Abstract
Glutamate transmission and activation of ionotropic glutamate receptors are the fundamental means by which neurons control their excitability and neuroplasticity 1. The N-methyl-D-aspartate receptor (NMDAR) is unique among all ligand-gated channels, requiring two ligands, glutamate, and glycine, for activation. These receptors function as hetero-tetrameric ion channels, with the channel opening dependent on the simultaneous binding of glycine and glutamate to the extracellular ligand-binding domains (LBDs) of the GluN1 and GluN2 subunits, respectively 2,3. The exact molecular mechanism for channel gating by the two ligands has been unclear, particularly without structures representing the open channel and apo states. Here, we show that the channel gate opening requires tension within the linker connecting the LBD and transmembrane domain (TMD) and rotation of the extracellular domain relative to the TMD. Using electron cryomicroscopy (cryo-EM), we captured the structure of GluN1–2B NMDAR in its open state bound to a positive allosteric modulator. This process rotates and bends the pore-forming helices in GluN1 and GluN2B, altering the symmetry of the TMD channel from pseudo-fourfold to twofold. Structures of GluN1–2B NMDAR in apo and single-liganded states showed that binding of either glycine or glutamate alone leads to distinct GluN1–2B dimer arrangements but insufficient tension in the LBD-TMD linker for channel opening. This mechanistic framework identifies a key determinant for channel gating and a potential pharmacological strategy for modulating NMDAR activity.
Keywords: NMDA receptor, ligand-gating, open channel, allosteric modulation, apo-states, single-particle cryo-EM
Excitatory neurotransmission and cellular signaling mediated by NMDARs are crucial for brain function, development, and health 1. NMDARs are ligand-gated cation-selective ion channels that assemble as hetero-tetramers comprising two GluN1 and GluN2 (A-D) subunits. A defining characteristic of NMDAR functionality lies in the unique requirement of simultaneous binding of glycine or D-serine to GluN1 and glutamate to GluN2 to activate the channel. This distinctive mechanism stands in stark contrast to the other members of ionotropic glutamate receptors (iGluRs), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR), and kainate receptors, where activation is mediated solely by glutamate 1–3. Opening of the NMDAR channels and subsequent relief of the Mg2+ blockade by membrane depolarization 4,5 can lead to an influx of sodium and calcium 6,7, which orchestrates neuroplastic signaling and is intimately associated with high-order brain functions, including learning and memory 8. Importantly, dysfunctional NMDARs have been implicated in a plethora of neurological diseases and disorders, such as schizophrenia, Alzheimer’s disease, depression, intellectual disability, autism spectrum disorder, epilepsy, and acute neuronal damage resulting from stroke or traumatic brain injury 1.
The inaugural structural analyses of the intact tetrameric GluN1–2 NMDAR channel in 2014 revealed a hetero-tetrameric subunit arrangement, showcasing a dimer of GluN1–2 heterodimers at both the amino-terminal domain (ATD) and the ligand-binding domain (LBD), whereas the transmembrane domain (TMD) displayed a pseudo-four-fold symmetry around the channel pore 9,10. The most recent study showed that the GluN1–3A NMDAR channel assembles as a dimer of GluN1–3A heterodimers at ATD and LBD yet exhibits a distinct conformational pattern from those of the GluN1–2 NMDARs 11. Subsequent studies expanded on these findings, presenting structures in various liganded states that prompted speculations on allosteric modulation 12–14, competitive inhibition 15,16, channel blockade 17–19, and allosteric mechanisms 12,16,20. The overarching interpretation from these structural studies, together with studies on the isolated domains, proposes that the bi-lobe architectures of GluN1 and GluN2 LBDs undergo closure upon agonist binding 21–23, and the GluN1–2 LBD heterodimers undergo rigid-body rotational movements 12,16,24, a process regulated by the ATDs 12,25–28. These motions are thought to collectively influence tension in the GluN2 LBD-TMD linker 29, ultimately impacting channel gating. Despite years of enthusiasm, understanding how the binding of the two agonists translates into the opening of the channel gate in the NMDAR has remained elusive due to the absence of a structure representing the open state. Rigorous endeavors in the past to structurally elucidate the open NMDAR channel have been met with challenges primarily stemming from a prevalence of mostly non-active and some pre-active (formerly referred to as active and active-SS) conformations where the receptors bind agonists, glycine, and glutamate, but the ion channel pore remains closed 12,16. Therefore, the pre-active state represents the state primed for channel opening, whereas the non-active state represents the more extensively closed state 16.
Here, we bridge these critical knowledge gaps by presenting the open channel structure of GluN1–2B NMDARs captured in the presence of glycine, glutamate, and a positive allosteric modulator (PAM), EU-1622–240. Additionally, we present a series of apo-states where both GluN1 and GluN2 are unoccupied with ligands, only GluN1 is occupied with an agonist, and only GluN2 is occupied with an agonist. Our findings show that the activation mechanism involves tension generation in the GluN2B LBD-TMD linkers, along with a robust rotation of the extracellular domain relative to the TMD channel. This action breaks the symmetry of the channel gate and pore from pseudo-four-fold into two-fold symmetry, introduces kinks in the GluN2B M3’ helices, and rotates GluN1 M3 helices to facilitate gate opening. The gate opening is stabilized by binding of EU-1622–240 to a novel site surrounded by GluN2B pre-M1’, M1’, and M4’ helices, which favors the bending of the GluN2B M3’. The apo-state structures demonstrate that glycine binding to GluN1 primes the NMDAR channel, resembling the pre-active state. Conversely, the absence of glycine rearranges the subunits into a position akin to the non-active1-like states, where the GluN1–2B LBD dimers are rotated downward toward the plane of the membrane, reducing the GluN2B LBD-TMD tension. The comprehensive scheme of conformational patterns and the newly identified PAM site outline potential therapeutic approaches for neurological diseases and disorders linked to NMDAR dysfunction.
Channel opening of GluN1–2B NMDAR involves linker stretching and global rotation.
To enhance the likelihood of capturing the open state, we introduced a PAM, EU-1622–240, which potentiates the macroscopic current when added with agonists (Fig. 1a). The single-particle cryo-EM on the GluN1–2B NMDAR in the presence of glycine, glutamate, and EU-1622–240 resulted in two major three-dimensional (3D) classes, one with a closed channel (non-active1 at 3.13 Å) and the other with an open channel (open state at 3.72 Å) (Fig. 1b, Extended Data Fig. 1). The quality of the TMD cryo-EM density was further improved by implementing focused refinement to facilitate reliable model building (Extended Data Fig. 1). Consequently, incorporating EU-1622–240 along with glycine and glutamate into the GluN1–2B NMDAR sample enabled the first-ever visualization of the open channel.
Figure 1. Structural analysis of GluN1–2B NMDAR in open state.
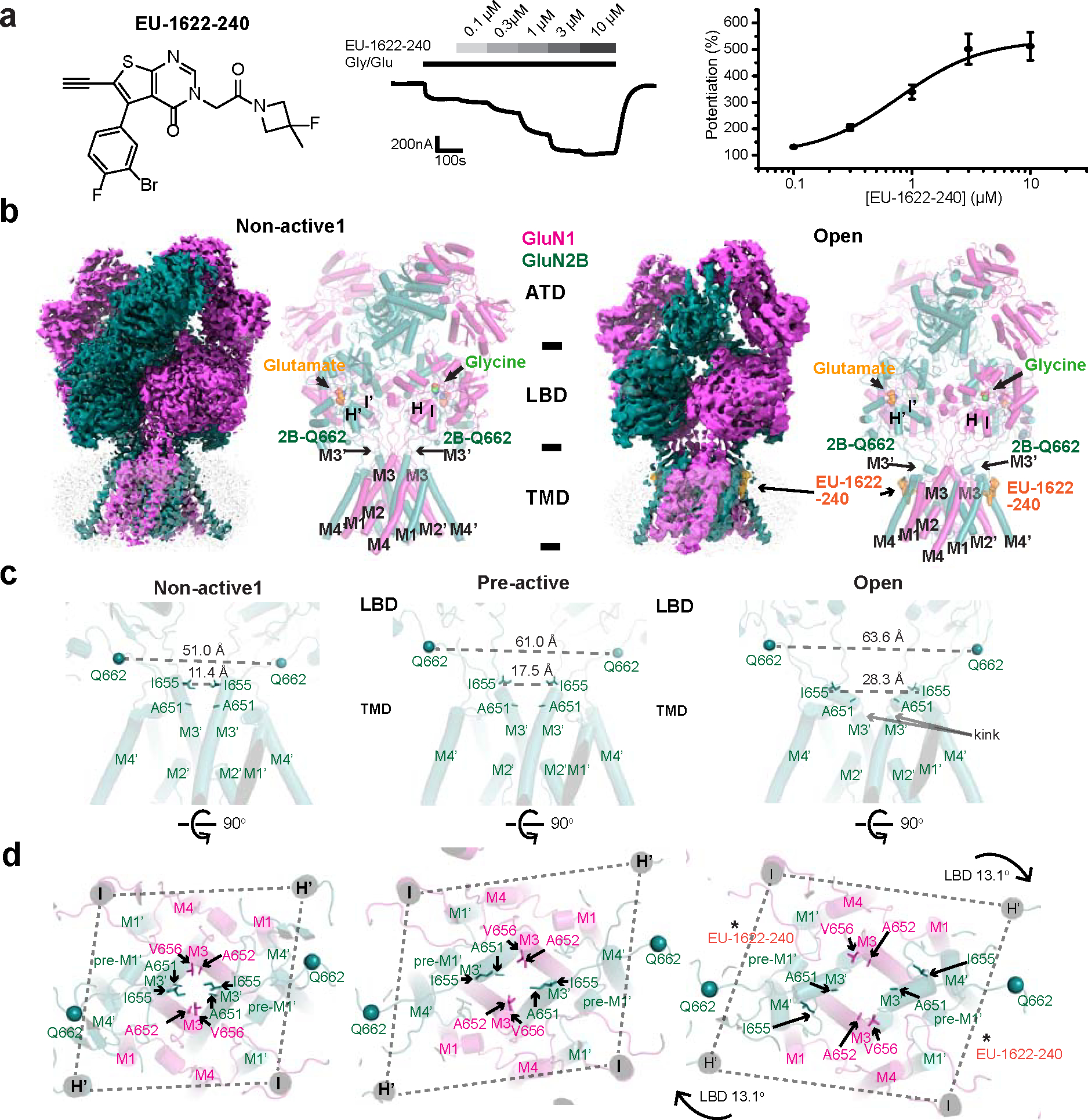
a, The chemical structure of EU-1622–240 alongside a representative two-electrode voltage clamp (TEVC) recording of rat GluN1–2B NMDAR expressed in Xenopus oocytes. Recordings were performed in the presence of 30 μM glycine, 100 μM glutamate, and various concentrations of EU-1622–240 at a holding potential of −40 mV. The concentration-response curve, calculated from TEVC recordings, revealed a 5.3±0.57-fold increase in maximum NMDAR current with an EC50 of 0.75±0.1 μM (n=6, Hill coefficient nH=1.55±0.05). Data are presented as mean ± SEM. n= number of oocytes measured. b, Cryo-EM density and modeled structure of GluN1–2B NMDAR in the non-active1 state (left) and open state complexed with EU-1622–240. The GluN1 and GluN2B subunits are colored in magenta and deep teal, respectively, with glycine and glutamate and Gln662 Cɑs shown as spheres. c, Comparison of GluN2B TMD in non-active1, pre-active (PDB code: 6WI1), and open states. d, Top view of the channel pores across various states, highlighting the alterations in the opening of the M3/M3’ gates clustered with hydrophobic residues (GluN1 Ala652 and Val656 and GluN2B Ile655 and Ala651). A notable 13.1° rotation between the LBD and TMD in the open state compared to the pre-active state is critical for gate opening.
The extracellular region of the open state structure resembles the pre-active state (formerly called Active-SS, PDB code: 6WI1) 12,16. This similarity is particularly evident in the arrangement of subunits within the LBD layer, where inter-subunit and inter-domain interactions occur. These interactions in the pre-active and open states are prominent at the interface of the GluN1–2B dimers (Extended Data Fig. 2a,b). Especially, the GluN1 loop2 (L2) and the GluN2B loop1 (GluN2B L1’) are proximal to the α4’ helix located in the GluN2B ATD (Extended Data Fig. 2b). In contrast, the non-active1 state features a downward rotation of the GluN1–2B LBD dimers towards the plane of the membrane compared to the open and pre-active states, leading to the movement of the GluN1 L2 away from the GluN2B α4’ helix (Extended Data Fig. 2c). This variance at the LBD layer correlates with changes at the GluN1–2B ATD dimer interface, where the open and pre-active states have reduced distances between the lower lobes (R2) of GluN1 and 2B ATDs compared to the non-active1 state (Extended Data Fig. 2d,e, 14.2, 12.3, and 19.2 Å for open, pre-active, and non-active1 states, respectively). Furthermore, the open and pre-active states are characterized by an open bi-lobe structure of GluN2B ATD, in contrast to the closed bi-lobe in the non-active1 state (Extended Data Fig. 2d,e) that resembles the inhibited state stabilized by a negative allosteric modulator (NAM), such as ifenprodil, Zn2+, or an inhibitory antibody 12,13,30,31.
In the open state, there is a notable increase in tension within the GluN2B LBD-M3’ loop as quantified by the distance between GluN2B Gln662 Cɑ atoms (Fig. 1c). The tension in the open state is significantly higher compared to the non-active1 (63.6 Å versus 51.0 Å) but similar to the pre-active state with a closed channel gate 16 (63.6 Å vs. 61.0 Å) (Fig. 1c). The critical step driving the opening of the channel gate from the pre-active state involves a 13.1° clockwise horizontal rotation of the extracellular domain relative to the TMD, which has not been predicted (Fig. 1d). This rotation, in turn, induces a corresponding rotation of the GluN1 M3 helices and a bending of the GluN2B M3’ helices (Fig. 1c,d). These structural changes transform the pseudo-four-fold symmetry of the TMD in the pre-active state into two-fold symmetry and result in gate-opening through the displacement of the hydrophobic residues at the pore entrance (Fig. 1d, Supplementary Video 1).
The non-active1 state had a closed channel where the agonist-bound GluN1–2B LBD dimers were not in a position to generate sufficient tension in the GluN2B LBD-M3’ loop for gating 12,16,29 (Fig. 1b,c; non-active1). Specifically, the lack of tension caused by the downward rotation of the GluN1–2B LBD dimers towards the plane of the membrane (described above, Extended Data Fig. 2c) leads to a clustering of hydrophobic residues (GluN1 Ala652 and Val656 and GluN2B Ala651 and Ile655) in GluN1 M3 and GluN2B M3’ helices around the channel entrance and gate, which prohibits ion passage (Fig. 1c,d; non-active1). Overall, our structural analysis delineated the cascade of conformational alterations coupling changes in the extracellular domains to the channel gate opening.
