ABSTRACT
The intestine is the largest organ in terms of surface area in the human body. It is responsible not only for absorbing nutrients but also for protection against the external world. The gut microbiota is essential in maintaining a properly functioning intestinal barrier, primarily through producing its metabolites: short-chain fatty acids, bile acids, and tryptophan derivatives. Ethanol overconsumption poses a significant threat to intestinal health. Not only does it damage the intestinal epithelium, but, maybe foremostly, it changes the gut microbiome. Those ethanol-driven changes shift its metabolome, depriving the host of the protective effect the physiological gut microbiota has. This literature review discusses the impact of ethanol consumption on the gut, the gut microbiota, and its metabolome, providing a comprehensive overview of the mechanisms through which ethanol disrupts intestinal homeostasis and discussing potential avenues for new therapeutic intervention.
KEYWORDS: Gut microbiome, gut microbiota, alcohol, ethanol, metabolome, short-chain fatty acids, bile acids, tryptophan derivatives, gut, intestine
Introduction
The intestines are the largest, surface-wise, barrier organ in humans. The intestinal barrier is a complex structure that acts as a selectively permeable shield responsible for absorbing nutrients while safeguarding against harmful external factors. It consists of several layers: the gut microbiota, the unstirred water layer, glycocalyx and the mucosal layer (containing antimicrobial products from both Paneth cells and enterocytes), and epithelial cells separated by junctions built by tight-junction proteins.1,2
The gut microbiota, encompassing diverse bacteria and microorganisms within the gastrointestinal tract, is integral in maintaining host health. Recognition of its significance is steadily increasing, as evidenced by a growing number of dedicated studies.3 It is fundamental for maintaining the proper function of the intestinal barrier, aiding in nutrient digestion and protecting against pathogens. This paper will focus on the bacteria that colonize the digestive tract, and for simplicity, we will refer to them as gut microbiota from this point on. The gut microbiota generates various metabolites that greatly influence the host’s metabolism. These include short-chain fatty acids (SCFAs), branch-chained amino acids (BCAAs), tryptophan metabolites, bile acids (BAs), trimethylamine N-oxide (TMAO) and many more. Physiologically, Firmicutes and Bacteroidetes phyla constitute over 90% of the total bacterial population. A shift in the number and type of species that form the gut microbiota may facilitate the onset of various diseases, promote malnutrition, and induce inflammatory responses.
Ethanol abuse is widely recognized as a significant public health concern. In 2016, ethanol use was identified as the seventh leading risk factor for deaths and disability-adjusted life-years globally.4 Its detrimental effects extend well beyond liver damage and addiction.5 Not only does it damage the intestinal barrier, but emerging research has unveiled the profound impact of ethanol consumption on the gut microbiome.6,7 Numerous studies have revealed that prolonged and excessive ethanol consumption can significantly alter the composition and diversity of the gut microbiota. These changes can impair its physiological function, potentially leading to various clinical consequences. Understanding the relationship between ethanol and the function of the gut microbiome may offer new therapeutic strategies in ethanol-related diseases of the gastrointestinal tract.
In this review, we will present ethanol’s detrimental effect on the gut. First, we will discuss the effects of its inherent toxicity on the intestines. Then, we will explore its impact on the gut microbiota and its metabolites, which appear to be crucial in maintaining gut barrier function and intestinal balance.
Main text
Toxic effect of ethanol on the intestine and the gut barrier
Most of the ingested ethanol is absorbed in the small intestine through diffusion.8,9 Ethanol has a direct toxic effect on the mucosal epithelium.10 It is known to disrupt cell membranes, increasing their fluidity and, thus, permeability.11 In experimental animals such as rodents and dogs, administration of ethanol at concentrations corresponding to those of commonly available alcoholic beverages leads to mucosal damage in the small intestine. This damage extends to the loss of epithelium at the tips of the villi, hemorrhagic erosions, and hemorrhage in the lamina propria.12 Barona et al. observed concentration-dependent morphological changes in the duodenum and jejunum of rats fed with ethanol, particularly evident at the villi tips where significant cell loss occurred.13 Broitman and Hoyumpa found similar changes in humans displaying villi damage in intestinal biopsies following ethanol consumption.8,14 The changes included reduced villus height, reduced mucosal surface area of villi, increases in the number of intra-epithelial mononuclear cells, goblet cell hyperplasia, and gastric metaplasia.15 Ethanol also increases the secretion of mucins by goblet cells and lowers the expression of heat shock proteins in a dose-dependent manner in murine models.16,17
Ethanol damages intestinal mucosa in a dose-dependent manner, compromising its barrier function by dismantling the microtubule skeleton and inducing oxidative stress.18 Recent experiments have suggested that the initial event of gut response to alcohol is an influx of leukocytes accompanied by enhanced release of toxic mediators such as reactive oxygen species, leukotrienes, and histamine by mast cells.19–21 However, duodenal or jejunal biopsies of actively drinking alcoholics taken after a few days of abstinence usually show no mucosal infiltrates of leukocytes or macrophages on routine histology or quantitative analysis compared to recently drinking alcohol-abusing subjects or controls.22,23
In the study by Palatino et al., ethanol increased the immunoreactivity of IL-6, MMP-9, and NF-κB in the jejunum.24 MMP-9, IL-6, and TNF-α are mediators playing a crucial role in the progression of inflammation and increase of intestinal permeability in patients with inflammatory bowel diseases.25,26
Ethanol lowers the expression of tight junction proteins, such as zonula occludens-1 and claudin-1, in the Caco-2 intestinal cell in vitro model.27 Ma et al. have shown that ethanol in Caco-2 monolayers can reversibly disrupt the intestinal epithelial tight junction integrity through myosin light chain kinase activation and subsequent modulation of perijunctional actin and myosin filaments.28 Furthermore, incubation of Caco-2 cells with ethanol for 24 hours has been shown to induce nuclear factor-kB activation, resulting in F-actin cytoskeleton instability and intestinal barrier dysfunction.29
Significantly increased intestinal permeability in actively drinking alcoholics has been confirmed for both micro- and macromolecules.30,31 It is supported by the observation of transient endotoxemia following acute alcohol consumption in healthy volunteers and in alcoholics with fatty liver.32 Chronic alcohol consumption in mice increases intestinal permeability and causes steatohepatitis, with the colon being the primary site of increased gut leakiness. While small intestinal permeability was unaffected, whole gut permeability was elevated, indicating colonic hyperpermeability as the main factor in alcohol-induced gut leakiness.33 In a study on healthy adults, acute binge drinking increased serum endotoxin and pro-inflammatory cytokine levels, proving that ethanol compromises the intestinal barrier, with this effect being more pronounced in women.34
Ethanol abuse induces alterations of the matrix network and increases the number of myofibroblast-like cells in the duodenal mucosa, findings compatible with the development of fibrosis of the intestinal mucosa.35
In a murine model of alcohol liver disease, ethanol feeding caused intestinal hyperpermeability represented by high concentrations of plasma LPS, though plasma LPS was significantly lower in Muc2(-/-) mice. Hartmann et al. postulated that the increased mucin secretion creates an environment favorable to microbial overgrowth that ultimately leads to liver injury.36 Germ-free mice fed with ethanol for seven days demonstrated only moderately higher intestinal permeability than control. In the same study, conventional mice had significantly higher intestinal permeability when exposed to the same level of ethanol consumption. This indicates that ethanol-induced alterations in the gut microbiome are the primary cause of increased intestinal permeability. Transferring these ethanol-modified gut microbes to germ-free mice resulted in high intestinal permeability and inflammation in the liver and intestines.37
We summarize the toxic effect of ethanol on intestine in Figure 1.
Figure 1.
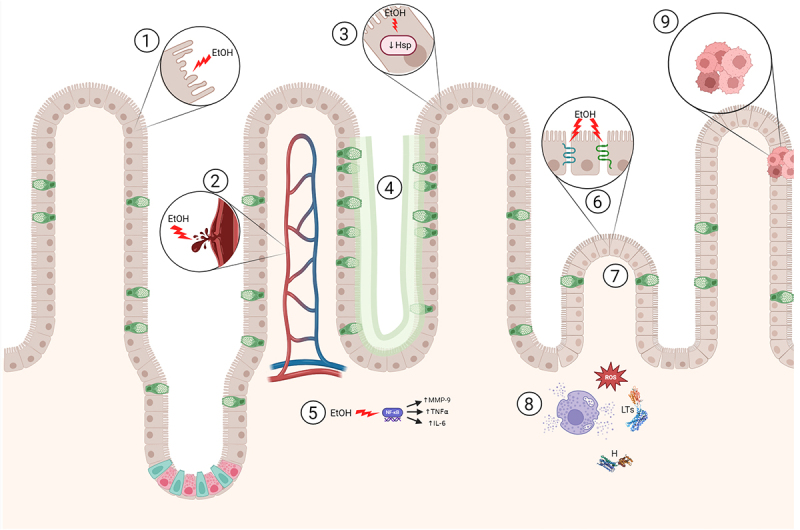
Detrimental effect of ethanol on intestine.
In this figure, we present the detrimental effect ethanol has on the intestinal tissue: 1) toxic damage of the intestinal cells, 2) hemorrhagic erosions and hemorrhage in the lamina propria, 3) lower expression of heat shock proteins (HSP), 4) increase in goblet cells and mucus production, 5) increased expression of nuclear factor kappa B (NF-κβ) and thus proinflammatory interleukin-6 (IL-6), matrix metalloproteinase-9 (MMP-9) and tumor necrosis factor α (TNF α), 6) decreased tight junction protein expression, 7) reduction of the height and surface of the villi, 8) influx of leukocytes which lead to release of toxic reactive oxygen species (ROS), leukotrienes (LTs) and histamine (H) by mast cells, 9) metaplasia.
Created with BioRender.com
Impact of ethanol on the gut microbiome
The difference in the gut microbiome between the population overusing ethanol and healthy controls has been shown in several studies. Only one systematic review was published by Litwinowicz et al.38 Changes in the gut microbiome observed in reviewed studies are presented in Table 1. Generally, there is a relatively higher abundance of Proteobacteria phylum in comparison to Firmicutes. Furthermore, there is a depletion of several bacterial genera. Almost all depleted genera are involved in producing SCFA, BCAAs, BAs and tryptophan derivatives. On the species level, the relative abundance of Akkermansia municiphila and Faecalibacterium prausnitzii was consistently lower in the population with alcohol use disorder.
Table 1.
Changes in the gut microbiota due to ethanol consumption based on the systematic review by Litwinowicz et al.
| Abundance | SCFA | BAs | Trp | BCAAs | TMA | Ref. | |
|---|---|---|---|---|---|---|---|
| PHYLUM | |||||||
| Proteobacteria | ↑ | – | – | – | – | + | 39,40 |
| Bacteroidetes | ↓ | + | + | + | – | + | 40 |
| FAMILY | |||||||
| Enterobacteriaceae | – | – | – | – | – | – | |
| Ruminococcaceae | ↑ | – | + | + | – | – | |
| GENUS | |||||||
| Akkermansia | ↓ | + | – | + | – | – | 41–43 |
| Alistipes | ↓ | + | – | + | – | – | 44 |
| Bacteroides | ↓ | + | + | + | + | – | 45–49 |
| Clostridium | ↓ | + | + | + | – | + | 40,45,50–52 |
| Collinsella | ↓ | + | – | – | – | – | 53 |
| Faecalibacterium | ↓ | + | – | – | + | – | 54–56 |
| Parabacteroides | ↓ | + | – | – | – | – | 57 |
| Paraprevotella | ↓ | – | – | – | – | – | |
| Prevotella | ↓ | + | – | – | + | – | 58,59 |
| Ruminococcus | ↓ | + | + | + | + | + | 60–66 |
A plus sign in the table indicates that the given bacteria are involved in the metabolism of the specified metabolites (in the case of bile acids, in the conversion of primary bile acids to secondary bile acids). References indicate that the bacteria in question are involved in the metabolism of the specified metabolites.
