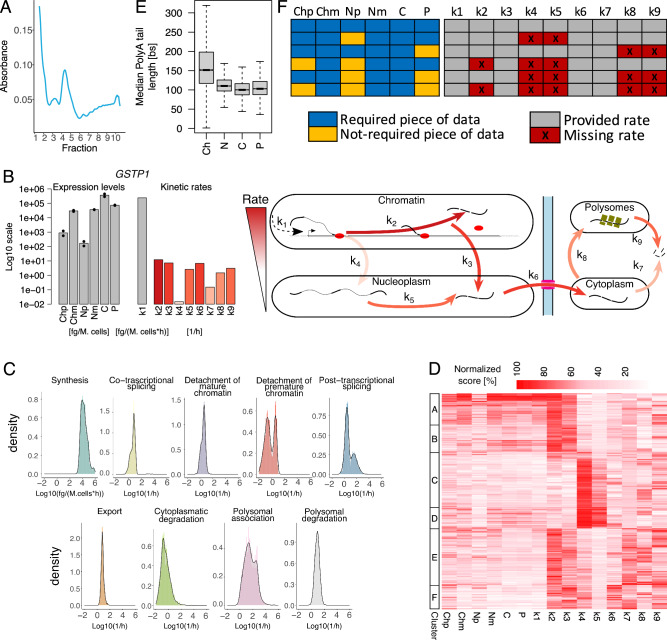
A Typical polysome trace for untreated SUM159 cells. B Schematic representation of the Nanodynamo framework, from input data to kinetic rates interpretation, for the gene GSTP1. Left, RNA species expression levels (bars and dots report mean and single replicate values respectively). Centre, kinetic rates coloured according to their relative values; the synthesis rate was reported in grey because of its different unit of measurement. Right, cartoon depicting the different efficiencies of the RNA life cycle steps. C Kinetic rates distributions in log10 scale. D Heatmap reporting for each modelled gene (rows) RNA species abundance and kinetic rates magnitude (columns). For each column, values are saturated between the 1st and 99th percentiles and normalised against the latter. The left bar indicates groups of genes identified by k-means clustering according to normalised abundances and kinetic rates. E Gene-level median polyA tail length distributions across RNA pools. F RNA species and kinetic rates for the simplified models (rows) implemented in Nanodynamo, compared to the complete model (1st row); input data required for each model are indicated in blue (not required in yellow) while the provided kinetic rates are indicated in grey (missing rates in red). In the figure, Ch, N, C, and P denote chromatin associated, nucleoplasmic, cytoplasmic, and polysomal RNA, respectively, while p and m additionally specify premature or mature forms. Analyses in C, D were performed on 1914 genes processed with the complete model while the analysis in E was performed on all the genes with at least one read with profiled polyA tail after replicates pooling (11774, 11957, 11760, 12739 genes for the chromatin, nucleoplasmic, cytoplasmic, and polysomal fractions respectively). For boxplots, the horizontal line represents the median value, the box edges represent the 25th (Q1) and 75th (Q3) percentiles, and the whiskers show the range of data excluding outliers (observations lower that Q1 − 1.5 * interquartile range or larger than Q3 + 1.5 * interquartile range). Source data are provided as a Source Data file.
To assess the reproducibility of Nanodynamo we separately analysed the two replicated datasets. In order to compare the results, we had to focus on the genes that could be analysed in both replicates (397 genes), the bottleneck being requiring the quantification of premature RNA species in all nuclear RNA pools. The Spearman correlation between the abundance of nascent, pre-existing and total RNA species between the two replicates ranged from 0.60 to 0.99 (median 0.95; Supplementary Fig. 7), with the smallest values associated with the lowest expressed species, while the correlation between the kinetic rates ranged from 0.37 to 0.95 (maximum p < 2.2e-16; median 0.67; Supplementary Fig. 8).
Jointly modelling the information coming from the two replicates, we were able to model 1914 genes. These were the genes that fulfilled the requirement of having at least one read for all the RNA species included in the model in at least one replicate. The minimum and median Spearman correlations between modelled expression levels and their experimental counterparts (replicates means) for these genes were 0.43 and 0.95 respectively, supporting the goodness of models’ fits (Supplementary Fig. 9). The median proportion of reads classified as nascent, upon 20’ 4sU metabolic labelling, ranged from 24% to 45% and followed the expected decreasing trend from chromatin to cytoplasmic RNA (Supplementary Fig. 10). The median proportion of reads classified as premature ranged from 7% to 15%, with the highest value detected in chromatin associated RNA (Supplementary Fig. 10), as expected. The dynamics of RNA metabolism for GSTP1, a representative gene involved in triple-negative breast cancer cells metabolism and pathogenicity29, are shown in Fig. 2B.
Among the nine inferred kinetic rates, RNA synthesis (k1) is the only one that is expressed in terms of polyA+ RNA produced per million cells, thus its absolute value (median of 15 pg Mcells−1 h−1) cannot be compared with the pure rates of the other RNA life-cycle steps (Fig. 2C). We extrapolated that this value is compatible with the expected yield of nascent RNA following a 20’ pulse30 (see the “Nascent RNA yield” methods section). Among the pure rates (k2-8), the fastest step was the association to actively translating polysomes, which had a median of 32.23 h−1 and spanned various orders of magnitude, suggesting that it could significantly shape gene expression programs. The remaining rates were slower with medians ranging from 0.25 h−1 (cytoplasmic decay) to 7.73 h−1 (polysomal decay). In terms of sequence features, the rate of RNA synthesis negatively associated with the size of transcriptional units and 5′ and 3′UTRs (Unpaired two-sided Wilcoxon test p < 1e-4), and genes with the fastest rates were involved in translation and focal adhesion (Unpaired two-sided Wilcoxon test p < 1e-10). All cytoplasmic rates (k7-9) were positively associated with the size of transcriptional units and 3′UTRs, and negatively associated with 5′UTR CG content (Supplementary Fig. 11; Unpaired two-sided Wilcoxon test p < 1e-4).
The rates involved in co-transcriptional splicing (k2-3) and those involved in post-transcriptional splicing (k4-5) had peculiar bimodal distributions (Fig. 2C). The genes characterised by low and high rates were conserved when analysing individual replicates (accuracy between 0.80 and 0.83, maximum two-sided exact Fisher test p < 0.0023; Supplementary Fig. 8). We excluded the numerical origin of these bimodal distributions which were not present in the simulated data (Supplementary Fig. 12A), nor did they depend on the initial conditions used for inference (see methods for details, Supplementary Fig. 12B). The clustering of genes based on the abundance of RNA species and magnitude of the kinetic rates revealed that 65% of the genes adopted co-transcriptional processing, in agreement with31, with a preference for genes with sustained transcriptional activity (Fig. 2D). Notably, distinct sets of genes relied on either co- or post-transcriptional processing pathways (i.e., genes with high k2-3 had low k4-5, and vice-versa). This was confirmed by the Spearman correlations between kinetic rates which were positive (>0.30) between synthesis and co-transcriptional processing rates, and negative (<−0.62) between co- and post-transcriptional processing rates. The same correlative analysis performed on simulated data provided 0.03 and −0.15 respectively (most extreme correlations), confirming that the structure of the model was not sufficient to generate similar results. Finally, these analyses also revealed that high expression levels were mainly mediated by high rates of synthesis, that genes efficiently translated tended to be efficiently degraded (Spearman correlations 0.28 and 0.26 against cytoplasmic and polysomal degradation, respectively), and that the rate of polysomal degradation was faster than the rate of cytoplasmic degradation (Unpaired two-sided Wilcoxon test p < 2.2e-16).
We took advantage that dRNA-seq data offer the possibility of quantifying the length of polyA tails. The median length of the tails for transcripts associated with chromatin was 152nt, which reduced to 105nt for transcripts retrieved from other RNA pools (Fig. 2E). This is in agreement with the recently reported rapid nuclear deadenylation of polyA tails occurring after transcription32. In addition, we found that transcripts with high rates of synthesis (Fig. 2D, cluster A) had particularly short tails (median 110 nt, Supplementary Fig. 13), derived from compact transcriptional units accounting for short 3′UTRs and a low number and size of exons and introns (Supplementary Fig. 14), and were often involved in translation (Hypergeometric test p < 8.85e-9).
Gathering all the expected data could be complicated due to various reasons. We thus developed alternative simplified models accommodating the lack of one or more RNA species (Fig. 2F, Supplemental Material and Supplementary Figs. 42–56). For instance, premature RNA might not be found in the nucleoplasm for genes that do not go through post-transcriptional processing, or due to insufficient sequencing depth, which could prevent the quantification of these low abundant species. We thus developed an alternative model in which RNA processing is only co-transcriptional. More in general, processing might be not applicable at all for certain genes like intron-less transcriptional units (~13% of the UCSC annotated genes). Modelling RNA processing could also be complicated for very compact genomes and short introns, such as yeast and A. thaliana. To accommodate these scenarios, we developed an alternative model in which transcripts are synthetized directly into their mature form. Finally, to deal with non-coding genes, we implemented a framework lacking the step of association with polysomes. These models could be useful also when technical reasons prevent the acquisition of polysomal RNA which typically requires dedicated instruments and specific expertise. Supplemental Material reports the reanalysis of untreated SUM159 cells with all these simplified models, and their comparison with the complete model.
Finally, we took advantage of the simplified models to compare RNA synthesis and cytoplasmic degradation rates returned by Nanodynamo against those determined with INSPEcT, which requires only metabolically labelled and total RNA sequencing data. Spearman correlations for the rates of RNA synthesis and degradation are 0.96 and 0.21 respectively. Noticeably, the modest score for RNA degradation likely reflects the differences in modelling between INSPEcT and Nanodynamo for this step of the RNA life cycle (Supplementary Fig. 1). We also extended the analysis including the rates estimations from nano-ID (Spearman correlation of 0.75 and 0.92 for synthesis and degradation, respectively).
Altogether, the application of Nanodynamo for analysing the dynamics of RNA metabolism in an untreated cell line revealed sets of genes characterised by substantially different combinations of kinetic rates and suggested the coordinated regulation of various steps of the RNA life cycle. We therefore set out to characterise how RNA dynamics are shaped by perturbations directed against specific steps of the RNA life cycle.
