Abstract
As the first identified multidrug efflux pump in Mycobacterium tuberculosis (Mtb), EfpA is an essential protein and promising drug target. However, the functional and inhibitory mechanisms of EfpA are poorly understood. Here we report cryo-EM structures of EfpA in outward-open conformation, either bound to three endogenous lipids or the inhibitor BRD-8000.3. Three lipids inside EfpA span from the inner leaflet to the outer leaflet of the membrane. BRD-8000.3 occupies one lipid site at the level of inner membrane leaflet, competitively inhibiting lipid binding. EfpA resembles the related lysophospholipid transporter MFSD2A in both overall structure and lipid binding sites and may function as a lipid flippase. Combining AlphaFold-predicted EfpA structure, which is inward-open, we propose a complete conformational transition cycle for EfpA. Together, our results provide a structural and mechanistic foundation to comprehend EfpA function and develop EfpA-targeting anti-TB drugs.
Subject terms: Permeation and transport, Cryoelectron microscopy, Computational biophysics, Pathogens, Membrane proteins
Multidrug efflux pump EfpA is an essential protein for M. tuberculosis. The authors determine the structures of MtEfpA bound to lipids or the inhibitor BRD-8000.3, and propose it may function as a lipid flippase with a defined inhibition mechanism.
Introduction
Tuberculosis (TB) is a major global infectious disease mainly caused by Mycobacterium tuberculosis (Mtb). It is estimated that over 10 million new cases of TB occur worldwide and causing more than 1.6 million deaths annually1. This made TB the second leading cause of death from a single infectious agent, following COVID-19, in 2021. Over recent decades, the emergence and continuous increase in drug-resistant and multidrug-resistant TB cases have made the global TB epidemic more severe and urgent1,2. One of the major mechanisms that causes Mtb drug resistance is genetic mutations in drug target-related genes, which render drugs ineffective3. In cases of isoniazid (INH), one of the most efficient drugs against Mtb, mutations in katG, a catalase-peroxidase responsible for the conversion of isoniazid to its active form, and target enzyme InhA were found4,5. Another mechanism of drug resistance is the activation and overexpression of efflux pumps, which reduce the effective drug concentration inside the cell6. In some drug-resistant clinical strains, genetic mutations have not been found in known drug targets, but multiple efflux pumps, including EfpA and MmpL7, have been observed with significant overexpression7–10. This suggests that these efflux pumps may contribute to drug resistance.
Efflux pumps are widely present in bacteria and can be classified into 5 families: the ATP-binding cassette (ABC) superfamily, the major facilitator superfamily (MFS), multidrug and toxic compound extrusion (MATE) family, resistance-nodulation-division (RND) family, and small multidrug resistance (SMR) family11. EfpA, a member of the QacA transporter family in the MFS superfamily12, was found to have higher expression level in Mtb upon exposure to INH, as demonstrated by DNA microarray13. RT‒qPCR analysis showed that EfpA was significantly upregulated in various clinical Mtb strains, including strains resistant to INH, rifampicin (RIF), ofloxacin (OFX), or multidrug14–16. Overexpression of EfpA from Mtb or M. bovis BCG in M. smegmatis (Ms) led to a significant increase in resistance to INH, RIF and amikacin by ten to hundreds of fold, and the resistance was further strengthened with a longer induction time17. Furthermore, a mutation of E520V was identified in MtEfpA of an INH-resistant strain but not in any of the susceptible clinical isolates18. These suggest EfpA plays a crucial role in drug resistance by effluxing multiple drugs in Mtb. Moreover, high-density mutagenesis and deep-sequencing studies revealed that EfpA is essential for Mtb growth in vitro, indicating that it may also efflux critical endogenous substrates19,20. However, the identity of the endogenous substrates of EfpA remains elusive.
The essential role of EfpA in Mtb and its exclusive presence in Actinomycetes make EfpA an attractive and promising drug target. Recently, new classes of Mtb inhibitors, the BRD-8000 series (BRD-8000s) and BRD-9327, were identified through large-scale chemical-genetics screening and have been demonstrated to specifically target EfpA21,22. Among them, the optimized inhibitor BRD-8000.3 exhibited the highest inhibitory activity against Mtb (MIC90 = 800 nM). BRD-8000.3 is effective against non-replicating, phenotypically drug-tolerant Mtb and Mycobacterium marinum (Mmar), but not Ms21,23. BRD-8000s exhibited non-competitive inhibition of the efflux of a known EfpA substrate, ethidium bromide (EtBr)24. Furthermore, expressing MtEfpA (WT) or BRD-8000s-resistant mutant MtEfpA (V319F) in Mtb does not affect the sensitivity of Mtb to INH21, which is mediated by EfpA efflux10,16. This suggests that BRD-8000s kill Mtb by inhibiting the endogenous function of MtEfpA. Although oral administration of BRD-8000.3 showed good plasma exposure in mice and a low risk of drug‒drug interactions21, further optimization is needed to enhance its inhibitory activity and improve its physicochemical properties for its use as a drug against Mtb. However, the binding site and inhibition mechanism of BRD-8000.3 on MtEfpA are not yet fully understood.
In this study, we expressed and purified MtEfpA and MsEfpA and determined their cryo-EM structures in an outward-open conformation. In the MtEfpA structure, three lipids are sequentially bound in head-to-head and tail-to-tail arrangements within a channel that traverses from the inner leaflet through the central pocket to the outer leaflet of the phospholipid bilayer. The cryo-EM structure of MtEfpA complex with BRD-8000.3 shows that BRD-8000.3 occupies the lipid binding pocket near the inner leaflet of the lipid bilayer, rather than the central pocket, suggesting that BRD-8000s competitively inhibit lipid binding to EfpA but non-competitively inhibit drug efflux. Finally, by comparing our structures with the inward-open structure of EfpA predicted by AlphaFold25, we proposed a “staircase-flips” model for the transport of lipids by EfpA. Our results provide structural insight into the endogenous function of EfpA and the mechanism of its inhibitors to help future development of anti-Mtb drugs.
Results
EfpA is in an outward-open conformation
EfpA is widely conserved among mycobacterial species, with some species having multiple paralogs17. Multiple sequence alignment shows high homology (> 50%) of EfpA homologs across different species, including both pathogenic and nonpathogenic Mycobacterium (Supplementary Fig. 1a). Specifically, MtEfpA and MsEfpA show a remarkable 79% sequence identity. To express MtEfpA with a C-terminal GFP and strep tag, a codon-optimized gene was integrated into the Expi293F cell genome using sleeping beauty transposase26, and stable cell lines with high expression were screened by flow cytometry. The protein purified using affinity and size-exclusion chromatography (Supplementary Fig. 1b, c) was used for single-particle cryo-EM study.