(SCFA – short-chain fatty acids, BAs – bile acids, Trp – tryptophan derivatives, BCAAs – branch-chained amino acids, TMA - trimethylamine).
Proteobacteria
Proteobacteria is currently the largest bacterial phylum. It is characterized by Gram-negative staining. Many recognized human pathogens, such as Helicobacter, Escherichia, Shigella, Salmonella and Yersinia, belong to this phylum. Researchers postulate that the higher abundance of Proteobacteria might be a marker of intestinal dysbiosis. It is also associated with metabolic disorders, such as obesity and type 2 diabetes mellitus.39
Higher Proteobacteria prevalence promotes pro-inflammatory conditions. In mice that were genetically modified to be susceptible to inflammation, Carvalho et al. observed higher levels of Proteobacteria, which seemed to trigger chronic colitis.67 IL-10− mice had a higher abundance of Proteobacteria and an onset and progression of chronic colonic inflammation.68 The excessive presence of Proteobacteria phylum characterizes inflammatory bowel disease patients and positively correlates with the severity of the inflammation.69,70 Researchers also observed the growth of the Proteobacteria population with aging accompanied by increased intestinal permeability and chronic inflammation.71 Low-grade chronic inflammation correlates with the development of age-related conditions and increased mortality.72
Akkermansia municiphila
Ethanol abuse leads to the depletion of Akkermansia municiphila. It is a mucin-degrading member of the Verrucomicrobium phylum, which can comprise up to 3% of the gut bacteria detected in human fecal samples.73 It produces SCFAs and lowers endotoxemia by maintaining intestinal barrier integrity.74,75 Extremely long-living people (i.e. over 100 years old) have a relatively higher abundance of Akkermansia.76 Its lower abundance is being linked with various metabolic diseases, such as obesity, type 2 diabetes mellitus and non-alcohol fatty liver disease. Many consider Akkermansia municiphila as a next-generation probiotic. Its supplementation was effective in the treatment of diet-induced obesity. It improved postprandial blood glucose, insulin sensitivity and total serum cholesterol. Furthermore, it seems to have a protective effect from cardiovascular diseases.77,78
Faecalibacterium prausnitzii
Faecalibacterium prausnitzii, a member of the Clostridium spp, is also depleted by excessive ethanol consumption. It is one of the most abundant butyrate-producing bacteria present in human fecal samples.79 It colonizes the mucus layer and plays a vital role in preserving the intestinal barrier.80,81 Faecalibacterium prausnitzii is often proposed as a marker of intestinal health. Patients with irritable bowel syndrome have a lower abundance of Faecalibacterium genus (including Faecalibacterium prausnitzii).82 In both in vitro and murine models of colitis, it exerts anti-inflammatory effects. Furthermore, a preoperative lower relative abundance of Faecalibacterium prausnitzii was associated with a higher risk of postoperative recurrence of ileal Crohn’s disease.55 Llopis et al. postulated that Faecalibacterium prausnitzii was a key species associated with protection from alcohol hepatitis in a murine model.83 Lower relative Faecalibacterium prausnitzii abundance was also found in patients with colorectal cancer.84
The gut microbiome metabolome and its response to ethanol consumption
Short-chain fatty acids
SCFAs are a group of fatty acids with a low number of carbon atoms predominantly produced in the colon through the bacterial fermentation of dietary fiber and other non-digestible carbohydrates.85,86 Generally, the Bacteroidetes phylum produces acetate and propionate, while the Firmicutes phylum produces butyrate.45 The majority of SCFAs is absorbed in the cecum and large intestine, with the remaining portion excreted in feces.87 In the following paragraphs, we will explore the diverse roles of SCFAs in intestinal health (Figure 2).
Figure 2.
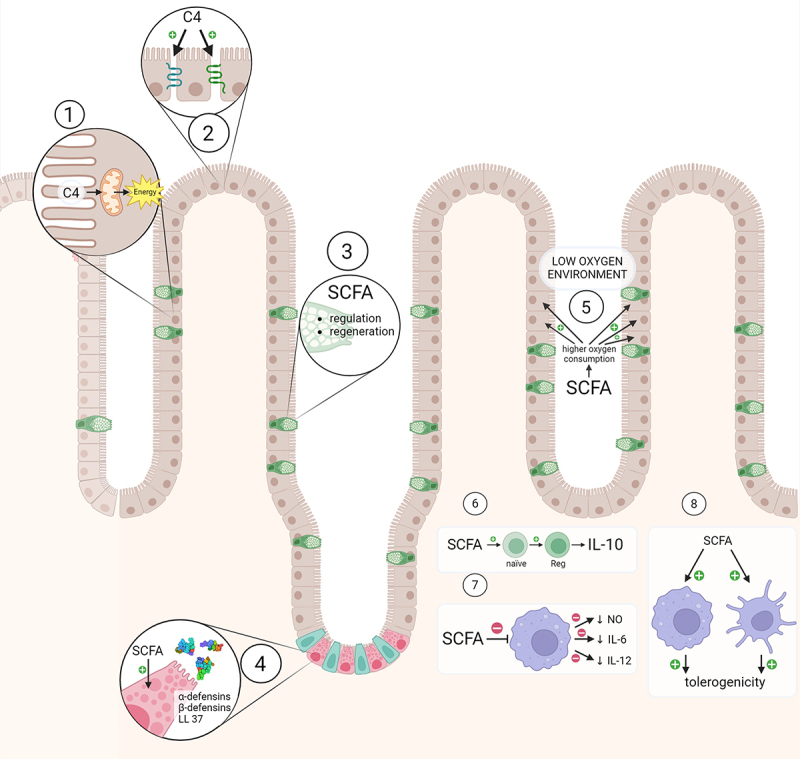
The impact of SCFAs on intestinal health.
SCFAs, particularly butyrate, serve as the primary energy source for colonocytes, fulfilling around 90% of their energy needs, and contribute approximately 5–10% of the total daily energy requirements in humans.88,89 Butyrate contributes to the production of tight-junction proteins, which play a critical role in maintaining the integrity of cell-to-cell junctions and regulating paracellular transport.90,91
The mucus forms a protective physical barrier that prevents harmful microorganisms and substances from reaching the epithelial surface.92 In vitro studies indicated that SCFAs can regulate the thickness of this mucus layer and are involved in restoring the mucus layer in instances of injury.93,94
Paneth cells, situated at the base of the small intestinal crypts, are responsible for secreting various antimicrobial peptides crucial for the intestine’s defense mechanisms.95 In vitro and in vivo studies indicate that SCFAs, especially butyrate, stimulate the production of antimicrobial peptides by Paneth cells, particularly alpha- and beta-defensins and LL-37.96–99
SCFAs participate in creating a favorable hypoxic environment that supports beneficial gut bacteria while inhibiting the growth of harmful pathogens. Butyrate stabilizes hypoxia-inducible factor transcription in intestinal epithelium cells, boosting oxygen consumption and fostering a physiologically advantageous hypoxic environment within the colon.100,101
SCFAs demonstrate potent immunomodulatory effects, particularly in the intestines, displaying notable anti-inflammatory properties. These effects operate through two primary pathways: G protein-coupled receptors (GPRs) and histone deacetylases (HDACs) inhibition.102 Dendritic cells (DCs) treated with SCFAs induced differentiation of naïve T-cells into Treg1 cells producing anti-inflammatory IL-10.103 Treatment of macrophages with n-butyrate led to the down-regulation of LPS-induced pro-inflammatory mediators, including nitric oxide, IL-6, and IL-12, while maintaining levels of TNF-α or MCP-1.104 Moreover, activation of GPR109a by butyrate enhanced the tolerogenic response of colonic macrophages and dendritic cells, reduced colonic inflammation and promoted homeostasis in mice.105 In a murine model of nonalcoholic steatohepatitis NASH, sodium butyrate treatment induced apoptosis of pro-inflammatory liver macrophages and promoted their differentiation into the anti-inflammatory M2 phenotype.106 SCFAs exert an anti-inflammatory and tolerogenic effect not only in intestinal epithelial cells but also in other organs.
In this figure we present the detrimental effect ethanol has on the intestinal tissue: 1) toxic damage of the intestinal cells, 2) hemorrhagic erosions and hemorrhage in the lamina propria, 3) reduction of the height and surface of the villi, 4) increase in goblet cells and mucus production, 5) metaplasia, 6) lower expression of heat shock proteins (HSP), 7) dismantling of cytoskeleton, 8) decreased tight junction protein expression, 9) increased activity of proinflammatory interleukin-6 (IL-6), matrix metalloproteinase-9 (MMP-9) and nuclear factor kappa B (NF-κβ), 10) influx of leukocytes which lead to release of toxic reactive oxygen species (ROS), leukotrienes (LTs) and histamine (H) by mast cells.
Created with BioRender.com
Impact of ethanol overconsumption on SCFA and SCFA-producing bacteria
Excessive ethanol consumption leads to a shift in SCFA-producing bacteria. There is a lesser abundance of Bacteroidetes on the phylum level and a lower Firmicutes/Proteobacteria ratio.39,107 As presented in Table 1. on genus level, there is less Akkermansia, Alistipes, Bacteroides, Clostridium, Collinsella, Faecalibacterium, Parabacteroides, Prevotella and Ruminococcus.
In mice, ethanol feeding causes colonic hyperpermeability, decreased butyrate to total SCFA ratio in the stool and steatohepatitis.33 It inhibits intestinal stem cells essential to maintain the continuous renewal of the intestinal epithelium.108 Ethanol feeding also suppressed Notch1 (the gene responsible for intestinal cell differentiation), resulting in gut leakiness, lower expression of tight-junction proteins and colonic inflammation.109,110 This effect can be attributed to lower butyrate production by the intestines.33 It is worth noting that the above effects happened only in the colon; none were observed in the jejunum. In an in vitro study of the intestinal cell model of ethanol abuse, treatment with a microbial synbiotic increased the relative abundance of SCFA-producing bacteria and butyrate and acetate production.111 Cresci et al. proved that tributyrin (butyrate and glycerol ester) supplementation in a chronic ethanol murine model resulted in higher expression of tight-junction proteins and lower intestinal permeability.112 Probiotic treatment with Pediococcus pentosaceus in chronic and binge ethanol murine models restored the relative abundance of SCFA-producing bacteria, improving the intestinal barrier function and reducing inflammation.113
Tryptophan and its derivatives
Tryptophan is one of the essential amino acids. Although most ingested tryptophan undergoes metabolism via kynurenine or serotonin pathways within the small intestine, the gut microbiome metabolizes a portion.114,115 Certain bacteria utilize tryptophan as an energy source, generating by-products that influence the immunological homeostasis within the intestine.116,117 In an experimental murine model, tryptophan deprivation led to a shift in the gut microbiome, notably an increase in Actinobacteria, Proteobacteria, and Firmicutes, alongside a decrease in Bacteroidetes.118
Tryptophan-derived microbial metabolites can function as ligands for the aryl hydrocarbon receptor (AHR). Within humans, AHRs are primarily situated at barrier sites, particularly in the intestines, contributing to maintaining immunological balance.119 Activation of AHRs exerts multifaceted effects. It contributes to preserving the proper function of the intestinal barrier, promoting the production of anti-inflammatory cytokines (especially IL-22, which plays a crucial role in early defense against bacterial pathogens) and inhibiting the production of pro-inflammatory cytokines (such as IL-17 or IFN-γ).120–124 AHR activation also helps regenerate the colitis model’s intestinal barrier.125
One of the better-studied gut microbiome-derived tryptophan metabolites is indole propionic acid (IPA).126 IPA is also involved in the maintenance of the intestinal barrier.127 It up-regulates the production of mucins and tight-junction proteins, reducing the intestinal epithelium’s permeability.128,129 IPA induces the expression of anti-inflammatory IL-10 while suppressing the pro-inflammatory NF-κβ signaling.130,131
Indole-3-lactic acid (ILA) is another bacterial tryptophan derivative that exhibits anti-inflammatory activity by activating AHR.132 Studies showed that it can reduce LPS-induced inflammation in intestinal epithelial cells and upregulate anti-inflammatory pathways in immature intestinal epithelial cells.133,134 In mice, it decreases the accumulation of pro-inflammatory macrophages, thereby reducing susceptibility to DSS-induced colitis.135 ILA increases the number of tryptophan-metabolizing bacteria, which results in increased production of indole-3-propionic acid and indole-3-acetic acid.136
A study by Li et al. demonstrated that indole-3-acetic acid, via an increase in the proportion of Treg cells, can alleviate DSS-induced colitis in an AHR-independent.137 Furthermore, by enhancing the sulfation of mucins, indole-3-acetic acid supports intestinal homeostasis and protects it from inflammation.138
We present the impact of microbial-derived tryptophan metabolites on intestines in Figure 3.