Analysis of channel gate and pore in multiple states
We next measured and compared channel diameters between the open, pre-active, and non-active1 states to assess which regions of the TMD undergo conformational alterations. Notably, we observed substantial diameter expansions in the open channel structure of the VIVI gate at the tip of the M3/M3’ helices and the conserved region, known as the SYTANLAAF motif, compared to pre-active and non-active1 structures (Fig. 2a–d). The hydrophobic cavity is more confined in the non-active1 structure than the pre-active and open state structures, while the Asn-rings at the entrance of the cation selectivity filter are equally narrow among all structures (Fig. 2a–d). Structurally, the VIVI gate is located next to the SYTANLAAF motif and is distinct from another gate at the M2 loop, which was suggested to exist recently 32.
Figure 2. Pore analysis on the structures of GluN1–2B NMDARs in various functional states.
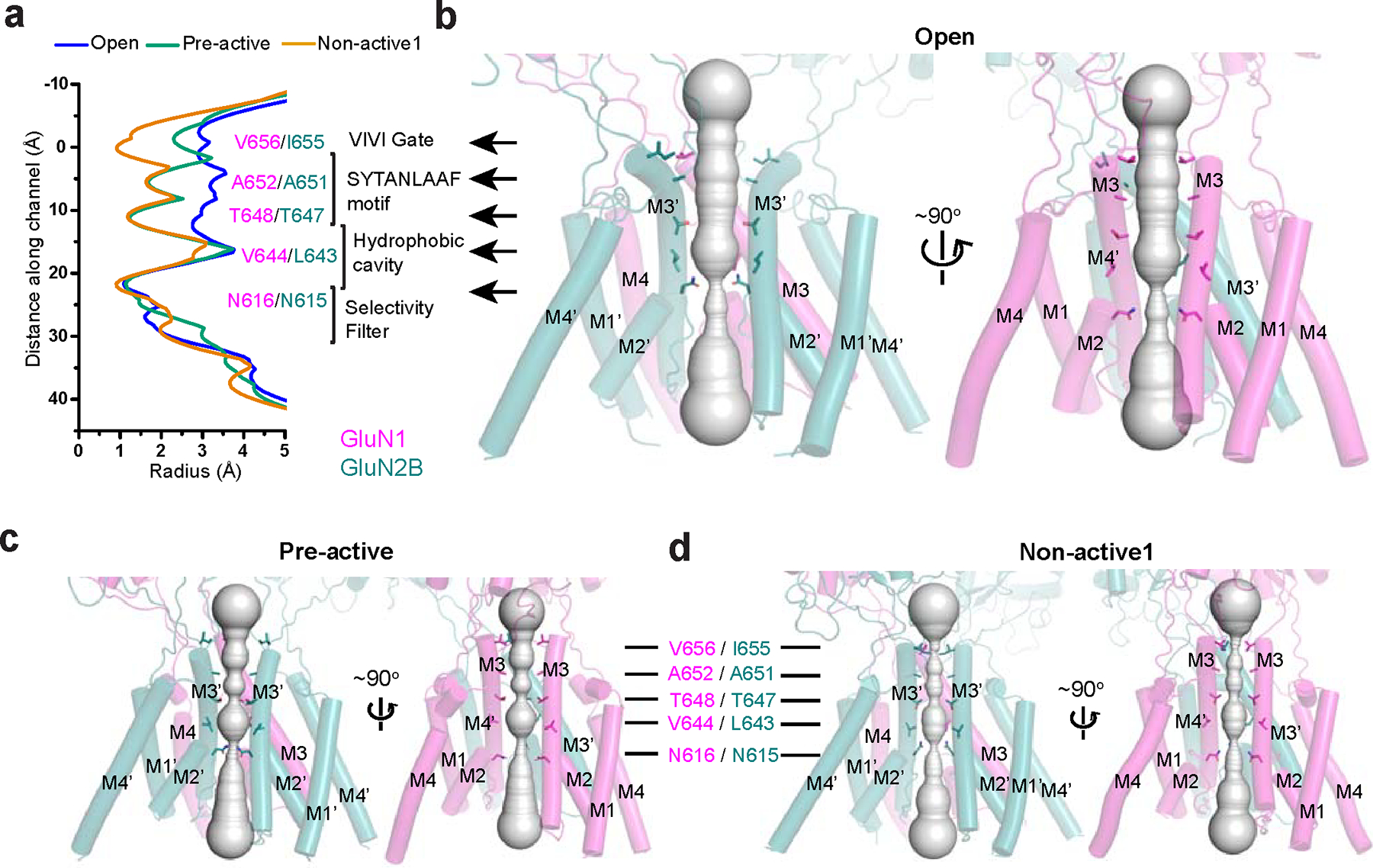
a-d, Pore radius measurement (a) and side views of the channel pores in open (b), pre-active (c), and non-active1 (d), as analyzed by the program, HOLE, which distinctively illustrate the gate widening and pore dilation specifically in the open state.
Consistent with the structural analysis above, Potential of Mean Force (PMF) calculations confirmed that only the open channel structure allows for the permeation of sodium ions (Extended Data Fig. 3). PMF calculations revealed substantial energy barriers at the VIVI-gate of 5 kcal mol−1 for non-activate1, and at the SYTANLAAF motif of 6.7 kcal mol−1 for non-active1 and 8 kcal mol−1 for the pre-active state, ruling out the possibility of ion passage (Extended Data Fig. 3). In contrast, the PMF at equivalent regions in the open channel structure hovers around 0 kcal mol−1, denoting substantially more favorable conditions for ion passage (Extended Data Fig. 3a). We also observed negative PMF for Na+ (−11.5 kcal mol−1) around the hydrophobic cavity in the non-active1, but not in the pre-active state or open state. While the closed gate prevents Na+ from permeating through the channel in the non-active1 conformation, its narrower hydrophobic cavity may accommodate some cation binding. Lastly, the PMF unequivocally established the impermeability of Cl−, consistently revealing a positive PMF throughout the channel (Extended Data Fig. 3a,b).
PAM manipulates channel gating determinants to stabilize open state in NMDAR
Our current structural analysis revealed an unprecedented binding pocket occupied by EU-1622–240 and crucial determinants for channel gating. The cryo-EM density, consistent with the approximate shape and size of EU-1622–240, was present at the juxtamembrane pocket, primarily formed by residues on GluN2B pre-M1’, M1’, and M4’, including, Phe550, Leu551, Trp559, and Met824 (Fig. 3a, blue mesh). This density was detected only when EU-1622–240 was added to the sample; however, it lacks sufficiently clear features to allow precise model fitting, likely stemming from multiple binding poses slightly differing from each other (Extended Data Fig. 1j). This observation aligns with the EU-1622–240 binding being almost exclusively facilitated through hydrophobic interactions. Similar to how NMDAR channel blockers like S-(+)-ketamine 17, it is probable that multiple binding modes of the EU-1622–240 binding result in an averaging effect on the cryo-EM density. The binding of EU-1622–240 induces the conformational shift in the GluN2B pre-M1’ and its preceding loop, pulling them away from the channel (Fig. 3b; asterisks and arrow). This movement consequently creates space, accommodating the bending of the GluN2B M3’, essential for the channel gate opening. We reasoned that the residues around the EU-1622–240 binding site (Fig. 3a) are critically involved in the channel gate control as EU-1622–240 manipulates them to favor the channel gate opening. To test this hypothesis, we performed site-directed mutagenesis and evaluated the channel open probability (Po) using two-electrode voltage clamp (TEVC) on Xenopus oocytes injected with cRNA encoding the receptors. Our methodology involved an indirect approach where the GluN1 Ala652Cys mutant was co-expressed with GluN2B mutants. These mutant receptors were then covalently modified by 2-aminoethylmethanethiosulphonatehydrobromide (MTSEA) at GluN1 Ala652Cys, a process designed to lock the channel in its open state 33. We assumed Po = 1.0 for MTSEA-modified receptors, implying that the maximal agonist response potentiation would inversely correlate with Po (Fig. 3c). Our analysis revealed that most mutations reduced Po, except GluN2B Pro547Ala, which had significantly increased Po, and GluN2B Ala827Trp, which produced no detectable change. The alanine mutation of GluN2B Trp559 on GluN2B M1’ resulted in no channel activity. These observations indicated that the EU-1622–240 binding region is crucial for gating control. The GluN2B pre-M1’ residue mutants (GluN2B Phe550Ala and Leu551Ala) had the largest enhancing effects on the PAM activity of EU-1622–240, validating the binding site observed in the cryo-EM structure (Fig. 3d). The mutations, including GluN2B Met824Trp and others around the binding pocket that do not directly interact, have minimal and statistically insignificant effects (Fig. 3d).
Figure 3. PAM site and channel gate determinants.
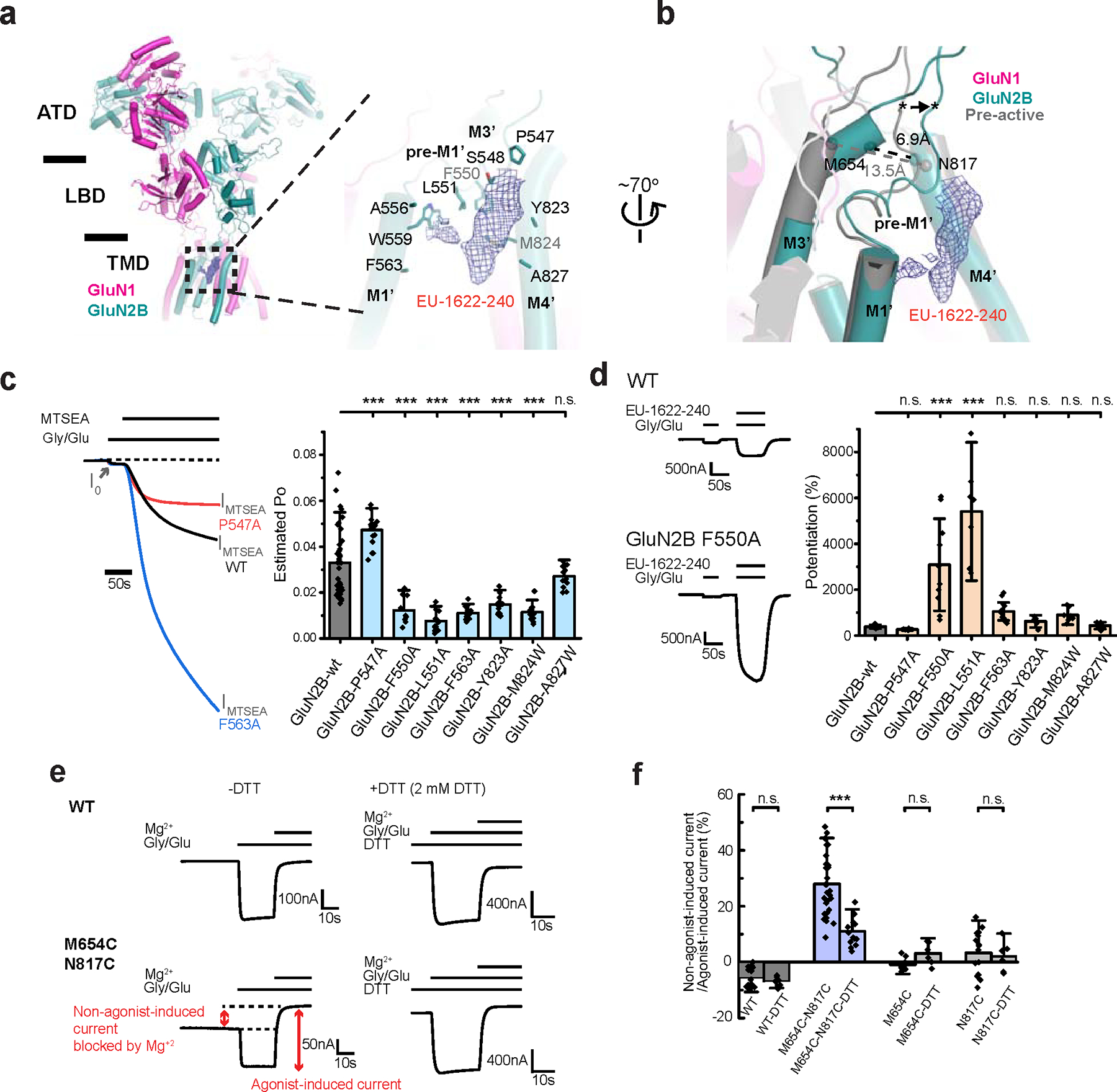
a, EU-1622–240 is exclusively bound to the open state as evident from the cryo-EM density (blue mesh) in a pocket formed by residues from GluN2B M1’, pre-M1’, and M4’. b, Comparison of the region around the PAM site between the open and pre-active states, where differences are shown by the Cα positions of Met654 on M3’ and Asn817 on M4’. The EU-1622–240 density is shown as blue mesh. c, Estimation of open probability (Po) by measurement of MTSEA potentiation in GluN1 Ala652Cys/GluN2B NMDARs. Representative TEVC traces (left) and the estimated Po for site-directed mutants based on MTSEA potentiation. (n from left to right = 41, 18, 10, 15, 19, 14, 22, and 12) d, Representative TEVC traces (left) and the changes in PAM activity ([EU-1622–240] = 3 μM) expressed as a percent of control for each mutant. The height of the bars represents the mean, and the whiskers represent ± 99% confidence interval (CI, n from left to right, 8, 6, 10, 7, 12, 7, 7, and 8). e-f, Disulfide crosslinking between Met654Cys and Asn817Cys locks a subset of receptors in the open state without agonists, which could be blocked by Mg2+ (red arrow). This current observed in the absence of agonists is diminished in the presence of DTT. WT or single-point mutants do not display this current (n in panel f from left to right = 16, 7, 30, 14, 10, 7, 16, and 6). For panels c and f, error bars represent mean ± SD. One-way ANOVA with post-hoc Dunnett’s (c, d) or Tukey’s test (f) was employed to determine the significant differences between the two experimental datasets. The asterisks denote p < 0.001 (***). n.s. denotes “not significant statistically”. TEVC recordings were conducted at a holding voltage of −60 mV. n = number of recordings from individual oocytes.
Next, we investigated if the open channel conformation observed in the presence of EU-1622–240 also occurs naturally, without the compound. To address this question, we sought to capture the open channel state by creating a disulfide bond through site-directed mutagenesis. Notably, a major structural change between the pre-active and open states is the bending of GluN2B M3’ helices towards the GluN2B M4’ helices (Fig. 3b). Based on this, we engineered a disulfide crosslinking by introducing cysteine residues at the GluN2B M3’ (Met654Cys) and M4’ (Asn817Cys). This modification was intended to stabilize the open channel during conformational transitions, thereby inducing some constitutive activity. Our TEVC recordings from oocytes expressing this mutant pair showed an increased current in the absence of applied co-agonists, which magnesium ions (2 mM) could block (Fig. 3e, left panel). This result implies that the crosslinking of these residues stabilizes open NMDAR channels, accounting for about 30% of the agonist-induced macroscopic NMDAR current. When we disrupted the disulfide bond using the reducing agent DTT, there was a marked decrease in the holding current (Fig. 3e, right panel). Furthermore, the double mutant effect was not observed in oocytes expressing the single point mutant (Fig. 3f; GluN2B Met654Cys or Asn817Cys), confirming that the current we measured in the nominal absence of agonists resulted from the formation of the engineered disulfide bond. Overall, these experiments collectively affirm that our EU-1622-240-bound structure highly resembles the authentic open channel.
