Abstract
Recent advances in high-throughput measurement technologies have enabled the analysis of molecular perturbations associated with disease phenotypes at the multi-omic level. Such perturbations can range in scale from fluctuations of individual molecules to entire biological pathways. Data-driven clustering algorithms have long been used to group interactions into interpretable functional modules; however, these modules are typically constrained to a fixed size or statistical cutoff. Furthermore, modules are often analyzed independently of their broader biological context. Consequently, such clustering approaches limit the ability to explore functional module associations with disease phenotypes across multiple scales. Here, we introduce AutoFocus, a data-driven method that hierarchically organizes biomolecules and tests for phenotype enrichment at every level within the hierarchy. As a result, the method allows disease-associated modules to emerge at any scale. We evaluated this approach using two datasets: First, we explored associations of biomolecules from the multi-omic QMDiab dataset (n = 388) with the well-characterized type 2 diabetes phenotype. Secondly, we utilized the ROS/MAP Alzheimer’s disease dataset (n = 500), consisting of high-throughput measurements of brain tissue to explore modules associated with multiple Alzheimer’s Disease-related phenotypes. Our method identifies modules that are multi-omic, span multiple pathways, and vary in size. We provide an interactive tool to explore this hierarchy at different levels and probe enriched modules, empowering users to examine the full hierarchy, delve into biomolecular drivers of disease phenotype within a module, and incorporate functional annotations.
Subject terms: Data integration, Functional clustering
AutoFocus is an interactive tool to explore hierarchical frameworks of multi-omics disease associations to probe modules and test for phenotype enrichment at every level within the hierarchy and incorporates functional annotations.
Introduction
The increasing availability of high-throughput measurement technologies has led to the generation of a large number of multi-omics datasets, providing molecular snapshots of biological systems at all -omic levels of regulation1,2. Such multi-omic datasets can be explored to infer molecular interactions3–5, or in the context of disease, to identify perturbations for a deeper understanding of pathophysiological mechanisms6–8. To this end, various computational methods have been developed to cluster multi-omic biomolecules into easier-to-interpret functional modules that attempt to describe alterations caused by a disease in a biological system5,9–13.
Functional modules generally consist of interacting biomolecules that are coordinated, coregulated, or otherwise involved in the same biological process14,15. Grouping molecules into such functional modules can often be achieved using existing functional annotations available in large databases comprised of experimentally derived interactions16,17. However, these types of annotations are constrained by research bias and are limited between -omic layers, for example those between metabolomics and transcriptomics18,19. Thus, while experimentally validated annotations promise to create well-supported functional modules, the lack of exhaustive annotations in a high-throughput context is often a severe limitation. Data-driven methods that infer interactions between biomolecules directly from the data are often a compelling alternative. Such methods include k-means clustering, hierarchical clustering, network approaches, principal component analysis (PCA), or other matrix factorization approaches12,20–23.
A significant challenge for these data-driven methods which statistically identify modules is determining the appropriate scale of a biological process that should be deemed a module. For instance, the catabolism of carbon units of cells can be studied at various levels, such as single-molecule level (glucose or pyruvate), pathway level (glycolysis), or functional pathway group level (central carbon metabolism, Fig. 1a)24. This exemplifies the concept that functional modules are not necessarily distinct processes, and that different hierarchical levels of super- and sub-modules exist25,26. In addition, previous work by our group has shown that phenotypes can impact biological system at a variety of levels; certain phenotypes, for example related to specific pathological perturbations, manifest at the level of a few molecules, while others, like sex effects, impact entire pathways or pathway groups12,27.