Figure 3.
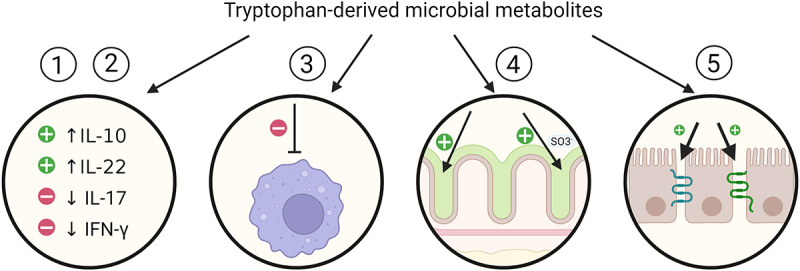
Impact of microbial-derived tryptophan metabolites on intestines.
Tryptophan-derived microbial metabolites: 1) partake in the creation of an anti-inflammatory environment by increasing the production of anti-inflammatory interleukin 10 (IL-10) and interleukin 22 (IL-22), 2) simultaneously inhibit the production of pro-inflammatory cytokines such as interferon γ (IFN-γ) and interleukin 17 (IL-17), 3) decrease the accumulation of pro-inflammatory macrophages, 4) up-regulate mucus production and its sulfation, 5) up-regulate tight junction proteins production.
Created with BioRender.com
Impact of ethanol abuse on tryptophan derivatives and the gut microbiota producing them
As presented in Table 1, ethanol abuse leads to a decrease in bacteria strains metabolizing tryptophan, namely Bacteroidetes phylum, Ruminococcaceae family and Akkermansia, Alistipes and Clostridium genus.
Ethanol consumption in mice led to intestinal dysbiosis, resulting in reduced levels of indole-3-acid – an AHR ligand involved in the regulation of the anti-inflammatory cytokine IL-22. Furthermore, patients with alcoholic hepatitis had lower fecal indole-3-acid compared to healthy controls. Feeding mice with Lactobacillus reuteri engineered to produce IL-22 reduced ethanol-driven liver damage, inflammation and bacterial translocation.139 Intervention with Lactiplantibacilus plantarum in a murine alcoholic liver injury model altered gut microbiome and increased indole-3-acetamide levels, elevating AHR expression and exhibiting anti-inflammatory properties. It up-regulated AHR expression, exerting anti-inflammatory effects.140 In a murine model of alcohol liver disease, prebiotic treatment leads to increased production of tryptophan-derived AHR ligands. It leads to a reduction in liver injury. However, this effect was absent in ahr knockout mice.141 Patients with alcoholic hepatitis and liver cirrhosis exhibited diminished levels of tryptophan metabolites in both serum and fecal samples.142
While plasma levels of indole-3-propionic acid are lowered in people who abuse ethanol, Mrdjen et al. did not observe lower levels of indole-3-acetic acid or indole-3-lactic acid.143
Summing up, bacterial tryptophan metabolites improve and regenerate the intestinal barrier while mitigating inflammation by activating diverse anti-inflammatory pathways. Ethanol consumption leads to gut microbiome changes, resulting in decreased levels of beneficial tryptophan derivatives.
Bile acids
Primary bile acids (BAs), chenodeoxycholic acid (CDCA) and cholic acid (CA) are derivatives of cholesterol. The liver is the only organ that can produce primary BAs.144 Once excreted into the intestine, more than 90% of BAs are absorbed in the ileum.145 In the colon, resident microbiome transform remaining primary bile acids into secondary bile acids, primarily lithocholic acid (LCA) and deoxycholic acid (DCA).146 This conversion involves deconjugation and dehydroxylation processes facilitated by specific enzymatic activity present in certain bacterial species. These include Gram-positive bacteria (mainly Firmicutes) and certain Gram-negative from the Bacteroides phylum.147,148 We will focus on the relationship between BAs and the gut microbiota and how BAs help maintain immunological homeostasis in the intestines.
Relation between bile acids and gut microbiome
BAs can modify the gut microbiome. High concentrations of hydrophobic bile acids (BAs) exhibit direct antimicrobial effects by damaging bacterial membranes. Gram-positive bacteria are more vulnerable to BAs than Gram-negative bacteria, with secondary BAs typically displaying higher toxicity to bacteria than primary BAs.146
Wang et al. observed a significant shift in the gut microbiome in mice fed with CA, which led to impaired intestinal barrier function and mild intestinal inflammation.149 Mice fed DCA displayed alterations in intestinal microbiome, alongside intestinal inflammation and the accumulation of fecal BAs.150 Supplementation of obeticholic acid induced changes in the gut microbiome composition in both mice and humans, reducing endogenous bile acid levels and augmenting the proportion of Firmicutes, notably in the small intestine.151 Interestingly, individuals with inflammatory bowel disease (IBD) often exhibit diminished primary bile acid conversion into secondary bile acids by their gut microbiome.152
Bile acids act as agonists for receptors found in various organs and cells. We will focus on receptors involved in preserving immunological balance within the intestines (Figure 4). Farnesoid-x-receptor (FXR) is a metabolic receptor expressed in several tissues, including the liver and the intestine.153 It is mainly activated by primary BAs – CDCA and CA.154 Its activation exerts several anti-inflammatory effects. It suppresses pro-inflammatory genes, reducing the expression of pro-inflammatory cytokines and leading to the attenuation of colitis in animal models.155,156 FXR agonists increase plasma levels of anti-inflammatory Il-10.157
Figure 4.
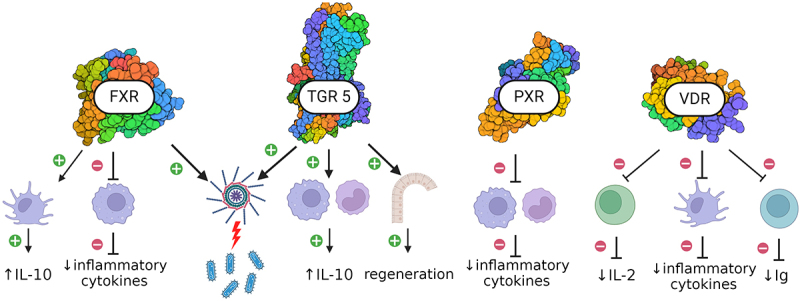
Bile acids receptors and intestinal health.
Secondary BAs, i.e. LCA and DCA, foremost activate the G protein bile acid receptor (TGR5). Primary BAs also activate TGR5 but at higher concentrations.154 It can be found in the intestines, monocytes and macrophages. Activation of TGR5 in intestinal macrophages and monocytes shifts their phenotypes into anti-inflammatory promoting expression of ll-10.158–160 BAs regenerate the intestinal epithelium via activation of TGR5 in intestinal stem cells.161
Both FXR and TGR5 promote inflammasome-mediated antimicrobial reactions in mice.156
The pregnane-x receptor (PXR) is expressed in the small intestine, colon and liver, as well as in CD4+ and CD8+ T lymphocytes, CD19+ B lymphocytes and CD14+ monocytes.162,163 LCA is the most potent PXR activator, although CDCA and DCA also activate it.164 PXR also takes part in the maintenance of intestinal homeostasis. Down-regulation of PXR by imidacloprid increased intestinal permeability, decreased the amounts of tight junction proteins and increased TNF-α and IL-1β levels, both in vitro and in vivo.165 PXR agonist protected mice from colitis, decreasing macrophage and monocyte infiltration and inhibiting the production of pro-inflammatory cytokines.166 Shah et al. observed that PXR agonists protected mice from colitis by reducing the mRNA expression of several NF-κβ target genes.167
The vitamin D receptor (VDR), located both in the liver and the intestine, is activated by LCA and DCA in addition to vitamin D.168 Activation of VDR in dendritic cells inhibits the production of inflammatory cytokines and reprograms them to become tolerogenic.169 Activation of VDR in CD8+ T lymphocytes reduces IL-2 production.170 Activation of VDR in B lymphocytes reduces the production of immunoglobulins.171
Bile acids work as agonists for various receptors found in the gastrointestinal tract.
Farnesoid-x-receptor (FXR) activation suppresses pro-inflammatory genes and increases plasma levels of anti-inflammatory interleukin 10 (IL-10).
G protein bile receptor (TGR5) activation shifts the phenotype of intestinal macrophages and monocytes into anti-inflammatory, resulting in higher production of IL-10. TGR5 activation in intestinal stem cells promotes regeneration of intestinal epithelium.
Both FXR and TGR promote antimicrobial inflammasome-mediated reactions.
Pregnane-x receptor activation inhibits the production of pro-inflammatory cytokines by intestinal monocytes and macrophages and reduces the expression of several NF-κβ genes.
Vitamin D receptor activation in 1) intestinal CD8+ T lymphocytes reduces the production of pro-inflammatory interleukin-2 (IL-2), 2) intestinal dendritic cells inhibits the production of inflammatory cytokines, 3) B lymphocytes reduce the production of immunoglobulins (Ig).
Created with BioRender.com
Ethanol consumption and bile acids
Ethanol stimulates bile acid synthesis in humans, possibly due to reduced feedback inhibition of bile acid synthesis caused by ethanol-driven diminished gall bladder contraction.172,173 However, in patients with alcohol hepatitis, despite high serum bile acid levels, de novo bile acid synthesis is significantly decreased.174 In mice, bile acid synthesis is regulated via ethanol-activated cannabinoid receptors in the liver.175 Within the cirrhotic population, currently drinking patients had significantly higher levels of secondary bile acids in stool.176
Muthiah et al. observed no significant difference in total fecal levels of both primary and secondary bile acids between drinking and non-drinking patients. However, individuals who consumed ethanol exhibited decreased levels of deoxycholate (DCA), along with alterations in the concentration of other specific bile acids.65 Ethanol increases the CDCA, DCA and LCA levels in the enterohepatic circulation.177 In a murine model of alcohol-induced liver injury, fiber feeding lowered the BA levels in plasma and liver. It increased their levels in the stool, probably through reshaping of the gut microbiome.178
Other metabolites
Branch-chained amino acids
BCAAs are essential amino acids involved in various metabolic processes. Leucine is a critical stimulator of protein synthesis, supporting albumin production, liver health, and muscle tissue growth.179 Elevated BCAA levels are associated with insulin resistance, type 2 diabetes risk, and hepatocellular carcinoma progression, but BCAA supplementation has shown benefits in liver diseases.180–183 The gut microbiota has a bidirectional relationship with BCAAs, affecting their levels and producing or utilizing them.184 Certain bacterial populations correlate with BCAA levels and insulin resistance.183,185,186 In the murine model, chronic ethanol consumption lowers concentrations of all three of BCAA in the gastrointestinal tract.187 However, as this review centers on exploring the relationship between gut microbiota and intestinal health, a detailed exploration of these aspects is beyond the scope of our focus.