Structural comparison of the open conformation between NMDAR and AMPAR
Prior to this study, the only open channel iGluR structures available were those of Stargazin complexed AMPARs 34–36. Our research presents the first opportunity to directly compare the channel-gating mechanisms of AMPARs and NMDARs, offering new insights into their functional differences. Given the differences in the ATD structures between NMDAR and AMPAR 26,37, our analysis compared the LBD and TMD along with their respective orientations. While the LBD architectures of NMDAR and AMPAR share broad similarities, with many of the secondary structures being conserved, notable differences are observed in the tetrameric arrangements. These are primarily attributed to the NMDAR-specific loops, L1 and L2, which are situated at the dimer-of-dimers interfaces of the LBD layer (Fig. 4a). The GluN1 L2, GluN2B L1’ from LBD, and GluN2B ɑ4’ from ATD are closely positioned, thereby influencing each other through steric effects (Fig. 4a). The involvement of ATD in these associations are pivotal in determining the degree of rotation in the GluN1–2B LBD dimers, thereby exerting critical control over the tension within the GluN2B M3’-LBD loop in NMDARs (Fig. 4a,b and Extended Data Fig. 2). Contrasting this, AMPARs and kainate receptors lack such motifs or interactions involving ATD, underscoring these features as distinctive to NMDARs, characterizing their unique motif and conformational dynamics.
Figure 4. Comparison between NMDAR and AMPAR open states.
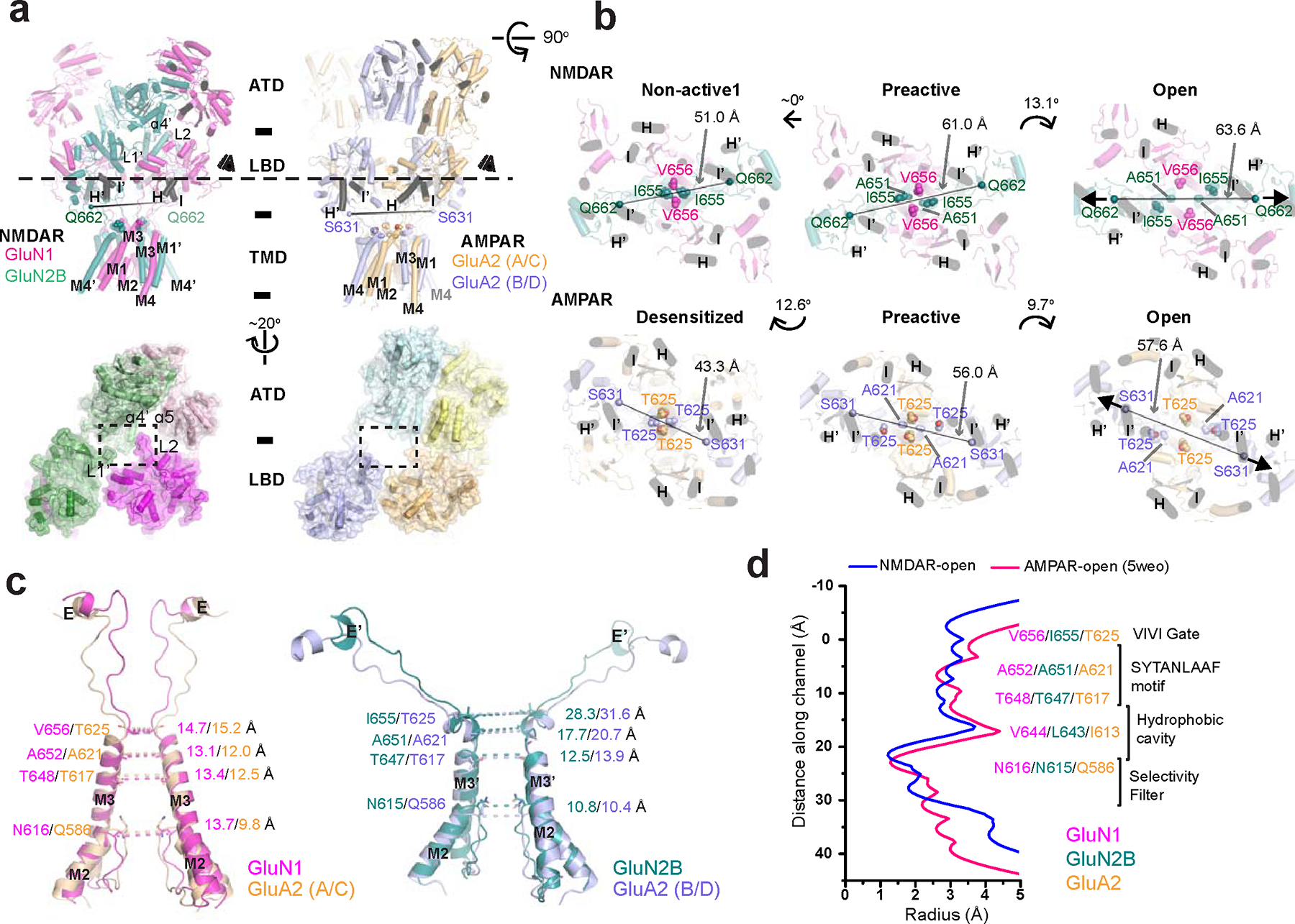
a, Side view of the open state GluN1–2B NMDAR and GluA2 AMPAR (PDB code: 5WEO) structures. The linker residues, GluN2B Gln662 and GluA2 Ser631 are shown as spheres. The ATD-LBD interaction modes are distinct between NMDAR and AMPAR, especially around the dimer of dimers interface, which involves the cluster of GluN1 L2, GluN2B L1’ and GluN2B ɑ4’ from the ATD. b, The NMDAR and AMPAR channel gates in different functional states are viewed from the LBD layer (‘eyes’ in panel a). The helices H and I are marked in black as references in both panel a and b. Gate residues are shown in spheres. The Cɑs of GluN2B Gln662 and GluA2 Ser631 in the LBD-M3 loop are connected with dotted lines to show the degree and direction of tensions necessary for channel gating. The angles between these connecting lines are shown with curved arrows between functional states. c, Structural comparisons of the pore-forming helices (M2 and M3) between the NMDAR and AMPAR open states. The distances between the Cαs of pore-forming residue pairs in the GluN1 and GluA2 A/C chains (left panel) and those in the GluN2B and GluA2 B/D chains (right panel) are shown. d, Radius measurements along the channel pores of the open NMDAR and open AMPAR structures.
In exploring the TMD channels of NMDARs and AMPARs, we find that the central pores, primarily formed by the M3 helices, exhibit similar structural architectures. The open states of both NMDAR and AMPAR feature a bending of two M3 helices, observed in the GluN2B subunits for NMDAR and the B/D subunits in GluA2 homotetramers 34,35 or GluA2 subunits in GluA1/2 heterotetramers 36. The bending of the M3 helices in both receptor types occurs at the first alanine residue within the SYTANLAAF gating motif conserved within the iGluR family. Despite these similarities, notable differences emerge in the domain orientation between the LBD and TMD. This is evident in the relative positioning of the helices in the LBD (Fig. 4a,b; Helix H, H’, I, and I’ in black) compared to the central M3/M3’ pore. Additionally, the directionality of the M3-LBD linkers, marked by the positions of the GluN2B Gln662 and GluA2 Ser631 residues, further distinguishes the two structures (Fig. 4b, Open, arrows). This suggests that the gating force emanating from the LBD layer, which involves pulling of the M3-LBD linker, is transduced in different directions for AMPAR and NMDAR. Further comparison of the pre-active, non-active1, and desensitized states reveals distinct LBD-TMD orientations. This rotation causes the GluN1 M3 helices in NMDAR to move Val656 away from the central pore, whereas, in AMPAR, the GluA2 (A/C) M3 helices move translationally, dilating the gate. Therefore, NMDAR and AMPAR have distinct ways to transduce LBD information to the channel-gate opening. The pore regions of NMDAR and AMPAR channels exhibit a few differences. Specifically, the VIVI gate region (TTTT in GluA2) is more open in GluA2 than in GluN1–2B. Conversely, the SYTANLAAF motif is marginally more expanded in GluN1–2B than in GluA2 (Fig. 4c,d).
Apo-state structures reveal the mechanism of dual-agonist requirement for NMDAR activation
We next sought to elucidate the mechanism underlying the dual-agonist requirement of GluN1–2 NMDAR channel activation. This unique functional feature of NMDAR was explored by comparing the agonist-bound structure with three apo-states: apo/apo (no ligand bound to GluN1/GluN2B) (Fig. 5), gly/apo (glycine only) (Fig. 6a–e), and apo/glu (glutamate only) (Fig. 6f–j). Using single-particle cryo-EM, we determined structures at 4.05, 3.69, and 3.90 Å for the apo/apo, gly/apo, and apo/glu states, respectively (Extended Data Fig. 4–6). Our analyses immediately revealed the closed channel gate in all structures as expected for the apo-states (Extended Data Fig. 7a,b,d). Here, we first compare these three apo-state structures with the pre-active state, which is bound to both glycine and glutamate but has not yet undergone further conformational changes for channel opening (Fig. 1c,d) 12,16.
Figure 5. Structural analysis of GluN1–2B NMDAR in apo/apo-state.
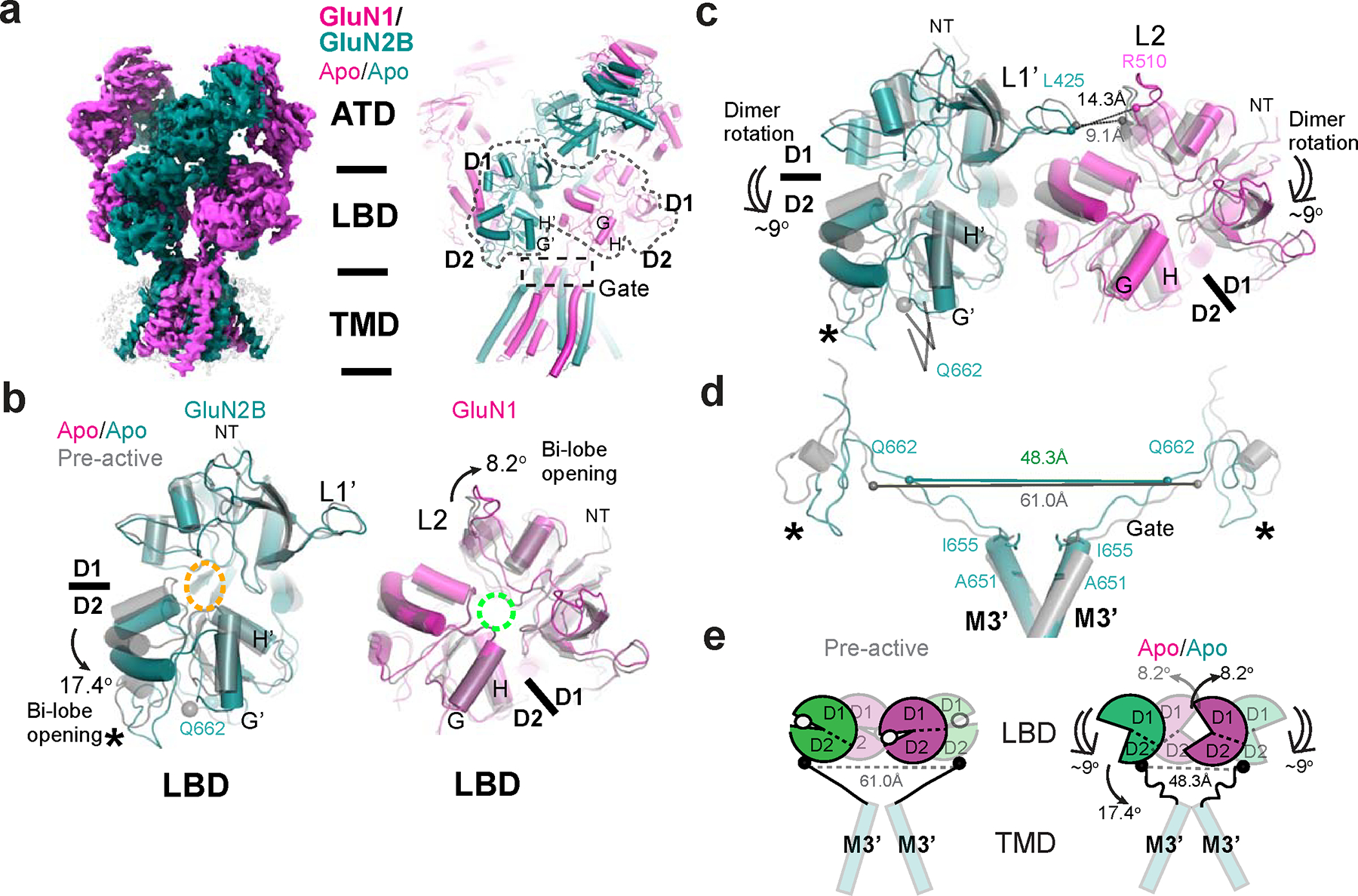
a, Cryo-EM density and modeled structure of GluN1–2B NMDAR in the apo/apo state. b, Comparison of LBDs in the apo/apo and pre-active (gray) states. Superposition of the apo/apo and pre-active structures at GluN2B D1 (upper lobe) and GluN1 D2 (lower lobe) demonstrates 17.4° and 8.2° domain openings, respectively, in the apo/apo state. Glutamate and glycine binding pockets are indicated by orange and cyan dotted ovals, respectively. c, In the apo/apo state, the GluN1–2B LBD dimers undergoes ~9° downward rotation toward the membrane plane compared to the pre-active state, which results in an increased distance between GluN2B L1’ and GluN1 L2 (represented by the distance between GluN1 Leu425 and Arg510). d-e, The tension in the GluN2B LBD-M3’ linkers is reduced in the apo/apo state compared to the pre-active state, which favors the closure of the channel gate. Asterisks in panels b-d indicate the location of the D2 loop in GluN2B.