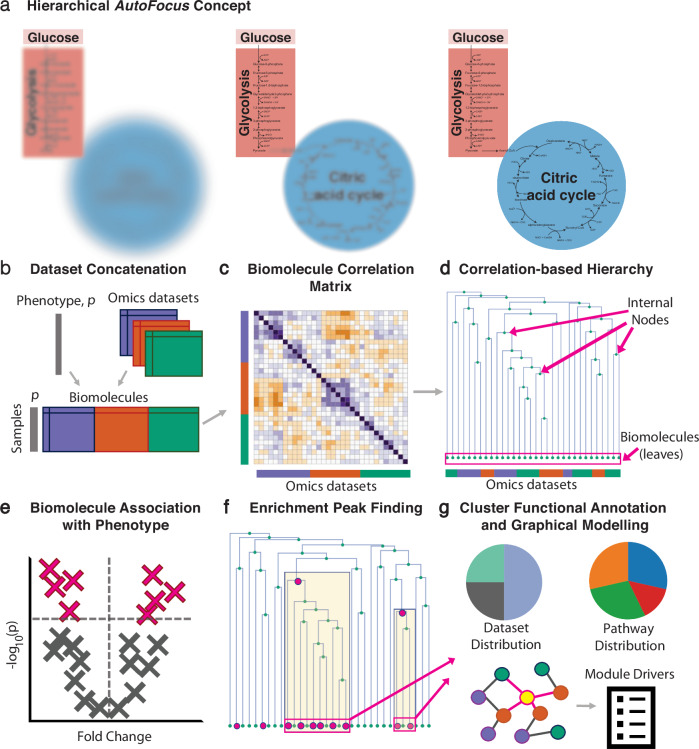
Fig. 1. AutoFocus method overview.
a Conceptual depiction of applying “focus” to the biological process of carbon metabolism at different hierarchical levels. b Multiple molecular datasets with biomolecules from the same n samples are concatenated into a single matrix, accompanied by sample phenotype information, p. c Correlation coefficients between molecules are calculated to generate a correlation matrix, d Correlation coefficients are converted to distances to create a hierarchical tree of biomolecules, e Biomolecules are univariately correlated with the phenotype of interest and filtered for statistical significance, f Enrichment “peaks” are detected by performing an enrichment analysis of the “leaves” descending from each internal node, i.e., the number of significantly correlated molecules in the respective cluster. g Functional annotation and module driver analysis is performed on each enriched module.
Despite the biological relevance of such hierarchies, current module identification algorithms are not designed to produce data-driven modules that can explore biological processes at multiple scales. Existing algorithms apply restrictive parameters, such as p-value cutoffs, network connectivity metrics, or desired module size, to demarcate modules at a fixed level, which are then further explored as standalone processes, disconnected from the larger biological context10,11,28–30. Thus, when analyzing the effects of a disease-phenotype on these modules, fixed-scale approaches do not reveal how a phenotype impacts different granularity levels within a module (e.g., single molecule versus pathway levels), and cannot determine how impacted modules relate to one another in the larger biological system. Further, fixed-scale modules restrict all phenotype associations to a single level, failing to capture the variety of scales that may exist among diverse phenotypes.
We here address the issue of identifying multi-level modules and allowing phenotype association to manifest at any scale by designing an interactive and adaptive hierarchical clustering and phenotype association approach. We introduce a new method, AutoFocus, that hierarchically structures molecular datasets, overlays phenotype association onto the hierarchy, and performs enrichment analysis to annotate functional modules within this system. The method is accompanied by an interactive application that allows a user to explore the hierarchy created by their data and provides functional insights through module annotation and the identification of module members driving phenotype association (Fig. 1). We then apply our method to two independent datasets to validate its ability to capture known disease signal and explore new findings: Type 2 Diabetes in The Qatar Metabolomics Study on Diabetes (QMDiab, n = 388)2, which contains 12 multi-omic datasets including metabolomics, proteomics, and glycomics; and the Alzheimer’s Disease in the Religious Orders Study/Memory and Aging Project (ROS/MAP, n = 500)31, which includes a metabolomics and proteomics platform and multiple clinical phenotypes. Finally, we examine how the clusters output by the AutoFocus method compare to those created by existing clustering algorithms.
Results
Description of AutoFocus framework
The AutoFocus tool enables fast clustering and phenotype association of multiple omics datasets, accompanied by an intuitive, interactive application for result exploration. Preprocessed, matched-sample omics datasets from any specimen, body fluid, or platform, are combined and pairwise correlated (Fig. 1b, c). These correlations are transformed into a distance metric that is used to structure all molecules into a single dependency tree based on well-established hierarchical clustering (Fig. 1d). Univariate associations of each molecule with a desired phenotype of interest are calculated, and significantly associated molecules, which are the “leaves” of the tree, are annotated at the bottom of the diagram (Fig. 1d–f).