Trimethylamine N-oxide
Trimethylamine N-oxide (TMAO) is another crucial microbial metabolite. The gut microbiota residing in the small intestine produces trimethylamine from choline and choline-containing compounds such as L-carnitine and betaine.188,189 It is mainly produced by the Firmicutes and Proteobacteria phyla.40 Then, in the liver, it is oxidized by the flavin monooxygenase family of enzymes forming TMAO.190 High TMAO serum levels are linked with several detrimental effects in various organs and systems. Li et al.‘s umbrella review found a positive association between circulating TMAO serum levels and all-cause mortality, cardiovascular diseases, major adverse cardiovascular events, hypertension, diabetes mellitus and a decrease in glomerular filtration rate.191 Furthermore, high TMAO serum levels were found in patients with breast, colorectal, gastric, liver and pancreatic cancers.192 TMAO levels are also positively associated with the development of NAFLD.193
Limited data exist on TMAO’s effects on the intestines. Still, it has been linked to the activation of NF-kB and NLRP3 pathways, contributing to inflammation and potentially the development of colorectal cancer. In vitro studies showed that TMAO activates NLRP3 inflammasomes and inhibits autophagy, suggesting a possible role in inflammatory bowel disease (IBD). However, one study found lower TMAO levels in IBD patients compared to a control group, indicating the need for further research.
There is insufficient data on the relationship between ethanol consumption and TMAO levels. Coulbalt et al. found no significant difference in serum TMAO levels between patients with alcohol use disorder and healthy controls, though the variability was greater in the ethanol group.194 Similarly, Haas et al. reported no difference in TMAO levels between men who moderately drank red wine and those who abstained for an equal period.195
Intestinal stem cells
Intestinal stem cells (ISCs) are undifferentiated, multipotent cells located in the crypts of the intestinal epithelium, responsible for the continuous renewal and repair of the intestinal lining.196,197 These cells give rise to the various cell types of the intestinal epithelium, including absorptive enterocytes, mucus-secreting goblet cells, hormone-producing enteroendocrine cells, and antimicrobial peptide-secreting Paneth cells.198–200 The proliferation and differentiation of ISCs are tightly regulated by signaling pathways such as Wnt, Notch, and BMP to maintain intestinal homeostasis and prevent uncontrolled cell growth and differentiation.201–203 The gut microbiota regulates ISC function and intestinal health through its metabolites. Butyrate, via the HDAC inhibition pathway, is a potent intestinal stem cell proliferation inhibitor. However, the unique architecture of the crypt structure usually shields intestinal stem cells from this effect, except during mucosal injury when they are exposed to butyrate.204 Propionate, and to a lesser extent acetate, significantly induces Reg3B and Reg3G expression both in vitro and in vivo, stimulating intestinal stem cells upon tissue injury.205 The probiotic bacterium Akkermansia muciniphila enhances ISCs function and epithelial development by producing SCFAs and activating the Wnt signaling pathway.206
At physiological levels, secondary bile acids act through FXR to regulate Wnt-β catenin signaling and ISCs proliferation, whereas lower levels can disrupt this regulation, potentially leading to carcinogenesis.207 Pai and colleagues found that low doses of DCA (5 to 50 µM) stimulate colonic cancer cell proliferation and migration via Wnt/β-catenin signaling, while higher concentrations (100 µM) inhibit these processes.208 Similar results were observed by Milovic et al. in an in vivo model.209 Sorrentino et al. demonstrated that bile acids (both isolated secondary bile acids as well as a mix) promote epithelial regeneration by activating the TGR5 receptor in intestinal stem cells. The BA-TGR5 axis was essential for reprogramming the intestinal epithelium into a proliferative, repairing tissue.161
Tryptophan metabolites influence intestinal stem cells (ISCs) by regulating the aryl hydrocarbon receptor (AHR), which controls WNT signaling and β-catenin levels, maintaining intestinal barrier integrity and preventing tumorigenesis.125 Park et al. showed that indole-3-carbinol, an AHR-dependent tryptophan metabolite, promotes goblet cell differentiation by regulating key transcription factors, underscoring its role in intestinal health and regeneration.210
Ethanol consumption and ISCs
There are still some conflicting data regarding ethanol’s effect on intestinal stem cells. Ethanol feeding in mice decreases Notch1 expression in the colon, associated with impaired cell integrity, decreased expression of tight junction proteins and colon inflammation.33,109,211 Ethanol exposure decreases ISC markers Lgr5 and Bmi1 by dysregulating β-catenin signaling, impairing small intestine stem cell proliferation and function in in vitro and in vivo models.212 On the other hand, in the NIAAA murine model, ethanol consumption alters small intestinal epithelium morphology by increasing crypt depth, proliferative activity, and migration of intestinal epithelial cells, upregulating ISC markers Lgr5 and Bmi1 and enhancing Wnt signaling-dependent ISC activity and IEC proliferation while leaving goblet cell numbers and Notch-1 pathway unchanged.213
The effect of probiotic treatment on the gut in ethanol abuse
Several clinical trials have explored the efficacy of probiotic treatment in ethanol-related diseases. They used predominantly animal models to simulate ethanol abuse. In mice, ethanol feeding increases the abundance of Proteobacteria and Actinobacteria while decreasing the abundance of Bacteroides and Firmicutes.214 In the following paragraphs, we will discuss how probiotic treatment can revert ethanol-induced changes to the gut microbiome and alleviate ethanol-induced intestinal injury.
Lactobacillus rhamnosus GG
Lactobacillus rhamnosus GG is Gram-positive, facultative anaerobic or microaerophilic bacteria. It preserves intestinal integrity, exerts anti-inflammatory effects in the intestines, and prevents dysbiosis.215 It is probably the most studied probiotic in the treatment of alcohol disease.
Treatment with Lactobacillus rhamnosus GG prevents ethanol-induced dysbiosis and increases the abundance of BSH-producing bacteria recovering ethanol-suppressed FXR activation.214,216 In vitro, Lactobacillus rhamnosus GG inhibits miR122a, increasing the occludin expression in Caco-2 intestinal monolayers treated with ethanol and restoring their barrier function.217 The same effect was observed in animal models where Lactobacillus rhamnosus GG recovered ethanol-induced damage to the intestinal villi and tight-junction proteins.218,219 It is, at least in part, mediated through exosome-like nanoparticles. They act through the AHR pathway, increasing the production of tight-junction proteins leading to improved intestinal barrier function.220 Furthermore, Lactobacillus rhamnosus GG reduces gut leakiness and decreases oxidative stress and inflammation in both the intestine and the liver, significantly ameliorating alcoholic steatohepatitis.221 In a murine model of alcohol-induced liver injury, Zhu et al. found that administration of Lactobacillus rhamnosus GG and inosine reversed ethanol-induced changes to the population of Treg and Th1 lymphocytes, alleviating inflammation in the intestines.218 Supernatant of Lactobacillus rhamnosus GG caused restoration of the intestinal barrier function and increased the expression of HIF 2α in mice.222
Other probiotics
Lactobacillus plantarum ST-III restored standard histological architecture of the intestine in ethanol-fed rats and mice.223 It has also restored the distribution of tight-junction proteins and the expression of P-gp protein (a protein that protects the intestine from harmful substances).224 In the DSS model of colitis treatment with Lactobacillus delbrueckii, it increased the expression of anti-inflammatory IL-22 and attenuated ethanol-induced exacerbation inflammation.225 In the small intestine, Lactobacillus fermentum inhibited inflammation by lowering the expression of IL-6 and transcription of TNF-α. Furthermore, it up-regulated the production of tight-junction proteins, restoring the intestinal barrier function.24
Administration of another probiotic – Kluyveromyces marxianus YG-4 – to ethanol-fed mice increased the number of tight-junction proteins and the number of goblet cells in the colon; furthermore, it reduced oxidative stress and inflammation in the liver.226
Pediococcus pentosaceus CGMC C7049 in mice restored ethanol-depleted abundance of SCFA-producing bacteria. It lead to increased SCFA levels in stool and increased expression of tight-junction proteins and the antimicrobial peptide Reg3B. Dissection of the murine livers showed lesser ethanol-induced injury in comparison with control and decreased levels of endotoxin and inflammatory cytokines.113
Interestingly, Jiang et al. engineered a recombinant strain of Lactococcus lactis that produced human alcohol dehydrogenase. Mice treated with the aforementioned probiotic not only showed greater tolerance for ethanol consumption but also were protected from ethanol-induced intestinal and liver damage.227
Compound probiotic (containing Lactobacillus, Bifidobacterium and Streptococcus) used together with metformin protected the intestine from ethanol-induced damage in vivo, in vitro and in silico. It has up-regulated tight junction proteins, reduced oxidative stress and alleviated inflammation.228
Concluding section
Ethanol’s detrimental impact on the intestines extends beyond its inherent toxicity. The gut microbiome, through the production of its metabolites, plays a vital part in maintaining intestinal homeostasis. Its modification caused by ethanol abuse creates a pro-inflammatory environment and compromises the intestinal barrier function. Several studies suggest that probiotic interventions have, at least partially, reversed these changes and restored immunological balance. Thus, further exploration of the gut microbiome in relation to ethanol consumption represents a promising research topic which may deliver new therapeutic options in ethanol-related gastrointestinal diseases.
Further studies are needed to address gaps in the current knowledge. In our opinion, data on the impact of ethanol abuse on the composition of the gut microbiota remain too sparse and too heterogeneous. Due to the ethical impossibility of conducting prospective randomized studies on the effects of ethanol on the microbiota in humans, we rely solely on observational studies. Therefore, more research on this topic is still needed to provide sufficient data for meta-analyses. The impact of ethanol on the gut microbiota in the small intestine, which constitutes the largest part of the digestive tract, is an almost unexplored area. Finding a method to non-invasively determine the composition of the microbiota there would be extremely valuable. The vast majority of studies assessing the impact of prebiotics, probiotics, and postbiotics on the gut microbiota in the context of ethanol abuse have been conducted on animals. Human studies are necessary.
Disclaimer
Views expressed in this paper are our own.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting this study’s findings are openly available in cited papers.