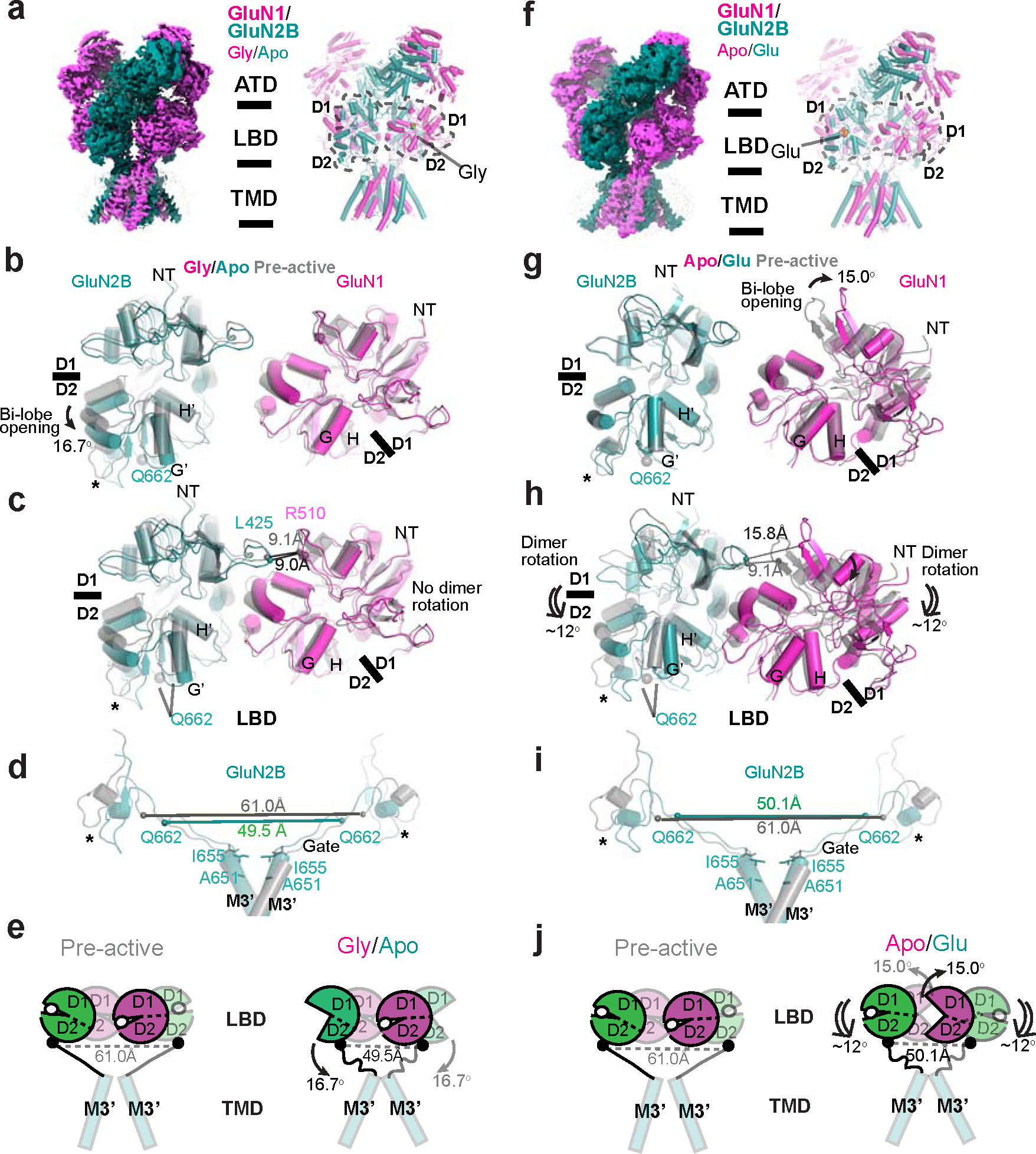
Figure 6. Structural analysis of GluN1–2B NMDAR in gly/apo and apo/glu states reveals distinct mechanisms for favoring channel closure.
a, Cryo-EM density and a structural model of the gly/apo state. b, Superposition of the gly/apo and pre-active structures at GluN2B D1 (upper lobe) and GluN1 D2 (lower lobe) demonstrates 16.7° opening of the GluN2B LBD bi-lobe and no change in the GluN1 LBD in the gly/apo state. c, There is no rotational movement in the GluN1–2B LBD dimers in this state relative to the pre-active state. d-e, The tension in the GluN2B LBD-M3’ linkers is reduced in the gly/apo state compared to the pre-active state as a result of the GluN2B LBD bi-lobe opening. f, Cryo-EM density and a structural model of the apo/glu state. g, Superposition of the apo/glu and pre-active structures at GluN2B D1 and GluN1 D2 demonstrates ~0° and ~15° domain openings, respectively, in the apo/glu state. h, In the apo/glu state, the GluN1–2B LBD dimers undergoes ~12° downward rotation compared to the pre-active state, which results in an increased distance between GluN2B L1’ and GluN1 L2. i-j, The tension in the GluN2B LBD-M3’ linker is reduced in the apo/glu state compared to the pre-active state as a result of the dimer rotation toward the membrane plane. Asterisks in panels b-d and g-i indicate the location of the D2 loop in GluN2B.
In the apo/apo state, two distinct types of conformational changes occur in the LBDs, which promote the closure of the channel gate (Fig. 5a–e). Firstly, there is an opening of the LBD bi-lobes: the GluN1 and GluN2B LBDs open by 8.2° and 17.4°, respectively, compared to the pre-active state where agonists are bound (Fig. 5b). Secondly, the GluN1–2B LBD dimers undergo a downward rotation of approximately 9° toward the membrane plane (Fig. 5c). These movements synergistically reduce tension in the GluN2B M3’-LBD linker, as measured by the shorter distance between the two GluN2B Gln662 residues, compared to the one in the pre-active state (Fig. 5d,e). The reduced GluN2B M3’-LBD linker tension favors the channel closure 29,38.
In the gly/apo state, the GluN1 LBD bi-lobes are in a closed conformation similar to the pre-active state, whereas the GluN2B LBD bi-lobes are open by 16.7° compared to the one in the pre-active state (Fig. 6a,b). Furthermore, the GluN1–2B LBD dimers are arranged similarly to the pre-active state (Fig. 6c). The resemblance in the orientation of the GluN1–2B LBD dimers is particularly noticeable in the similar arrangement of GluN2B L1’ and GluN1 L2 (Fig. 6c). In this state, the 16.7° opening of the GluN2B LBD bi-lobe serves as a direct factor for channel closing by relaxing the GluN2B M3’-LBD linkers (Fig. 6c–e). The inter-residue distance between the GluN2B Gln662 sites is reduced to 49.5 Å in the gly/apo state, down from 61.0 Å in the pre-active state (Fig. 6d,e, Supplementary Video 2).
The apo/glu state represents a theoretical functional state where glutamate is transmitted from the presynaptic neuron, yet the co-agonist, glycine or D-serine, is available in restricted quantities within the synaptic cleft. AMPAR and kainate receptors are activated in this scenario, while NMDAR remains inactive. In this state, the GluN1 LBD bi-lobes open by 15° relative to the pre-active state (Fig. 6g), a more significant bi-lobe opening than the apo/apo structure (Fig. 5c). This is accompanied by a ~12° downward rotation of the GluN1–2B LBD dimers (Fig. 6h), a feature also observed in the apo/apo and non-active1 states. Despite the closing of the GluN2B LBD bi-lobes due to glutamate binding, this downward rotation relaxes the GluN2B M3’-LBD linkers (Fig. 6h–j), hindering the opening of the channel gate. Notably, the downward rotation of the GluN1–2B LBD dimers is linked to the dimeric arrangement of the GluN1–2B ATDs, resulting in a greater distance between the R2 lobes (Extended Data Fig. 7c, Supplementary Video 3). This structural interplay between the ATDs and LBDs enables ATD-binding compounds, like ifenprodil, to control channel gating indirectly, a function unique to NMDARs 12.
The structural comparison of the gly/apo and apo/glu states with the apo/apo state provides insights into changes that occur in the LBD layer upon agonist binding. The glycine binding to the apo/apo state closes the GluN1 LBD bi-lobe by ~8.1° (Extended Data Fig. 8a), which in turn, results in ~4° rotation of the GluN1–2B LBD dimers upward relative to the membrane plane (Extended Data Fig. 8b). These rotations prime the gly/apo state glutamate binding away from transitioning to the pre-active state; thus, the GluN2B LBD-M3’ linker tension remains similarly loose compared to the apo/apo state (Extended Data Fig. 8c,d). The glutamate binding to the apo/apo state closes the GluN2B LBD bi-lobe by 16.1° and further opens the GluN1 LBD by 10.7°, which is coupled to the 8° downward rotation of the GluN1–2B LBD dimers relative to the membrane plane (Extended Data Fig. 8e,f). These rotations prevent the GluN2B LBD-M3’ linkers from forming tension, thereby ensuring channel closure (Extended Data Fig. 8g,h).
Overall, NMDARs exhibit a distinctive mechanism to prevent channel opening in response to a single ligand, varying between the GluN1 and GluN2 subunits. Furthermore, these three apo-state structures demonstrate that the binding of glycine, or another co-agonist, D-serine, to GluN1 primes the NMDAR channel to become activated following synaptic transmission, transitioning from pre-active to the open state.
Discussion
In summary, we demonstrate that NMDAR channel gating involves the generation of tension in the GluN2B M3’-LBD linker by agonist bindings followed by the clockwise rotations of the extracellular domain relative to the TMD channel. These movements facilitate the bending of the GluN2B M3’ helix and rotation of the GluN1 M3, which steer the hydrophobic residues away from the gate entrance, allowing for ion permeation. The extracellular regions and TMD channels are coupled distinctly between AMPAR and NMDAR due to robust differences in the relative orientation of LBD and TMD, indicating that NMDAR and AMPAR undergo distinct gating mechanisms. Furthermore, the NMDAR becomes primed for channel gating by the presence of tonic concentrations of co-agonists glycine or D-serine bound to GluN1 LBD. In the absence of co-agonist binding to GluN1, regardless of the occupancy of GluN2B LBD by glutamate, a downward rotation of the GluN1–2B LBD dimers occurs, reducing the GluN2B M3’ tension necessary for channel gating (Supplementary Video 4). This unique requirement of dual-ligands for NMDAR activation depends on glutamate availability via synaptic transmission and co-agonists, glycine or D-serine, controlled by serine racemase 39 as well as ASCT1 and 2 40 and Asc-1 amino acid transporters 41. Lastly, our study unveils a previously unidentified PAM site in the juxtamembrane region of GluN2B that controls channel gating. When occupied by the compound EU-1622–240, this site promotes bending of the GluN3B M3’ segment, a critical action necessary for channel gating. The gained insights into the dual-ligand-gated ion channel opening and the novel PAM site hold the potential to open new avenues for developing therapeutic approaches in conditions like cognitive impairment 42 and schizophrenia 43, where enhancing NMDAR activity has demonstrated potential benefits.
Methods
Protein expression and purification
The expression and purification methods of apo, glycine-bound, glutamate-bound, and EU-1622–240-bound (open) GluN1-GluN2B NMDARs were adopted from Earlybac insect expression system 44 and the previously established purification protocols 45. In brief, the Sf9 insect cells were infected by the baculovirus harboring GluN1 and GluN2B genes at a cell density of 4 × 106 cells/ml. The cells were harvested at 48 hours post-infection, resuspended in the purification buffer (20 mM HEPES-Na pH 7.5, 150 mM NaCl (apo), with 1 mM glycine (glycine-bound), 1 mM glutamate (glutamate-bound), or 1 mM glycine and glutamate (open)) supplemented with 1 mM phenylmethylsulphonyl fluoride (PMSF), and lysed by the high-pressure cell homogenizer (EmulsiFlex-C5, Avestin). The cell debris was removed by centrifugation at 5,000g, and the membrane fraction was collected by ultracentrifugation (40k rpm at 4°C) of the supernatant. The pelleted membrane was then solubilized by 0.5% LMNG in the purification buffer with gentle stirring for 2 hours at 4°C followed by ultracentrifugation. The supernatant was subjected to Strep-tactin Sepharose by gravity flow followed by washes with the purification buffer and the purification buffer with 3 mM Mg-ATP. The protein was eluted by the purification buffer with 3 mM desthiobiotin in the purification buffer. The eluted proteins were subjected to size-exclusion chromatography (SEC). For open state, 100 μM EU-1622–240, 1 mM glycine, and 1 mM glutamate were added to the sample. For the apo and glutamate-bound NMDARs, additional buffer wash of the corresponding purification buffers with glycine-specific antagonist 1 μM L689,560 was introduced to complete away any bound glycine. L689,560 was removed by extensive buffer wash before elution.
Single-particle cryo-EM
The purified and concentrated (4 mg/ml) proteins were vitrified on the glow-discharged UltrAufoil holey gold grids and Holey carbon grids (Quantifoil). Glow discharge took place in PELCO easiGlowTM glow discharge cleaning system (Ted Pella) for 35 sec under 15 mA. The grids were blotted using FEI Vitrobot Mark IV (ThermoFisher Scientific) at 12°C with 85% humidity with a blot time of 3–5 sec under level 7 blot force. The electron micrographs were acquired by FEI Titan Krios G3 (ThermoFisher Scientific), operating at 300 kV with the GIF quantum energy filter (Gatan Inc.) under 105k magnification. Micrographs were recorded as dose-fractionated movie frames by the K3 direct electron detector (Gatan Inc.) for apo and open NMDARs and the K2 direct electron detector (Gatan Inc.) for gly/apo and apo/glu NMDARs at electron counting mode. The applied defocus was set with a range from −1.4 to −2.8 μm. The semi-automated data acquisition was executed by EPU. Micrographs were taken at 105k magnification. The images were fractionated into 30 frames between 0.06–0.07 seconds of exposure. Total exposure times between 1.8–2.1 seconds were accumulated under the electron flux of 1.96–2.27 e/Å2/frame, yielding total doses between 58.8–68.1 e−/Å2 on the specimens. The movie alignment, CTF estimation, particle picking, 2D classification, ab-initio 3D map generation, 3D classification, and non-uniform refinement were done using the program cyoSPARC3.1.0. 46 Model fitting and building were done using UCSF Chimera 47 and COOT 48. The final models were refined against the cryo-EM maps using Phenix real space refinement 49 with secondary structure and Ramachandran restraints. The FSCs were calculated by phenix.mtriage. A summary of data collection and refinement statistics is shown in Extended Data Table 1. The presentations of the structures were illustrated by the program PyMOL2.5 (Schrödinger, LLC.).