Blocking the spliceosomal machinery leads to a switch from co- to post-transcriptional processing
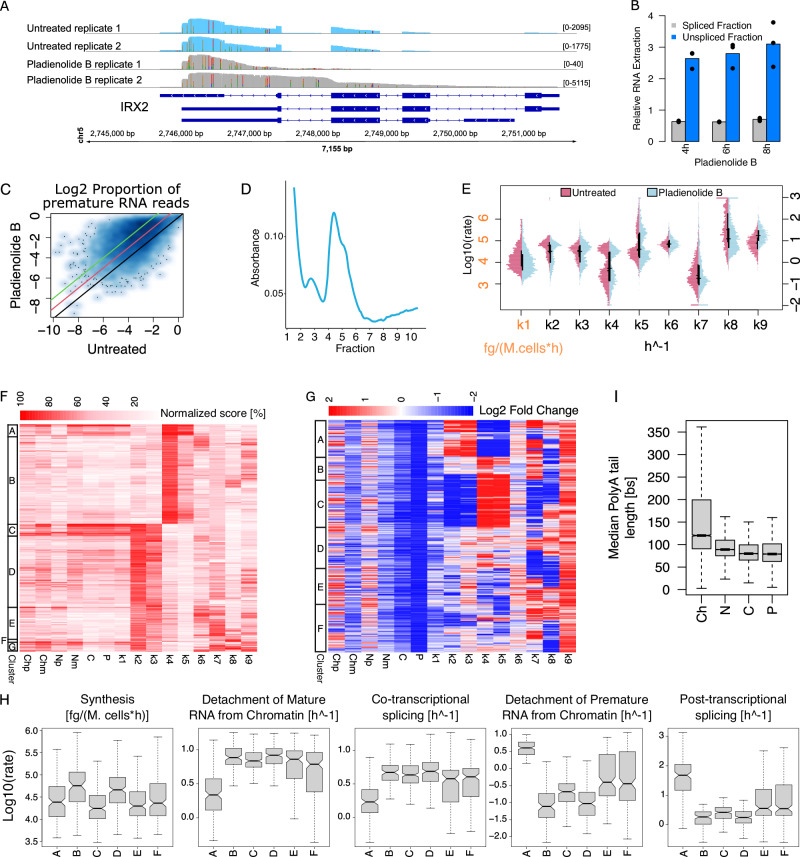
After characterising the transcriptional programs of untreated SUM159 cells, we investigated the response of the same cell system to splicing perturbation mediated by Pladienolide B. This drug targets the splicing factor SF3B1, resulting in an increase of intronic signal33. We confirmed the expected accumulation of intronic signal by RT-PCR and by inspecting various genes following dRNA-seq (Fig. 3A, B and Supplementary Figs. 15, 16). Globally, the proportion of premature reads increased by at least 4-fold for 30% of genes, compared to untreated cells (Fig. 3C). We also observed a significant reduction of nascent RNA for all nuclear fractions (between 17% and 27%, two-sided Kolmogorov-Smirnov test p = 0; Supplementary Fig. 17) suggesting a reduction in RNA synthesis. Finally, polysome fractionation revealed a marked reduction in the presence of polysome-bound RNAs (Fig. 3D, Supplementary Fig. 6B), suggesting a reduced association with the polysome.
Fig. 3. Blocking the spliceosomal machinery with Pladienolide B leads to a switch from co- to post-transcriptional processing.
A Coverage for IRX2 in untreated (blue) and treated (grey) cells. B Expression, relative to RPLPO, of CNBP spliced (black) and unspliced (blue) reads over time after treatment. Control is set to 1 (dashed line), 3 technical replicates and their mean values are reported. C Scatterplot comparing, for each gene, the proportion of intron-containing reads in untreated and treated cells (identity line in black, 2 and 4 folds increases lines compared to untreated cells in red and green). D Typical polysome trace for treated cells. E Distributions of log10 kinetic rates for untreated (purple) and treated (light blue) cells. Distributions medians and interquartile ranges as horizontal and vertical black lines respectively. F Heatmap reporting for each modelled gene (rows) RNA species abundance and kinetic rates magnitude (columns). For each column, values are saturated between the 1st and 99th percentiles and normalised against the latter. The left bar indicates gene sets similar in abundances and kinetic rates (k-means clustering). G Heatmap depicting abundances and kinetic rates modulations in response to treatment (log2 fold changes saturated between −2 and +2). The left bar indicates gene sets similar in abundances and kinetic rates modulations (k-means clustering). H Distributions of log10 synthesis and processing rates in untreated cells for genes clusters defined in G. I Gene-level median polyA tail length distributions across RNA pools. In the figure, Ch, N, C, and P denote chromatin associated, nucleoplasmic, cytoplasmic, and polysomal RNA while p and m specify premature and mature forms. Analyses reported in E, F were performed on 1950 genes processed with the complete model in the treated condition. Analyses reported in C, G, H were performed on 996 genes processed with the complete model also in untreated condition. The analysis reported in I was performed on genes with at least one profiled polyA tail after replicates pooling (11319, 11385, 11262, 11168 genes for Ch, N, C, and P). For boxplots, the horizontal line represents the median value, the box edges represent the 25th and 75th percentiles, and the whiskers show the range of data excluding outliers. Source data are provided as a Source Data file.
Following the same approach presented for the untreated condition, we checked the reproducibility of our results with the independent analysis of the two replicates (Supplementary Figs. 18 and 19). We were able to model 1950 genes with Nanodynamo following the treatment of SUM159 cells with Pladienolide B (Fig. 3E; minimum and median Spearman correlations between modelled and experimental data 0.51 and 0.92, respectively; Supplementary Fig. 20). Pladienolide B treatment led to a significant albeit mild reduction in RNA synthesis (Fig. 3F, Unpaired one-sided Wilcoxon test p < 5.51e-25). The rates that presented bimodal distribution in the untreated cells confirmed their bimodality following the drug treatment (Fig. 3F). We observed the reduction in co-transcriptional splicing and detachment of mature RNA from chromatin (Unpaired one-sided Wilcoxon test p < 8.6e-5), matching an increase in premature RNA associated with chromatin (Fig. 3F, G). The slowdown in co-transcriptional RNA processing is an expected consequence of Pladienolide B treatment and reassures about the rates modelled with Nanodynamo. In addition, the reduction in the co-transcriptional steps were accompanied by a slight increase in the detachment of premature RNA from chromatin and in the processing of premature nucleoplasmic transcripts. These data suggested a switch from co- to post-transcriptional processing pathways. Indeed, for 26% of the 966 genes that could be modelled in both the untreated and Pladienolide B treated cells, the drug treatment led to reduced co-transcriptional rates (k2-3) and increased nuclear post-transcriptional rates (k4-5), compared to untreated cells. Rather, 15% of genes switched in the opposite direction. Eventually, the reduction in co-transcriptional processing prevailed, since the net result of these opposite modulations resulted in a marked reduction of mature RNA in the nucleoplasm (Fig. 3G). Noticeably, genes repressed in co-transcriptional processing (Fig. 3G cluster C) were the least expressed among the most efficiently co-transcriptionally spliced (low k1 and high k2, Fig. 3H). Similarly, genes repressed in post-transcriptional processing (Fig. 3G cluster A) were the most efficiently post-transcriptionally spliced and were also particularly low expressed (high k5 and low k1, Fig. 3H).
We also observed an increase in nucleoplasmic RNA export following Pladienolide B treatment, which might represent an attempt to compensate for the aforementioned shortage of nucleoplasmic transcripts (Fig. 3F, G). However, in the cytoplasm, the rate of association with actively translating polysomes decreased, as a consequence of the strong reduction in the yield of the polysomal transcripts (Fig. 3G). This was accompanied by a marked increase in polysomal degradation, suggesting the need of removing transcripts that could not be properly translated, potentially through the nonsense-mediated decay pathway34. Transcripts containing Terminal Oligo Pyrimidine motifs at their 5′ end (5′ TOP) encode proteins that are essential for protein synthesis, whose translation is decreased under stress conditions. The vast majority of 5′ TOP factors that we could assess had indeed a markedly reduced rate of polysomal association, suggesting their involvement in the broad reduction of this RNA life cycle step (Supplementary Fig. 21A)35.
Finally, we found that the length of polyA tails was reduced following Pladienolide B treatment and that their shortening after transcription was less pronounced, compared to untreated cells (Fig. 3I). Changes in polyA tails length were positively correlated with changes in the rate of RNA export (Spearman correlation 0.25, p < 8.29e-15), suggesting that the tails length could impact export efficiency36. Rather, polyA tails length for genes in cluster E are more similar to those of untreated cells (Supplementary Fig. 22). Genes in this cluster are characterised by a marked increase in RNA export, and a mild reduction in RNA synthesis, potentially as an attempt to compensate for the drug effects.
Altogether, Nanodynamo revealed that treating cells with a drug directed against the splicing machinery has broad consequences that go beyond the expected repression of RNA processing, involving a switch from co- to post-transcriptional RNA processing and impacting the export and translational machineries.
Perturbation of the export machinery leads to downstream alterations of translation and degradation machineries
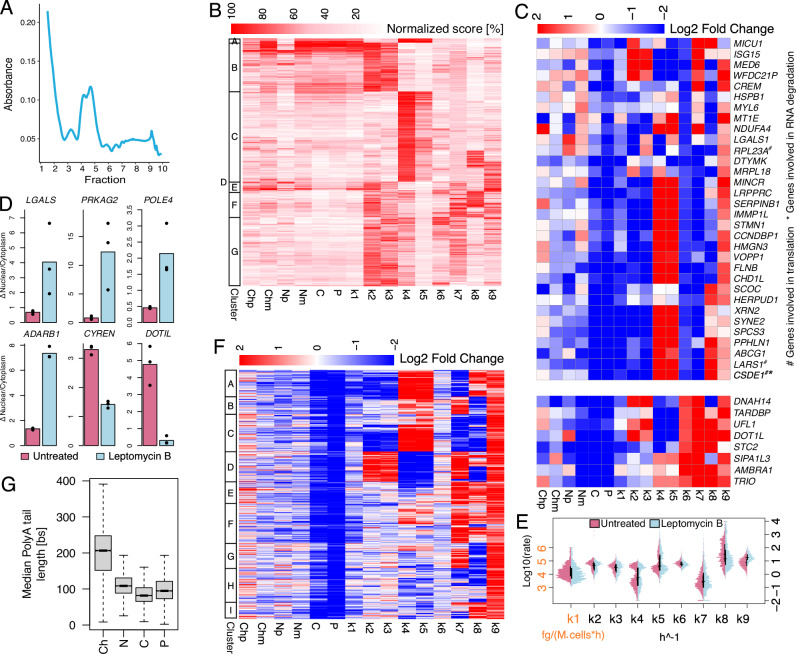
For the block of RNA export, we opted for treating SUM159 cells for 16 h with Leptomycin B, an inhibitor of CRM1, a major receptor for the export of RNA and proteins to cytoplasm37. Polysome fractionation revealed a marked reduction in the presence of polysome-bound RNAs (Fig. 4A, Supplementary Fig. 6D), suggesting a reduced rate of association with the polysome. After a check on the reproducibility of the modelling across two independent replicates (Supplementary Figs. 23 and 24), we performed the inference of the complete model as described above resulting in 1371 genes (Fig. 4B).
Fig. 4. Perturbation of the export machinery with Leptomycin B leads to downstream alterations of translation and degradation machineries.