References
- 1.Farhadi A, Banan A, Fields J, Keshavarzian A.. Intestinal barrier: an interface between health and disease. J Gastro Hepatol. 2003;18(5):479–23. doi: 10.1046/j.1440-1746.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- 2.Camilleri M, Madsen K, Spiller R, Greenwood-Van Meerveld B, Verne GN. Intestinal barrier function in health and gastrointestinal disease. Neurogastro Motil. 2012;24(6):503–512. doi: 10.1111/j.1365-2982.2012.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. 2020;113(12):2019–2040. doi: 10.1007/s10482-020-01474-7. [DOI] [PubMed] [Google Scholar]
- 4.GBD . Alcohol collaborators. Alcohol use and burden for 195 countries and territories, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2016;392(2018):1015–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dguzeh U, Haddad NC, Smith KTS, Johnson JO, Doye AA, Gwathmey JK, Haddad GE. Alcoholism: a multi-systemic cellular insult to organs. Int J Environ Res Public Health. 2018;15(6):1083. doi: 10.3390/ijerph15061083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engen PA, Green SJ, Voigt RM, Forsyth CB, Keshavarzian A. The gastrointestinal microbiome: alcohol effects on the composition of intestinal microbiota. Alcohol Res. 2015;37(2):223–236. [PMC free article] [PubMed] [Google Scholar]
- 8.Broitman SA, Gottlieb LS, Vitale JJ. Augmentation of ethanol absorption by mono- and disaccharides. Gastroenterology. 1976;70(6):1101–1107. doi: 10.1016/S0016-5085(76)80319-8. [DOI] [PubMed] [Google Scholar]
- 9.Ch H, Ea R, E M. Distribution of ethanol in the human gastrointestinal tract. The Am J Clin Nutr. 1973;26(8):831–834. doi: 10.1093/ajcn/26.8.831. [DOI] [PubMed] [Google Scholar]
- 10.Bode JC. Alcohol and the gastrointestinal tract. Ergeb Inn Med Kinderheilkd. 1980;45:1–75. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein DB. Effect of alcohol on cellular membranes. Ann Emerg Med. 1986;15(9):1013–1018. doi: 10.1016/S0196-0644(86)80120-2. [DOI] [PubMed] [Google Scholar]
- 12.Beck IT, Dinda PK. Acute exposure of small intestine to ethanol: effects on morphology and function. Dig Dis Sci. 1981;26(9):817–838. doi: 10.1007/BF01309614. [DOI] [PubMed] [Google Scholar]
- 13.Barona E, Pirola RC, Leiber CS. Small intestinal damage and changes in cell population produced by ethanol ingestion in the rat. Gastroenterology. 1974;66(2):226–234. doi: 10.1016/S0016-5085(74)80106-X. [DOI] [PubMed] [Google Scholar]
- 14.Hoyumpa AM, Breen KJ, Schenker S, Wilson FA. Thiamine transport across the rat intestine. II. Effect of ethanol. J Lab Clin Med. 1975;86(5):803–816. [PubMed] [Google Scholar]
- 15.Persson J. Alcohol and the small intestine. Scand J Gastroenterol. 1991;26(1):3–15. doi: 10.3109/00365529108996478. [DOI] [PubMed] [Google Scholar]
- 16.Lee SW, Choi DW, Park SC, Kim HJ, Nam YH, Choi DH, Kang CD, Lee SJ, Chun WJ, Ryu Y-J, et al. Expression of heat shock proteins and cytokines in response to ethanol induced damage in the small intestine of ICR mice. Intest Res. 2014;12(3):205–213. doi: 10.5217/ir.2014.12.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grewal RK, Mahmood A. Ethanol induced changes in glycosylation of mucins in rat intestine. Ann Gastro. 2009;22(3):178–183. [Google Scholar]
- 18.Banan A, Choudhary S, Zhang Y, Fields JZ, Keshavarzian A. Ethanol-induced barrier dysfunction and its prevention by growth factors in human intestinal monolayers: evidence for oxidative and cytoskeletal mechanisms. J Pharmacol Exp Ther. 1999;291(3):1075–1085. [PubMed] [Google Scholar]
- 19.Dinda PK, Kossev P, Beck IT, Buell MG. Role of xanthine oxidase-derived oxidants and leukocytes in ethanol-induced jejunal mucosal injury. Dig Dis Sci. 1996;41(12):2461–2470. doi: 10.1007/BF02100144. [DOI] [PubMed] [Google Scholar]
- 20.Beck IT, Boyd AJ, Dinda PK. Evidence for the involvement of 5-lipoxygenase products in ethanol-induced intestinal plasma protein loss. Am J Physiol. 1988;254(4):G483–G488. doi: 10.1152/ajpgi.1988.254.4.G483. [DOI] [PubMed] [Google Scholar]
- 21.Dinda PK, Leddin DJ, Beck IT. Histamine is involved in ethanol-induced jejunal microvascular injury in rabbits. Gastroenterology. 1988;95(5):1227–1233. doi: 10.1016/0016-5085(88)90355-1. [DOI] [PubMed] [Google Scholar]
- 22.Bode JC, Knüppel H, Schwerk W, Lorenz-Meyer H, Dürr HK. Quantitative histomorphometric study of the jejunal mucosa in chronic alcoholics. Digestion. 1982;23(4):265–270. doi: 10.1159/000198760. [DOI] [PubMed] [Google Scholar]
- 23.Maier A. Effects of chronic alcohol abuse on duodenal mononuclear cells in man. Dig Dis Sci. 1999;44(4):691–696. doi: 10.1023/A:1026697305769. [DOI] [PubMed] [Google Scholar]
- 24.Paladino L, Rappa F, Barone R, Macaluso F, Zummo FP, David S, Szychlinska MA, Bucchieri F, Conway de Macario E, Macario AJL, et al. Nf-kB regulation and the chaperone system mediate restorative effects of the probiotic lactobacillus fermentum LF31 in the small intestine and cerebellum of mice with ethanol-induced damage. Biol (Basel). 2023;12(11):1394. doi: 10.3390/biology12111394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinecker HC, Steffen M, Witthoeft T, Pflueger I, Schreiber S, MacDERMOTT RP, Raedler A. Enhand secretion of tumour necrosis factor-alpha, IL-6, and IL-1β by isolated lamina ropria monouclear cells from patients with ulcretive cilitis and crohn’s disease. Clin Exp Immunol. 1993;94(1):174–181. doi: 10.1111/j.1365-2249.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruder B, Atreya R, Becker C. Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int J Mol Sci. 2019;20(8):1887. doi: 10.3390/ijms20081887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Tong J, Chang B, Wang B, Zhang D, Wang B. Effects of alcohol on intestinal epithelial barrier permeability and expression of tight junction-associated proteins. Mol Med Rep. 2014;9(6):2352–2356. doi: 10.3892/mmr.2014.2126. [DOI] [PubMed] [Google Scholar]
- 28.Ma TY, Nguyen D, Bui V, Nguyen H, Hoa N. Ethanol modulation of intestinal epithelial tight junction barrier. Am J Physiol. 1999;276(4):G965–G974. doi: 10.1152/ajpgi.1999.276.4.G965. [DOI] [PubMed] [Google Scholar]
- 29.Banan A, Keshavarzian A, Zhang L, Shaikh M, Forsyth CB, Tang Y, Fields JZ. Nf-κB activation as a key mechanism in ethanol-induced disruption of the F-actin cytoskeleton and monolayer barrier integrity in intestinal epithelium. Alcohol. 2007;41(6):447–460. doi: 10.1016/j.alcohol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 30.The leaky gut of alcoholism: possible route of entry for toxic compounds - PubMed. https://pubmed.ncbi.nlm.nih.gov/6141332/. [DOI] [PubMed]
- 31.Parlesak A, Schäfer C, Schütz T, Bode JC, Bode C. Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J Hepatol. 2000;32(5):742–747. doi: 10.1016/S0168-8278(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 32.Fukui H, Brauner B, Bode JC, Bode C. Plasma endotoxin concentrations in patients with alcoholic and non-alcoholic liver disease: reevaluation with an improved chromogenic assay. J Hepatol. 1991;12(2):162–169. doi: 10.1016/0168-8278(91)90933-3. [DOI] [PubMed] [Google Scholar]
- 33.Forsyth CB, Shaikh M, Bishehsari F, Swanson G, Voigt RM, Dodiya H, Wilkinson P, Samelco B, Song S, Keshavarzian A, et al. Alcohol feeding in mice promotes colonic hyperpermeability and changes in colonic organoid stem cell fate. Alcohol Clin Exp Res. 2017;41(12):2100–2113. doi: 10.1111/acer.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bala S, Marcos M, Gattu A, Catalano D, Szabo G, Feng W. Acute binge drinking increases serum endotoxin and bacterial DNA levels in healthy individuals. PLOS ONE. 2014;9(5):e96864. doi: 10.1371/journal.pone.0096864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casini A, Galli A, Calabro’ A, Di Lollo S, Orsini B, Arganini L, Jezequel AM, Surrenti C. Ethanol-induced alterations of matrix network in the duodenal mucosa of chronic alcohol abusers. Virchows Arch. 1999;434(2):127–135. doi: 10.1007/s004280050316. [DOI] [PubMed] [Google Scholar]
- 36.Hartmann P, Chen P, Wang HJ, Wang L, McCole DF, Brandl K, Stärkel P, Belzer C, Hellerbrand C, Tsukamoto H, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58(1):108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M C C C, Nl L, Cm F, Jl G, D A, C G, G C, Sh P, C M, Fs M, et al. Comparing the effects of acute alcohol consumption in germ-free and conventional mice: the role of the gut microbiota. BMC Microbiol. 2014;14(1):240. doi: 10.1186/s12866-014-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litwinowicz K, Choroszy M, Waszczuk E. Changes in the composition of the human intestinal microbiome in alcohol use disorder: a systematic review. Am J Drug Alcohol Abuse. 2020;46(1):4–12. doi: 10.1080/00952990.2019.1669629. [DOI] [PubMed] [Google Scholar]
- 39.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:1–7. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romano KA, Vivas EI, Amador-Noguez D, Rey FE, Blaser MJ. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6(2):e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Huang S, Li T, Li N, Han D, Zhang B, Xu ZZ, Zhang S, Pang J, Wang S, et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome. 2021;9(1):184. doi: 10.1186/s40168-021-01115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Z, Pei W, Shen Y, Wang L, Zhu J, Zhang Y, Fan S, Wu Q, Li L, Zhang Z, et al. Akkermansia muciniphila and its outer protein Amuc_1100 regulates tryptophan metabolism in colitis. Food Funct. 2021;12(20):10184–10195. doi: 10.1039/D1FO02172A. [DOI] [PubMed] [Google Scholar]
- 43.van Beek AA, Hugenholtz F, Meijer B, Sovran B, Perdijk O, Vermeij WP, Brandt RMC, Barnhoorn S, Hoeijmakers JHJ, de Vos P, et al. Frontline science: tryptophan restriction arrests B cell development and enhances microbial diversity in WT and prematurely aging Ercc1−/Δ7 mice. J Leukoc Biol. 2017;101(4):811–821. doi: 10.1189/jlb.1HI0216-062RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62(1):67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 46.Ovadia C, Perdones‐Montero A, Spagou K, Smith A, Sarafian MH, Gomez‐Romero M, Bellafante E, Clarke LCD, Sadiq F, Nikolova V, et al. Enhanced microbial bile acid deconjugation and impaired ileal uptake in pregnancy repress intestinal regulation of bile acid synthesis. Hepatology. 2019;70(1):276–293. doi: 10.1002/hep.30661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res. 2015;56(6):1085–1099. doi: 10.1194/jlr.R054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaga S, Lee S, Ji B, Andreasson A, Talley NJ, Agréus L, Bidkhori G, Kovatcheva-Datchary P, Park J, Lee D, et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci Rep. 2020;10(1):14977. doi: 10.1038/s41598-020-71939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook KL, Rothrock MJ, Loughrin JH, Doerner KC. Characterization of skatole-producing microbial populations in enriched swine lagoon slurry. FEMS Microbiol Ecol. 2007;60(2):329–340. doi: 10.1111/j.1574-6941.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- 50.Coleman JP, Hudson LL. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl Environ Microbiol. 1995;61(7):2514–2520. doi: 10.1128/aem.61.7.2514-2520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme a transferases from clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J Lipid Res. 2012;53(1):66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, Alves JMP, Ridlon JM. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol. 2018;84(10):e00235–18. doi: 10.1128/AEM.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin P, Zou Y, Dai Y, Luo G, Zhang X, Xiao L. Characterization a novel butyric acid-producing bacterium collinsella aerofaciens subsp. shenzhenensis subsp. Nov. Microorganisms. 2019;7(3):78. doi: 10.3390/microorganisms7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moran-Ramos S, Macias-Kauffer L, López-Contreras BE, Villamil-Ramírez H, Ocampo-Medina E, León-Mimila P, Del Rio-Navarro BE, Granados-Portillo O, Ibarra-Gonzalez I, Vela-Amieva M, et al. A higher bacterial inward BCAA transport driven by faecalibacterium prausnitzii is associated with lower serum levels of BCAA in early adolescents. Mol Med. 2021;27(1):108. doi: 10.1186/s10020-021-00371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y-J, Xu X-J, Zhou N, Sun Y, Liu C, Liu S-J, You X. Parabacteroides acidifaciens sp. nov. isolated from human faeces. Int J Syst Evol Microbiol. 2019;69(3):761–766. doi: 10.1099/ijsem.0.003230. [DOI] [PubMed] [Google Scholar]