Two-electrode voltage clamp
cRNAs encoding rat GluN1–1a or GluN1–4a and rat GluN2B were injected into defolliculated Xenopus laevis oocytes at 1:1 ratio (total of 0.3–1 ng). Some oocytes were then incubated in recovery medium (0.5X L-15 medium (Hyclone) buffered by 15 mM Na-HEPES at a final pH of 7.4), supplemented with 100 μg/ml streptomycin, and 100 U/ml penicillin at 18°C. Two-electrode voltage clamp (TEVC; Axoclamp-2B or Warner 725B/C) recordings were performed between 24 to 48 hours after injection. Crosslinking experiments used an extracellular solution containing 5 mM HEPES, 100 mM NaCl, 0.3 mM BaCl2, 10 mM Tricine at final pH 7.4 (adjusted with KOH). The current was measured using agarose-tipped microelectrode (0.4–0.9 MΩ) at the holding potential of −60 mV. Maximal response currents were evoked by 100 μM of glycine and 100 μM of L-glutamate. Data was acquired by the program PatchMaster (HEKA) or EasyOocyte (Emory) and analyzed by Origin 8 (OriginLab Corp) or custom software. Experiments to assess the potency and efficacy of EU-1622–240 and NMDAR mutations were performed in solution containing (in mM) 90 mM NaCl, 1 mM KCl, 0.5 mM BaCl2, 10 mM HEPES at pH 7.4 (adjusted with NaOH) using borosilicate microelectrodes (2–10 MΩ) filled with 0.3–3 M KCl at the holding potential of −40 mV. The channel open probability was estimated from the fold Potentiation observed in MTSEA using:
where and were the single channel chord conductance values estimated from GluN1/GluN2A receptors ( was 0.67 50) and fold was defined as the ratio of current in the presence of MTSEA to current in the absence of MTSEA. All recordings were made at room temperature.
Molecular dynamics simulation
Missing atoms on initial Cryo-EM models were fixed with MODELLER version 9.12 51 prior to simulation, with only the TMDs used to improve sampling. Umbrella sampling with the Gromacs weighted histogram analysis method (WHAM) was used to determine a one-dimensional potential of mean force (PMF). Initial configurations for each window were generated by positioning the monovalent ion in question at the center of geometry of the Cα carbons of the Asn-ring and then manually adjusting the position of said ion by 0.5 Å to 1 Å increments along the z-dimension of the channel pore perpendicular to the phospholipid bilayer, between ± 3.5 nm of this position. Each window was energy minimized using the steepest descent until converged, with protein-heavy atom restraints of 1000 kJ/mol/nm2 and a 100000 kj/mol/nm2 restraint on the monovalent PMF ion. Each window was then subjected to 2 ns of equilibration in the isothermal isobaric ensemble with umbrella harmonic restraints on. A 1000 kj/mol/nm2 umbrella restraint was applied to most windows, with a 2000 or 3000 kj/mol/nm2 restraint applied on windows occurring at steep energy barriers/saddles. For the Open state Na+ system, a total of 220 windows of 6 ns in length for a total of 1.32 μs sampling was obtained. The Open state Cl− PMF was comprised of 144 windows of 14 ns each for a total of 2.016 μs. The pre-active Na+ PMF possessed 71 windows of 18 ns each for a total of 1.278 μs of sampling, and the nonactivate1 Na+ PMF was comprised of 80 windows of 18 ns each, giving 1.44 μs sampling. The AMBER ff99sb-ildn forcefield 52 was used to model the protein with TIP3P water 53 and AMBER Slipids for POPC 54. 150 mM NaCl was added to the resulting system with additional ions to neutralize the overall system charge. Electrostatics were calculated according to Particle Mesh Ewald (PME) 55 and LINCS algorithm was used for holonomic constraints to heavy atom H-bonds 56. The time step chosen was 2 femtoseconds. The system was coupled to a heat bath via the Nosé-Hoover temperature coupling algorithm 57 at 298 K, with a TT of 0.5. Pressure was maintained at 1 bar with a 4.5e−5 isothermal compressibility using the Parrinello-Rahman algorithm 58 with semi-isotropic pressure coupling type and TP value of 1. Protein Cα atoms were restrained with a force constant of 1000 kj/mol/nm2. Simulations were performed in Gromacs version 2021.3. The inbuilt Gromacs weighted histogram analysis (WHAM) method 59 was used to calculate the PMF, and statistical uncertainty measured by using five rounds of bootstrapping. Convergence of PMFs were assessed with block analysis, whereby 2 ns increments were added to each successive WHAM calculation to determine if the overall change in energy between successive profiles were within thermal energy. Open Na+, open Cl−, pre-active Na+, and non-activate1 Na+ achieved convergence after 8 ns, 6 ns, 6 ns, and 6 ns, respectively, with the final successive 2 ns blocks within thermal energy for all systems. The results of this block analysis are included in Extended Data Fig. 3b.
Quantification and Statistical Analysis
Data processing and statistical analysis of electrophysiology data were conducted using the software Origin. ANOVA was implemented for analyses and comparison of all site-directed mutants with Dunnett’s post hoc test. The n values in these experiments represent the numbers of Xenopus oocytes from which electrophysiological experiments were conducted. The data points are represented as mean ± SEM or mean ± SD, as indicated. No method was applied to determine whether the data met the assumptions of the statistical approach. The resolution of the cryo-EM maps was estimated by the FSC = 0.143 criteria, calculated from two half maps with a soft mask. Details of data processing statistics and map quantifications are listed in Extended Data Table 1.
Extended Data
Extended Data Figure 1. Single-particle cryo-EM on glycine-, glutamate-, and EU-1622–240-bound rat GluN1–2B NMDAR.
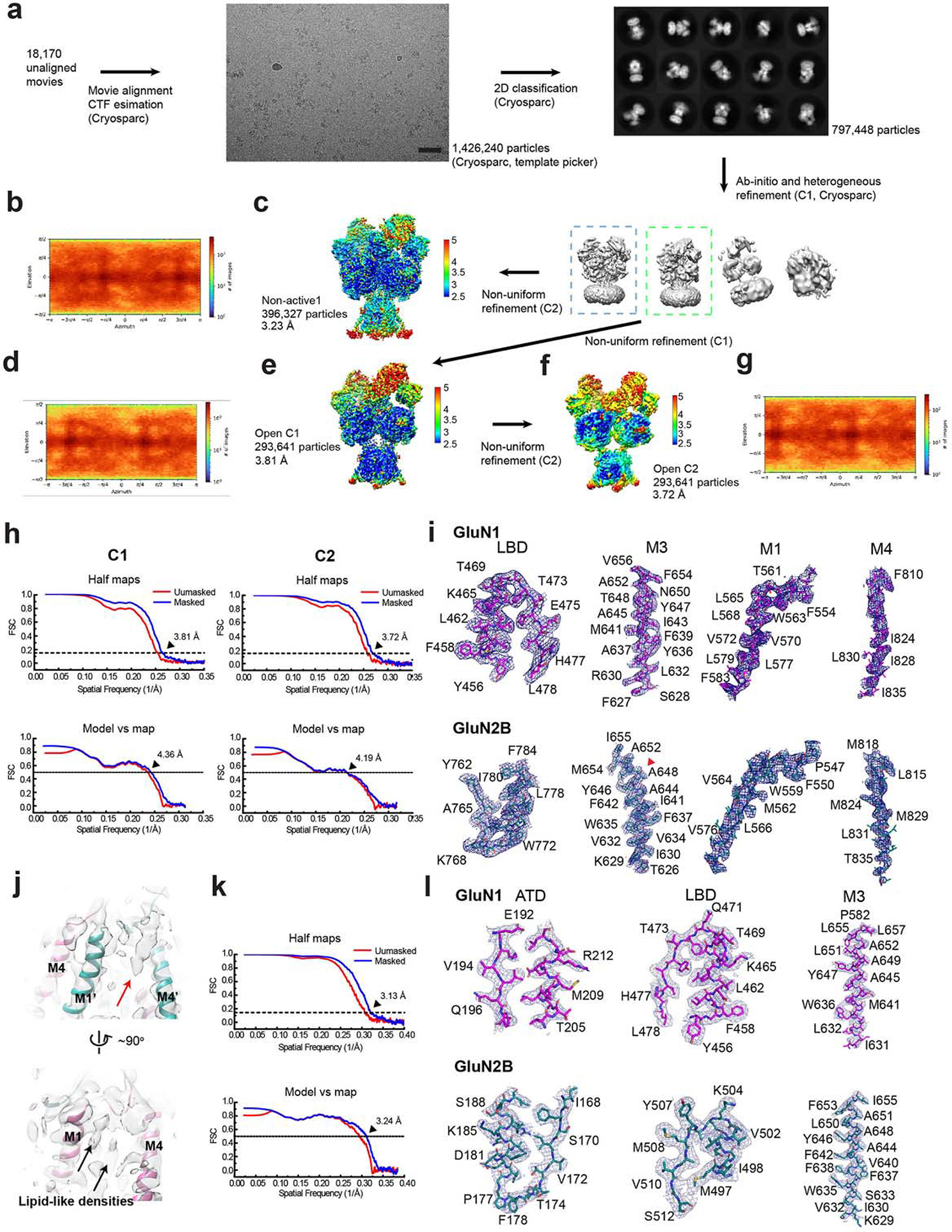
a, A representative EM micrograph, 2D classes, and the 3D classification and refinement workflow. The scale bar on the micrograph equates to 49.5 nm. b, d, and g, Orientation distribution maps of the particles used in reconstructing the final map of the non-active1 (b), open C1 (d), and open C2 (g) structures. c, e, and f, Local resolution estimation calculated by ResMap for the non-active1 (c), open C1 (e), and open C2 (f) structures. h and k, Post-processing analysis of open (h) and non-active1 (k) state structures. The masked (blue) and unmasked (red) Fourier shell correlation (FSC) curves of two half maps (top), map vs. model (bottom). i and l, Representative zoom-in views of the cryo-EM density in conserved regions for both open (i) and non-active1 (l) states fitted with molecular models. A red arrowhead indicates the starting residue of the GluN2B M3’ helix bending in the open state structure. j, A zoom-in view of the cryo-EM density of the bound EU-1622–240 compound in the open state structure (red arrow).
Extended Data Figure 2. Structural comparison of open state, pre-active, and non-active1 states.
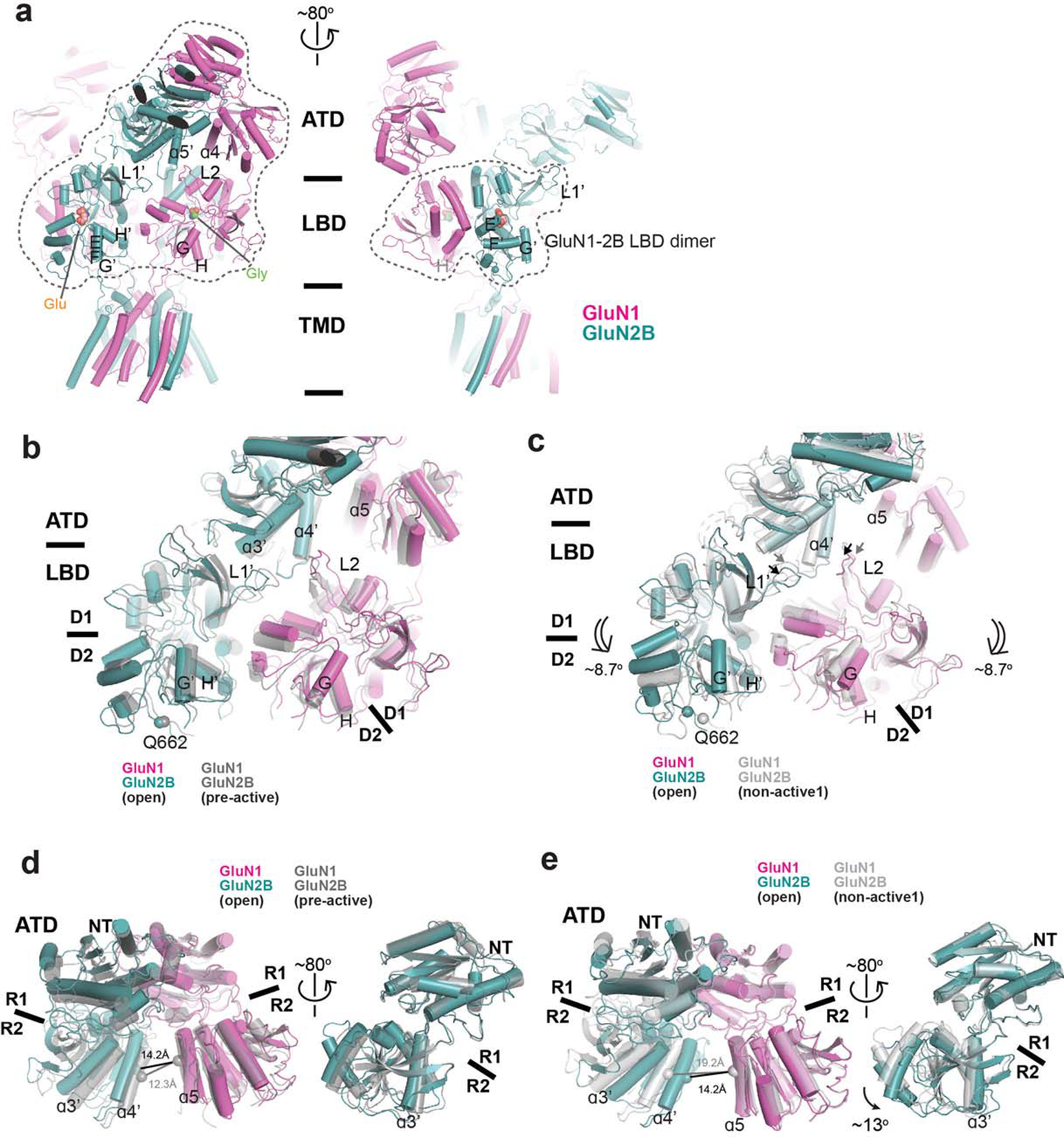
a, Cartoon representation of GluN1–2B NMDARs in the open state. Dotted lines on the left panel enclose one GluN1–2B ATD dimer and two GluN1–2B LBD dimers, whereas the ones on the right panel enclose the GluN1–2B LBD heterodimer. The color codes are as in Figure 1. b-c, Comparison of the LBD dimer arrangements and the interfaces involving GluN2B ATD, GluN2B L1’, and GluN1 L2 (arrows) between the open and pre-active (gray) states (b) and the open and non-active1 (gray) states (c). The arrangements are similar between the open and pre-active states but show divergence between the open and non-active states, especially the positionings of the L1’ and L2 due to the dimer rotation (double-line arrows). d-e, Comparison of the GluN1–2B ATD dimers and GluN2B ATD bi-lobe structures between the open and pre-active (gray) states (d) and between the open and non-active1 states (e). Open and pre-active states exhibit similar conformations, whereas substantial changes are evident between the open and non-active1 states, as highlighted by the differences in the ɑ4’-ɑ5 distances (panel e, left). GluN2B ATD bi-lobe structure is ~13° more open in the open state than the non-active1 state (e, right).
Extended Data Figure 3. PMF calculations.