A Typical polysome trace for treated cells. B Heatmap reporting for each modelled gene (rows) RNA species abundance and kinetic rates magnitude (columns). For each column, values are saturated between the 1st and 99th percentiles and normalised against the latter. The left bar indicates gene sets similar in abundances and kinetic rates (k-means clustering). C Heatmap depicting genes (rows) abundances and kinetic rates modulations (columns - log2 fold changes saturated between −2 and +2) in response to Leptomycin B treatment for genes reduced or induced in export (k6 log2 fold change < −1.5 or > 1.5 respectively). D Nuclear vs cytoplasmic RNA expression, relative to RPLPO, for the indicated genes in untreated cells (purple) and after 16 h of Leptomycin B treatment (light blue); 3 technical replicates and their mean values are reported. E Kinetic rates distributions in log10 scale for untreated (purple) and treated (light blue) cells; horizontal and vertical black lines represent medians and interquartile ranges of the distributions, respectively. F As in C for 1371 genes modelled both in untreated and treated cells. The left bar indicates gene sets similar in abundances and kinetic rates modulations (k-means clustering). G Gene-level median polyA tail length distributions across RNA pools. In the figure, Ch, N, C, and P denote chromatin associated, nucleoplasmic, cytoplasmic, and polysomal RNA while p and m specify premature and mature forms. Analyses reported in B, E were performed on 1371 genes processed with the complete model in the treated condition. The analysis reported in F was performed on 778 genes processed with the complete model in both untreated and treated conditions. The analysis reported in G was performed on genes with at least one profiled polyA tail after replicates pooling (12620, 11369, 10138, 12028 genes for Ch, N, C, and P). For boxplots, the horizontal line represents the median value, the box edges represent the 25th (Q1) and 75th (Q3) percentiles, and the whiskers show the range of data excluding outliers (observations lower that Q1 − 1.5 * interquartile range or larger than Q3 + 1.5 * interquartile range). Source data are provided as a Source Data file.
According to a recent report, overnight Leptomycin B treatment impacted RNA export only for selected genes. Consistently, we found 40 genes that were markedly impacted in their export rates (k6 ratio > 2.5 compared to untreated cells), which were reduced for 32 of them (Fig. 4C). RT-PCR validation in an independent experiment confirmed the altered ratio of nuclear vs cytoplasmic RNA for 6 genes previously reported to be affected by Leptomycin B. Specifically, the transcripts for 4 genes (LGALS, PRKAG2, POLE4, ADARB1) were accumulated in the nucleus and depleted in the cytoplasm, while RNAs for 2 genes (CYREN, DOTIL) were accumulated in the cytoplasm and depleted in the nucleus, as previously described38. We additionally validated 2 genes that were not previously reported being altered in RNA export (DOTIL and LGALS) (Fig. 4D) and two genes that we identified as not impacted in their RNA export (Supplementary Fig. 25). Genes up-regulated in k6 were enriched in transcriptional units down-regulated in nuclear mature RNA (Two-sided exact Fisher test p < 4.0e-4) and vice versa for genes down-regulated in k6 (Two-sided exact Fisher test p < 1.5e-3—see the “Differential RNA species” methods section for the definition of differentially expressed genes. Notably, genes modulated in opposite directions in RNA export were differently affected in other RNA life cycle steps. In particular, RNAs whose export was reduced were more likely to be reduced in RNA synthesis and co-transcriptional events (while being promoted in post-transcriptional nuclear events), whilst those that were promoted in the export were more likely to be increased in cytoplasmic degradation and association with polysomes.
Comparing all the modelled genes to untreated cells revealed that 48% of these were polarised in terms of co- or post-transcriptional processing (Fig. 4E, F). Indeed, these genes were either promoted in co-transcriptional processing and detachment of mature RNA from chromatin, while being hampered in the detachment of premature RNA from chromatin and post-transcriptional processing (Fig. 4F cluster D), or subjected to the opposite modulation (Fig. 4F clusters A-C). Even more strikingly and consistently, most of the modelled genes had altered rates of polysomal association and degradation (Fig. 4E, F). In particular, we observed a marked increase in polysomal degradation (98% of the genes). Similarly to Pladienolide B, the polysomal association of transcripts encoding for 5’ TOP factors was markedly reduced, suggesting their involvement in the broad reduction of this RNA life cycle step (Supplementary Fig. 21B)35. Analysis of sequence features for the modelled genes indicated that transcripts repressed in RNA export (Fig. 4C top) had shorter 5′ UTRs and fewer, shorter exons compared to genes unaffected in RNA export and, even more prominently, compared to genes with increased RNA export (Supplementary Fig. 26).
Finally, the analysis of polyA tails length following Leptomycin B treatment revealed that, while they were shortened following the detachment from chromatin as discussed for the untreated cells, they had a marked increase in length for the transcripts associated with chromatin (Fig. 4G). Similarly to Pladienolide B, changes in polyA tails length were positively correlated with changes in the rate of RNA export (Spearman correlation 0.11, p < 0.0023).
Eventually, given the length of the Leptomycin B treatment and the impact on the export of transcripts encoding for proteins involved in RNA decay and translation (Fig. 4C), it is possible that the observed major alterations in polysomal association and decay could be attributed to indirect downstream effects of the alteration of those Leptomycin B targets. Altogether, Nanodynamo revealed that treating the cells with a commonly used drug against RNA and protein export has consequences that are broader and more complex than expected, possibly due to nonspecific effects following the prolonged drug treatment.
Blocking the translational machinery hampers RNA export and cytoplasmatic degradation
The block of translation was obtained by treating SUM159 cells for 1 h with Harringtonine, an inhibitor of translation initiation. The net result of this treatment was a clear block of translation, as determined through Polysome fractionation profiles (Fig. 5A, Supplementary Fig. 6C). As a result, we could not isolate enough RNA associated with polysomes for the sequencing (Fig. 5B). For this reason, we applied Nanodynamo based on a simplified model which neglects RNA translation and has only one step of cytoplasmic degradation (Supplementary Fig. 27). After a check on the reproducibility of the modelling across two independent replicates (Supplementary Figs. 28 and 29), we succeeded in modelling 1616 genes (Fig. 5C). The block of translation had positive consequences on RNA synthesis for 17% of the modelled genes, and negative consequences for the remaining ones (Fig. 5D, E). 40% of the genes switched from co- to post-transcriptional processing or vice-versa. These changes were accompanied by a reduction in RNA export (71% of the genes). The slowdown in RNA export was also associated with a marked reduction in cytoplasmic degradation (77% of the genes), leading to an accumulation of transcripts in the cytoplasm.
Fig. 5. Blocking the translational machinery hampers RNA export and cytoplasmatic degradation.
A Typical polysome trace for Harringtonine treated cells. B Yield of polysomal RNA in untreated and Harringtonine treated cells; two biological replicates per condition reported. C Heatmap reporting for each of the modelled genes (rows) RNA species abundance and kinetic rates magnitude (columns). For each column, values are saturated between the 1st and 99th percentiles and normalised against the latter. The left bar indicates gene sets similar in abundances and kinetic rates (k-means clustering). D Heatmap depicting genes (rows) abundances and kinetic rates modulations (columns) in response to Harringtonine treatment (log2 fold changes saturated between −2 and +2). The left bar indicates gene sets similar in abundances and kinetic rates modulations (k-means clustering). E Kinetic rates distributions in log10 scale for untreated (purple) and Harringtonine treated (light blue) cells; horizontal and vertical black lines represent medians and interquartile ranges of the distributions, respectively. F Gene-level median polyA tail length distributions across RNA pools. In the figure, Ch, N, and C denote chromatin associated, nucleoplasmic, and cytoplasmic RNA while p and m specify premature and mature forms. Analyses performed in C, E were performed on 1616 genes processed with a model accounting for all the steps of the RNA life cycle except for association with polysomes and polysomal degradation in the Harringtonine treated condition. The analysis reported in D was performed on 896 genes processed with the same model used for C, E in both untreated and Harringtonine treated conditions. The analysis reported in F was performed on genes with at least one profiled polyA tail after replicates pooling (11832, 12242, 11646 genes for Ch, N, and C). For boxplots, the horizontal line represents the median value, the box edges represent the 25th (Q1) and 75th (Q3) percentiles, and the whiskers show the range of data excluding outliers (observations lower that Q1 − 1.5 * interquartile range or larger than Q3 + 1.5 * interquartile range). Source data are provided as a Source Data file.
Finally, similarly to the Leptomycin B treatment, also in the case of Harringtonine treatment the length of polyA tails showed a marked increase for the transcripts associated with chromatin (Fig. 5F).
Altogether, Nanodynamo revealed that blocking RNA translation has major consequences on all steps of the RNA life cycle, leading to an accumulation of transcripts in both the nucleus and the cytoplasm.
Coupling of RNA life cycle steps markedly influence the coordinated response of RNA metabolism to perturbations
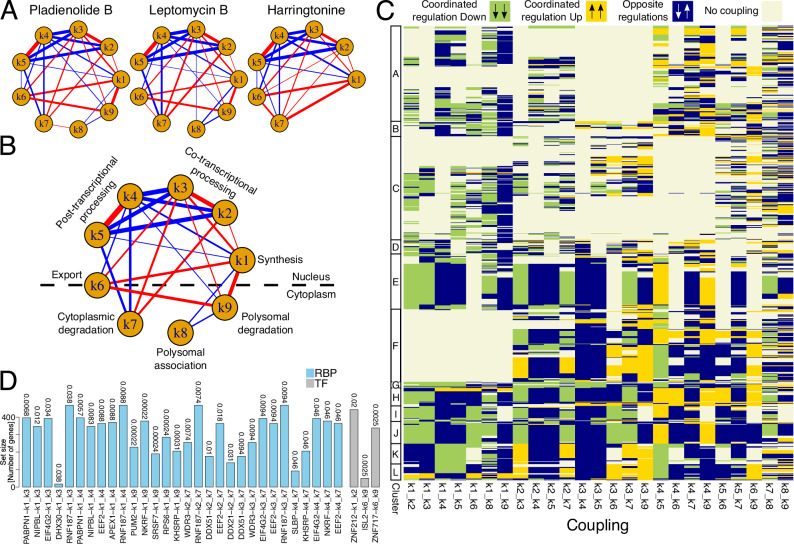
The comprehensive characterisation of RNA dynamics in untreated cells and how they are impacted by a set of perturbations is well suited for studying the coupling between steps of the RNA life cycle. We reasoned that we could study the coordination of the corresponding machineries by determining whether changes in the kinetic rates following the described drug treatments are correlated. We then determined, for each pair of kinetic rates and for each drug treatment, the correlation between kinetic rates log2 Fold Changes to the untreated condition. Significant correlations (p < 1e-4) upon treatment with Pladienolide B were displayed as edges in a graph, whose width and colour identify strength and direction of coupling, respectively. The same analysis was repeated upon perturbation with Leptomycin B and Harringtonine (Fig. 6A). Importantly, the latter is only partially informative to this regard, as it derives from the implementation of a simplified model lacking RNA life cycle steps associated with polysome. In addition, while the readout of the Leptomycin B treatment could be complicated by potentially prevalent indirect effects, this was not relevant for the identification of couplings. Indeed, what mattered for these analyses is that whichever the perturbation was, we could determine how RNA metabolism adapted to it. Eventually, 92% of the couplings identified upon Pladienolide B treatment were shared with and had the same direction of those identified upon Leptomycin B treatment. Similarly, the couplings were shared and had consistent direction with those reported for the Harringtonine treatment (Fig. 6A).
Fig. 6. Coupling of RNA life cycle steps markedly influence the coordinated response of RNA metabolism to perturbations.