- 58.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, Escobar JS. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid–producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 59.de la Rubia Ortí JE, Moneti C, Serrano-Ballesteros P, Castellano G, Bayona-Babiloni R, Carriquí-Suárez AB, Motos-Muñoz M, Proaño B, Benlloch M. Liposomal epigallocatechin-3-gallate for the treatment of intestinal dysbiosis in children with autism spectrum disorder: a comprehensive review. Nutrients. 2023;15(14):3265. doi: 10.3390/nu15143265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gui Q, Li H, Wang A, Zhao X, Tan Z, Chen L, Xu K, Xiao C. The association between gut butyrate-producing bacteria and non-small-cell lung cancer. J Clin Lab Anal. 2020;34(8):e23318. doi: 10.1002/jcla.23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki M, Schwab C, Ramirez Garcia A, Li Q, Ferstl R, Bersuch E, Akdis CA, Lauener R, Frei R, Roduit C, et al. The abundance of ruminococcus bromii is associated with faecal butyrate levels and atopic dermatitis in infancy. Allergy. 2022;77(12):3629–3640. doi: 10.1111/all.15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhakan DB, Maji A, Sharma AK, Saxena R, Pulikkan J, Grace T, Gomez A, Scaria J, Amato KR, Sharma VK, et al. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience. 2019;8(3):giz004. doi: 10.1093/gigascience/giz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon - PubMed. https://pubmed.ncbi.nlm.nih.gov/23729502/. [DOI] [PMC free article] [PubMed]
- 64.Paik D, Yao L, Zhang Y, Bae S, D’Agostino GD, Zhang M, Kim E, Franzosa EA, Avila-Pacheco J, Bisanz JE, et al. Human gut bacteria produce τη17-modulating bile acid metabolites. Nature. 2022;603(7903):907–912. doi: 10.1038/s41586-022-04480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muthiah MD, Smirnova E, Puri P, Chalasani N, Shah VH, Kiani C, Taylor S, Mirshahi F, Sanyal AJ. Development of alcohol-associated hepatitis is associated with specific changes in gut-modified bile acids. Hepatol Commun. 2022;6(5):1073–1089. doi: 10.1002/hep4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams BB, Van Benschoten A, Cimermancic P, Donia M, Zimmermann M, Taketani M, Ishihara A, Kashyap P, Fraser J, Fischbach M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16(4):495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carvalho FA, Koren O, Goodrich J, Johansson MV, Nalbantoglu I, Aitken J, Su Y, Chassaing B, Walters W, González A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12(2):139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, Plevy S, Sartor RB, Carroll IM. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4(4):316–324. doi: 10.4161/gmic.25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, Nie Y, Li M, Zhi F, Liu S, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3(1):e00188–17. doi: 10.1128/msystems.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He X-X, Li Y-H, Yan P-G, Meng X-C, Chen C-Y, Li K-M, Li J-N. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative colitis. World J Gastroenterol. 2021;27(28):4722–4737. doi: 10.3748/wjg.v27.i28.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maynard C, Weinkove D. The gut microbiota and ageing. Subcell Biochem. 2018;90:351–371. [DOI] [PubMed] [Google Scholar]
- 72.Baechle JJ, Chen N, Makhijani P, Winer S, Furman D, Winer DA. Chronic inflammation and the hallmarks of aging. Mol Metab. 2023;74:101755. doi: 10.1016/j.molmet.2023.101755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The mucin degrader akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74(5):1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110(22):9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe −/− mice. Circulation. 2016;133(24):2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 76.Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, Quercia S, Scurti M, Monti D, et al. Gut Microbiota and Extreme Longevity. Curr Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Zhai Q, Feng S, Arjan N, Chen W. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr. 2019;59(19):3227–3236. doi: 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 78.Yan J, Sheng L, Li H. Akkermansia muciniphila: is it the holy grail for ameliorating metabolic diseases? Gut Microbes. 2021;13(1):1984104. doi: 10.1080/19490976.2021.1984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hold GL, Schwiertz A, Aminov RI, Blaut M, Flint HJ. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microbiol. 2003;69(7):4320–4324. doi: 10.1128/AEM.69.7.4320-4324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Velasquez-Manoff M. Gut microbiome: the peacekeepers. Nature. 2015;518(7540):S3–11. doi: 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- 81.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof F-M, Van de Wiele T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7(5):949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P. Gut microbiota in patients with irritable bowel syndrome—A systematic review. Gastroenterology. 2019;157(1):97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 83.Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65(5):830–839. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 84.Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastro Hepatol. 2008;23(8pt1):1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 85.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106(14):5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruppin H, Bar-Meir S, Soergel KH, Wood CM, Schmitt MG. Absorption of short-chain fatty acids by the colon. Gastroenterology. 1980;78(6):1500–1507. doi: 10.1016/S0016-5085(19)30508-6. [DOI] [PubMed] [Google Scholar]
- 88.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39(2):338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 89.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 90.Wang H-B, Wang P-Y, Wang X, Wan Y-L, Liu Y-C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein claudin-1 transcription. Dig Dis Sci. 2012;57(12):3126–3135. doi: 10.1007/s10620-012-2259-4. [DOI] [PubMed] [Google Scholar]
- 91.Yan H, Ajuwon KM, Koval M. Butyrate modifies intestinal barrier function in IPEC-J2 cells through a selective upregulation of tight junction proteins and activation of the akt signaling pathway. PLOS ONE. 2017;12(6):e0179586. doi: 10.1371/journal.pone.0179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McShane A, Bath J, Jaramillo AM, Ridley C, Walsh AA, Evans CM, Thornton DJ, Ribbeck K. Mucus. Curr Biol. 2021;31(15):R938–R945. doi: 10.1016/j.cub.2021.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaudier E, Jarry A, Blottière HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol. 2004;287(6):G1168–G1174. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- 94.Finnie IA, Dwarakanath AD, Taylor BA, Rhodes JM. Colonic mucin synthesis is increased by sodium butyrate. Gut. 1995;36(1):93–99. doi: 10.1136/gut.36.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sankaran-Walters S, Hart R, Dills C. Guardians of the gut: enteric defensins. Front Microbiol. 2017;8:647. doi: 10.3389/fmicb.2017.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beisner J, Filipe Rosa L, Kaden-Volynets V, Stolzer I, Günther C, Bischoff SC. Prebiotic inulin and sodium butyrate attenuate obesity-induced intestinal barrier dysfunction by induction of antimicrobial peptides. Front Immunol. 2021;12:678360. doi: 10.3389/fimmu.2021.678360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun. 2002;70(2):953–963. doi: 10.1128/IAI.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raqib R, Sarker P, Bergman P, Ara G, Lindh M, Sack DA, Nasirul Islam KM, Gudmundsson GH, Andersson J, Agerberth B, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci USA. 2006;103(24):9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, Xiao Y, Huang X, Eaves-Pyles TD, Golovko G, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11(3):752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365(6):537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- 101.Kelly CJ, Zheng L, Campbell E, Saeedi B, Scholz C, Bayless A, Wilson K, Glover L, Kominsky D, Magnuson A, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17(5):662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martin-Gallausiaux C, Marinelli L, Blottière HM, Larraufie P, Lapaque N. SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc. 2021;80(1):37–49. doi: 10.1017/S0029665120006916. [DOI] [PubMed] [Google Scholar]
- 103.Kaisar MMM, Pelgrom LR, van der Ham AJ, Yazdanbakhsh M, Everts B. Butyrate conditions human dendritic cells to prime type 1 regulatory T cells via both histone deacetylase inhibition and G protein-coupled receptor 109A signaling. Front Immunol. 2017;8:1429. doi: 10.3389/fimmu.2017.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111(6):2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad P, Manicassamy S, Munn D, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sarkar A, Mitra P, Lahiri A, Das T, Sarkar J, Paul S, Chakrabarti P. Butyrate limits inflammatory macrophage niche in NASH. Cell Death Dis. 2023;14(5):332. doi: 10.1038/s41419-023-05853-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Litwinowicz K, Gamian A. Microbiome alterations in alcohol use disorder and alcoholic liver disease. Int J Mol Sci. 2023;24(3):2461. doi: 10.3390/ijms24032461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 109.Min X-H, Yu T, Qing Q, Yuan Y-H, Zhong W, Chen G-C, Zhao L-N, Deng N, Zhang L-F, Chen Q-K, et al. Abnormal differentiation of intestinal epithelium and intestinal barrier dysfunction in diabetic mice associated with depressed Notch/NICD transduction in Notch/Hes1 signal pathway. Cell Biol Int. 2014;38(10):1194–1204. doi: 10.1002/cbin.10323. [DOI] [PubMed] [Google Scholar]
- 110.Dahan S, Rabinowitz KM, Martin AP, Berin MC, Unkeless JC, Mayer L. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology. 2011;140(2):550–559. doi: 10.1053/j.gastro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tierney BT, Van den Abbeele P, Al-Ghalith GA, Verstrepen L, Ghyselinck J, Calatayud M, Marzorati M, Gadir AA, Daisley B, Reid G, et al. Capacity of a microbial synbiotic to rescue the in vitro metabolic activity of the gut microbiome following perturbation with alcohol or antibiotics. Appl Environ Microbiol. 2023;89(3):e01880–22. doi: 10.1128/aem.01880-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cresci GA, Glueck B, McMullen MR, Xin W, Allende D, Nagy LE. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J Gastro Hepatol. 2017;32(9):1587–1597. doi: 10.1111/jgh.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang X-W, Li Y-T, Ye J-Z, Lv L-X, Yang L-Y, Bian X-Y, Wu W-R, Wu J-J, Shi D, Wang Q, et al. New strain of pediococcus pentosaceus alleviates ethanol-induced liver injury by modulating the gut microbiota and short-chain fatty acid metabolism. World J Gastroenterol. 2020;26(40):6224–6240. doi: 10.3748/wjg.v26.i40.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith EA, Macfarlane GT. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol. 1997;33(3):180–188. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- 115.Yokoyama MT, Carlson JR. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am J Clin Nutr. 1979;32(1):173–178. doi: 10.1093/ajcn/32.1.173. [DOI] [PubMed] [Google Scholar]
- 116.Jin U-H, Lee S-O, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85(5):777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zelante T, Iannitti R, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 118.Sonner JK, Keil M, Falk-Paulsen M, Mishra N, Rehman A, Kramer M, Deumelandt K, Röwe J, Sanghvi K, Wolf L, et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat Commun. 2019;10(1):4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32(1):403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 120.Sun M, Ma N, He T, Johnston LJ, Ma X. Tryptophan (trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr. 2020;60(10):1760–1768. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- 121.Cella M, Miller H, Song C. Beyond NK cells: the expanding universe of innate lymphoid cells. Front Immunol. 2014;5:282. doi: 10.3389/fimmu.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 123.The role of intestinal microbes on intestinal barrier function and host immunity from a metabolite perspective - PubMed. https://pubmed.ncbi.nlm.nih.gov/37876938/. [DOI] [PMC free article] [PubMed]