The tree is then scanned from top to bottom. For each internal node of the tree, the leaves descending from that node create a cluster (see highlighted parts of Fig. 1f). An enrichment analysis of significant hits is performed on the molecules within that cluster. If a user-defined enrichment threshold is reached, that internal node is labeled as an “enrichment peak” (Fig. 1f). Finally, functional annotation is performed on the modules associated with each peak along with a phenotype “driver” analysis (Fig. 1g). Drivers are defined as module members sharing a direct, unconfounded correlation edge with the disease phenotype based on a mixed-distribution graphical model.
All AutoFocus functionalities are available as an R package at https://github.com/krumsieklab/autofocus. As input, the method accepts Excel sheets of preprocessed measurements from multiple omics datasets along with dataset-specific molecular annotations and sample-specific annotations, including phenotype(s) of interest and covariate information. Accompanying the workflow in Fig. 1 is an interactive Shiny application that allows a user to set an enrichment threshold and easily explore the resulting functional modules.
Intra- and inter-dataset relationships in the 12-dataset multi-omics QMDiab study
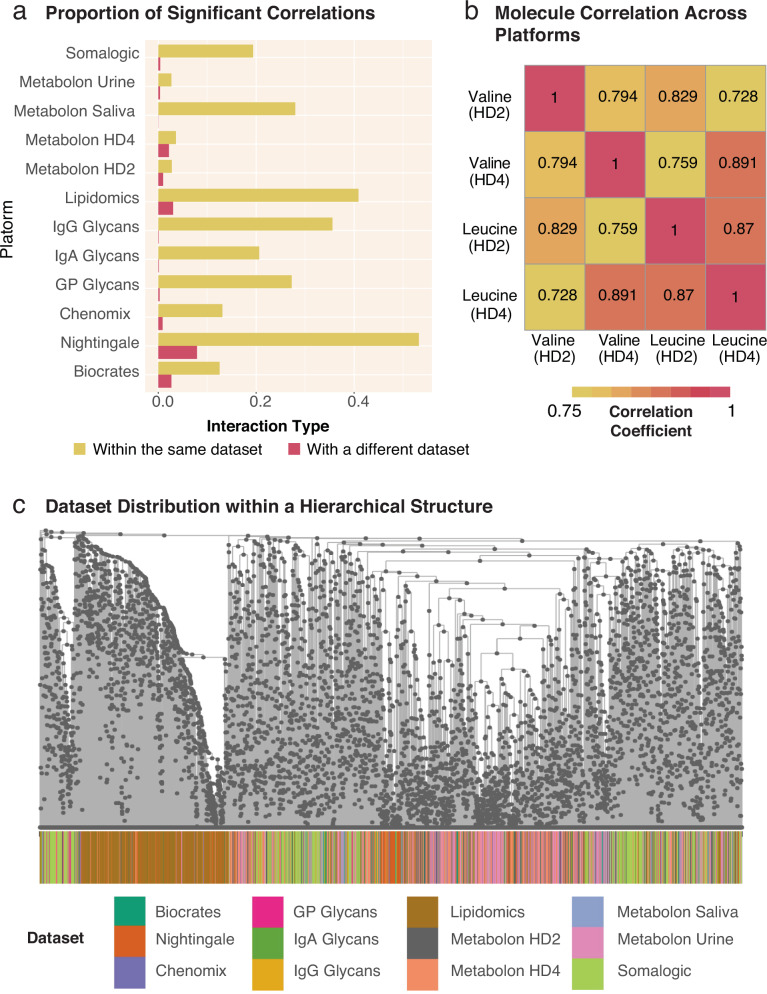
As all data-driven clustering methods depend on the similarity relationships between the measured variables, we first explored the correlation structure of a dataset with various omics layers to get an overview of the highly complex underlying statistical structures. The QMDiab dataset consists of 5135 biomolecules from 8 metabolomics datasets (5 different platforms performed on plasma, 2 on urine, and 1 on saliva), 3 blood glycomics datasets, and 1 blood proteomics dataset (Table 1). These 12 datasets were combined and the pairwise biomolecule correlations were calculated. A systemic correlation bias was detected across the various assays: Intra-dataset correlations were systematically higher than inter-dataset correlations (Fig. 2a). This bias persisted even in instances where the same molecule was measured on different platforms. For example, when analyzing two specific molecules, valine and leucine, measured on two almost identical plasma metabolomics platforms, we observed that valine had a higher correlation with leucine measured on the same platform than its correlation with itself measured on a different platform (Fig. 2b). As a consequence of this bias, molecules from the same dataset tended to be in close proximity in a hierarchical structure (Fig. 2c). This poses a problem when using correlation networks to statistically extract interactions between these molecules, a common approach to inferring biological relationships. We systematically probed the QMDiab correlation network for the optimal statistical cutoff to create a network whose edge set best models ground truth interactions. We found that this optimal cutoff differs between ground truth annotations for intra-dataset edges (metabolite pathways) and ground truth annotations for inter-dataset edges (KEGG and Recon3D-based gene-metabolite edges, Supplementary Fig. 1)32. This indicates the inability of a single statistical cutoff to recover biologically relevant interactions in a multi-dataset context.