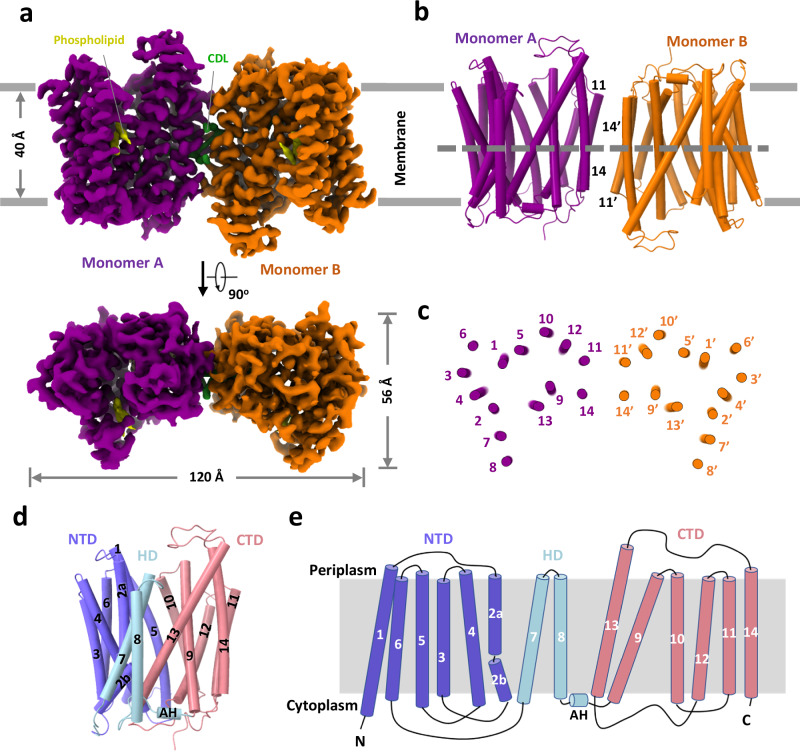
The cryo-EM map of MtEfpA at 3.1-Å resolution revealed a dimeric assembly with a total of 28 transmembrane segments (TMs) in the micelles, as well as two loops and two terminal tails extending out of the micelles (Fig. 1a and Supplementary Fig. 2c). The well-resolved TMs with clear α-helical and sidechain features allowed us to build a model of MtEfpA from residues 45 to 519 (Supplementary Fig. 2g). The EfpA dimer is in an antiparallel configuration, with TM11 and TM14 mediating the interactions between the two monomers (Fig. 1b, c). These interactions are mainly hydrophobic, except for a pi-bond formed by two Y378 residues from each monomer (Supplementary Fig. 1g). Mutation of Y378 to alanine disrupts dimer formation (Supplementary Fig. 3b, c). To determine the topology of MtEfpA in the membrane, we incubated MtEfpA-cGFP expressing cells, before and after permeabilization, with an allophycocyanin (APC) conjugated anti-GFP antibody, and then analyzed GFP and APC signals using flow cytometry. APC signals were observed in permeabilized cells, but not in nonpermeabilized cells (Supplementary Fig. 1d). This indicates that the C-terminus of MtEfpA in the membrane is inside the cytosol. Therefore, the antiparallel MtEfpA dimer was likely formed during solubilization and purification. Interestingly, a similar phenomenon has also been reported in other transporters27. MFS family lipid transporters, MFSD2A28,29 and LtaA30, function as monomers, so EfpA is likely to function as a monomer as well.
Fig. 1. Overall structure of MtEfpA.
a 3D reconstruction map of dimeric MtEfpA in sideview (top) and topview (bottom). Two MtEfpA monomers are colored purple and orange. Phospholipids are colored yellow, and cardiolipin (CDL) is in green. b Cartoon model of MtEfpA dimer shown as in (a). c Cross sectional view of MtEfpA at the level indicated as the dotted line in (b). d Structure of monomeric MtEfpA. The hinge domain (HD) TM7-8 (light blue) connects the N-terminal domain (NTD) TM1-6 (Blue) and C-terminal domain (CTD) TM9-14 (pink). The second transmembrane fragment breaks into two half helixes (2a and 2b). An amphiphilic helix (AH) between TM8 and TM9, is located on the cytoplasmic surface of the membrane. e Topology schematic of MtEfpA.
In each monomer, MtEfpA contains 14 transmembrane helices (TMs 1-14) (Fig. 1d), instead of 12 TMSs in canonical MFS transporters31, with both termini located on the cytoplasmic side (Fig. 1e). Different from canonical MFS fold, which comprises two domains with pseudo-two-fold symmetry connected by a long loop27,32–35, MtEfpA has a hinge domain (HD), which contains a long loop (229-239), two TMs (TM7-8), and an amphiphilic helix (AH), to connect the N-terminal domain (NTD) consisting of TMs 1-6 and the C-terminal domain (CTD) consisting of TMs 9-14 (Fig. 1d, e). Furthermore, the root-mean-square deviation (RMSD) value between NTD and CTD is 11.5 Å for 96 Cα atoms, indicating a large structural difference and lack of internal pseudo-two-fold symmetry in EfpA.
Unlike the formation of dimeric MtEfpA resulting from solubilizing membrane with lauryl maltose neopentyl glycol (LMNG) and cholesteryl hemisuccinate (CHS), dodecyl-β-D-maltoside (DDM) and CHS generated predominantly monomeric EfpA (Supplementary Figs. 1b and 3a). To investigate the conformation of monomeric EfpA, we determined the structure of MsEfpA in DDM-CHS at 3.7 Å resolution (Supplementary Fig. 4). The structure of monomeric MsEfpA is essentially identical to MtEfpA, with an RMSD value of 0.868 Å for 425 Cα atoms (Supplementary Fig. 4g). Thus, both structures of dimeric MtEfpA and monomeric MsEfpA are in the outward-open conformation (Supplementary Figs. 1f and 4g).
Three lipids within EfpA define an internal pathway
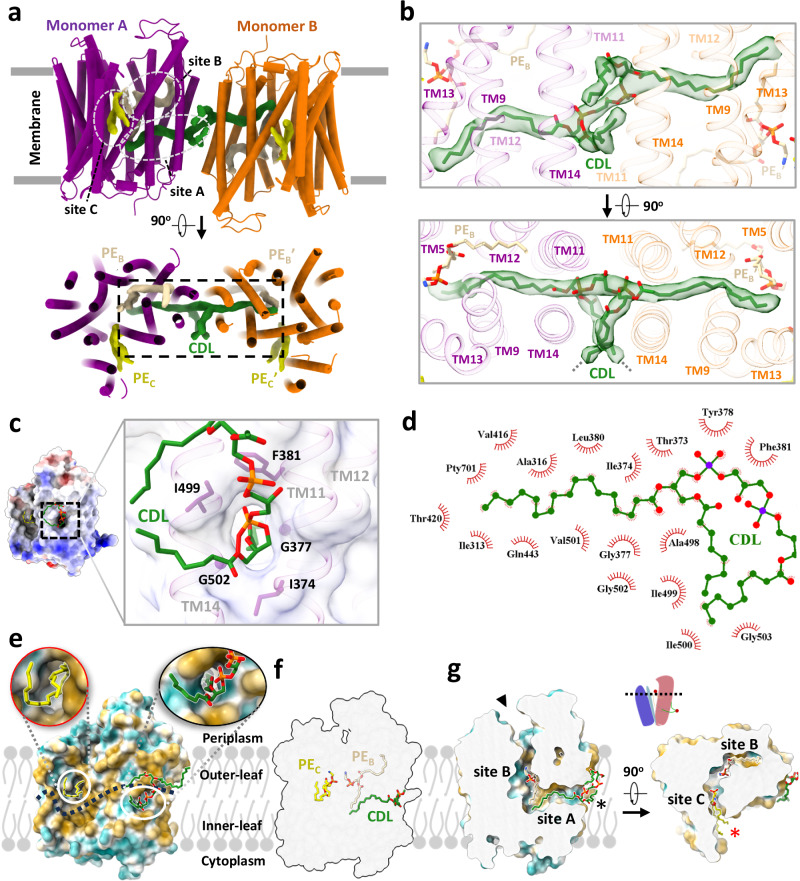
In the cryo-EM map of dimeric MtEfpA, strong nonproteineous densities were identified at three adjacent sites (sites A-C) within MtEfpA (Fig. 2a). These densities exhibit two or four elongated tails which do not correspond to the shapes of detergent molecules (Fig. 2a and Supplementary Fig. 5, b and c). In addition, the density at site A with four tails has two long tails inserted into the hydrophobic pockets of each MtEfpA monomer (Fig. 2b). These features suggest that the additional densities are lipids. Untargeted lipidomic analysis showed that the amount of cardiolipin in MtEfpA dimer is four times higher than that in monomeric MtEfpA purified with DDM-CHS (Supplementary Fig. 1e). Therefore, the four-tail density linking two monomers is likely cardiolipin which indeed fits well into this density (Fig. 2b). Since there is no lipid density in this pocket of monomeric MsEfpA (Supplementary Fig. 4f), cardiolipin likely binds to this pocket during solubilization and purification in detergents, forming an artificial dimer. Therefore, it is unlikely to modulate EfpA’s physiological functions. The other two lipids are likely glycerophospholipids, although the resolution is not sufficient to confidently identify the headgroups. Nevertheless, phosphatidylethanolamine (PE), the most abundant lipid identified by lipidomics in dimeric MtEfpA, fits well into each of these two densities and was used in atomic model building (Supplementary Fig. 5, b and c). Molecular dynamic simulation was utilized to analyze the binding mode and stability of these cardiolipin and PE molecules bound to MtEfpA, showing stable binding of cardiolipin in site A pocket for up to 500 ns (RMSD < 4.0 Å) and more motion for PE at site B (PEB, RMSD < 10.0 Å) and PE at site C (PEC, RMSD < 7.0 Å) (Supplementary Fig. 6a). Considering different glycerophospholipids in mycobacteria36 compared to HEK293 cells37, future investigation is needed to identify the exact lipids bound in the three pockets of EfpA in mycobacterial membranes.