A Networks depicting, for each treatment, coupling among steps of the RNA life cycle identified through Spearman correlation of rates modulations in treated versus untreated cells (log2 fold changes). Nodes and edges represent steps of the RNA life cycle and significant correlations among them respectively (p < 1e-4 - significance estimated with the AS89 algorithm of the cor.test R function). Edges thicknesses and colours represent magnitude and sign of correlations respectively (negative and positive values in blue and red respectively). B As in A for couplings conserved between Pladienolide B and Leptomycin B treated cells (edges thicknesses given by correlations mean across treatments). C Heatmap depicting, for the Pladienolide B treatment, the couplings (columns—see panel A) exploited by each gene (rows). Colours represent specific regulations: beige indicates that at least one of the two rates for a given coupling is not regulated, yellow denotes their coordinated up-regulation, green denotes their coordinated down-regulation, and blue indicates their opposite regulation. A rate is considered modulated given an absolute log2 fold change larger than 0.5. D Bar plot reporting the 5 most significant proteins (RNA Binding Proteins—RBPs—in light blue and Transcription Factors—TF—in grey), according to a GSEA analysis, for each coupling identified in Pladienolide B treated cells (see panel A). For a pair of rates defining a coupling, the GSEA ranking was defined according to the product of their modulations in Pladienolide B treated versus untreated cells (log2 fold changes) times the sign of their Spearman correlation. GSEA adjusted p-values (Benjamini-Hochberg) are reported above each bar. Analyses in (A - “Pladienolide B”) and C, D were performed on 996 genes processed with the complete model in both untreated and Pladienolide B treated conditions. Analysis in (A - “Leptomycin B”) was performed on 778 genes processed with the complete model in both untreated and Leptomycin B treated conditions. Analysis in (A - “Harringtonine”) was performed on 896 genes processed with a model accounting for all RNA life cycle steps except for association with polysomes and polysomal degradation in both untreated and Harringtonine treated conditions. Source data are provided as a Source Data file.
The final network of couplings shared between Pladienolide B and Leptomycin B revealed couplings linking processes within and between nucleus and cytoplasm (Fig. 6B). RNA synthesis (k1) turned out to be positively coordinated with co-transcriptional processing and detachment of mature RNA from chromatin (k2-3). Detachment of premature transcripts and post-transcriptional processing (k4-5) were also positively associated. Rather, transcription and co-transcriptional processing steps were anti-correlated with the post-transcriptional processing counterparts. Additionally, the rate of synthesis (k1) was positively coupled with RNA export (k6) and polysomal degradation (k9), while both k1 and k9 were negatively coupled with polysomal association (k8). k1 and k9 were globally down- and up- regulated in response to the Pladienolide B (Fig. 3G). Consequently, their positive coupling denoted that strong regulations of the former were associated with weak down-regulations of the latter, and vice-versa (Supplementary Fig. 30). Finally, these analyses revealed that co-transcriptional processing steps were positively coupled with several downstream steps, including export and both degradation rates. The same holds true for post-transcriptional processing steps, while this occurred through negative couplings. These observations suggested the relevance of RNA processing in shaping gene expression responses.
We next wondered how the response of individual genes is shaped by the reported couplings. For each of the identified global couplings, and for each modelled gene we determined whether that gene exploited it, and whether the coupling involved the coordinated increase or decrease of the kinetic rates or their opposite modulation (Fig. 6C for Pladienolide B response and Supplementary Fig. 31 for the other drug treatments).
Genes responding to Pladienolide B exploited a remarkable fraction of the possible mechanisms (14 couplings for 50% of the genes) typically involving the coordination between rates in different cellular compartments. Half of the genes (clusters E-L) were pervasively regulated by couplings, involving most of the interactions reported for Pladienolide B (Fig. 6C). Among these, only those in cluster F differed since they did not exploit couplings stemming from the synthesis step, while they relied on the coordinated response between co- and post-transcriptional processing steps (k2-5) and between these and cytoplasmic steps. We characterized the clusters of genes in terms of structural features, CG content, and polyA tail length. Genes within cluster G, which were down-regulated in synthesis and in both co- and post- transcriptional processing rates, were the only that had longer tails compared to untreated cells in contrast with the trend observed for the other gene sets (compare Supplementary Fig. 32 with Figs. 2E and 3I). In terms of size and genomic complexity, genes in clusters I, which did not exploit the coupling to polysomal degradation, were characterized by the longest transcriptional units. Rather, genes in the aforementioned cluster F, which did not exploit the coupling with RNA synthesis, were characterized by the shortest transcriptional units (Supplementary Fig. 33). These results were largely confirmed by the analysis of Leptomycin B response.
We used the Hamming distance to compare the coupling profiles of the genes modelled upon both Pladienolide B and Leptomycin B treatments. Using as a reference a null distribution of Hamming distances, obtained through the shuffling of the heatmaps columns (Supplementary Fig. 34—see methods section “Couplings” for details), we identified 97 genes whose coupling profiles were consistent following the two treatments. This analysis indicated the conservation of gene-level coupling for a sizable fraction of the analysed genes, involving genes with either a high or a low number of couplings (Supplementary Fig. 35).
Finally, we sought to identify regulatory factors potentially responsible for each of the detected couplings. To this end, we took advantage of ENCODE data to search for RNA Binding Proteins (RBPs) and Transcription Factors (TFs) targeting genes supported by a given coupling. Specifically, we computed the product of log2 Fold Changes for each pair of coupled rates, and we used this quantity to perform Gene Set Enrichment Analyses (see methods section “Couplings” for further details). Overall, we identified 100 and 114 factors for Pladienolide B and Leptomycin B respectively (GSEA adjusted p < 0.05 for at least one edge), the vast majority deriving from the k1,9 edge. 93 of these factors were shared further supporting the conservation of coupling mechanisms. The 5 most significant factors for each coupling were reported in Fig. 6D for the Pladienolide B treatment, and in Supplementary Fig. 36 for the Leptomycin B one. Three proteins involved in gene expression regulation emerged as top candidates for implementing the couplings between RNA synthesis and processing (k3-5) in both the treatments: NIPBL, APEX1, and PABPN1. Interestingly, the latter takes part in RNA polyadenylation which might suggest the involvement of this regulatory layer in mediating transcriptional couplings39,40.
Discussion
The Nanodynamo model, limitations and potential extensions
We developed Nanodynamo, a method combining an experimental and computational approach for the quantification of the kinetic rates governing the dynamics of RNA metabolism, and we used it to unravel coupling interactions between steps of the RNA life cycle. Nanodynamo combines the profiling of native metabolically labelled transcripts from various RNA pools, including cell fractions and polysomes, with mathematical modelling. Compared to available methods, Nanodynamo significantly expands the considered steps of the RNA life cycle, which is integrally modelled from the birth of the transcripts within chromatin until their decay in the cytoplasm. The adopted model does not only account for a chain of steps, but also includes two branch points, where the transcripts can be directed towards co- or post-transcriptional processing in the nucleus, or towards degradation or association to polysomes in the cytoplasm (Fig. 1).
This model relies on the following assumptions: (i) RNA degradation only occurs in the cytoplasm, i.e., nuclear RNA is not degraded, (ii) premature RNA is not exported into the cytoplasm, (iii) ribosome-bound RNA that is not in active translation can be degraded.
Nanodynamo complete model disregards nuclear degradation. We tested extending the model to incorporate degradation of either premature or mature nucleoplasmic RNA (see Supplemental Material). Even though the models including these extra steps are globally identifiable, we could not infer the corresponding nucleoplasmic decay rates (Spearman correlation coefficients based on simulated data 0.05 and 0.08, respectively), possibly due to a lack of sensitivity for these parameters compared to the other rates. Importantly, the presence or lack of these extra steps had no impact on the other kinetic rates (Supplementary Figs. 37, 38). It could be interesting to further investigate this aspect searching for rates configurations more dependent on nuclear degradation, which could reveal the importance of this step of the RNA life cycle for specific subsets of genes, as suggested in ref. 21.
Regarding the second assumption, there are two ways to avoid it. The first possibility is by assuming that premature and mature RNA can be exported, subjected to cytoplasmic degradation, polysomal association and subsequent degradation with the same rates as the mature species. We tested this possibility and confirmed that the main conclusions drawn by this study are maintained, see for example Supplementary Fig. 39 for the impact of Pladienolide B. The second possibility is to markedly expand the model incorporating a number of steps which act specifically on premature species. This would substantially complicate the model and likely require additional data. For all these reasons we deemed reasonable to maintain this assumption.
Regarding the third assumption, avoiding it would likely imply to extend the model. RNA translation is a multistep process including the association of multiple ribosomes to coding transcripts, initiation, elongation and detachment of the translational machinery. Nanodynamo currently only considers the association of mRNAs with polysomes and the subsequent polysomal degradation. In fact, our modelling does not assume nor depend on the fact that translation actually occurred. In addition, we have no information on whether RNAs profiled in the cytoplasm were previously associated with polysomes or not. Further development of the method could in the future be dedicated to the inclusion of additional steps for a more thorough modelling of the translation machinery.
The same perspective could be applied also to improve our characterization of RNA synthesis by explicitly modelling key steps of RNA polymerase activity27 (e.g., initiation, pause-release, elongation, and termination). More generally, the Nanodynamo framework could be extended to include rates describing the transition of RNA molecules across a large set of states defined according to a specific feature of the transcripts (e.g., retention of intronic signal) and/or their localization. In this regard, we foresee an interesting extension of our model based on the isolation of biomolecular condensates (e.g., stress granules and P-bodies).
Moreover, we anticipate the possibility of extending the Nanodynamo framework to incorporate information about key determinants of gene expression programs and kinetic rates couplings, such as the level of RNA modifications, RBPs, and TFs. For the latter two classes of regulatory factors, gene expression levels and/or the rate of association with polysomes are potential proxies for those factors protein abundance. For example, the models of genes targeted by specific factors could be coupled with the equations describing those factors’ life cycle.
All these potential extensions are feasible until the ODEs system parameters are globally identifiable, and the required RNA pools can be isolated for: dRNA-seq library preparation and RNA yield measurement. Noticeably, these two steps of the Nanodynamo framework are decoupled and they can be performed on independent samples providing more flexibility in the experimental design and allowing for the collection of various replicates of RNA yields without incurring additional sequencing costs and waiting time. This is particularly important because RNA yield quantification is crucial for inference purposes, and it can potentially introduce systematic biases affecting the absolute kinetic rates quantifications.
The adopted experimental workflow, which relies on the sequencing of two independent replicates through Nanopore dRNA-seq, allowed us to apply the complete model quantifying the kinetic rates for 9 steps of the RNA life cycle of 1.5–2 thousand genes, depending on the condition. This is mostly under the control of the sequencing throughput and could be significantly improved by switching from MinION/GridION to PromethION Nanopore flow cells, which provide a substantial increase in the throughput. As an alternative, PromethION flow cells could also be adopted to combine the profiling of the various RNA pools within the same sequencing run, by setting up a barcoding strategy to track the RNAs within each pool. An increase in throughput would also improve the inference performance for genes already profiled. Indeed, we observed a mild overfit for low-expressed species (i.e., higher correlation between inferred and expression data than between replicates for nascent nucleoplasmic premature RNA) which was significantly reduced selecting highly expressed genes (top 10% genes in nucleoplasmic premature RNA - Supplementary Fig. 40). Similarly, the inference performance would benefit from the profiling of a higher number of replicates, as suggested by the modest yet clear trend observed in simulated datasets (Supplementary Fig. 4). Clearly, the drawback of all these improvements is the significant increase in experimental costs; for this reason, we suggest the experimental design used in this study as a reasonable compromise.