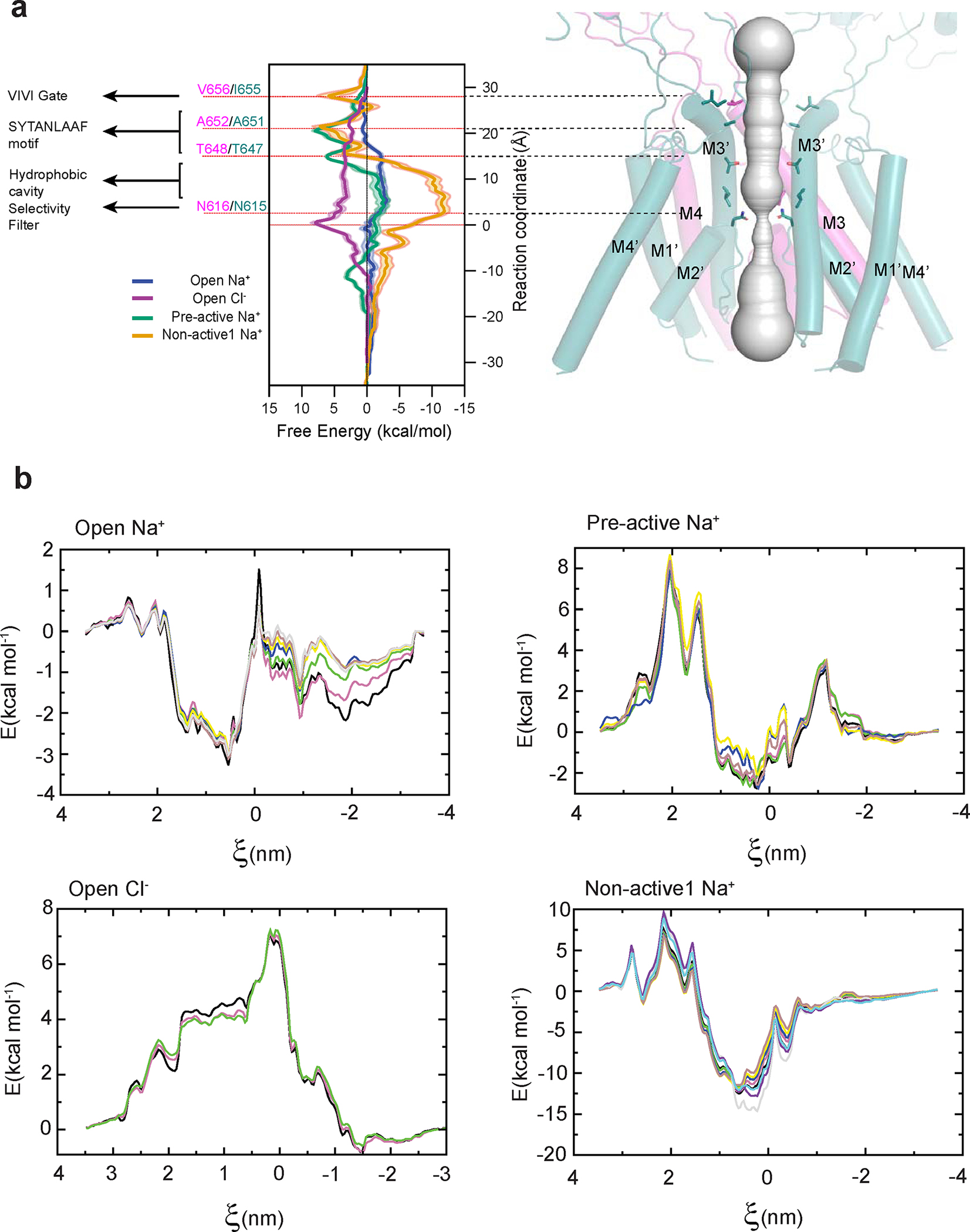
a, All-atom Potential of Mean Force (PMF) calculations for the TMD channel highlight a more favorable free energy for Na+ ions around the VIVI gate and SYTANLAAF motif in the open state (blue), as opposed to the pre-active (green) and non-active1 (red) states, consistent with the gate opening and pore dilation in the open state structure. The placement of Cl− is shown to be unfavorable, indicated by the positive free energy level (purple), consistent with the cation selectivity of the NMDAR channel. b, Block analysis of the PMF calculation. Each color represents an additional block where the PMF was rerun with two ns of additional data. The two final PMF blocks for all systems were within thermal energy, demonstrating convergence.
Extended Data Figure 4. Single-particle analysis on rat GluN1–2B NMDAR in apo/apo state.
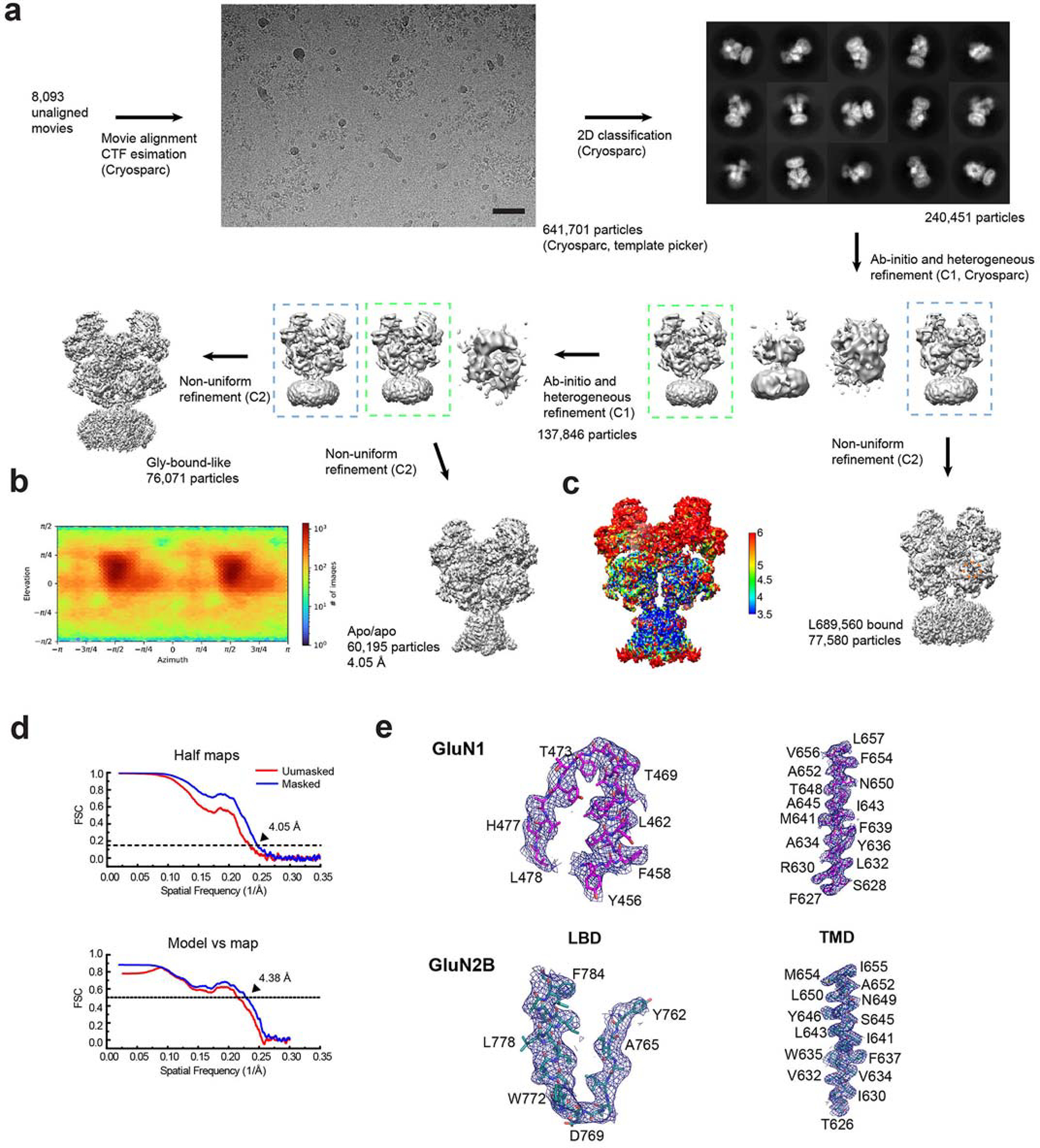
a, A representative EM micrograph, 2D classes, and the 3D classification and refinement workflow. The scale bar on the micrograph equates to 49.5 nm. b, An orientation distribution map of the particles used to reconstruct the final map. c, Local resolution estimation calculated by ResMap. d, Post-processing analysis. The masked (blue) and unmasked (red) Fourier shell correlation (FSC) curves of two half maps (top), map vs. model (bottom). e, Representative zoom-in views of the cryo-EM density in different conserved regions fitted with molecular models.
Extended Data Figure 5. Single-particle analysis on rat GluN1–2B NMDAR in gly/apo state.
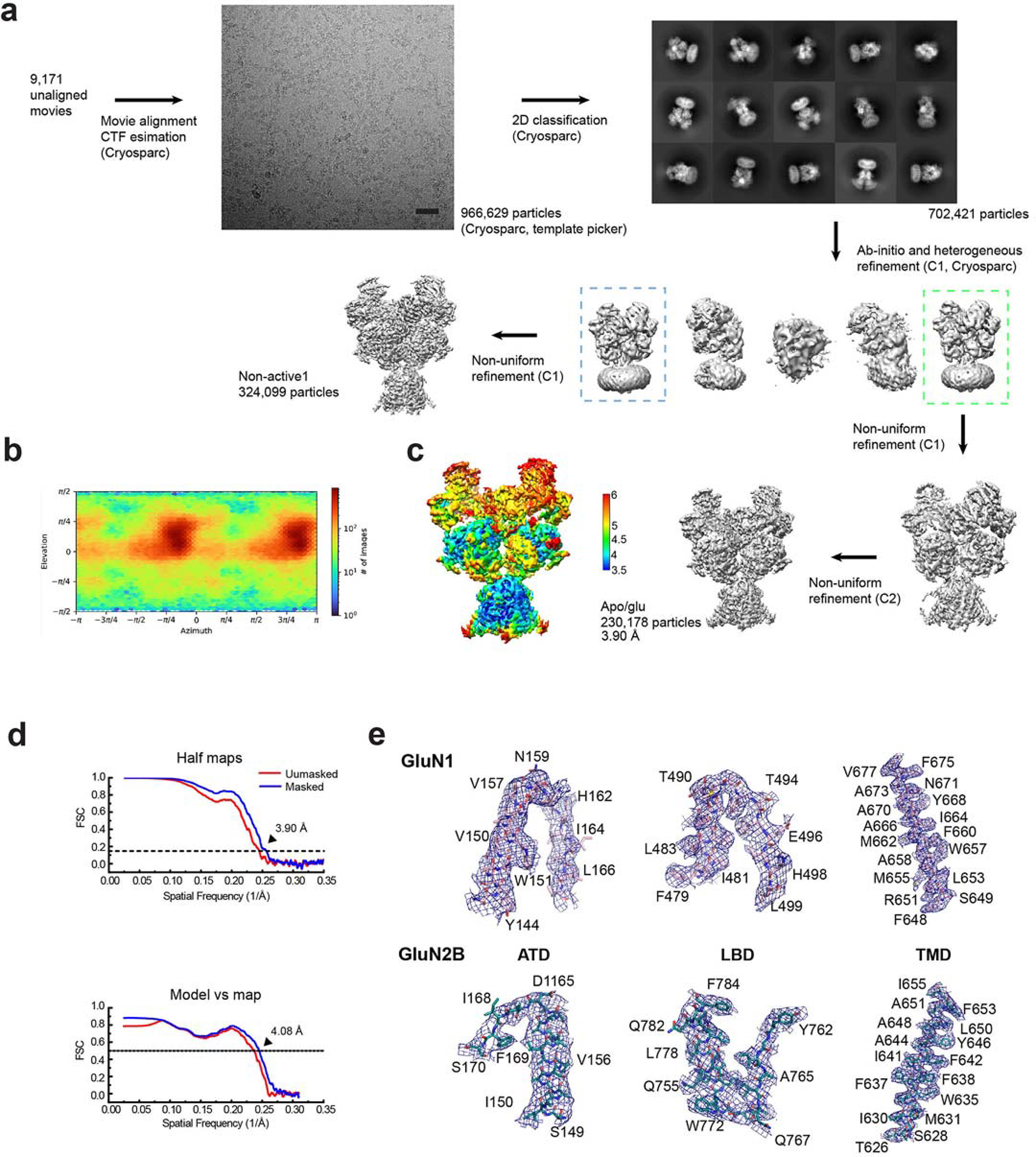
a, A representative EM micrograph, 2D classes, and the 3D classification and refinement workflow. The scale bar on the micrograph equates to 40.5 nm. b, An orientation distribution map of the particles used to reconstruct the final map. c, Local resolution estimation calculated by ResMap. d, Post-processing analysis. The masked (blue) and unmasked (red) Fourier shell correlation (FSC) curves of two half maps (top), map vs. model (bottom). e, Representative zoom-in views of the cryo-EM density in different conserved regions fitted with molecular models.
Extended Data Figure 6. Single-particle analysis on rat GluN1–2B NMDAR in apo/glu state.