Fig. 2. Three lipids binding in a channel in MtEfpA.
a The model of MtEfpA with densities of cardiolipin (CDL, green) and 2 molecules of phosphatidylethanolamine (PE, colored wheat at site B and yellow at site C). b Superimposition of the MtEfpA model and the cardiolipin density (5σ) at site A, shown in side view (upper) and top view (bottom). The gray dashed lines represent the missing ends of two tails extending inward toward the membrane. c The surface and residues at the lateral gate of cardiolipin insert into MtEfpA. d The residues of MtEfpA that interact with cardiolipin within 4 Å. e The two lateral gates of a channel in MtEfpA open separately towards the inner and outer leaflets of the membrane. The dashed line represents the boundary between the inner leaflet and outer leaflet of membrane. f Three lipids bind to MtEfpA in “staircase”-shaped tunnel from a side view. g Cross-sectional perspectives of the lipids-binding channel in MtEfpA. Left, view from side. Right, view from top. The two lateral gates are indicated by black and red stars, while the funnel opens toward the periplasm, marked with a black triangle. The cartoon on top represents the cross-sectional position from a lateral perspective.
Within our cryo-EM structure of MtEfpA, the three lipids are generally parallel, instead of perpendicular, to the membrane surface. The cardiolipin molecule has its hydrophilic head group at the dimer interface, two acyl chains inserted into the crevice formed by TM11-TM12 and TM9-TM14 of each monomer, and the other two acyl chains extending into the membrane (Fig. 2b, bottom). The residues I374, G377, F381, I499, and G502 at the dimeric interface of EfpA formed a lateral gate to accommodate the insertion of the hydrophobic cardiolipin acyl chain (Fig. 2c). Single mutations of G377D and G502D, which are located at dimeric interface as well as the lateral gate of cardiolipin acyl chains, weakened EfpA dimer formation (Supplementary Fig. 3, b and c). However, mutations of L195D and T324D, situated at lipid-binding site B, exhibited no discernible impact on dimer formation. The hydrophobic tails of cardiolipin that insert into EfpA interact with I313, A316, V501, A498, T373, I412, V416, T420, and Q443 from TM9, TM14, TM11, TM12, and TM13 (Fig. 2b, d). The lipid at site B (PEB), adjacent to cardiolipin, is surrounded by TM9, TM14, TM11, TM12, TM10, and TM5, with its hydrophobic acyl chains interacting with M320, L323, I327, L491, M384, P404, I405, F349, and F346 (Supplementary Fig. 5b). The lipid at site C (PEC) binds in the gap between the hinge domain and TM9-TM13 from CTD, with its acyl chains extending into the membrane (Supplementary Fig. 5c). Conservation analysis using ConSurf38 on EfpA from different species of Mycobacterium revealed that lipid-binding site A exhibits variability, while sites B and C are highly conserved (Supplementary Fig. 7). In addition, a hydrophilic cavity is located at the center of MtEfpA, formed by the head groups of PEB and PEC, as well as TM1, TM4, TM2a, TM13 and TM5. The central pocket exhibits a width of approximately 6–10 Å and possesses a negatively charged inner surface with residues Y97, S155, S69, T66, Q62, S191, Q443, and S444 (Supplementary Fig. 7b). The size and charged surface of this central pocket align well with the hydrophilic substrates of EfpA, EtBr and INH, both of which contain primary amino groups. Remarkably, among these residues, Y97, which is solvent-exposed and faces the periplasm, is conserved in all species (Supplementary Fig. 7b), indicating its essential role in the function of EfpA.
A solvent-accessible pathway within MtEfpA was identified using the MOLEonline program39, consisting of three openings oriented toward the periplasm, the inner and outer leaflets of the phospholipid bilayer (Fig. 2e). Intriguingly, this pathway accommodates the sequential binding of three lipids, forming a “staircase”-shaped pathway that starts from the inner leaflet, traverses the central pocket, and reaches the outer leaflet of the phospholipid bilayer (Fig. 2f, g). The lipids bind within the pathway in a head-to-head and tail-to-tail arrangement (Fig. 2g, Supplementary Fig. 5d, and Supplementary Movie 1).
BRD inhibitors occupy lipid binding sites in EfpA
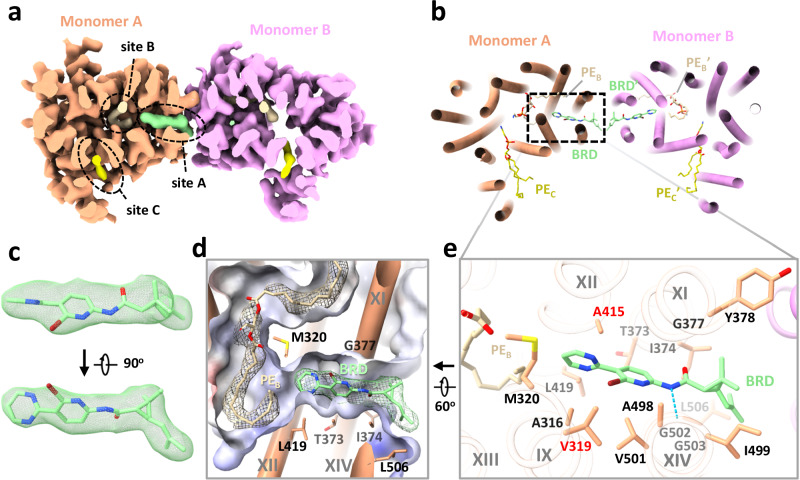
Two series of EfpA-targeting Mtb inhibitors, BRD-8000s and BRD-9327, were recently identified through high-throughput chemical-genetic screening21,22. These two series of compounds inhibit EfpA efflux in a non-competitive manner, and display potentiation when used in combination22. To reveal their inhibitory mechanism, we supplemented BRD-8000.3, which exhibited the highest inhibitory activity, during MtEfpA purification and determined the cryo-EM structure of MtEfpA-BRD-8000.3 complex at 3.3 Å resolution (Supplementary Fig. 8). In this cryo-EM map, BRD-8000.3 fits well into the density at lipid-binding site A, displacing the cardiolipin observed in our cryo-EM map of MtEfpA without compound (Fig. 3a, c). The density of BRD-8000.3 is distinct from that of cardiolipin (Supplementary Fig. 9b, c). Molecular dynamic simulations show that BRD-8000.3 can stably bind to the site A pocket for up to 500 ns with an RMSD within 2 Å (Supplementary Fig. 6b).