Table 1.
Overview of -omic types, sample types, platforms, collection methods, and analyte count for the QMDiab and ROS/MAP datasets
| Dataset | Omic type | Sample type | Platform | Method | # of analytes |
|---|---|---|---|---|---|
|
QMDiab (n = 410) |
Metabolomics | Plasma | Metabolon (HD2) | UHPLC/GC-MS/MS | 466 |
| Plasma | Metabolon (HD4) | UHPLC/GC-MS/MS | 843 | ||
| Plasma | Nightingale | NMR | 224 | ||
| Plasma | Biocrates p150 | FIA-MS/MS | 161 | ||
| Saliva | Metabolon | UHPLC/GC-MS/MS | 251 | ||
| Urine | Metabolon | UHPLC/GC-MS/MS | 695 | ||
| Urine | Chenomx | NMR | 32 | ||
| Proteomics | Plasma | SOMAscan | SOMAmer + DNA microarray | 1141 | |
| Lipidomics | Plasma | Metabolon (Lipidyzer) | LC-MS | 1133 | |
| Glycomics | Plasma | Genos | IgG | 39 | |
| Plasma | Genos | Total N-glycans | 60 | ||
| Plasma | Leiden University | IgA | 90 | ||
|
ROS/MAP (n = 500) |
Metabolomics | Brain Tissue | Metabolon | UHPLC/GC-MS/MS | 667 |
| Proteomics | Brain Tissue | ACQUITY UPLC + TSQ-Vantage MS | UHPLC-MS/MS | 7526 |
Fig. 2. Correlation values within and across datasets.
a Proportion of significant correlations between biomolecules within and across datasets. For every dataset, the proportion of significant correlation coefficients within each dataset is substantially larger than across datasets. Consequently, statistical methods that depend on correlations will be biased towards intra-dataset interactions in a multi-omics setting. b Example correlations between two molecules measured on the sample blood samples using two similar metabolomics platforms, Metabolon Plasma HD2 and Metabolon Plasma HD4. Valine on the HD2 platform correlated stronger with Leucine measured on the same platform than with Valine on the HD4 platform. This further illustrates the tendency for stronger correlations within a dataset than between datasets. c Dataset distribution in the correlation-based hierarchical structure formed on the QMDiab dataset. Strong intra-dataset correlations can be seen for lipids (brown) and to a lesser extent for proteomics (light green), as these two datasets have dense regions where they segregate from the other -omics datasets which are otherwise thought to be well integrated.
Notably, this observation of higher correlations within the same omics layers appears to be a natural feature of multi-omics datasets. Neither AutoFocus, nor any other clustering-based method can sufficiently remove this bias, as all clustering methods rely on the associations between measured molecules. However, unlike existing methods, the hierarchical framework of AutoFocus does not apply a fixed statistical cutoff to correlations between analytes, allowing any intra- and inter- dataset relationships to naturally emerge from the data structure. As the AutoFocus method evaluates clusters at every internal node of a hierarchy, clusters formed by nodes closer to the root of the tree will encompass molecules spanning different -omics, fluids, and datasets whose relationships would have been excluded when using statistical significance-based cutoffs (Fig. 1c). The resulting clusters are more representative of multi-omic, multi-fluidic, and multi-dataset biological interactions as compared to cutoff-dependent clustering methods.