We anticipate that an alternative and effective workaround to reduce the experimental cost of Nanodynamo would be shifting from the Nanopore to the Illumina RNA sequencing platform, leveraging protocols for the chemical conversion of incorporated nucleotides for nascent RNA profiling17,21. This would provide better control over sequencing depth (i.e., cost) and a higher ratio of detected genes per million sequenced bases. On the other hand, this approach would not benefit from key features of long-read direct RNA-seq, such as the ability to better discriminate expressed isoforms and intronic signal, as well as the intrinsic profiling of important determinants of gene expression programs like RNA modifications and polyA tails.
dRNA-seq requires the presence of polyA tails, and the profiling of the various RNA pools in this study was performed by selecting polyA+ transcripts. With this approach, we could have lost premature transcripts not yet polyadenylated. To assess this, we considered depleting ribosomal species as an alternative to polyA selection. We have previously shown that the profiling of premature and mature RNA through Illumina short reads sequencing and the subsequent quantification of RNA dynamics are consistent across various library preparation and RNA selection methods16. To further assess this, we performed dRNA-seq through ribo depletion - following in vitro polyadenylation to comply with dRNA-seq requirements of polyA tails - and compared it to polyA selection (both experiments performed with K562 cells). Ribo-depletion resulted in a slightly higher yield of premature reads (4.0%), compared to polyA selection (3.2%), which, however, were associated with a lower proportion of genes (48%), compared to polyA selection (64%). Eventually, this alternative approach proved not so reliable in our hands and often resulted in reduced sequencing throughput. Given the limited amount of extra information gained, we considered that the increased number of steps and costs required for this alternative approach was not worth it, and we decided to rely on polyA selection.
The pervasive role of post-transcriptional RNA processing
The analysis of the dynamics of RNA metabolism in SUM159 TNBC cells (Fig. 2) revealed that a substantial number of genes adopt the post-transcriptional processing pathway, which is still largely under-investigated compared to its co-transcriptional counterpart. The consistent association of specific gene sets to co- and post-transcriptional pathways in different biological replicates, suggests that the assignment of a given gene to either pathway is not the consequence of stochastic leakage of premature transcripts between chromatin and nucleoplasmatic fractions. In addition, the lack of structural and sequence features differentiating these gene sets suggests that they are unlikely to be originated by chemical or physical properties leading to contamination between fractions (Supplementary Fig. 14).
Our data indicate that co- and post-transcriptional processing pathways are often mutually exclusive, suggesting the existence of different machineries, or that mechanisms exist that select whether the same spliceosomal machinery has to be recruited within chromatin or in the nucleoplasm. All the perturbations considered in this study caused for many genes a switch between these alternative processing pathways (Figs. 3–5); which is confirmed between treatments biological replicates. In particular, the treatment with a drug inhibiting splicing, Pladienolide B, which impacted both processing pathways, revealed the widespread transition of genes from co- to post-transcriptional processing and vice-versa (Fig. 3G clusters A and C) specifically affecting low expressed, efficiently spliced genes (Fig. 3H). The impact of the drug on poorly expressed genes is reasonable because, assuming a uniform distribution of Pladienolide B, they would be those with the highest proportion of drug molecules per transcript. On the other hand, the up-regulation of post-transcriptional processing rates in response to the impairment of the co-transcriptional processing ones—and vice-versa—suggests the existence of either active or passive compensatory mechanisms. These mechanisms could be based, for instance, on the coordinated modulation of kinetic rates mediated by RBPs. Additionally, these mechanisms could originate from the release of spliceosomal resources which would promote the re-equilibrium between the two mutually exclusive processing pathways. Importantly, regardless of the biological explanation, the increase in co-transcriptional processing as a consequence of switching was facilitated by the low magnitude of the co-transcriptional rate for genes in cluster A, and the same applied to genes in cluster C for the switch to post-transcriptional processing.
A comprehensive analysis of coupling among RNA life cycle steps
The quantification of RNA dynamics with Nanodynamo enabled us to identify coupling interactions between steps of the RNA life cycle (Fig. 6) which do not emerge from properties of the data or of the adopted modelling. The fact that these interactions are consistent across perturbations not only reinforces our conclusions, but also suggests that these coupling mechanisms are robust and could not be easily perturbed.
While coupling interactions are unable to predict differential gene expression programs resulting from perturbations of RNA life cycle steps, they should be able to shape how steps of the RNA life cycle adapt to the perturbation consequences. Depending on the extent and length of the perturbation, only the steps directly coupled to the perturbed one(s) could have enough time to reshape the RNA life cycle. If enough time is given, also indirect coupling connections could be activated and shape a new steady-state gene expression program. Minimizing the length of a given treatment is important to minimize the occurrence of indirect effects. However, distinguishing between direct or indirect effects, or avoiding the latter, is not relevant for the purpose of identifying coupling interactions.
The method that we devised for the identification of coupling interactions aims at identifying global coupling mechanisms that are supported by large sets of genes. Nevertheless, it is possible that different coupling modes exist, or that the coupling interactions that we revealed have the opposite sign, for specific subset of genes. This is especially true for the weakest global coupling interactions.
Discrete modules of coupling interactions emerged, involving co-transcriptional (k1,2,3) and post-transcriptional (k4,5) processing steps. Each module relied on positive interactions. This could be due to the fact that positive coupling interactions are effective for steps in a chain of events directed towards the goal of maximising an output, such as the production of processed (mature) transcripts. Rather, co- and post-transcriptional processing modules were linked by negative couplings, in agreement with the aforementioned mutual exclusivity of the two processes. A set of additional modules based on positive couplings stemmed from the co-transcriptional module, involving export and both degradation steps (k1,6,9 and k2,3,7). The positive coordination between transcription and degradation is likely instrumental for RNA production buffering8. On the other hand, modules stemming from the post-transcriptional processing module negatively coordinate these steps with the degradation machinery (k4,5,7 and k4,1,9).
The positive coupling between RNA synthesis and co-transcriptional processing was expected and is supported by various studies5. However, our study documented the intricacies of these connections, revealed the prevalence of an alternative post-transcriptional processing pathway, and how this is connected with RNA synthesis and co-transcriptional processing machineries.
RNA processing was also previously shown being coupled with RNA export, apparently favoured by the co-localization of the two machineries41. Indeed, we found a conserved positive association between the detachment of mature RNA from chromatin and RNA export.
The coupling between the translational and the degradation machineries was also previously documented and includes a variety of mechanisms linked to the quality control steps associated with ribosomes. One of the most important coupling mechanisms between translation and degradation is the decay of transcripts that cannot be properly translated11,12. In agreement with this mechanism, we revealed a negative coupling between polysomal association and degradation, following both Pladienolide B and Leptomycin B treatments.
Final perspectives
We developed Nanodynamo as a framework for the comprehensive analysis of the dynamics of RNA metabolism. This approach could facilitate studying the functional role of poorly characterized RNA binding proteins or RNA modifications, by determining how their loss or depletion impacts RNA fate. We illustrated the application of Nanodynamo in the identification of couplings between steps of the RNA life cycle. Follow up studies might be dedicated to shedding light on specific coupling mechanisms. For example by studying RNA modifications, RNA binding proteins or transcription factors acting as coupling factors, including those that we proposed here to be associated with specific couplings (Fig. 6D). The data generated in this study and the described approaches could also be instrumental for studying system-level properties emerging from the combined regulation of the different machineries of the RNA life cycle. Finally, studying whether RNA dynamics and coupling mechanisms are aberrant in disease conditions might point to unexpected vulnerabilities of RNA metabolism.
Methods
Cell culture
SUM159PT (Asterand, RRID:CVCL_5423, Female) -CRISPRi cells (PB TRE dCAS9-KRAB), from here on and in the text named SUM159, were cultured in HAM’s-F12 medium (Thermofisher 11765054) with 5% TET-free serum (Thermofisher A4736301), insulin 5 μg/ml (Lonza BE02-033E20), hydrocortisone 1 μg/ml (MERK H0888), HEPES 10 mM (Thermofisher,15630080) and Hygromycin B 100 μg/mL (Thermofisher, 10453982). Cells were grown at 37 °C and 10% carbon dioxide. In absence of doxycycline, we checked that these cells showed no difference with SUM159 parental cells in terms of Cas9 expression. To inhibit splicing, SUM159 cells were treated with 100 nM of Pladienolide B (Santa Cruz CAS 445493-23-2) for 4 h. To inhibit nuclear export, cells were treated with 20 ng/ml of Leptomycin B (Santa Cruz CAS 87081-35-4) for 18 h. One hour of Harringtonine (1ug/ml, Abcam 26833-85-2) was used to inhibit translation. For metabolic labelling SUM159 cells were treated with 500uM of 4-Thiouridine (Santa Cruz CAS 13957-31-8) for 20 min.
K562 (Kristian Helin lab, RRID:CVCL_0004, Female)-MycER cells - from here on, and in the text, named K562 - were grown and maintained in RPMI-1640 medium (Invitrogen Corporation; Cat No. 23400-021) containing 5% FBS in a 5% CO2 incubated at 37 °C. In absence of OHT, K562-MycER showed no difference with K562 parental cells in terms of expression of MycER and endogenous MYC levels. For metabolic labelling, K562 cells were treated with 500 μM 5-Ethynyl Uridine (Jena Bioscience CLK-N002-10) for 60 min.
Western Blot
Proteins from the different fractions were extracted from Qiazol (Qiagen 79306) and quantified using BCA Protein Assay Kit (Thermofisher 23227). The absorbance was read at 562 nm with Glomax Explorer. Precast TGX Stain-Free 4-15% gradient SDS-PAGE gels (Criterion 5678084) were used for protein separation. Proteins were then transferred onto nitrocellulose membranes, which were blocked with BSA 5% and incubated with antibodies against 1:10000 Vinculin (SIGMA V9131), 1:1000 LaminB1 (SantaCruz sc-374015), 1:1000 H3 (Abcam ab1791). Membranes were then incubated with peroxidase-labelled goat anti-rabbit IgG 1:10000 (Cell signaling 7074P2) or goat anti-mouse IgG 1:10000 (Cell signaling 7076P2) for 60 min. Antigen detection was achieved using the Clarity Western ECL substrate (Biorad 1705061). See Supplementary Fig. 6E and Supplementary Fig. 57.
Retrotranscription and qRT-PCR
SUM159 cells treated with 100 nM of Pladienolide B (Santa Cruz CAS 445493-23-2) and total RNA was extracted from the different fractions using Qiazol (Qiagen 79306). 1ug of purified RNA was retrotranscribed using Superscript III Reverse Transcriptase (Thermofisher 18080093), following manufacturer’s protocol. Briefly, The reaction mix included 1 µL of oligo(dT)20 (50 µM), 1 µL of 10 mM dNTP Mix, and sterile, distilled water to a final volume of 13 µL. The mix was heated to 65 °C for 5 minutes and chilled on ice for 1 minute. After brief centrifugation, 4 µL of 5× First-Strand Buffer, 1 µL of 0.1 M DTT, 1 µL of RNaseOUT, and 1 µL of SuperScript III RT were added. The reaction was incubated at 50 °C for 30–60 minutes and terminated by heating at 70 °C for 15 minutes. Working stock was diluted at 5 ng/ul and stored at −20 °C. Complementary DNA was analysed by qPCR using SYBR® Green Master Mix (Biorad 1725150) with the primers reported in Table S1.