Fig. 3. Structure of MtEfpA bound to the inhibitor BRD-8000.3.
a Top view of BRD-8000.3 density (light green) at the lipid binding site A in one MtEfpA monomer (light orange). b Cartoon model of MtEfpA bound to BRD-8000.3 (BRD, light green) and two phosphatidylethanolamine (PE) molecules (wheat at site B and yellow at site C). c Superimposition of the BRD-8000.3 model and density (5σ). d Electrostatic surface of the BRD-8000.3 binding pocket. e Residues of MtEfpA (light orange) interacting with BRD-8000.3 (light green) within 4 Å. The hydrogen bond between A489’s O3 of MtEfpA and N9 of BRD-8000.3 is shown in a blue dashed line. Resistant mutation residues are labeled in red.
The binding of BRD-8000.3 to MtEfpA caused only minor structural changes, as compared to the structure of MtEfpA without compound (RMSD = 0.462 Å for 475 Cα atoms) (Supplementary Fig. 9a). Similar to the hydrophobic tail of cardiolipin inserted in MtEfpA, BRD-8000.3 binds in the gap between TM11-TM12 and TM9-TM14, interacting with M320, A316 and V319 on TM9, V501, A498, I499, G502, G503 and L506 on TM14, T373, I374, G377, Y378 on TM11, and A415 on TM12 (Fig. 3b, e). A previous study identified several BRD-8000.3 resistant mutants, including V319F in MtEfpA and V319M, A415V in MmEfpA, which has 90% identity to MtEfpA and is same at these residues22, corroborating our structural finding of BRD-8000.3 bound in site A (Fig. 3e and Supplementary Fig. 6c). The large side chains resulting from these mutations likely cause steric hindrance with BRD-8000.3 and prevent its binding to MtEfpA. In addition to hydrophobic interactions, a hydrogen bond between N9 of BRD-8000.3 and O3 of MtEfpA A498 may facilitate the binding of BRD-8000.3 and displacement of lipid from the site A pocket (Fig. 3e).
In contrast, the MmEfpA mutants resistant to BRD-9327 were identified with mutations G328C, G328D, F346L, and A339T22, which are located at lipid-binding site B (Supplementary Fig. 6d, left). AutoDock Vina40 shows that BRD-9327 can fit into the site B pocket, thus preventing lipid binding (Supplementary Fig. 6d, right). BRD-9327 and BRD-8000s binding to distinct sites in EfpA is consistent with the reported synergistic inhibition of M. marinum when combining BRD-8000.3 and BRD-932722. Thus, our structural analyses indicate that BRD-8000s and BRD-9327 compete with lipids for binding to two distinct sites in EfpA, thus inhibiting the endogenous functions of EfpA and ultimately leading to the death of Mycobacterium.
EfpA resembles related lipid transporters
The presence of a lipid-occupying pathway within EfpA and the displacement of these lipids by small-molecule Mtb inhibitors imply a connection between these lipids and EfpA’s essential functions for Mtb. To gain further insights into EfpA function, we conducted structure-based functional predictions using COFACTOR41 against BioLip protein function database and Dali server42 against the Protein Data Bank (PDB). Both searches consistently identified MFSD2A, a lysophospholipid transporter in the MFS superfamily, as the top hit. MFSD2A transports long-chain unsaturated fatty acids with a zwitterionic lysophosphatidylcholine headgroup, such as docosahexaenoic acid (DHA) and α-linolenic acid (ALA), across the blood brain barrier43,44. MFSD2A is the best studied MFS lipid transporter with well-resolved substrate lipid densities and high-resolution structures in both outward- and inward-open conformations28,29,43–45.
Upon superimposing our structure of MtEfpA and the outward-open structure of MFSD2A from Mus musculus (MmusMFSD2A)29, we found that, despite low sequence homology (11%), they exhibit high-level structural similarity, except for the additional hinge domain of MtEfpA (Supplementary Fig. 10a). Furthermore, the density of the substrate, lysophospholipid, in the outward-open conformation of MmusMFSD2A corresponds to the lipid bound to site C in MtEfpA, albeit with a more vertical orientation (Supplementary Fig. 10c). Additionally, we compared the AlphaFold-predicted structure of MtEfpA, which adopts an inward-open conformation, with the recently reported inward-open structure of MFSD2A from Gallus gallus (GgMFSD2A)28. Similar to their outward-open counterparts, the inward-open conformations of MtEfpA and GgMFSD2A also exhibit analogous folding patterns (Supplementary Fig. 10b). Moreover, in the inward-open conformation of GgMFSD2A, the substrate lysophosphatidylcholine (LPC) is situated in a pocket corresponding to site B in MtEfpA (Supplementary Fig. 10d). The remarkable structural similarity in both inward and outward conformations between MFSD2A and MtEfpA, together with their highly analogous lipid-binding sites, strongly supports the notion that MtEfpA shares a similar function with MFSD2A as a lipid flippase, facilitating the translocation of substrate lipids between the inner and outer leaflets of the Mtb membrane. However, identifying the exact endogenous substrate and comprehending the lipid transport cycle of EfpA require further studies.