AutoFocus analysis on QMDiab reveals impact of type 2 diabetes at multiple levels of molecular interactions
Systems-level analysis of type 2 diabetes
The phenotype of interest used for this analysis was Type 2 Diabetes (T2D) diagnosis. After correcting for age, sex, and BMI, 188 of the 5135 molecules were found to be significantly associated with Type 2 Diabetes (p < 0.05, Bonferroni adjusted), covering 10 of the 12 omics datasets. The IgG and IgA glycomics datasets showed no significant associations with T2D. We observed a broad distribution of signal across the hierarchical tree (Fig. 3a), suggesting a system wide T2D effect across omics and body fluids. Certain regions of the tree had substantially denser distributions of significantly associated biomolecules, suggesting hotspots of T2D perturbation.
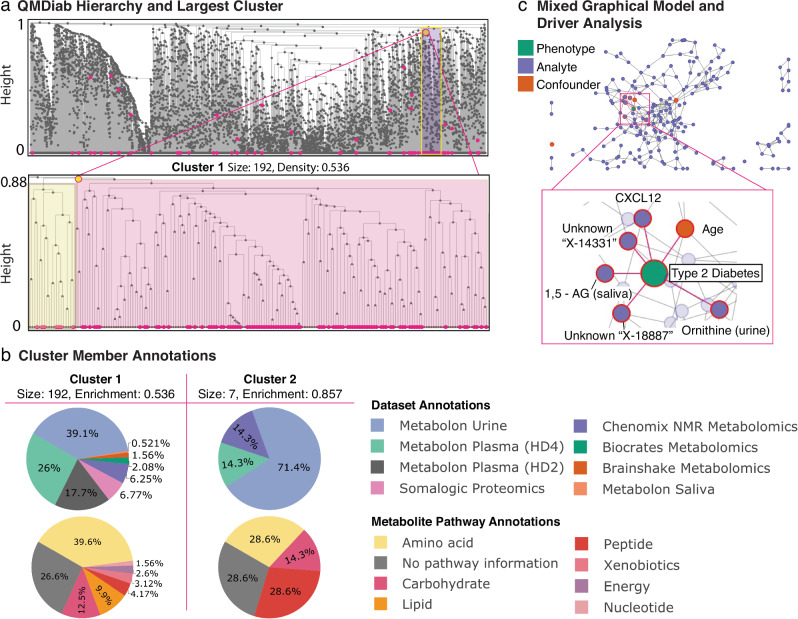
Fig. 3. AutoFocus on the QMDiab dataset.
The dataset included a total of 388 samples and 5135 biomolecules from 12 datasets: 5 metabolomics platforms on plasma, 2 on urine, and 1 on saliva, 3 blood glycomics datasets and 1 blood proteomics dataset. a View of the full hierarchical structure created from the QMDiab dataset. Magenta circles at the bottom of the tree indicate significant molecules, circles within the tree indicate modules that passed the enrichment threshold. Significant molecules were dispersed throughout the leaves of the tree and enriched modules were scattered throughout the hierarchy at a wide range of heights. The high-density region of significant molecules towards the right corresponds to the largest enriched module at the highest height. Below is a zoomed view of this module, with the left sub-tree in yellow and the right sub-tree in pink. b Pie charts of the dataset and pathway makeup of the two largest modules along with their size and significant-node enrichment fraction. Pathway annotations were only available for the metabolites measured by Metabolon. c Confounder-corrected mixed graphical model of the molecules in the largest module with phenotype. The zoomed-in view is of nodes with edges to the Type 2 Diabetes phenotype which include 1,5-AG in saliva, ornithine in urine, and the CXCL12 protein, along with the confounder age and 2 unknown molecules. As these molecules are directly connected to the T2D phenotype, we mark them as statistical “drivers” of the disease in this module.
Type 2 diabetes modules
To identify T2D modules for the QMDiab dataset, we applied a “majority vote” enrichment threshold of 0.5, where at least 50% of a cluster’s members must be significantly associated with T2D for it to be designated as a T2D module. The AutoFocus method identified 21 modules, ranging in size from 2 to 192 biomolecules (Supplementary Fig. 2 and Supplementary Data 1). In addition, there were 33 single-molecule modules, identified as T2D-associated molecules that did not belong to any of the 21 modules. The identified T2D modules substantially ranged in scale (Fig. 3a), from very high correlation near the leaves at tree height 0 to low correlations near the root at tree height 1. This shows that Type 2 Diabetes manifests at various levels of the biological hierarchy, from closely connected molecules to larger pathways.