Fractionation and mRNA extraction
SUM159 cells were resuspended in PBS, counted, centrifugated at 110 g for 5 min and washed with ice cold PBS containing RNAse inhibitor (New England Biolabs M0307L). The pellet was resuspended in a cytoplasmic lysis buffer for the subsequent fractionation, following the protocol described in ref. 42. Briefly, cell pellets were rinsed with 1× PBS/1 mM EDTA and lysed in ice-cold NP-40 lysis buffer (10 mM Tris-HCl [pH 7.5], 0.05% NP-40, 150 mM NaCl) for 5 min. The lysate was then placed on 2.5 volumes of chilled sucrose cushion (24% sucrose in lysis buffer) and centrifuged at 16,000 g for 10 minutes at 4 °C, the supernatant containing the cytoplasmic fraction was collected. The nuclear pellet was gently rinsed with ice-cold 1× PBS/1 mM EDTA and resuspended in prechilled glycerol buffer (20 mM Tris-HCl [pH 7.9], 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, 50% glycerol) by gently flicking the tube. An equal volume of cold nuclei lysis buffer (10 mM HEPES [pH 7.6], 1 mM DTT, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M UREA, 1% NP-40) was then added. The mixture was gently vortexed for 2 × 2 s, incubated on ice for 2 min, and centrifuged again at 16,000 g for 2 min at 4 °C. The supernatant, containing the nuclear fraction, was collected. Pellet of chromatin fraction was resuspended in the homogenization buffer. The RNA in the three subcellular fractions was extracted using Maxwell RSC miRNA tissue Kit (Promega AS1460). Total RNA was quantified, and mRNA purification was performed with up to 200ug of Total RNA using µMACS™ mRNA Isolation Kit (Miltenyi Biotec 130-075-201) following the manufacturer’s protocol. The mRNA was quantified with Qubit and used as an input for Nanopore Direct RNA sequencing libraries preparation. Fractions yields are reported in Table S2.
Polysome Profiling
SUM159 cells were labelled with 500uM of 4-Thiouridine (Santa Cruz CAS 13957-31-8) for 20 min and treated with cycloheximide 100 µg/ml (Merk C4859) for 10 min before being lysed and washed twice with ice-cold PBS containing cycloheximide 10 µg/ml. Cells were lysed in a buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.1% NP-40, 100 µg/ml cycloheximide, 1 mM DTT, and 0.2 mg/ml heparin and let sit 10 minutes on ice. After centrifugation at 14,000 g for 10 min at 4 °C, cytoplasmic extracts were loaded on a 15–50% sucrose gradient and centrifuged at 4 °C in a SW41Ti Beckman rotor for 3 h 30 min at 180,000 g Absorbance at 254 nm was recorded by BioLogic LP software (BioRad). RNA from polysome fraction was extracted using Qiazol (Qiagen 79306). mRNA purification was performed using µMACS™ mRNA Isolation Kit (Miltenyi Biotec 130-075-201) and used as an input for Nanopore Direct RNA sequencing.
Ribo depletion and PolyA tailing
10ug of total RNA purified from K562 cells were Ribo depleted using RiboMinus™ Eukaryote Kit (Thermofisher Scientific A1083708) following manufacturer’s protocol. Two rounds of Ribo depletion were performed for each sample. Around 1ug of ribo depleted RNA were obtained. Subsequently, Ribo depleted RNA was denatured, and Poly(A) tailed in vitro using E. coli Polymerase NEB (NEB# M0276). 1ug of RNA was incubated 30 min at 37 °C. RNA was cleaned using RNAClean XP beads (Beckman Coulter A63987) and then loaded into µMACS™ columns for mRNA purification. 100 ng of mRNA were used as an input for Nanopore library preparation.
dRNA-seq
Between 100 ng and 500 ng of Poly(A)-selected RNA were used as input for Nanopore direct RNA sequencing kit (SQK-RNA002). RNA was prepared following the manufacturer’s protocol. Sequencing was carried out on an Oxford Nanopore GridION Mk1 using R9.4.1 flow cells for ∼72 h. Table S3 reports statistics on input material and output data for the samples sequenced in this study.
Reads Alignment
Direct RNA nanopore sequencing reads were obtained for each cellular fraction (chromatin, nucleoplasm, cytoplasm), and for polysomes. FAST5 files were basecalled using Guppy6 (v6.2.1 + 6588110) with the following parameters: --fast5_out -c rna_r9.4.1_70bps_hac.cfg --num_callers 20 --gpu_runners_per_device 1 --trim_strategy ‘rna’ --disable_qscore_filtering. Subsequently, FASTQ were filtered based on the average read quality score (qvalue = 7) with NanoFilt (v2.8.0)43, and the selected reads were aligned to the human genome assembly GRCh38 extended with ERCC sequences (provided by ThermoFisher web documentation) and yeast ENO2 gene (assembly R64-1-1, Ensembl version 109) with minimap2 (v0.1 - minimap2 -ax splice –k14)44,45. Subsequently, samtools (v1.6)46 was used to filter out unmapped reads, reads with not primary alignments and with supplementary alignments.
Quantification of gene expression levels
Premature reads profiling
For each gene, we retrieved regions annotated as exonic in at least one isoform from the R object TxDb.Hsapiens.UCSC.hg38.knownGene (Bioconductor v3.17)47 through the R package GenomicFeatures (v1.48.1)48, and we defined gaps between these portions as intronic regions. We loaded samples BAM files in R, and we associated each read to the corresponding gene according to their overlap (findOverlaps method of the R package GenomicAlignments v1.32.1)48. Genes overlapping the intronic regions of the gene were classified as premature, the others as mature. We considered overlaps of at least 10 bases and reads overlapping more than one gene were discarded. Premature reads were discarded for cytoplasmic and polysomal samples.
Nascent reads profiling
Transcripts containing 4sU were profiled with nano-ID26 (reference GitHub repository: https://github.com/birdumbrella/nano-ID) relying on a neural network trained to distinguish between reads from unlabelled and fully-labelled samples; the latter obtained after 8 hours of 4sU metabolic labelling (500 nM) followed by nascent transcripts pull-down. Briefly, total RNA was extracted with Trizol and the isolated RNA was treated with biotin that specifically reacts with 4sU. To purify biotinylated RNA we used Dynabeads MyOne Streptavidin T1 (Invitrogen 65601). The isolated newly synthesised RNA labelled with 4sU was converted into libraries used for direct RNA sequencing with Nanopore. We processed the training samples by extracting all the features required by nano-ID with a custom Nextflow pipeline (Manuscript in preparation) that performs: reads alignment (minimap2 -ax splice -k14), BAM sorting and filtering (samtools view -F 2308 -q 20), and executes all the R scripts provided by Maier and colleagues in the original nano-ID publication. The dataset was then sub-sampled to balance the amount of labelled and unlabelled reads per gene, and splitted in training and test sets (70% and 30% of the reads, respectively) which were finally used for the training of the neural network and its evaluation. The same pipeline was used to profile nascent RNA in SUM159 untreated and treated samples. In this case, after feature extraction, we used the previously trained instance of nano-ID to estimate a modification probability for each read. Reads with modification probability greater or smaller than 0.5 were classified as nascent or pre-existing respectively. Spike-ins and Yeast – Reads mapping to spike-ins and S. cerevisiae ENO2 gene were retrieved for each sample from BAM files in R. Total reads - The total number of reads was retrieved for each sample from the FASTQ files through the bash command cat file.fastq | seqtk seq -A - | grep \“^>\“ | wc -l”.
Counts normalization
For each sample, the following factor was computed to account for the amount of polyA RNA extracted from each fraction; a crucial step to move from relative to absolute gene expression levels.
| 1 |
Then, for each RNA species and sequenced sample, we estimate genes counts and we use them to split the yield of the corresponding fraction (e.g., given 100 fg/Millions of cells of chromatin RNA, if 7% of the reads were annotated as nascent premature the yield of Chpn would be 7 fg/Millions of cells - see the methods section “Fractionation and mRNA extraction”). Then, we applied DESeq2 independently to each RNA species to estimate the parameters of gene-specific negative binomial distributions: dispersion values, and replicate-specific mean values. After that, we randomly sampled each distribution and we used the resulting counts to split the yield of each RNA species across genes (same principle applied above for fractions). Iterating this sampling scheme 1000 times, we got a normalized expression level distribution for each RNA species at the single gene and single replicate resolution. The means of these distributions were used as input data for rates inference. Importantly, the profiling of RNA yield and the acquisition of transcriptional profiles through dRNA-seq were performed on independent samples for all the fractions except for polysomes.
Gene expression quantification was performed in R 4.2 using the Bioconductor package DESeq249 (v1.38.3).
Differential RNA species
The normalized counts distributions previously described allow to identify RNA species modulations between samples. We used this approach to get genes modulated in nuclear mature RNA in response to Leptomycin B. Specifically, we considered a gene up-regulated when comparing a Leptomycin B sample against an untreated one if the mean of the treated distribution was larger than the 97.5% quantile of the untreated one while the mean of the untreated distribution was lower than the 2.5% of treated one. Vice versa, we classified it as down-regulated if the mean of the untreated distribution was larger than the 97.5% quantile of the treated one while the mean of the treated distribution was lower than the 2.5% of the untreated one. We repeated this analysis for each pair of untreated and treated samples and we decided to classify a gene as up- or down- regulated if it was coherently modulated in at least 2 of the 4 possible combinations (configurations characterized by the same number of opposite regulations were discarded). Noticeably, the fraction of genes classified as up- or down-regulated comparing replicates from the same treatment was remarkably low, reassuring about the precision of our procedure (Supplementary Fig. 41).
Transcripts characterization
PolyA tails
We applied the polya routine of Nanopolish (v0.13.3)50 to profile transcripts polyA tail length; only esteems flagged as “PASS” in the polya_results.tsv file were used for our analyses.
UTRs structural features
3’UTRs genomic coordinates were extracted from the R object TxDb.Hsapiens.UCSC.hg38.knownGene (Bioconductor v3.17)47 through the R routine threeUTRsByTranscript. Standard chromosomes were kept (keepStandardChromosomes routine), and the longest 3’UTR for each gene was selected for further analyses. Their coordinates were saved in BED format and used to extract the corresponding genomic sequence with bedtools getfasta (v2.30.0)51. The same was done for 5’UTRs (fiveUTRsByTranscript). Custom R scripts were used to estimate UTRs GC content and Shannon Entropy (Entropy function of the package DescTools - v 0.99.49).
ODEs model solution
The complete model equations presented in Fig. 1 were numerically solved (see the dedicated method section) except for their steady state solution which was obtained analytically setting the time derivatives to zero. Since pre-existing RNA species are equal to their steady state values before labelling, these equations were used to estimate their initial conditions:
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
The very same approach was used for all the simplified frameworks; see Supplemental material for the corresponding equations.