Conformational transition of EfpA during substrate efflux
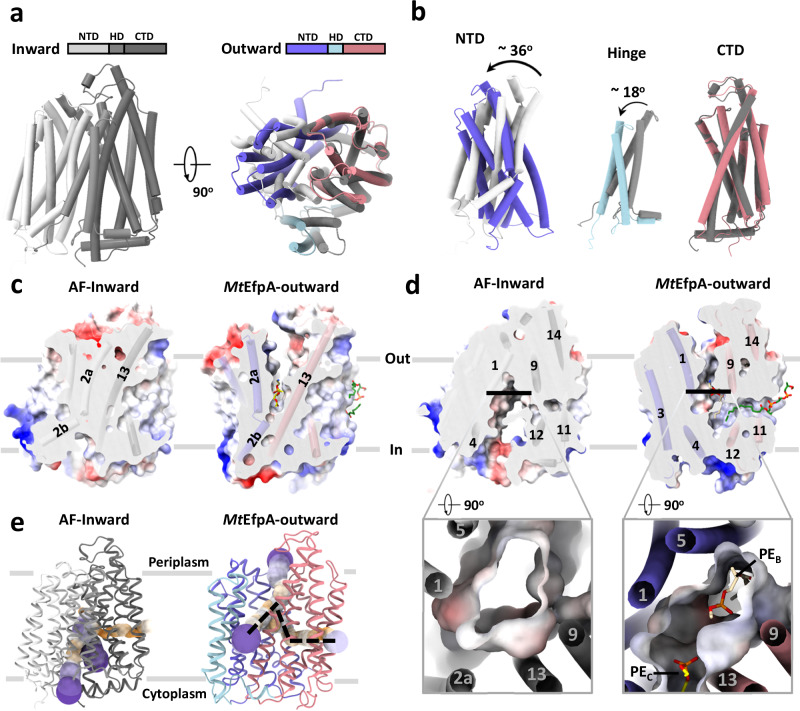
The structures of EfpA from different Mycobacterium species were predicted using AlphaFold25, and all of them, including MtEfpA, exhibited an inward-open conformation (Fig. 4a and Supplementary Fig. 11a, b). Compared to the outward-open conformation of our MtEfpA structures, the CTD in the predicted inward-open structure exhibits minimal variation (Fig. 4b, right), while the NTD and HD of the inward-open conformation rotate by approximately 36° and 18°, respectively (Fig. 4b, left and middle). In the inward-open conformation, the rotation of HD and the movement of TM2a toward TM13 result in the occlusion of lipid-binding site C (Fig. 4c). Additionally, the opening of the central hydrophilic pocket shifts from the cytoplasm to the periplasm (Fig. 4d). Despite the rotations and movements, the structures of individual domains of EfpA remain largely unchanged between the outward- and inward-open conformations (Supplementary Fig. 11c). This indicates that EfpA may employ the canonical rocker-switch mechanism46 for the transport of hydrophilic drugs bound at the central pocket (Supplementary Movie 2).
Fig. 4. Comparison of inward and outward facing states of MtEfpA.
a Superimposition of inward state of MtEfpA (lightgray-gray-darkgray), predicted by AlphaFold, with outward state of MtEfpA (blue-lightblue-lightpink). b Movement of MtEfpA’s NTD and hinge domain from an inward state to an outward state. c The PE binding cavity at site C in outward state (right) is blocked by TM2a moving near TM13 in inward state (left). d The binding cavity at site B changes from inward open (left) to outward open (right). e The tunnel in inward and outward states of MtEfpA.
The solvent-accessible pathway in the inward-open conformation of EfpA differs from that in the outward-open conformation (Fig. 4e). The pathway in the inward-open conformation constricts at lipid-binding site A, and the lateral gate opens to the middle of membrane, while the other two funnels of the pathway open toward the cytoplasm and the inner leaflet of membrane (Fig. 4e left). These observations suggest a mechanism for EfpA to efflux the drug molecules from the cytoplasm: the compounds bind to the central hydrophilic pocket of EfpA in its inward-open conformation and are subsequently released to the periplasm when EfpA transitions to the outward-open conformation. Such conformational changes may also cause the association and dissociation of lipid molecules, as well as the movement of lipids between different binding sites within EfpA.
Discussion
EfpA was the first identified efflux pump in Mtb12 and has been implicated in multidrug resistance, particularly against INH10,14–17. The essential role of EfpA in the in vitro growth of Mtb19,20 indicates that, in addition to export drugs, EfpA may have other crucial, unknown endogenous functions. In this study, the presence of a lipid-binding tunnel in MtEfpA that spans from the inner leaflet to the outer leaflet of the membrane, along with inhibitors targeting the lipid-binding sites and the significant structural and lipid-binding site similarity to the lysophospholipid transporter MFSD2A, strongly suggest that EfpA functions as a lipid transporter.
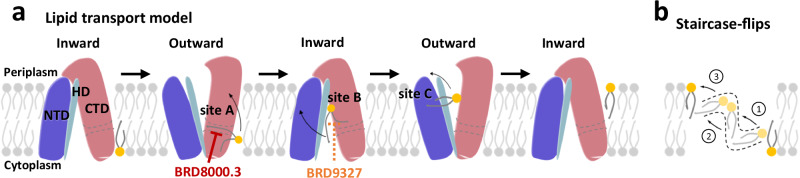
Two general models have been proposed to describe lipid translocation by lipid transporters: the trap-and-flip model30,47 and the credit-card model48–50. While the only characterized structures of MFS lipid transporters, MFSD2A28,29 and LtaA51, suggest the possibility of MFS lipid transporters employing the rocker-switch30,45 or trap-and-flip30 mechanism, the rotational dynamics of lipids within the transmembrane region remain unclear. Based on the lipid-binding pathway observed in MtEfpA, we propose a “staircase-flips” model to elucidate how EfpA translocate lipids from the inner membrane leaflet to the outer leaflet (Fig. 5a, b). In this model, the transition of EfpA from an inward-open to an outward-open conformation, expanding lipid-binding site A and facilitating the binding of substrate lipids from the inner-leaf of the membrane. Subsequently, in multiple conformational changes of EfpA, the substrate lipid gradually flips from lipid-binding site A to site B and site C before ultimately being released to the outer-leaf of the membrane (Fig. 5a). Notably, the inhibitor BRD-8000.3 prevents lipid entry into the pathway, while BRD-9327 likely disrupts lipid flipping from site A to site B.
Fig. 5. Proposed model for lipid transport by MtEfpA and small-molecule inhibition.
a Lipid transport model for MtEfpA and distinct target sites of different inhibitors. NTD, N-terminal domain. HD, hinge domain. CTD, C-terminal domain. The substrate lipid flips to adjacent binding site or releases to the outer-leaf of membrane during each conformational transition. BRD-8000.3 targets lipid-binding site A and BRD-9327 was supposed to target lipid-binding site B to inhibit MtEfpA function. b Lipids undergo a stepwise flipping, resembling a staircase, from the inner leaflet to the outer leaflet of the membrane.
Despite these findings, the exact substrate lipid of MtEfpA remains elusive and requires further investigation. Additionally, the lack of lipid-bound EfpA in the inward-open conformation hinders our understanding of the exact transport cycles required to transport a lipid from the inner leaflet to the outer leaflet. Moreover, although BRD-8000s and BRD-9327 have been reported to non-competitively inhibit the efflux of EtBr, a hydrophilic substrate of EfpA21,22, the relationship between the functions of EfpA (drug efflux and lipid transport) remains unclear. In conclusion, our work sheds light on the endogenous functions of EfpA, elucidates inhibitor mechanisms, and proposes a comprehensive lipid transport model. These findings provide a foundation for the structure-based design and optimization of novel EfpA-targeting drugs against Mtb, and advance our understanding of the mechanisms of MFS lipid transporters.