As expected, most of the smaller, highly correlated modules tended to contain molecules from only one dataset, due to the aforementioned within-dataset correlation biases, most notably within the lipidomic and proteomics datasets. However, AutoFocus identified six T2D modules that contained molecules from multiple omics or fluids (Supplementary Fig. 2). The smaller of these modules included molecules that were measured multiple times but on different platforms, e.g., one module which was made up of pyroglutamine measured on the Metabolon HD2 and HD4 platforms. The largest module with 192 molecules (Fig. 3a), comprising of the bulk of the T2D-associated analytes in the QMDiab dataset, brought together molecules from both metabolomic and proteomic datasets and all three body fluids in QMDiab (Fig. 3b).
This 192-analyte module contained two sub-modules, each with substantially different functional components. The larger, right-hand “child” tree (Fig. 3a, pink) contained molecules involved in energy metabolism, including various carbohydrates, such as mannose, glucose, and 1,5-anhydroglucitol, which are known biomarkers of diabetes33,34, as well as TCA cycle metabolites like pyruvate and lactate in plasma. In addition, this module showed significant changes in the abundance of ketone bodies acetoacetate and 3-hydroxybutyrate in urine, supporting the prevalence of ketosis and ketone body secretion in T2D patients35.
The left sub-tree (Fig. 3a, yellow) in this module contained biomolecules related to bone growth, mineralization, and degradation, as well as some chemokines and endothelial cell proteins. The bone degradation molecules included the proteins Osteomodulin (OMD), Integrin-binding sialoprotein (IBSP), and C-type lectin domain protein (Clec11A) and the metabolite prolylhydroxyproline in both plasma and urine36–39. Osteoporesis has a well-documented relationship to T2D, and although the mechanisms are not established, hypotheses for the link include inflammation and microangiopathy40. The presence of chemokines Stromal cell-derived factor 1 (CXCL12) and C-C motif chemokine 22 (CCL22), as well as Endothelial cell-specific molecule 1 (ESM1) in this sub-module presented potential osteoporosis links to inflammation and microangiopathy, respectively.
Type 2 diabetes module driver analysis
We further analyzed this module using a mixed graphical model (MGM) approach, which allowed us to differentiate direct correlations between biomolecules and T2D from indirect, statistically confounded correlations. We identified and labeled as drivers those molecules that had a direct correlation with T2D diagnosis, signified by sharing an edge in the MGM network. The MGM identified 5 biomolecules showing direct correlations with T2D, including CXCL12 and ornithine in urine, 1, 5-anhydroglucitol in saliva, as well as 2 unknown urine metabolites (Fig. 3c). The variety of drivers likely reflects the multiple functional components associated with T2D (such as hyperglycemia and inflammation).
Stability of QMDiab hierarchy
The modules identified by the AutoFocus algorithm are highly dependent on the hierarchical structure of the data. A bootstrapping method was used to assess the stability of the hierarchical structure in the QMDiab dataset. Briefly, this involved generating bootstrap samples by randomly sampling with replacement and constructing hierarchical trees from these samples. The cophenetic correlation between each bootstrapped tree and the original tree was calculated, and the procedure was repeated 100 times. The results showed an average cophenetic correlation of 0.924 with a standard deviation of 0.006, indicating high stability of the QMDiab hierarchical structure.
Taken together, the AutoFocus analysis on this large T2D dataset showed the benefits of exploring multi-omics datasets with a hierarchical algorithm: First, AutoFocus was able to cluster and draw links between multiple omics and fluids into functional modules in T2D at a variety of scales within the hierarchy. Second, the granularity of the hierarchical structure allowed us to explore the functional sub-modules of an identified enrichment peak separately and in detail. For the largest QMDiab cluster associated with T2D, we were able to identify that one sub-module was enriched for energy metabolism molecules and the other for bone growth and degradation, while the peak annotations showed us how these two processes interacted together at a larger biological scale. Finally, mixed graphical models allowed us to perform a driver analysis on each module, indicating which molecules had direct statistical links to the T2D phenotype in each module, and which ones were confounded correlations.