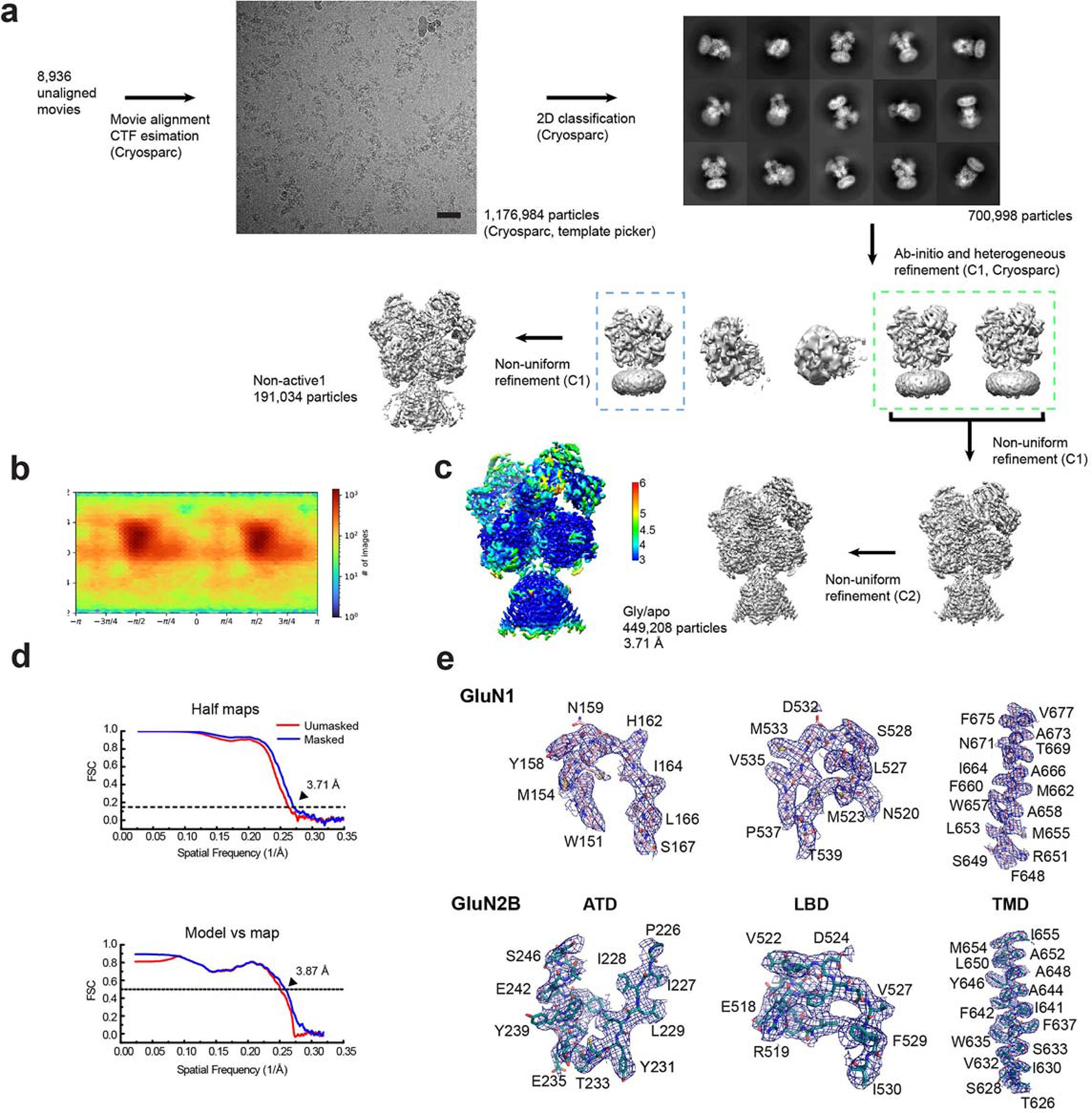
a, A representative EM micrograph, 2D classes, and the 3D classification and refinement workflow. The scale bar on the micrograph equates to 40.5 nm. b, An orientation distribution map of the particles used to reconstruct the final map. c, Local resolution estimation calculated by ResMap. d, Post-processing analysis. The masked (blue) and unmasked (red) Fourier shell correlation (FSC) curves of two half maps (top), map vs. model (bottom). e, Representative zoom-in views of the cryo-EM density in different conserved regions fitted with molecular models.
Extended Data Figure 7. Structural comparisons between apo and pre-active states.
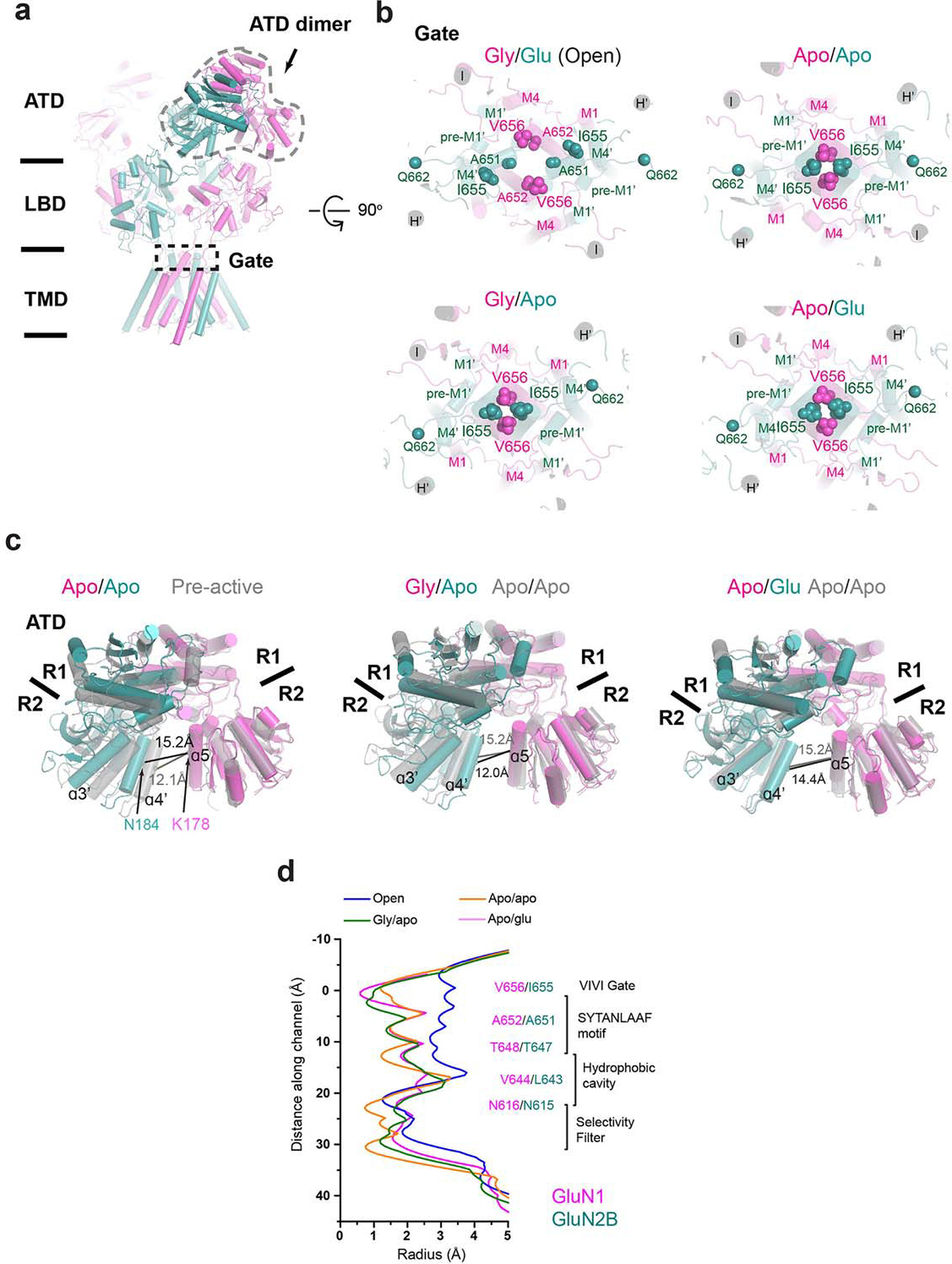
a, Cartoon representation of GluN1–2B NMDAR in the apo/apo state. The GluN1–2B ATD heterodimer and the channel gate are highlighted with dotted lines. b, A top-down view of the channel gate in the open, apo/apo, gly/apo, and apo/glu states. The gate residues are shown in spheres. c, Structural comparisons of GluN1–2B ATD heterodimers in different functional states. Distances between the GluN1 α5 and GluN2B α4’ in each state are shown for each functional state. d, Measurement of the central pore radii of the apo/apo, gly/apo, and apo/glu state structures.
Extended Data Figure 8. Structural comparisons of GluN1–2B NMDAR in gly/apo and apo/glu states with apo/apo state.
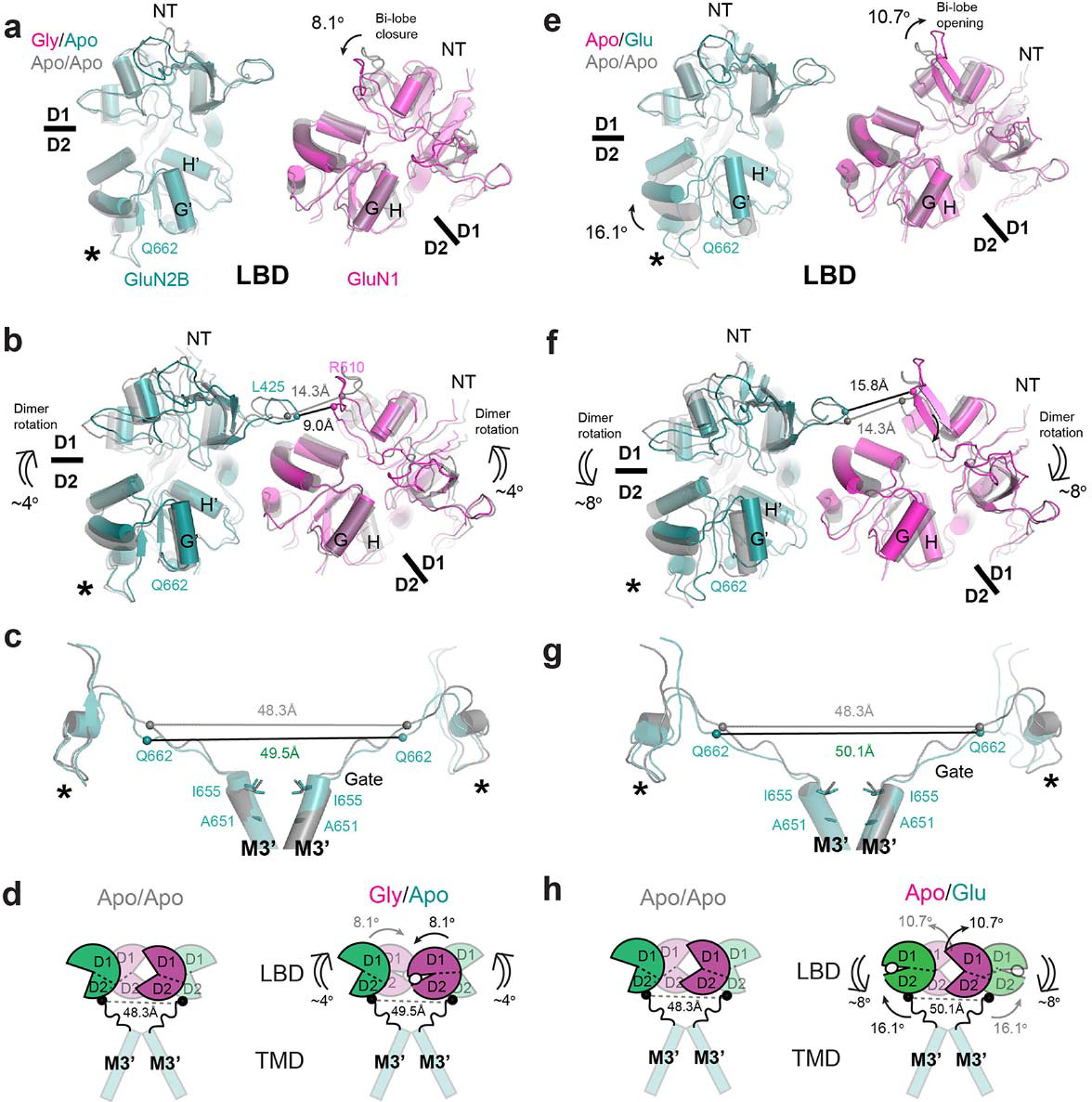
a, Superposition of the gly/apo and apo/apo structures at GluN2B D2 (lower lobe) and GluN1 D1 (upper lobe) demonstrates no change in the bi-lobe orientation for GluN2B LBD and an 8.1° domain closure for GluN1 LBD (single-line arrow) in the gly/apo state. b, The GluN1 LBD bi-lobe closure is coupled to the 4° upward rotational movement of GluN1–2B LBD dimers relative to the membrane plane from the apo/apo to gly/apo (double-line arrows). c-d, These rotational movements are insufficient to create tension in the GluN2B LBD-M3’ linker for channel gating as measured by the distance between the GluN2B Gln662 residues. e, Superposition of the apo/glu and apo/apo structures at GluN2B D2 (lower lobe) and GluN1 D1 (upper lobe) displays 16.1° closure of the GluN2B LBD bi-lobe and 10.7° opening of the GluN1 LBD bi-lobe compared to the apo/apo state. f, These LBD-bi-lobe movements are coupled to an 8° downward rotational movement relative to the membrane plane compared to the apo/apo state (double-line arrows). g-h, The GluN2B LBD-M3’ linkers in the apo/glu state do not have sufficient tension for channel gating as in the gly/apo and apo/apo states. Asterisks indicate the location of the D2 loop.
Extended Data Table 1.