Methods
Protein overexpression and purification
The DNA coding sequences of full-length MtEfpA were optimized for expression in Expi293F™ Cells (Cat. No.: A14527, ThermoFisher Scientific) and cloned into pSBbi vector with GFP and Strep-Tag II at C-terminal (pSBbi-MtEfpA-CGS)26. The primers sequences are provided in Supplementary Data 1. pSBbi-MtEfpA-CGS was co-transfected with pCMV(CAT)T7-SB100, which contains gene of sleeping beauty transposase, to Expi293F cell line. After a week of screening with 2 mg/L puromycin, a cell line with high expression of MtEfpA was obtained. The cells with MtEfpA were grown at 37 °C with 8% CO2 to 4 × 106 cells per milliliter and harvested after centrifugation at 1000 g for 10 min. Cells pellet were resuspended with buffer A (20 mM Hepes pH 7.0, 300 mM NaCl), then lysed by passing through a microfluidizer (ATS Scientific Inc) at 200 bars after adding protease inhibitor cocktail. Cell debris was removed by centrifugation at 10,000 g for 10 min followed by collecting membrane from the supernatant at 150,000 g for 1 h at 4 °C. Membrane was resuspended with buffer B (20 mM Hepes pH 7.0, 300 mM NaCl, 10% (w/v) glycerol), and supplemented with final concentration is 1% LMNG-CHS (5:1). After solubilized at 4 °C for 2 hours, the solution was centrifuged at 40,000 g for 30 min, and the supernatant was loaded to Strep-Tactin®XT resin. The resin bound with protein was rinsed with buffer B containing 0.04% LMNG-CHS, and the target protein was eluted from the resin with buffer B containing 50 mM biotin. The crude protein was further purified by gel filtration with column superose 6. After gel filtration, the collections were used for preparing negative stains or cryo-EM grids. MtEfpA mutants are purified according to the same protocol as the wide type. BRD-8000.3 was added to the buffer for membrane solubilization and subsequent steps to acquire the MtEfpA-BRD-8000.3 complex. The monomeric MtEfpA and MsEfpA were purified with 1% DDM-CHS (5:1), instead of LMNG-CHS, and exchanged to amphipol PMAL-C8 before gel filtration for freezing cryo-EM grids as previously described52.
Flow cytometry assay
Cells with MtEfpA constitutive expression, with empty Expi293F as control, were collected, resuspended in PBS and aliquoted into three. One aliquot was fixed with 1% Paraformaldehyde (BioLegend). Another was incubated with APC anti-GFP antibody (# 338010, BioLegend, Cat. No. 338010, Clone FM264G) followed by fixation. The third one was incubated with APC anti-GFP antibody, fixed, and permeabilized followed by incubating with APC anti-GFP antibody. All samples were made at least three parallel. Cell samples were assayed for FITC and APC with a CytoFLEX flow cytometer (Beckman). Data were analyzed with FlowJo Software (BD Biosciences).
Negative stain analysis
All negative-stain EM grids were made by applying 4 μL protein samples at 10 μg ml-1 to glow-discharged homemade grids. Negative-stain EM images were collected on a Tecnai T12 microscope (ThermoFisher) with pixel size of 1.68 Å. Automated particle picking from 3x binned images and 2D classification were performed with SAMUEL and SamViewer53.
Cryo-EM specimen preparation and data collection
All cryo-EM grids were prepared with Vitrobot Mark IV (Thermo Fisher Scientific) by application of 3 μL protein at 10 mg ml−1 to glow-discharged holey gold grid (Quantifoil, R1.2/1.3 μm, 300 mesh) and blotted 4 s with force −1 at 100% humidity and 4 °C before plunged into liquid ethane. Micrographs of MtEfpAs were recorded on Talos Arctica or Titan Krios (ThermoFisher) operated at 200 kVe or 300 kVe with K3 direct electron detector (Gatan). SerialEM was used for automatic data collection with one shot per hole and nine holes per movement. The MtEfpA dataset was collected with a pixel size of 0.87 Å, a total dose of 54 e Å–2, and 50 frames in total. The MtEfpA-BRD8000.3 dataset was acquired in a super-resolution pixel size of 0.394 Å, 50 frames and total dose 49 e Å–2 with specimen stage tilt 20 degree. For monomeric MsEfpA, two datasets were collected on Talos Arctica and Titan Krios with pixel size of 1.1 Å and 0.6632 Å, respectively. The defocus of all micrographs was set to range from −1.2 μm to −2.5 μm.
Cryo-EM data processing
3465 movies of MtEfpA were collected and were drift-corrected by MotionCorr254. CTF information were estimated using CTFFind455. 2,424,800 particles were picked and extracted from 3031 selected micrographs with binning 4. Two rounds of 2D classification and deep2D classification were performed using SAMUEL53. After selection of good classes from deep2D, 450,607 particles are used for 3D classification and 3D refinement with C2 symmetry in Relion56. In the process of 3D classification and 3D refinement, as the resolution improved, we gradually replaced the binned particles with less binned or original dataset. Finally, a 3.1 Å resolution map of MtEfpA in dimer was reconstructed by 49,783 particles with C2 symmetry after local refinement and postprocess.
For MtEfpA complexed with BRD8000.3, the dataset was processed using cryoSPARC57. Briefly, after motion correction and CTF estimation, 2,014,853 particles were auto picked and extracted from 6,550 micrographs with binned 4-fold. Junk particles are removed by 2D classification, and 1,072,224 particles are re-extracted from micrographs with bin 2. After two rounds of ab-initio with setting maximum resolution were 6 Å and 4 Å, respectively, re-extraction selected particles without bin and supplement of sideview and top-view particles. Then the third round of ab-initio was performed with setting maximum resolution is 4 Å, following with non-uniform refinement58 and local refinement with applying C2 symmetry, a 3.3 Å map of mtEfpA-BRD8000.3 in outward state reconstructed by 52,099 particles are acquired finally.
For the two datasets of monomeric MsEfpA, 6,546,828 particles and 3,142,841 particles were picked from two datasets with 3280 and 3554 micrographs, respectively, after motion correction and CTF estimation by cryoSPARC. After 2D classifications, 2,205,382 particles are selected and re-extracted with both pixel sizes are 1.1 Å before being combined. Then one round of ab-initio and two rounds of joint non-uniform refinement and non-alignment classification were performed with the combined particles. A 3.7 Å map of MsEfpA was achieved in reconstructed by 52,150 particles after local refinement and post-process.
Gold standard Fourier Shell Correlation (FSC) = 0.14359 was applied to estimate the resolution of the final maps. Local resolutions of maps were calculated by RasMap60.
Structural model building and determination
To build MtEfpA models, the AlphaFold-predicted model of MtEfpA was cleaved into domains and docked into the 3.1 Å map of MtEfpA in Chimera61. Then the model was adjusted with COOT62 and refined against the cryo-EM maps using real-space refinement in Phenix63. The structures and restraints of lipids and BRD8000.3 were generated by eBLOW64, then fitted into the map of MtEfpA or MtEfpA-BRD, respectively, following with real-space refinement and validated by MolProbity65. The MsEfpA model was determined by molecular replacement with Phaser using MtEfpA model as searching template before real-space refinement. All figures and movies were generated with Chimera or ChimeraX66.