Parameters identifiability
To assess model’s parameters identifiability, we used the Julia implementation of software for structural identifiability analysis of ODE models (SIAN)48,52,53 which takes as input the system of ODEs and the parameters of the model and returns a summary of globally, locally, and not identifiable parameters. Briefly, the software performs a Taylor expansion of the ODEs to obtain a polynomial representation of the systems. Polynomials are then truncated in such a way to obtain the minimal system containing all identifiability information. The problem is solved for each parameter, chosen in a random order, up to a correctness level p (by default 0.99). Last, the algorithm uses the results of the third step to distinguish between locally, globally, and not identifiable parameters.
Data simulation
General Framework
For the in-silico data simulation, we started from the median rates of synthesis, processing and degradation estimated by INSPEcT in27: 12 RPKM/h, 30 1/h and 1.3 1/h respectively. We linked these data to the rates of our extended model with the following numerical coefficients defined to maintain a biologically reasonable ranking between the processes: synthesis = synthesis, co-transcriptional processing = processing, detachment of mature chromatin RNA = processing, detachment of premature chromatin RNA = 0.1*processing, post-transcriptional processing = 0.25*processing, export = 1.5*processing, association to polysomes = 0.5*degradation, cytoplasmic degradation = degradation, polysomal degradation = degradation, and nucleoplasmic degradation = 0.5*degradation. Each initial rate was used as mean of a normal distribution with variation coefficient equal to 5 sampled to simulate different genes; this produced a set of simulated rates. For each set of rates, we solved the ODE system at given time-points defined in input (deSolve R package54) to retrieve the corresponding expression levels for all the RNA species involved in the model. To simulate the impact of noise, we employed each value as the mean of a normal distribution. The variation coefficient was set equal to the median CV of the normalized expression levels of the corresponding RNA species in the untreated condition. Noticeably, CVs for missing species (e.g., nascent RNA at 0 hours) were set to 0.5. The resulting values were used as RNA normalized counts for the RNA species of interest. We set expression levels lower than 1e-10 in module to this value, and we filtered out genes with negative expression levels because the corresponding set of rates is not biologically meaningful. Finally, we retrieved the first 1000 genes for further analyses. The same data generation routine was used to simulate all the different configurations and models presented in the article. In this regard, it was designed to select a specific ODE model according to the initial input rates.
Temporal Design
To define the optimal temporal design, we investigated nascent RNA accumulation simulating 1000 genes without noise. We noticed a very rapid saturation of nascent premature RNA associated to chromatin which, on average, reached its steady state value around 20 minutes as expected for an RNA species actively transcribed. We observed slower dynamics for other nuclear RNA species which bring the nascent RNA between 83% and 95% of the steady state value in 20 minutes. We also noticed that around 20 minutes most of the RNA species are far both from the corresponding steady state, and from the initial exponential regime where small fluctuations in labelling time and/or efficiency could result in large deviations of nascent gene expression levels. This is particularly important to avoid further increasing the variance for RNA species that are already noisy due to their poor abundance, such as nuclear premature RNAs. As previously mentioned in the main text, all these considerations suggest that a labelling time of 20 minutes could be a good trade-off for all the RNA species. Data simulation was always performed with R 4.2.
Rates inference
Nanodynamo
We designed a routine for kinetic rates inference suitable to process both real and simulated data. According to the expression levels in input and the initial rates, the function selects the proper model defining the correct ODE system. Then, for each gene, we performed the minimization of the following cost function (optim method from the R package stats) seeking for the set of rates which better recapitulates the expression levels:
| 8 |
Notice that each replicate contributes independently to the cost function, therefore, a gene can be analysed with a given model if all the RNA species required are defined at least once. This approach accommodates missing datapoints fully exploiting the available information. We performed each minimization with three different algorithms (“L-BFGS-B”,“BFGS”,“Nelder-Mead”), and three different initial values equal to the order of magnitude of ceiling, round, and floor of the initial rates values. The rationale behind these initial conditions was to exploit what we expected in terms of order of magnitudes of the rates without providing too much information about real rates mean values. We also tested a less informative initial condition with all the rates equal to 1 finding a minimum impact on the inferred rates. For each gene, the rates’ configuration minimizing the cost function was finally selected. The regularization strength lambda was set to 0.05. Rates below 1e-6 and above 1e4 were penalized with a fixed cost function value of 1e8; the upper bound was set to 1e6 for real data to accommodate high rates of synthesis. The parameters optimization was performed in the logarithmic space. Rates inference was performed with R 4.2.
INSPEcT
To analyse the Untreated SUM159 dataset with the first version of INSPEcT, we had to merge gene expression levels to mimic the standard input data of the tool. Specifically, for each replicate and each gene, we started excluding polysomal RNA species from the normalized counts matrix. Then, we simulated 4sU-seq data summing all nascent RNA species to generate the exonic signal, and all the premature nascent RNA species to estimate the intronic one. For genes lacking premature RNA, we set the intronic signal to 0. Similarly, to simulate Total RNA-seq data, we summed all the RNA species to generate the exonic signal, and all the premature RNA species to estimate the intronic one. After this preliminary step, we applied to the Total and Nascent RNA datasets the INSPEcT method quantifyExpressionsFromTrAbundance merging, in this way, the information from the two replicates. The resulting objects were finally used as input for the newINSPEcT method which computed the rates estimation. This analysis was performed in R 4.2 with INSPEcT 1.28.
Nascent RNA yield
Nascent transcripts after 15 minutes of metabolic labelling are expected to represent the 1.5% of the total RNA yield30. According to our quantifications of RNA cumulative yield in untreated SUM159 cells (~180 ng - see the methods section “Fractionation and mRNA extraction”), this corresponds to ~11 ng/(Millions of cells * hour). This value is well recapitulated by Nanodynamo which returned a median rate of synthesis per gene of ~15 pg/(Millions of cells * h). Indeed, assuming 103−104 expressed genes per cell, this sums up to 15-150*103 pg/(Millions of cells * h), which corresponds to 15-150 ng/(Millions of cells * h). Notably, this analysis was conducted using a limited set of highly expressed genes, which may account for the slight overestimation observed.
Couplings
Couplings conservation
To evaluate the conservation of the coupling mechanisms in response to Pladienolide B and Leptomycin B at single gene resolution, we started defining, independently for each gene and each treatment, a couplings vector. This object comprised an element for each treatment-specific coupling (Fig. 6A). The values were set as follows: 0 if at least one of the two rates defining the coupling was not modulated compared to the untreated counterpart (|log2 Fold Change | <0.5), 1.25 or 0.75 if both rates were up- or down-regulated (log2 Fold Change > 0.5 and < −0.5 respectively), or −1 if both rates were regulated but in opposite directions. The Hamming distance between the Pladienolide B and Leptomycin B coupling vectors of the same gene, restricted to the common elements, provided a quantitative estimation of coupling conservation. To identify distances significantly lower than expected by chance (i.e., significantly conserved genes), we computed a null distribution of Hamming distances by shuffling Pladienolide B and Leptomycin B coupling vectors (Supplementary Fig. 34). Specifically, we retrieved coupling vectors from the Pladienolide B treatment for half of the genes and coupling vectors from the Leptomycin B treatment for the remaining ones, generating a first set of vectors. Then, we created a second complementary set composed of vectors from Leptomycin B and Pladienolide B for the first and second half of the genes respectively. Finally, for 1000 iterations, we shuffled gene names independently within each set of vectors and computed gene Hamming distances. The threshold for identifying significantly conserved genes was set at the 5% level of the resulting distribution. Notably, the coupling vectors for a given treatment were clustered according to the Hamming distance and depicted as heatmaps in Fig. 6C and Supplementary Fig. 30.
Enrichment analysis
We performed a GSEA-based analysis to identify enrichments in RBPs and TFs targets among genes supporting a given coupling. Proteins binding sites were retrieved from the ENCODE web portal55,56 downloading ChIP-seq and eCLIP BED files respectively (GRCh38 - K562 and HepG2 not perturbed cell lines). RBPs binding sites were annotated according to the overlap with genes exonic regions (see the methods section “Premature reads profiling”); only genes with at least 25 binding sites for a given RPB were considered targets. TFs binding sites were annotated according to the overlap with promoters which were defined as regions 2000 bases upstream and 1000 bases downstream genes transcription start sites (TSSs) in a strand-aware manner. TSSs were retrieved from genes exonic regions (see the methods section “Premature reads profiling”) taking the lower coordinate for genes on the positive strand and the larger coordinate otherwise. The rankings for the GSEA analyses were defined according to the product of rates Log2 Fold Changes compared to the untreated condition times the sign of their Spearman correlation. In this way, the top genes for positive or negative couplings were characterised by strong coherent or opposite modulations respectively. Noticeably, this score accommodated also the peculiar positive coupling between synthesis and polysomal degradation where the first rate was globally down-regulated while the second one was globally up-regulated. Indeed, genes strongly supporting the coupling were characterised by a modulation in k1 and not in k9 or vice-versa, which resulted in scores small in modules that guaranteed a top ranking since the scores were mainly negative. All these analyses were performed in R 4.2 using the GSEA function of the Bioconductor package clusterProfiler (v4.6.2)57,58 with default options (p cut off = 0.05, p-value adjusted with Benjamini Hochberg method).
Statistics & Reproducibility
The optimum sample size (i.e., number of replicates and labelling time-points) was determined based on simulated data analyses. No data were excluded from the study. Data and kinetic rate reproducibility were confirmed through correlative analyses across biological replicates. Sample allocation and randomization, as well as blinding, were not relevant for this study. Statistical analyses were performed using R, nano-ID and Excel. Spearman correlation significance was estimated using the default options of the R 4.2 cor.test function from the stats package (algorithm AS 89 for n < 1290 or via the asymptotic t approximation otherwise). Details regarding other statistical tests and analyses are indicated in dedicated methods sections, as well as in the main text and figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank Stefano Campaner, Francesco Nicassio, Logan Mulroney, Stefano de Pretis, Gianluca Mastrantonio, Luca Rotta, Giorgia Garbeglio, Iris Tanaka, Björn Schwalb, Roberto Albanese, and members of the iRNA@IIT initiative for insightful discussions. We would like to acknowledge that the research activity herein was carried out using the IIT HPC infrastructure and through the support of the IEO Genomic Unit. V.F. is a PhD student within the European School of Molecular Medicine (SEMM). This work was supported by grants from the Italian Association for Cancer Research (AIRC) - project IG 2020 (ID. 24784) to M.P., the Giorgio Boglio fellowship from AIRC (ID. 26611) to M.F., and an AIRC fellowship (ID. 25399) to L.C.T, NextGenerationEU (PNRR M4C2-Investimento 1.4 -CN00000041-PNRR_CN3RNA_SPOKE2) to G.D. – and through the Translacore COST Action (CA21154).
Author contributions
L.C.T. developed the Nanodynamo experimental workflow, designed the experiments and performed the majority of the experiments; V.F. and A.d.P. contributed to conducting the experiments; V.F. and M.F. developed the Nanodynamo computational workflow and performed the majority of the analyses; S.M. contributed to the analyses; G.D. and S.B. developed the experimental workflow for the analysis of polysomal RNA; G.D. and L.C.T conducted the experiments for the profiling of polysomal RNA; M.F. and M.P. conceived and supervised the project; L.C.T., V.F., M.F., and M.P. wrote the paper.