Cryo-EM data collection and refinement statistics
| GluN1-GluN2B in complex with glycine, glutamate, and EU-1622-A in open conformation. C2 (EMDB-43779) (PDB 9ARE) |
GluN1-GluN2B in complex with glycine, glutamate, and EU-1622-A in open conformation. C1 (EMDB-44586) (PDB 9BIB) |
GluN1-GluN2B in complex with glycine, glutamate in non-active1 conformation (EMDB-43780) (PDB 9ARF) |
GluN1-GluN2B in apo conformation (apo/apo) (EMDB-43781) (PDB 9ARG) |
GluN1-GluN2B in complex with glycine (gly/apo) (EMDB-43782) (PDB 9ARH) |
GluN1-GluN2B in complex with glutamate (apo/glu) (EMDB-43783) (PDB 9ARI) |
|
|---|---|---|---|---|---|---|
|
| ||||||
| Data collection and processing | ||||||
| Microscope | Titan Krios | Titan Krios | Titan Krios | Titan Krios | Titan Krios | Titan Krios |
| Camera | K3/counting | K3/counting | K3/counting | K3/counting | K2/counting | K2/counting |
| Magnification | 105,000 | 105,000 | 105,000 | 105,000 | 130,000 | 130,000 |
| Energy filter | Gatan | Gatan | Gatan | Gatan | Gatan | Gatan |
| Energy filter slit width (eV) | 8 | 8 | 8 | 20 | 20 | 20 |
| Collection software | EPU | EPU | EPU | EPU | EPU | EPU |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Cumulative exposure (e−/Å2) | 66.3–68.1 | 66.3–68.1 | 66.3–68.1 | 59.7 | 63.9–65.4 | 58.8–67.2 |
| Exposure rate (e−/Å2/frame) | 2.21–2.27 | 2.21–2.27 | 2.21–2.27 | 1.99 | 2.13–2.18 | 1.96–2.24 |
| Defocus range (pm) | 1.4–2.8 | 1.4–2.8 | 1.4–2.8 | 1.4–2.8 | 1.6–2.8 | 1.6–2.8 |
| Pixel size (Å) | 0.861 | 0.861 | 0.861 | 0.856 | 1.07 | 1.07 |
| Symmetry imposed | C2 | C1 | C2 | C2 | C2 | C2 |
| Number of micrographs | 18,170 | 18,170 | 18,170 | 8,093 | 8,936 | 9,171 |
| Initial particle images (no.) | 1,426,240 | 1,426,240 | 1,426,240 | 641,701 | 1,176,984 | 966,629 |
| Final particle images (no.) | 293,641 | 293,641 | 396,327 | 60,195 | 449,208 | 230,178 |
| 0.143 FSC half map masked (Å) | 3.72 | 3.81 | 3.13 | 4.05 | 3.69 | 3.90 |
| 0.143 FSC half map unmasked(Å) | 3.88 | 3.96 | 3.42 | 4.26 | 3.80 | 4.40 |
| Refinement | ||||||
| Refinement package | Phenix | Phenix | Phenix | Phenix | Phenix | Phenix |
| Initial model used (PDB code) | 7SAA | 9ARE | 7SAA | 7SAA | 6WHR | 6WHR |
| 0.5 FSC model resolution masked (Å) | 4.04 | 4.18 | 3.24 | 4.50 | 3.85 | 4.14 |
| 0.5 FSC model resolution unmasked (Å) | 4.19 | 4.36 | 3.41 | 4.76 | 3.99 | 4.30 |
| Model resolution range (Å) | 3–6 | 3–6 | 2.5–6 | 3–6 | 3–6 | 3–6 |
| Map sharpening B factor (Å2) | 150.2 | 150.2 | 122.6 | 120.9 | 152.0 | 164.6 |
| CC (model vs map, mask) | 0.65 | 0.64 | 0.76 | 0.66 | 0.74 | 0.71 |
| Model composition | ||||||
| Non-hydrogen atoms | 19,280 | 19,327 | 23,034 | 21,008 | 22,792 | 20,756 |
| Protein residues | 3,172 | 3,172 | 3,166 | 3,166 | 3,182 | 3,178 |
| Ligands | 6 | 6 | 10 | 0 | 8 | 2 |
| Rms. deviations | ||||||
| Bond lengths (Å) | 0.007 | 0.004 | 0.010 | 0.002 | 0.013 | 0.008 |
| Bond angles (°) | 1.027 | 0.629 | 1.148 | 0.558 | 1.579 | 1.125 |
| Validation | ||||||
| MolProbity score | 2.44 | 1.81 | 2.32 | 2.00 | 2.97 | 2.79 |
| Clashscore | 10.67 | 3.25 | 8.31 | 3.93 | 18.66 | 12.43 |
| Poor rotamers (%) | 5.24 | 4.65 | 5.07 | 4.82 | 3.48 | 9.27 |
| Ramachandran plot Favored (%) | 95.07 | 96.91 | 95.32 | 95.67 | 92.26 | 92.95 |
| Allowed (%) | 4.80 | 2.99 | 4.56 | 4.33 | 7.36 | 6.80 |
| Disallowed (%) | 0.13 | 0.10 | 0.12 | 0.00 | 0.38 | 0.25 |
| C-beta deviations (%) | 1.22 | 0.41 | 1.42 | 0.00 | 2.49 | 1.48 |
| CaBLAM outliers (%) | 3.12 | 3.99 | 3.95 | 3.21 | 4.87 | 5.25 |
Supplementary Material
Acknowledgment
We thank N. Simorowski for technical support. D. Thomas and M. Wang are thanked for managing the cryo-EM facility and computing facility at Cold Spring Harbor Laboratory, respectively. A. Sobolevsky is thanked for critical comments on this work. This work was funded by the NIH (NS111745 and MH085926 to H.F and NS111619 to S.F.T), Austin’s purpose (to H.F and S.F.T), Robertson funds at CSHL, Doug Fox Alzheimer’s fund, Heartfelt Wing Alzheimer’s fund, and the Gertrude and Louis Feil Family Trust (to H.F). The computational work was performed with assistance from an NIH grant (S10OD028632-01)
Footnotes
Conflict of Interest
SFT is a member of the medical advisory boards for the CureGRIN Foundation and the GRIN2B Foundation, is a member of the scientific advisory boards for Sage Therapeutics, Eumentis Therapeutics, and Neurocrine, is a Senior Advisor for GRIN Therapeutics, is co-founder of NeurOp, Inc and AgriThera, Inc., and is on the Board of Directors for NeurOp Inc. D.C.L. is on the Board of Directors for NeurOp Inc. Multiple authors are co-inventors on Emory-owned IP involving NMDA receptor modulators (RGF, NSA, SP, SFT, DCL). The remaining authors declare no competing interests.
Data Availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes, EMD-43779 (GluN1–2B, EU-1622–240 bound, open conformation, C2 symmetry), EMD-44586 (GluN1–2B, EU-1622–240 bound, open conformation, C1 symmetry), EMD-43780 (GluN1–2B, non-active1 conformation), EMD-43781 (GluN1–2B, apo conformation), EMD-43782 (GluN1–2B, glycine/apo), EMD-43783 (GluN1–2B, apo/glutamate). The structural coordinates have been deposited in the RCSB Protein Data Bank (PDB) under accession codes, 9ARE(GluN1–2B, EU-1622–240 bound, open conformation, C2 symmetry), 9BIB (GluN1–2B, EU-1622–240 bound, open conformation, C1 symmetry), 9ARF (GluN1–2B, non-active1 conformation), 9ARG (GluN1–2B, apo conformation), 9ARH (GluN1–2B, glycine/apo), 9ARI (GluN1–2B, apo/glutamate). The structure of pre-active GluN1–2B NMDAR is available in the PDB under the accession code 6WI1. The structural coordinates of pre-active, open, and desensitized GluA2 AMPAR are available in the PDB under the accession codes 4U5C, 5WEO, and 7RZA, respectively.
References
- 1.Hansen KB et al. Structure, Function, and Pharmacology of Glutamate Receptor Ion Channels. Pharmacol Rev 73, 298–487, doi: 10.1124/pharmrev.120.000131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mony L & Paoletti P Mechanisms of NMDA receptor regulation. Curr Opin Neurobiol 83, 102815, doi: 10.1016/j.conb.2023.102815 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Zhou C & Tajima N Structural insights into NMDA receptor pharmacology. Biochem Soc Trans 51, 1713–1731, doi: 10.1042/BST20230122 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer ML, Westbrook GL & Guthrie PB Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Nowak L, Bregestovski P, Ascher P, Herbet A & Prochiantz A Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307, 462–465, doi: 10.1038/307462a0 (1984). [DOI] [PubMed] [Google Scholar]
- 6.Jahr CE & Stevens CF Glutamate activates multiple single channel conductances in hippocampal neurons. Nature 325, 522–525, doi: 10.1038/325522a0 (1987). [DOI] [PubMed] [Google Scholar]
- 7.MacDermott AB, Mayer ML, Westbrook GL, Smith SJ & Barker JL NMDA-receptor activation increases cytoplasmic calcium concentration in cultured spinal cord neurones. Nature 321, 519–522, doi: 10.1038/321519a0 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Kauer JA, Malenka RC & Nicoll RA NMDA application potentiates synaptic transmission in the hippocampus. Nature 334, 250–252, doi: 10.1038/334250a0 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Karakas E & Furukawa H Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997, doi: 10.1126/science.1251915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CH et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511, 191–197, doi: 10.1038/nature13548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalski K & Furukawa H Structure and function of GluN1–3A NMDA receptor excitatory glycine receptor channel. Sci Adv 10, eadl5952, doi: 10.1126/sciadv.adl5952 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajima N et al. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68, doi: 10.1038/nature17679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jalali-Yazdi F, Chowdhury S, Yoshioka C & Gouaux E Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor. Cell 175, 1520–1532 e1515, doi: 10.1016/j.cell.2018.10.043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, Du J, Goehring A & Gouaux E Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355, doi: 10.1126/science.aal3729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S et al. Mechanism of NMDA Receptor Inhibition and Activation. Cell 165, 704–714, doi: 10.1016/j.cell.2016.03.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou TH, Tajima N, Romero-Hernandez A & Furukawa H Structural Basis of Functional Transitions in Mammalian NMDA Receptors. Cell 182, 357–371 e313, doi: 10.1016/j.cell.2020.05.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou TH et al. Structural insights into binding of therapeutic channel blockers in NMDA receptors. Nat Struct Mol Biol 29, 507–518, doi: 10.1038/s41594-022-00772-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y et al. Structural basis of ketamine action on human NMDA receptors. Nature, doi: 10.1038/s41586-021-03769-9 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Song X et al. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 556, 515–519, doi: 10.1038/s41586-018-0039-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou T-H, Kang H, Simorowski N, Traynelis SF & Furukawa H Structural insights into assembly and function of GluN1–2C, GluN1–2A-2C, and GluN1–2D NMDARs. Molecular Cell 82, 4548–4563.e4544, doi: 10.1016/j.molcel.2022.10.008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa H, Singh SK, Mancusso R & Gouaux E Subunit arrangement and function in NMDA receptors. Nature 438, 185–192, doi: 10.1038/nature04089 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC & Furukawa H Structural insights into competitive antagonism in NMDA receptors. Neuron 81, 366–378, doi: 10.1016/j.neuron.2013.11.033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa H & Gouaux E Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. The EMBO journal 22, 2873–2885, doi: 10.1093/emboj/cdg303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esmenjaud JB et al. An inter-dimer allosteric switch controls NMDA receptor activity. EMBO J, doi: 10.15252/embj.201899894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karakas E, Simorowski N & Furukawa H Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475, 249–253, doi: 10.1038/nature10180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karakas E, Simorowski N & Furukawa H Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. The EMBO journal 28, 3910–3920, doi: 10.1038/emboj.2009.338 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan MC et al. Structural elements of a pH-sensitive inhibitor binding site in NMDA receptors. Nat Commun 10, 321, doi: 10.1038/s41467-019-08291-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroebel D et al. A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol Pharmacol 89, 541–551, doi: 10.1124/mol.115.103036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazi R, Dai J, Sweeney C, Zhou HX & Wollmuth LP Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nature neuroscience 17, 914–922, doi: 10.1038/nn.3724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tajima N et al. Development and characterization of functional antibodies targeting NMDA receptors. Nat Commun 13, 923, doi: 10.1038/s41467-022-28559-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero-Hernandez A, Simorowski N, Karakas E & Furukawa H Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain. Neuron 92, 1324–1336, doi: 10.1016/j.neuron.2016.11.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin JB et al. Two gates mediate NMDA receptor activity and are under subunit-specific regulation. Nat Commun 14, 1623, doi: 10.1038/s41467-023-37260-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones KS, VanDongen HM & VanDongen AM The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci 22, 2044–2053 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature 549, 60–65, doi: 10.1038/nature23479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S et al. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246 e1214, doi: 10.1016/j.cell.2017.07.045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herguedas B et al. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science 364, doi: 10.1126/science.aav9011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin R et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. The EMBO journal 28, 1812–1823, doi: 10.1038/emboj.2009.140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y et al. Mechanism of glutamate receptor desensitization. Nature 417, 245–253, doi: 10.1038/417245a (2002). [DOI] [PubMed] [Google Scholar]
- 39.Mothet JP et al. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc Natl Acad Sci U S A 97, 4926–4931, doi: 10.1073/pnas.97.9.4926 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribeiro CS, Reis M, Panizzutti R, de Miranda J & Wolosker H Glial transport of the neuromodulator D-serine. Brain Res 929, 202–209, doi: 10.1016/s0006-8993(01)03390-x (2002). [DOI] [PubMed] [Google Scholar]
- 41.Fukasawa Y et al. Identification and characterization of a Na(+)-independent neutral amino acid transporter that associates with the 4F2 heavy chain and exhibits substrate selectivity for small neutral D- and L-amino acids. J Biol Chem 275, 9690–9698, doi: 10.1074/jbc.275.13.9690 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Hill MD et al. SAGE-718: A First-in-Class N-Methyl-d-Aspartate Receptor Positive Allosteric Modulator for the Potential Treatment of Cognitive Impairment. J Med Chem 65, 9063–9075, doi: 10.1021/acs.jmedchem.2c00313 (2022). [DOI] [PubMed] [Google Scholar]
Method References
- 43.Hanson JE et al. Therapeutic potential of N-methyl-D-aspartate receptor modulators in psychiatry. Neuropsychopharmacology 49, 51–66, doi: 10.1038/s41386-023-01614-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furukawa H, Simorowski N & Michalski K Effective production of oligomeric membrane proteins by EarlyBac-insect cell system. Methods Enzymol 653, 3–19, doi: 10.1016/bs.mie.2020.12.019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regan MC et al. Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron 98, 521–529 e523, doi: 10.1016/j.neuron.2018.03.034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Pettersen EF et al. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan H, Erreger K, Dravid SM & Traynelis SF Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J Biol Chem 280, 29708–29716, doi: 10.1074/jbc.M414215200 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Eswar N et al. Comparative Protein Structure Modeling Using Modeller. Current Protocols in Bioinformatics 15, doi: 10.1002/0471250953.bi0506s15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindorff-Larsen K et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics 78, 1950–1958, doi: 10.1002/prot.22711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 79, 926–935, doi: 10.1063/1.445869 (1983). [DOI] [Google Scholar]
- 54.Jämbeck JPM & Lyubartsev AP Derivation and Systematic Validation of a Refined All-Atom Force Field for Phosphatidylcholine Lipids. The Journal of Physical Chemistry B 116, 3164–3179, doi: 10.1021/jp212503e (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darden T, York D & Pedersen L Particle mesh Ewald: An Nꞏlog(N) method for Ewald sums in large systems. The Journal of Chemical Physics 98, 10089–10092, doi: 10.1063/1.464397 (1993). [DOI] [Google Scholar]
- 56.Hess B, Bekker H, Berendsen HJC & Fraaije JGEM LINCS: A Linear Constraint Solver for molecular simulations. Journal of Computational Chemistry 18, 1463–1472, doi: (1997). [DOI] [Google Scholar]
- 57.Nosé S A unified formulation of the constant temperature molecular dynamics methods. The Journal of Chemical Physics 81, 511–519, doi: 10.1063/1.447334 (1984). [DOI] [Google Scholar]
- 58.Parrinello M & Rahman A Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 52, 7182–7190, doi: 10.1063/1.328693 (1981). [DOI] [Google Scholar]
- 59.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH & Kollman PA THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. Journal of Computational Chemistry 13, 1011–1021, doi: 10.1002/jcc.540130812 (1992). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes, EMD-43779 (GluN1–2B, EU-1622–240 bound, open conformation, C2 symmetry), EMD-44586 (GluN1–2B, EU-1622–240 bound, open conformation, C1 symmetry), EMD-43780 (GluN1–2B, non-active1 conformation), EMD-43781 (GluN1–2B, apo conformation), EMD-43782 (GluN1–2B, glycine/apo), EMD-43783 (GluN1–2B, apo/glutamate). The structural coordinates have been deposited in the RCSB Protein Data Bank (PDB) under accession codes, 9ARE(GluN1–2B, EU-1622–240 bound, open conformation, C2 symmetry), 9BIB (GluN1–2B, EU-1622–240 bound, open conformation, C1 symmetry), 9ARF (GluN1–2B, non-active1 conformation), 9ARG (GluN1–2B, apo conformation), 9ARH (GluN1–2B, glycine/apo), 9ARI (GluN1–2B, apo/glutamate). The structure of pre-active GluN1–2B NMDAR is available in the PDB under the accession code 6WI1. The structural coordinates of pre-active, open, and desensitized GluA2 AMPAR are available in the PDB under the accession codes 4U5C, 5WEO, and 7RZA, respectively.