Shotgun lipidomic and small-molecule analysis
Lipids and small molecule analytes were extracted from ~20 μL samples of protein at 0.1 mg/mL and containing 5 μL of SPLASH LIPIDOMIX mass spectrometry standard (Avanti Polar Lipids) with the addition of methyl tert-butyl ether (Sigma Aldrich) and methanol (Sigma Aldrich) under 7:2:1.5 (v/v/v) mixing ratio of methyl tert-butyl ether, methanol and purified proteins, respectively67. Samples were mixed and incubated on shaker for 60 min at 4 °C with 1000 rpm agitating speed. After extraction, samples were centrifuged at 10,000 g for 5 min at 4 °C. Lipid-containing organic phase of each sample were transferred to new tubes and dried down under vacuum concentrator. Extracted lipids were resolubilized using a 2:1 (v/v) mixing ratio of chloroform and methanol prior to mass spectrometry (MS) analysis. MS analysis of extracted lipids were performed using an Orbitrap ID-X Tribrid mass spectrometer (Thermo Scientific), equipped with an automated Triversa Nanomate nanospray interface (Advion Bioscience) for lipid solution delivery. Direct infusion spray was achieved using 0.95 psi of gas pressure and 1.4 kV of delivery voltage for 6 min in each polarity. All full scan mass spectra (m/z 400–1100) were acquired in positive and negative modes, respectively, using mass resolution of 500,000 (FWHM at m/z 200). Data independent acquisition of MS/MS scans were performed at mass resolution of 30,000 (FWHM at m/z 200) Analysis of mass spectrometry data was performed using LipidXplorer 1.2.8 (CBG-MPI) software68,69. Duplicate lipids were excluded. One biological repeat was performed for this experiment. All the raw data, mzmL files, and mfql files of the shotgun lipidomic and small-molecule analysis are included in the Supplementary Data 2.
All-atom molecular dynamics (MD) simulations and docking
The resolved monomeric and dimeric structures of MtEfpA and MtEfpA-BRD were utilized as initial structures for MD simulations. The enveloping POPC lipid bilayer around MtEfpAs was constructed using packmol-memgen program70 in AmberTools2271. The system was solvated in TIP3P water model with addition of 150 mM Na+ and Cl- ions. The protein and POPC were modeled by AMBER ff14SB force field72 and Lipid21 force field73, respectively. The ligand molecules, including CDL, PE, and BRD, have their hydrogen atoms added by PyMOL. Then they were assigned GAFF2 parameters and AM1-BCC charge using the antechamber and parmchk2 modules in AmberTools 22 package. After an initial energy minimization, the systems were initially heated to 100 K with fixed volume and further heated to 300 K, followed by pre-equilibration under isothermal-isobaric (NPT) conditions with decreasing positional restraints. Subsequently, a 100-ns NPT simulation without any restraints was performed, followed by a 500-ns production run. The temperature was maintained at 300 K using Langevin dynamics with a collision frequency of 2.0 ps−1, and the pressure was regulated using Monte Carlo barostat. The van der Waals and electrostatic interactions in real space were truncated at 10 Å, and the long-range electrostatic interaction was calculated using the particle mesh Ewald method. SHAKE was employed, and a time step of 2 fs was used in the above simulations, except for the final production simulation, in which hydrogen mass repartitioning74 was employed to enable a larger time step of 4 fs. All MD simulations were performed on GPUs using the pmemd.cuda module in the AMBER22. Four independent replicates of the simulations were performed using the same coordinates but different velocities. The topology files, initial structure files, simulation parameter control files, and result data files of molecular dynamics trajectories are provided in Supplementary Data 3.
BRD9327 was docked into MtEfpA at the lipid binding site B using Smina75. The docking was carried out with the option exhaustiveness set to 96, and the top five poses returned by Smina were saved.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Z. Li, S. Sterling and R. Walsh at Harvard cryo-EM center for training and assistance in data collection of MsEfpA monomer, and M. Farias from Pacific Northwest Center for Cryo-EM for collecting the dataset of MtEfpA-BRD. We are grateful to A. J Baublis for assistance in mass spectrometry experiments. We thank all the members in Liao laboratory and S. Shao for their help and support on this project. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. This work was supported in part by National Natural Science Foundation of China (32330054 to M.L.). M. Liao is an investigator of SUSTech Institute for Biological Electron Microscopy.
Author contributions
M.L. and S.W. conceived the project. S.W. conducted molecular cloning, protein purification, mutagenesis assay, grid preparation, EM data collection and processing, and model building. S.W. and D.L. cultured the cell lines. Y.S. and S.W. carried out the flow cytometry assay. K.S. and C.X. helped on cryo-EM data collection. K.W. and Z.L. performed the mass spectrometry analysis. P.L. and Y.M. conducted the MD simulation and docking. S.W., M.L., and K.W. contributed to drafting the manuscript. All authors participated in data analysis and manuscript preparation.
Peer review
Peer review information
Nature Communications thanks Russell Bishop, Johanna Striesow and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The cryo-EM density maps have been deposited into the Electron Microscopy Data Bank under accession numbers EMD-37641 (MtEfpA with PE and cardiolipin), EMD-42204 (MtEfpA with PE and BRD8000.3) and EMD-42205 (MsEfpA with PE). The atomic coordinates are deposited in the Protein Data Bank under accession codes PDB 8WM5 (MtEfpA with PE and cardiolipin), 8UFD (MtEfpA with PE and BRD8000.3) and 8UFE (MsEfpA with PE). Source data are provided with this paper.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shuhui Wang, Email: shuhui.wang@yale.edu.
Maofu Liao, Email: liaomf@sustech.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51948-9.
References
- 1.Organization, W. H. Global tubersulosis report 2022. (2022).
- 2.Somoskovi, A., Parsons, L. M. & Salfinger, M. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res.2, 164–168 (2001). 10.1186/rr54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossini, N. O. & Dias, M. V. B. Mutations and insights into the molecular mechanisms of resistance of Mycobacterium tuberculosis to first-line. Genet Mol. Biol.46, e20220261 (2023). 10.1590/1678-4685-gmb-2022-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, Y., Heym, B., Allen, B., Young, D. & Cole, S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature358, 591–593 (1992). 10.1038/358591a0 [DOI] [PubMed] [Google Scholar]
- 5.Banerjee, A. et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science263, 227–230 (1994). 10.1126/science.8284673 [DOI] [PubMed] [Google Scholar]
- 6.De Rossi, E., Ainsa, J. A. & Riccardi, G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev.30, 36–52 (2006). 10.1111/j.1574-6976.2005.00002.x [DOI] [PubMed] [Google Scholar]
- 7.Machado, D. et al. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One7, e34538 (2012). 10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, H. et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet.45, 1255–1260 (2013). 10.1038/ng.2735 [DOI] [PubMed] [Google Scholar]
- 9.Calgin, M. K. et al. Expression analysis of efflux pump genes among drug-susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates and reference strains. Diagn. Microbiol. Infect. Dis.76, 291–297 (2013). 10.1016/j.diagmicrobio.2013.02.033 [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues, L., Machado, D., Couto, I., Amaral, L. & Viveiros, M. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect. Genet. Evol.12, 695–700 (2012). 10.1016/j.meegid.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 11.Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs64, 159–204 (2004). 10.2165/00003495-200464020-00004 [DOI] [PubMed] [Google Scholar]