- 124.J Y, Qiu J, Bostick JW, Ueda A, Schjerven H, Li S, Jobin C, Chen ZME, Zhou L. The aryl hydrocarbon receptor preferentially marks and promotes gut regulatory T cells. Cell Rep. 2017;21(8):2277–2290. doi: 10.1016/j.celrep.2017.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, Stockinger B, et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49(2):353–362.e5. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jiang H, Chen C, Gao J. Extensive summary of the important roles of indole propionic acid, a gut microbial metabolite in host health and disease. Nutrients. 2022;15(1):151. doi: 10.3390/nu15010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jennis M, Cavanaugh CR, Leo GC, Mabus JR, Lenhard J, Hornby PJ. Microbiota-derived tryptophan indoles increase after gastric bypass surgery and reduce intestinal permeability in vitro and in vivo. Neurogastro Motil. 2018;30(2). doi: 10.1111/nmo.13178. [DOI] [PubMed] [Google Scholar]
- 128.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet A, Qiu Z, Maher L, Redinbo M, Phillips R, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41(2):296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li J, Zhang L, Wu T, Li Y, Zhou X, Ruan Z. Indole-3-propionic acid improved the intestinal barrier by enhancing epithelial barrier and mucus barrier. J Agric Food Chem. 2021;69(5):1487–1495. doi: 10.1021/acs.jafc.0c05205. [DOI] [PubMed] [Google Scholar]
- 130.Zh Z, Xin F-Z, Xue Y, Hu Z, Han Y, Ma F, Zhou D, Liu X-L, Cui A, Liu Z, et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp Mol Med. 2019;51(9):1–14. doi: 10.1038/s12276-019-0304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alexeev EE, Lanis JM, Kao DJ, Campbell EL, Kelly CJ, Battista KD, Gerich ME, Jenkins BR, Walk ST, Kominsky DJ, et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am J Pathol. 2018;188(5):1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ehrlich AM, Pacheco AR, Henrick BM, Taft D, Xu G, Huda MN, Mishchuk D, Goodson ML, Slupsky C, Barile D, et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020;20(1):357. doi: 10.1186/s12866-020-02023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, Zhu W, Walker WA. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020;88(2):209–217. doi: 10.1038/s41390-019-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang W, Cho KY, Meng D, Walker WA. The impact of indole-3-lactic acid on immature intestinal innate immunity and development: a transcriptomic analysis. Sci Rep. 2021;11(1):8088. doi: 10.1038/s41598-021-87353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yu K, Li Q, Sun X, Peng X, Tang Q, Chu H, Zhou L, Wang B, Zhou Z, Deng X, et al. Bacterial indole-3-lactic acid affects epithelium–macrophage crosstalk to regulate intestinal homeostasis. Proc Natl Acad Sci USA. 2023;120(45):e2309032120. doi: 10.1073/pnas.2309032120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang G, Fan Y, Zhang G, Cai S, Ma Y, Yang L, Wang Y, Yu H, Qiao S, Zeng X, et al. Microbiota-derived indoles alleviate intestinal inflammation and modulate microbiome by microbial cross-feeding. Microbiome. 2024;12(1):59. doi: 10.1186/s40168-024-01750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li M, Han X, Sun L, Liu X, Zhang W, Hao J. Indole-3-acetic acid alleviates dss-induced colitis by promoting the production of R-equol from bifidobacterium pseudolongum. Gut Microbes. 2024;16(1):2329147. doi: 10.1080/19490976.2024.2329147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li M, Ding Y, Wei J, Dong Y, Wang J, Dai X, Yan J, Chu F, Zhang K, Meng F, et al. Gut microbiota metabolite indole-3-acetic acid maintains intestinal epithelial homeostasis through mucin sulfation. Gut Microbes. 2024;16(1):2377576. doi: 10.1080/19490976.2024.2377576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.T H, Duan Y, Wang Y, Oh J-H, Alexander LM, Huang W, Stärkel P, Ho SB, Gao B, Fiehn O, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68(8):1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang M, Feng X, Zhao Y, Lan Y, Xu H. Indole-3-acetamide from gut microbiota activated hepatic AhR and mediated the remission effect of Lactiplantibacillus plantarum P101 on alcoholic liver injury in mice. Food Funct. 2023;14(23):10535–10548. doi: 10.1039/D3FO03585A. [DOI] [PubMed] [Google Scholar]
- 141.Wrzosek L, Ciocan D, Hugot C, Spatz M, Dupeux M, Houron C, Lievin-Le Moal V, Puchois V, Ferrere G, Trainel N, et al. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut. 2021;70(7):1299–1308. doi: 10.1136/gutjnl-2020-321565. [DOI] [PubMed] [Google Scholar]
- 142.Gao B, Duan Y, Lang S, Barupal D, Wu T-C, Valdiviez L, Roberts B, Choy YY, Shen T, Byram G, et al. Functional microbiomics reveals alterations of the gut microbiome and host Co-metabolism in patients with alcoholic hepatitis. Hepatol Commun. 2020;4(8):1168–1182. doi: 10.1002/hep4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mrdjen M, Huang E, Pathak V, Bellar A, Welch N, Dasarathy J, Streem D, McClain CJ, Mitchell M, Radaeva S, et al. Dysregulated meta-organismal metabolism of aromatic amino acids in alcohol-associated liver disease. Hepatol Commun. 2023;7(11):e0284. doi: 10.1097/HC9.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72(1):137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 145.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50(12):2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 147.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 148.Yao L, Seaton, SC, Ndousse-Fetter, S, Adhikari, AA, DiBenedetto, N, Mina, AI, Banks, AS, Bry, L., Devlin, AS. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife. 2018;7:e37182. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang S, Dong W, Liu L, Xu M, Wang Y, Liu T, Zhang Y, Wang B, Cao H. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol Carcinog. 2019;58(7):1155–1167. doi: 10.1002/mc.22999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu M, Cen M, Shen Y, Zhu Y, Cheng F, Tang L, Hu W, Dai N. Deoxycholic acid-induced gut dysbiosis disrupts bile acid enterohepatic circulation and promotes intestinal inflammation. Dig Dis Sci. 2021;66(2):568–576. doi: 10.1007/s10620-020-06208-3. [DOI] [PubMed] [Google Scholar]
- 151.Friedman ES, Li Y, Shen TCD, Jiang J, Chau L, Adorini L, Babakhani F, Edwards J, Shapiro D, Zhao C, et al. FXR-Dependent modulation of the human small intestinal microbiome by the bile acid derivative obeticholic acid. Gastroenterology. 2018;155(6):1741–1752.e5. doi: 10.1053/j.gastro.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. 2019;7(1):75. doi: 10.1186/s40168-019-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jiang L, Zhang H, Xiao D, Wei H, Chen Y. Farnesoid X receptor (FXR): structures and ligands. Comput Struct Biotechnol J. 2021;19:2148–2159. doi: 10.1016/j.csbj.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chiang JYL. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183(10):6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 156.Kang J-H, Kim M, Yim M. FXR/TGR5 mediates inflammasome activation and host resistance to bacterial infection. Biochem Biophys Rep. 2021;27:101051. doi: 10.1016/j.bbrep.2021.101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Massafra V, Ijssennagger N, Plantinga M, Milona A, Ramos Pittol JM, Boes M, van Mil SWC. Splenic dendritic cell involvement in fxr-mediated amelioration of DSS colitis. Biochim Biophys Acta. 2016;1862(2):166–173. doi: 10.1016/j.bbadis.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 158.Biagioli M, Carino A, Cipriani S, Francisci D, Marchianò S, Scarpelli P, Sorcini D, Zampella A, Fiorucci S. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol. 2017;199(2):718–733. doi: 10.4049/jimmunol.1700183. [DOI] [PubMed] [Google Scholar]
- 159.Haselow K, Bode JG, Wammers M, Ehlting C, Keitel V, Kleinebrecht L, Schupp A-K, Häussinger D, Graf D. Bile acids pka-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol. 2013;94(6):1253–1264. doi: 10.1189/jlb.0812396. [DOI] [PubMed] [Google Scholar]
- 160.Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume MT, Higuchi H, Matsuoka K, Watanabe M, Itoh H, Kanai T, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology. 2012;136(2):153–162. doi: 10.1111/j.1365-2567.2012.03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, Pellicciari R, Schoonjans K. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology. 2020;159(3):956–968.e8. doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
- 162.Zhang B, Xie W, Krasowski MD. PXR: a xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics. 2008;9(11):1695–1709. doi: 10.2217/14622416.9.11.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dubrac S, Elentner A, Ebner S, Horejs-Hoeck J, Schmuth M. Modulation of T lymphocyte function by the pregnane X receptor. J Immunol. 2010;184(6):2949–2957. doi: 10.4049/jimmunol.0902151. [DOI] [PubMed] [Google Scholar]
- 164.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98(6):3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao G-P, Wang X-Y, Li J-W, Wang R, Ren F-Z, Pang G-F, Li Y-X. Imidacloprid increases intestinal permeability by disrupting tight junctions. Ecotoxicol Environ Saf. 2021;222:112476. doi: 10.1016/j.ecoenv.2021.112476. [DOI] [PubMed] [Google Scholar]
- 166.Uehara D, Tojima H, Kakizaki S, Yamazaki Y, Horiguchi N, Takizawa D, Sato K, Yamada M, Uraoka T. Constitutive androstane receptor and pregnane X receptor cooperatively ameliorate DSS-induced colitis. Dig Liver Dis. 2019;51(2):226–235. doi: 10.1016/j.dld.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 167.Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. Pregnane X receptor activation ameliorates dss-induced inflammatory bowel disease via inhibition of NF-κB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292(4):G1114–G1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 168.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296(5571):1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 169.Széles L, Keresztes G, Töröcsik D, Balajthy Z, Krenács L, Póliska S, Steinmeyer A, Zuegel U, Pruenster M, Rot A, et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol. 2009;182(4):2074–2083. doi: 10.4049/jimmunol.0803345. [DOI] [PubMed] [Google Scholar]
- 170.Bhalla AK, Amento EP, Krane SM. Differential effects of 1,25-dihydroxyvitamin D3 on human lymphocytes and monocyte/macrophages: inhibition of interleukin-2 and augmentation of interleukin-1 production. Cell Immunol. 1986;98(2):311–322. doi: 10.1016/0008-8749(86)90291-1. [DOI] [PubMed] [Google Scholar]
- 171.Lemire JM, Adams JS, Sakai R, Jordan SC. 1 alpha,25-dihydroxyvitamin D3 suppresses proliferation and immunoglobulin production by normal human peripheral blood mononuclear cells. J Clin Invest. 1984;74(2):657–661. doi: 10.1172/JCI111465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Axelson M, Mörk B, Sjövall J. Ethanol has an acute effect on bile acid biosynthesis in man. FEBS Lett. 1991;281(1–2):155–159. doi: 10.1016/0014-5793(91)80382-D. [DOI] [PubMed] [Google Scholar]
- 173.Marin GA, Ward NL, Fischer R. Effect of ethanol on pancreatic and biliary secretions in humans. Am J Dig Dis. 1973;18(10):825–833. doi: 10.1007/BF01073332. [DOI] [PubMed] [Google Scholar]
- 174.Brandl K, Hartmann P, Jih LJ, Pizzo DP, Argemi J, Ventura-Cots M, Coulter S, Liddle C, Ling L, Rossi SJ, et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol. 2018;69(2):396–405. doi: 10.1016/j.jhep.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chanda D, Kim Y-H, Li T, Misra J, Kim D-K, Kim JR, Kwon J, Jeong W-I, Ahn S-H, Park T-S, et al. Hepatic cannabinoid receptor type 1 mediates alcohol-induced regulation of bile acid enzyme genes expression via CREBH. PLOS ONE. 2013;8(7):e68845. doi: 10.1371/journal.pone.0068845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gong X, Zhang Q, Ruan Y, Hu M, Liu Z, Gong L. Chronic alcohol consumption increased bile acid levels in Enterohepatic circulation and reduced efficacy of irinotecan. Alcohol Alcohol. 2020;55(3):264–277. doi: 10.1093/alcalc/agaa005. [DOI] [PubMed] [Google Scholar]