Peer review
Peer review information
Nature Communications thanks Takayuki Nojima and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequencing data generated in this study have been deposited in the SRA database under the accession code PRJNA1023045. The re-analysed sequencing data of K562 cells are publicly available in the GEO database under the accession code GSM4663623. Abundance of RNA species and the corresponding kinetic rates for untreated cells and the drug treatments are available as SupplementaryData 1. Source data are provided with this paper.
Code availability
The Nanodynamo source code, as well as the scripts used for the analyses and figures included in this study, have been uploaded to GitHub [https://github.com/mfurla/Nanodynamo.git]59.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lucia Coscujuela Tarrero, Valeria Famà.
These authors jointly supervised this work: Mattia Furlan, Mattia Pelizzola.
Contributor Information
Mattia Furlan, Email: mattia.furlan@iit.it.
Mattia Pelizzola, Email: mattia.pelizzola@iit.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51917-2.
References
- 1.Cramer, P. Eukaryotic Transcription Turns 50. Cell179, 808–812 (2019). 10.1016/j.cell.2019.09.018 [DOI] [PubMed] [Google Scholar]
- 2.Komili, S. & Silver, P. A. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Genet9, 38–48 (2008). 10.1038/nrg2223 [DOI] [PubMed] [Google Scholar]
- 3.Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature416, 499–506 (2002). 10.1038/416499a [DOI] [PubMed] [Google Scholar]
- 4.Dahan, O., Gingold, H. & Pilpel, Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet.27, 316–322 (2011). 10.1016/j.tig.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 5.Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet15, 163–175 (2014). 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, K. A. & Young, E. T. Coupling mRNA Synthesis and Decay. Mol. Cell. Biol.34, 4078–4087 (2014). 10.1128/MCB.00535-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slobodin, B. et al. Transcription Dynamics Regulate Poly(A) Tails and Expression of the RNA Degradation Machinery to Balance mRNA Levels. Mol. Cell78, 434–444.e5 (2020). 10.1016/j.molcel.2020.03.022 [DOI] [PubMed] [Google Scholar]
- 8.Timmers, H. T. M. & Tora, L. Transcript Buffering: A Balancing Act between mRNA Synthesis and mRNA Degradation. Mol. Cell72, 10–17 (2018). 10.1016/j.molcel.2018.08.023 [DOI] [PubMed] [Google Scholar]
- 9.García-Moreno, J. F. & Romão, L. Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay. IJMS21, 9424 (2020). 10.3390/ijms21249424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, P., James, R. S., Mee, C. J. & Morozov, I. Y. mRNA levels are buffered upon knockdown of RNA decay and translation factors via adjustment of transcription rates in human HepG2 cells. RNA Biol.16, 1147–1155 (2019). 10.1080/15476286.2019.1621121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris, C., Cluet, D. & Ricci, E. P. Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny. WIREs RNA12, e1658 (2021). 10.1002/wrna.1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, L. Y., Mugler, C. F., Heinrich, S., Vallotton, P. & Weis, K. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. eLife7, e32536 (2018). 10.7554/eLife.32536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang, K. & Bartel, D. P. The molecular basis of coupling between poly(A)-tail length and translational efficiency. eLife10, e66493 (2021). 10.7554/eLife.66493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, C. et al. Structural basis of transcription-translation coupling. Science369, 1359–1365 (2020). 10.1126/science.abb5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercier, B. C. et al. Translation-dependent and -independent mRNA decay occur through mutually exclusive pathways defined by ribosome density during T cell activation. Genome Res. genome;gr.277863.123v2 10.1101/gr.277863.123 (2024). [DOI] [PMC free article] [PubMed]
- 16.Furlan, M. et al. Genome-wide dynamics of RNA synthesis, processing, and degradation without RNA metabolic labeling. Genome Res30, 1492–1507 (2020). 10.1101/gr.260984.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furlan, M., de Pretis, S. & Pelizzola, M. Dynamics of transcriptional and post-transcriptional regulation. Brief. Bioinforma.22, bbaa389 (2021). 10.1093/bib/bbaa389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiler, P., Van Den Berge, K., Street, K. & Tiberi, S. A Guide to Trajectory Inference and RNA Velocity. In Single Cell Transcriptomics (eds. Calogero, R. A. & Benes, V.) 2584 269–292 (Springer US, New York, NY, 2023). [DOI] [PubMed]
- 19.Chen, T. & Van Steensel, B. Comprehensive analysis of nucleocytoplasmic dynamics of mRNA in Drosophila cells. PLoS Genet13, e1006929 (2017). 10.1371/journal.pgen.1006929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingolia, N. T., Hussmann, J. A. & Weissman, J. S. Ribosome Profiling: Global Views of Translation. Cold Spring Harb. Perspect. Biol.11, a032698 (2019). 10.1101/cshperspect.a032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smalec, B. M. et al. Genome-Wide Quantification of RNA Flow across Subcellular Compartments Reveals Determinants of the Mammalian Transcript Life Cycle. http://biorxiv.org/lookup/doi/10.1101/2022.08.21.504696 (2022) 10.1101/2022.08.21.504696. [DOI] [PMC free article] [PubMed]
- 22.Ren, J. et al. Spatiotemporally resolved transcriptomics reveals the subcellular RNA kinetic landscape. Nat. Methods20, 695–705 (2023). 10.1038/s41592-023-01829-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Pretis, S. et al. INSPEcT: a computational tool to infer mRNA synthesis, processing and degradation dynamics from RNA- and 4sU-seq time course experiments. Bioinformatics31, 2829–2835 (2015). 10.1093/bioinformatics/btv288 [DOI] [PubMed] [Google Scholar]
- 24.de Pretis, S., Furlan, M. & Pelizzola, M. INSPEcT-GUI Reveals the Impact of the Kinetic Rates of RNA Synthesis, Processing, and Degradation, on Premature and Mature RNA Species. Front. Genet.11, 759 (2020). 10.3389/fgene.2020.00759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer, A. & Churchman, L. S. Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nat. Protoc.11, 813–833 (2016). 10.1038/nprot.2016.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier, K. C., Gressel, S., Cramer, P. & Schwalb, B. Native molecule sequencing by nano-ID reveals synthesis and stability of RNA isoforms. Genome Res30, 1332–1344 (2020). 10.1101/gr.257857.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Pretis, S. et al. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res27, 1658–1664 (2017). 10.1101/gr.226035.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uvarovskii, A., Naarmann-de Vries, I. S. & Dieterich, C. On the optimal design of metabolic RNA labeling experiments. PLoS Comput Biol.15, e1007252 (2019). 10.1371/journal.pcbi.1007252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louie, S. M. et al. GSTP1 Is a Driver of Triple-Negative Breast Cancer Cell Metabolism and Pathogenicity. Cell Chem. Biol.23, 567–578 (2016). 10.1016/j.chembiol.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biasini, A. & Marques, A. C. A Protocol for Transcriptome-Wide Inference of RNA Metabolic Rates in Mouse Embryonic Stem Cells. Front. Cell Dev. Biol.8, 97 (2020). 10.3389/fcell.2020.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girard, C. et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun.3, 994 (2012). 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- 32.Alles, J., Legnini, I., Pacelli, M. & Rajewsky, N. Rapid nuclear deadenylation of mammalian messenger RNA. iScience26, 105878 (2023). 10.1016/j.isci.2022.105878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drexler, H. L., Choquet, K. & Churchman, L. S. Splicing Kinetics and Coordination Revealed by Direct Nascent RNA Sequencing through Nanopores. Mol. Cell77, 985–998.e8 (2020). 10.1016/j.molcel.2019.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishigaki, Y., Li, X., Serin, G. & Maquat, L. E. Evidence for a pioneer round of mRNA translation. Cell106, 607–617 (2001). 10.1016/S0092-8674(01)00475-5 [DOI] [PubMed] [Google Scholar]
- 35.Cockman, E., Anderson, P. & Ivanov, P. TOP mRNPs: molecular mechanisms and principles of regulation. Biomolecules10, 969 (2020). 10.3390/biom10070969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart, M. Polyadenylation and nuclear export of mRNAs. J. Biol. Chem.294, 2977–2987 (2019). 10.1074/jbc.REV118.005594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdullah, A. et al. Nucleocytoplasmic translocation of UBXN2A is required for apoptosis during DNA damage stresses in colon cancer cells. J. Cancer6, 1066–1078 (2015). 10.7150/jca.12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel, K. L. et al. Analysis of subcellular transcriptomes by RNA proximity labeling with Halo-seq. Nucleic Acids Res.50, e24–e24 (2022). 10.1093/nar/gkab1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonelig, M. PABPN1 shuts down alternative poly(A) sites. Cell Res.22, 1419–1421 (2012). 10.1038/cr.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang, L. et al. The polyA tail facilitates splicing of last introns with weak 3′ splice sites via PABPN1. EMBO Rep.24, e57128 (2023). 10.15252/embr.202357128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valencia, P., Dias, A. P. & Reed, R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA105, 3386–3391 (2008). 10.1073/pnas.0800250105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatt, D. M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell150, 279–290 (2012). 10.1016/j.cell.2012.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics34, 2666–2669 (2018). 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics34, 3094–3100 (2018). 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics37, 4572–4574 (2021). 10.1093/bioinformatics/btab705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience10, giab008 (2021). 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bioconductor Core Team, B. P. M. O. [Cre. TxDb.Hsapiens.UCSC.hg38.knownGene. [object Object] 10.18129/B9.BIOC.TXDB.HSAPIENS.UCSC.HG38.KNOWNGENE (2017).
- 48.Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput Biol.9, e1003118 (2013). 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loman, N. J., Quick, J. & Simpson, J. T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods12, 733–735 (2015). 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- 51.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong, H., Ovchinnikov, A., Pogudin, G. & Yap, C. SIAN: software for structural identifiability analysis of ODE models. Bioinformatics35, 2873–2874 (2019). 10.1093/bioinformatics/bty1069 [DOI] [PubMed] [Google Scholar]
- 53.Linden, N. J., Kramer, B. & Rangamani, P. Bayesian parameter estimation for dynamical models in systems biology. PLoS Comput Biol.18, e1010651 (2022). 10.1371/journal.pcbi.1010651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soetaert, K., Petzoldt, T. & Setzer, R. W. Solving Differential Equations in R: Package deSolve. J. Stat. Soft. 33, 1–25 (2010).
- 55.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed]
- 56.Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res.48, D882–D889 (2020). 10.1093/nar/gkz1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol.16, 284–287 (2012). 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.mfurla. mfurla/Nanodynamo: Nanodynamo_PublicationRelease_v1.0. Zenodo10.5281/ZENODO.12784887 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing data generated in this study have been deposited in the SRA database under the accession code PRJNA1023045. The re-analysed sequencing data of K562 cells are publicly available in the GEO database under the accession code GSM4663623. Abundance of RNA species and the corresponding kinetic rates for untreated cells and the drug treatments are available as SupplementaryData 1. Source data are provided with this paper.
The Nanodynamo source code, as well as the scripts used for the analyses and figures included in this study, have been uploaded to GitHub [https://github.com/mfurla/Nanodynamo.git]59.