- 12.Doran, J. L. et al. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab Immunol.4, 23–32 (1997). 10.1128/cdli.4.1.23-32.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson, M. et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl Acad. Sci. USA96, 12833–12838 (1999). 10.1073/pnas.96.22.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kardan-Yamchi, J. et al. Expression analysis of 10 efflux pump genes in multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis clinical isolates. J. Glob. Antimicrob. Resist.17, 201–208 (2019). 10.1016/j.jgar.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 15.Machado, D. et al. Interplay between Mutations and Efflux in Drug Resistant Clinical Isolates of Mycobacterium tuberculosis. Front. Microbiol.8, 711 (2017). 10.3389/fmicb.2017.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, G. et al. Efflux pump gene expression in multidrug-resistant Mycobacterium tuberculosis clinical isolates. PLoS One10, e0119013 (2015). 10.1371/journal.pone.0119013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai, D. & Mehra, S. The Mycobacterial Efflux Pump EfpA Can Induce High Drug Tolerance to Many Antituberculosis Drugs, Including Moxifloxacin, in Mycobacterium smegmatis. Antimicrob. Agents Chemother.65, e0026221 (2021). 10.1128/AAC.00262-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, M. et al. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol.43, 5477–5482 (2005). 10.1128/JCM.43.11.5477-5482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol.48, 77–84 (2003). 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 20.Griffin, J. E. et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog.7, e1002251 (2011). 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, E. O. et al. Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature571, 72–78 (2019). 10.1038/s41586-019-1315-z [DOI] [PubMed] [Google Scholar]
- 22.Johnson, E. O., Office, E., Kawate, T., Orzechowski, M. & Hung, D. T. Large-Scale Chemical-Genetic Strategy Enables the Design of Antimicrobial Combination Chemotherapy in Mycobacteria. ACS Infect. Dis.6, 56–63 (2020). 10.1021/acsinfecdis.9b00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, S. S. et al. Identification of novel inhibitors of nonreplicating Mycobacterium tuberculosis using a carbon starvation model. ACS Chem. Biol.8, 2224–2234 (2013). 10.1021/cb4004817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X. Z., Zhang, L. & Nikaido, H. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother.48, 2415–2423 (2004). 10.1128/AAC.48.7.2415-2423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowarz, E., Loscher, D. & Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J.10, 647–653 (2015). 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- 27.Ravera, S. et al. Structural insights into the mechanism of the sodium/iodide symporter. Nature612, 795–801 (2022). [DOI] [PMC free article] [PubMed]
- 28.Cater, R. J. et al. Structural basis of omega-3 fatty acid transport across the blood-brain barrier. Nature595, 315–319 (2021). 10.1038/s41586-021-03650-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood, C. A. P. et al. Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A. Nature596, 444–448 (2021). 10.1038/s41586-021-03782-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert, E., Mehdipour, A. R., Schmidt, A., Hummer, G. & Perez, C. Evidence for a trap-and-flip mechanism in a proton-dependent lipid transporter. Nat. Commun.13, 1022 (2022). 10.1038/s41467-022-28361-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler, B. D. & Kaatz, G. W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist Updat27, 1–13 (2016). 10.1016/j.drup.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 32.Niu, Y. et al. Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter. Nature601, 280–284 (2022). 10.1038/s41586-021-04212-9 [DOI] [PubMed] [Google Scholar]
- 33.Han, L. et al. Structure and mechanism of the SGLT family of glucose transporters. Nature601, 274–279 (2022). 10.1038/s41586-021-04211-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, N. et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell184, 370–383.e13 (2021). 10.1016/j.cell.2020.11.043 [DOI] [PubMed] [Google Scholar]
- 35.Debruycker, V. et al. An embedded lipid in the multidrug transporter LmrP suggests a mechanism for polyspecificity. Nat. Struct. Mol. Biol.27, 829–835 (2020). 10.1038/s41594-020-0464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson, M. The mycobacterial cell envelope-lipids. Cold Spring Harb Perspect Med. 4, a021105 (2014). [DOI] [PMC free article] [PubMed]
- 37.Casares, D., Escriba, P. V. & Rossello, C. A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int J Mol Sci. 20, 2167 (2019). [DOI] [PMC free article] [PubMed]
- 38.Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pravda, L. et al. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 (2018). 10.1093/nar/gky309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput Chem.31, 455–461 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, C., Freddolino, P. L. & Zhang, Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 45, W291–W299 (2017). 10.1093/nar/gkx366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm, L., Laiho, A., Toronen, P. & Salgado, M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci.32, e4519 (2023). 10.1002/pro.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen, L. N. et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature509, 503–506 (2014). 10.1038/nature13241 [DOI] [PubMed] [Google Scholar]
- 44.Ben-Zvi, A. et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature509, 507–511 (2014). 10.1038/nature13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen, C. et al. Lipid flipping in the omega-3 fatty-acid transporter. Nat. Commun.14, 2571 (2023). 10.1038/s41467-023-37702-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quistgaard, E. M., Low, C., Guettou, F. & Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol.17, 123–132 (2016). 10.1038/nrm.2015.25 [DOI] [PubMed] [Google Scholar]
- 47.Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature549, 233–237 (2017). 10.1038/nature23649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature524, 433–438 (2015). 10.1038/nature14953 [DOI] [PubMed] [Google Scholar]
- 49.Hiraizumi, M., Yamashita, K., Nishizawa, T. & Nureki, O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science365, 1149–1155 (2019). 10.1126/science.aay3353 [DOI] [PubMed] [Google Scholar]
- 50.Pomorski, T. & Menon, A. K. Lipid flippases and their biological functions. Cell Mol. Life Sci.63, 2908–2921 (2006). 10.1007/s00018-006-6167-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, B. et al. Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis. Nat. Struct. Mol. Biol.27, 561–569 (2020). 10.1038/s41594-020-0425-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui, X. et al. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature581, 323–328 (2020). 10.1038/s41586-020-2289-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ru, H. et al. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell163, 1138–1152 (2015). 10.1016/j.cell.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 58.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). 10.1038/s41592-020-00990-8 [DOI] [PubMed] [Google Scholar]
- 59.Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy135, 24–35 (2013). 10.1016/j.ultramic.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods11, 63–65 (2014). 10.1038/nmeth.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–1612 (2004). 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 62.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D. Biol. Crystallogr65, 1074–1080 (2009). 10.1107/S0907444909029436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. Biol. Crystallogr66, 12–21 (2010). 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matyash, V., Liebisch, G., Kurzchalia, T. V., Shevchenko, A. & Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008). 10.1194/jlr.D700041-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herzog, R. et al. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One7, e29851 (2012). 10.1371/journal.pone.0029851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herzog, R. et al. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol.12, R8 (2011). 10.1186/gb-2011-12-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schott-Verdugo, S. & Gohlke, H. PACKMOL-Memgen: A Simple-To-Use, Generalized Workflow for Membrane-Protein-Lipid-Bilayer System Building. J. Chem. Inf. Model59, 2522–2528 (2019). 10.1021/acs.jcim.9b00269 [DOI] [PubMed] [Google Scholar]
- 71.Case, D. A. et al. Amber22, University of California, San Francisco. (2022).
- 72.Maier, J. A. et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput.11, 3696–3713 (2015). 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickson, C. J., Walker, R. C. & Gould, I. R. Lipid21: Complex Lipid Membrane Simulations with AMBER. J. Chem. Theory Comput.18, 1726–1736 (2022). 10.1021/acs.jctc.1c01217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Compu.t11, 1864–1874 (2015). 10.1021/ct5010406 [DOI] [PubMed] [Google Scholar]
- 75.Koes, D. R., Baumgartner, M. P. & Camacho, C. J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model53, 1893–1904 (2013). 10.1021/ci300604z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The cryo-EM density maps have been deposited into the Electron Microscopy Data Bank under accession numbers EMD-37641 (MtEfpA with PE and cardiolipin), EMD-42204 (MtEfpA with PE and BRD8000.3) and EMD-42205 (MsEfpA with PE). The atomic coordinates are deposited in the Protein Data Bank under accession codes PDB 8WM5 (MtEfpA with PE and cardiolipin), 8UFD (MtEfpA with PE and BRD8000.3) and 8UFE (MsEfpA with PE). Source data are provided with this paper.