- 178.Ciocan D, Spatz M, Trainel N, Hardonnière K, Domenichini S, Mercier-Nomé F, Desmons A, Humbert L, Durand S, Kroemer G, et al. Modulation of the bile acid Enterohepatic cycle by intestinal microbiota alleviates alcohol liver disease. Cells. 2022;11(6):968. doi: 10.3390/cells11060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tajiri K. Branched-chain amino acids in liver diseases. World J Gastroenterol. 2013;19(43):7620–7629. doi: 10.3748/wjg.v19.i43.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.van den Berg EH, Flores-Guerrero JL, Gruppen EG, de Borst MH, Wolak-Dinsmore J, Connelly MA, Bakker SJL, Dullaart RPF. Non-alcoholic fatty liver disease and risk of incident type 2 diabetes: role of circulating branched-chain amino acids. Nutrients. 2019;11(3):705. doi: 10.3390/nu11030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nezami Ranjbar MR, Luo Y, Di Poto C, Varghese RS, Ferrarini A, Zhang C, Sarhan NI, Soliman H, Tadesse MG, Ziada DH, et al. GC-MS based plasma metabolomics for identification of candidate biomarkers for hepatocellular carcinoma in Egyptian cohort. PLOS ONE. 2015;10(6):e0127299. doi: 10.1371/journal.pone.0127299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ishikawa T. Branched-chain amino acids to tyrosine ratio value as a potential prognostic factor for hepatocellular carcinoma. World J Gastroenterol. 2012;18(17):2005–2008. doi: 10.3748/wjg.v18.i17.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vilstrup H, Amodio P, Bajaj J, Cordoba J, Ferenci P, Mullen KD, Weissenborn K, Wong P. Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. Hepatology. 2014;60(2):715–735. doi: 10.1002/hep.27210. [DOI] [PubMed] [Google Scholar]
- 184.Jc K, De H, Garsin DA. Branching out: alterations in bacterial physiology and virulence due to branched-chain amino acid deprivation. mBio. 2018;9(5). doi: 10.1128/mBio.01188-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park JG, Tak WY, Park SY, Kweon YO, Chung WJ, Jang BK, Bae SH, Lee HJ, Jang JY, Suk KT, et al. Effects of branched-chain amino acid (BCAA) supplementation on the progression of advanced liver disease: a Korean nationwide, multicenter, prospective, observational, cohort study. Nutrients. 2020;12(5):1429. doi: 10.3390/nu12051429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xie G, Zhong W, Zheng X, Li Q, Qiu Y, Li H, Chen H, Zhou Z, Jia W. Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J Proteome Res. 2013;12(7):3297–3306. doi: 10.1021/pr400362z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zeisel SH, Warrier M. Trimethylamine N-Oxide, the microbiome, and heart and kidney disease. Annu Rev Nutr. 2017;37(1):157–181. doi: 10.1146/annurev-nutr-071816-064732. [DOI] [PubMed] [Google Scholar]
- 189.Stremmel W, Schmidt KV, Schuhmann V, Kratzer F, Garbade SF, Langhans C-D, Fricker G, Okun JG. Blood trimethylamine-N-Oxide originates from microbiota mediated breakdown of phosphatidylcholine and absorption from small intestine. PLOS ONE. 2017;12(1):e0170742. doi: 10.1371/journal.pone.0170742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li D, Lu Y, Yuan S, Cai X, He Y, Chen J, Wu Q, He D, Fang A, Bo Y, et al. Gut microbiota–derived metabolite trimethylamine-N-oxide and multiple health outcomes: an umbrella review and updated meta-analysis. Am J Clin Nutr. 2022;116(1):230–243. doi: 10.1093/ajcn/nqac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhou Y, Zhang Y, Jin S, Lv J, Li M, Feng N. The gut microbiota derived metabolite trimethylamine N-oxide: its important role in cancer and other diseases. Biomed Pharmacother. 2024;177:117031. doi: 10.1016/j.biopha.2024.117031. [DOI] [PubMed] [Google Scholar]
- 193.Dai X, Hou H, Zhang W, Liu T, Li Y, Wang S, Wang B, Cao H. Microbial metabolites: critical regulators in NAFLD. Front Microbiol. 2020;11:567654. doi: 10.3389/fmicb.2020.567654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Coulbault L, Laniepce A, Segobin S, Boudehent C, Cabé N, Pitel AL. Trimethylamine N-Oxide (TMAO) and indoxyl sulfate concentrations in patients with alcohol use disorder. Nutrients. 2022;14(19):3964. doi: 10.3390/nu14193964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Haas EA, Saad MJ, Santos A, Vitulo N, Lemos WJ, Martins AM, Picossi CR, Favarato D, Gaspar RS, Magro DO, et al. A red wine intervention does not modify plasma trimethylamine N-oxide but is associated with broad shifts in the plasma metabolome and gut microbiota composition. Am J Clin Nutr. 2022;116(6):1515–1529. doi: 10.1093/ajcn/nqac286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.El-Hashash A. Chapter 6 - application of new approaches for intestinal repair and regeneration via stem cell–based tissue engineering. In: El-Hashash AHK, and Meguid EA. editors. The intestine. Vol. 2. London: Academic Press; 2021. p. 87–99. [Google Scholar]
- 197.Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastro Hepatol. 2019;16(1):19–34. doi: 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 198.Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer NF, Bourgaux J-F, Pignodel C, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192(5):767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Luo H, Li M, Wang F, Yang Y, Wang Q, Zhao Y, Du F, Chen Y, Shen J, Zhao Q, et al. The role of intestinal stem cell within gut homeostasis: focusing on its interplay with gut microbiota and the regulating pathways. Int J Biol Sci. 2022;18(13):5185–5206. doi: 10.7150/ijbs.72600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rees WD, Tandun R, Yau E, Zachos NC, Steiner TS. Regenerative intestinal stem cells induced by acute and chronic injury: the saving grace of the epithelium? Front Cell Dev Biol. 2020;8:583919. doi: 10.3389/fcell.2020.583919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17(14):1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Palikuqi B, Rispal J, Klein O. Good neighbors: the niche that fine tunes mammalian intestinal regeneration. Cold Spring Harb Perspect Biol. 2022;14(5):a040865. doi: 10.1101/cshperspect.a040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Okamoto R, Tsuchiya K, Nemoto Y, Akiyama J, Nakamura T, Kanai T, Watanabe M. Requirement of notch activation during regeneration of the intestinal epithelia. Am J Physiol Gastrointest Liver Physiol. 2009;296(1):G23–35. doi: 10.1152/ajpgi.90225.2008. [DOI] [PubMed] [Google Scholar]
- 204.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165(7):1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bajic D, Niemann A, Hillmer A-K, Mejias-Luque R, Bluemel S, Docampo M, Funk MC, Tonin E, Boutros M, Schnabl B, et al. Gut microbiota-derived propionate regulates the expression of Reg3 mucosal lectins and ameliorates experimental colitis in mice. J Crohns Colitis. 2020;14(10):1462–1472. doi: 10.1093/ecco-jcc/jjaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Duan C, Wu J, Wang Z, Tan C, Hou L, Qian W, Han C, Hou X. Fucose promotes intestinal stem cell-mediated intestinal epithelial development through promoting akkermansia-related propanoate metabolism. Gut Microbes. 2023;15(1):2233149. doi: 10.1080/19490976.2023.2233149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Markandey M, Bajaj A, Ilott NE, Kedia S, Travis S, Powrie F, Ahuja V. Gut microbiota: sculptors of the intestinal stem cell niche in health and inflammatory bowel disease. Gut Microbes. 2021;13(1):1990827. doi: 10.1080/19490976.2021.1990827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates β-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15(5):2156–2163. doi: 10.1091/mbc.e03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Milovic V, Teller IC, Faust D, Caspary WF, Stein J. Effects of deoxycholate on human colon cancer cells: apoptosis or proliferation. Eur J Clin Invest. 2002;32(1):29–34. doi: 10.1046/j.0014-2972.2001.00938.x. [DOI] [PubMed] [Google Scholar]
- 210.Park J-H, Lee J-M, Lee E-J, Hwang W-B, Kim D-J. Indole-3-carbinol promotes goblet-cell differentiation regulating wnt and notch signaling pathways AhR-dependently. Mol Cells. 2018;41(4):290–300. doi: 10.14348/molcells.2018.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mathern DR, Laitman LE, Hovhannisyan Z, Dunkin D, Farsio S, Malik TJ, Roda G, Chitre A, Iuga AC, Yeretssian G, et al. Mouse and human notch-1 regulate mucosal immune responses. Mucosal Immunol. 2014;7(4):995–1005. doi: 10.1038/mi.2013.118. [DOI] [PubMed] [Google Scholar]
- 212.Lu R, Voigt RM, Zhang Y, Kato I, Xia Y, Forsyth CB, Keshavarzian A, Sun J. Alcohol injury damages intestinal stem cells. Alcohol Clin Exp Res. 2017;41(4):727–734. doi: 10.1111/acer.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Park J-H, Jung IK, Lee Y, Jin S, Yun HJ, Kim BW, Kwon HJ. Alcohol stimulates the proliferation of mouse small intestinal epithelial cells via wnt signaling. Biochem Biophys Res Commun. 2021;534:639–645. doi: 10.1016/j.bbrc.2020.11.028. [DOI] [PubMed] [Google Scholar]
- 214.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of lactobacillus rhamnosus GG treatment. PLOS ONE. 2013;8(1):e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Capurso L. Thirty years of lactobacillus rhamnosus GG: a review. J Clin Gastro. 2019;53(Suppl 1):S1–S41. doi: 10.1097/MCG.0000000000001170. [DOI] [PubMed] [Google Scholar]
- 216.Jiang M, Li F, Liu Y, Gu Z, Zhang L, Lee J, He L, Vatsalya V, Zhang H-G, Deng Z, et al. Probiotic-derived nanoparticles inhibit ALD through intestinal miR194 suppression and subsequent FXR activation. Hepatology. 2023;77(4):1164–1180. doi: 10.1002/hep.32608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhao H, Zhao C, Dong Y, Zhang M, Wang Y, Li F, Li X, McClain C, Yang S, Feng W, et al. Inhibition of miR122a by lactobacillus rhamnosus GG culture supernatant increases intestinal occludin expression and protects mice from alcoholic liver disease. Toxicol Lett. 2015;234(3):194–200. doi: 10.1016/j.toxlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 218.Zhu Y, Wang X, Zhu L, Tu Y, Chen W, Gong L, Pan T, Lin H, Lin J, Sun H, et al. Lactobacillus rhamnosus GG combined with inosine ameliorates alcohol-induced liver injury through regulation of intestinal barrier and Treg/Th1 cells. Toxicol Appl Pharmacol. 2022;439:115923. doi: 10.1016/j.taap.2022.115923. [DOI] [PubMed] [Google Scholar]
- 219.Bruch-Bertani JP, Uribe-Cruz C, Pasqualotto A, Longo L, Ayres R, Beskow CB, Barth AL, Lima-Morales D, Meurer F, Tayguara Silveira Guerreiro G, et al. Hepatoprotective effect of probiotic lactobacillus rhamnosus GG through the modulation of gut permeability and inflammasomes in a model of alcoholic liver disease in zebrafish. J Am Coll Nutr. 2020;39(2):163–170. doi: 10.1080/07315724.2019.1627955. [DOI] [PubMed] [Google Scholar]
- 220.Gu Z, Li F, Liu Y, Jiang M, Zhang L, He L, Wilkey DW, Merchant M, Zhang X, Deng Z-B, et al. Exosome-like nanoparticles from lactobacillus rhamnosusGG protect against alcohol-associated liver disease through intestinal aryl hydrocarbon receptor in mice. Hepatol Commun. 2021;5(5):846–864. doi: 10.1002/hep4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 2009;43(2):163–172. doi: 10.1016/j.alcohol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang Y, Liu Y, Sidhu A, Ma Z, McClain C, Feng W. Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am J Physiol Gastrointest Liver Physiol. 2012;303(1):G32–41. doi: 10.1152/ajpgi.00024.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Arora S, Kaur IP, Chopra K, Rishi P. Efficiency of double layered microencapsulated probiotic to modulate proinflammatory molecular markers for the management of alcoholic liver disease. Mediators Inflamm. 2014;2014:1–11. doi: 10.1155/2014/715130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xu F, Chen, Z, Xie, L, Yang, S, Li, Y, Wu, J, Wu, Y, Li, S, Zhang, X, Ma, Y., Liu, Y. Lactobacillus plantarum ST-III culture supernatant protects against acute alcohol-induced liver and intestinal injury. Aging (Albany NY). 2023;15:2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cannon AR, Shim EH, Kuprys PV, Choudhry MA. IL-22 and Lactobacillus delbrueckii mitigate alcohol-induced exacerbation of dss-induced colitis. J Leukoc Biol. 2022;112(6):1471–1484. doi: 10.1002/JLB.4A0122-068R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cui Y, Guo P, Ning M, Yue Y, Yuan Y, Yue T. Kluyveromyces marxianus supplementation ameliorates alcohol-induced liver injury associated with the modulation of gut microbiota in mice. Food Funct. 2023;14(21):9920–9935. doi: 10.1039/D3FO01796F. [DOI] [PubMed] [Google Scholar]
- 227.Jiang X, Yan C, Zhang H, Chen L, Jiang R, Zheng K, Jin W, Ma H, Liu X, Dong M, et al. Oral probiotic expressing human ethanol dehydrogenase attenuates damage caused by acute alcohol consumption in mice. Microbiol Spectr. 2023;11(3):e0429422. doi: 10.1128/spectrum.04294-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Patel F, Parwani K, Rao P, Patel D, Rawal R, Mandal P. Prophylactic treatment of probiotic and metformin mitigates ethanol-induced intestinal barrier injury. In Vitro, In Vivo, and In Silico Approaches. Mediators Inflamm. 2021;2021:1–32. doi: 10.1155/2021/5245197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this study’s findings are openly available in cited papers.