AutoFocus on ROS/MAP dataset shows Alzheimer’s disease phenotype impact at different levels of the biological hierarchy
As another use case, AutoFocus was applied to an Alzheimer’s disease (AD) dataset of brain samples from the Religious Order Study (ROS) and Rush Memory and Aging Project (MAP) cohorts31. This dataset consisted of 8,193 biomolecules from one metabolomics and one proteomics platform, both performed on brain tissue from post-mortem samples (Table 1). For this analysis, we examined the association between the biomolecules and two clinical AD phenotypes simultaneously: 1) Neurofibrillary tangles (NFT), defined by the immunohistochemistry-based overall paired helical filament tau tangles load from post-mortem pathology, and 2) cognitive decline (CD), defined by the rate of change in global cognition over lifetime. These phenotypes were chosen because they represent two distinct effects of AD, molecular and cognitive.
Systems-level analysis of AD phenotypes
Of the 8193 molecules in the ROS/MAP dataset, 887 molecules significantly associated with NFT and 763 molecules significantly associated with CD (p < 0.05, adjusted p-values). All statistical models were corrected for age at death, sex, BMI, post-mortem interval, years of education, and APOE genotype. To maintain consistency with previous studies published on the ROS/MAP dataset41, the FDR p-value correction method was used instead of Bonferroni, leading to a dense distribution of significant hits across the tree (Fig. 4a). Both phenotypes had robust metabolic associations, as metabolites made up 20% and 26% of significant hits in NFT and CD, respectively, even though metabolites only made up 8.14% of the underlying dataset. There were 358 overlapping molecules significantly associated with both phenotypes.
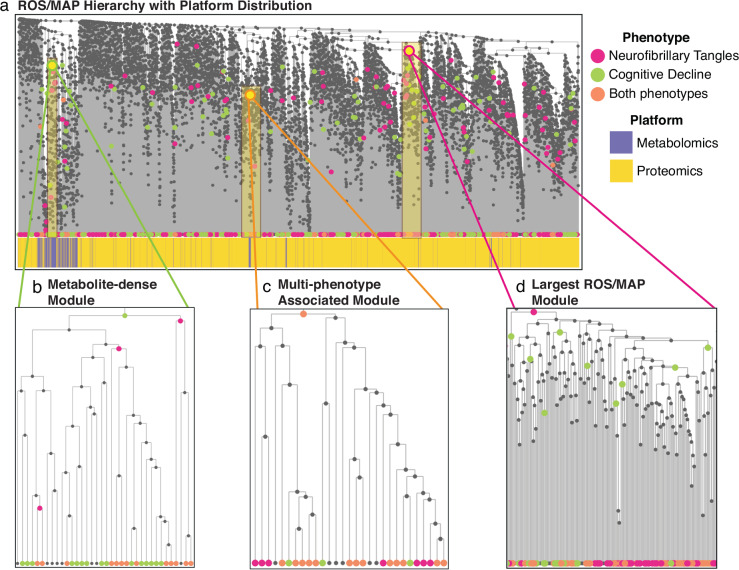
Fig. 4. Results of running the AutoFocus method on the ROS/MAP dataset.
The dataset included a total of 500 samples, which contained 8193 biomolecules from a metabolomics platform a proteomics platform performed on post-mortem brain tissue. a View of the full hierarchical structure created from the ROS/MAP dataset with two phenotypes annotated and dataset distribution below. Magenta circles represent the neurofibrillary tangles phenotype, green circles represent cognitive decline, and orange circles are overlaps between the two. Significant molecules are dispersed densely throughout the tree and enriched modules are scattered throughout the hierarchy at a large range of heights. b Zoomed-in view of a metabolomics module enriched for significant hits associated with cognitive decline. This module contained metabolites related to oxidative stress and lipid peroxidation. c Zoomed in view of the largest module found in the dataset which was enriched for metabolites and proteins significantly associated with neurofibrillary tangles. d Zoomed-in view of the largest module enriched for both phenotypes with the left sub-tree (yellow) enriched for mitochondrial proteins and the right sub-tree (pink) enriched for proteins related to synaptic vesicle exocytosis and inhibitory neurotransmission.
AD modules
A total of 171 modules were identified with a “majority vote” enrichment threshold of 0.5, with 83 modules unique to the NFT phenotype, 82 unique to the CD phenotype, and 6 modules associated with both phenotypes. There were 466 single-molecule modules that did not belong to any of the 171 modules. The multi-molecule modules ranged in size from 2 to 165 biomolecules (Supplementary Fig. 3, Supplementary Data 2). Similar to the QMDiab dataset, modules associated with both phenotypes ranged drastically in tree height across the tree, from 0 to 0.83 (Fig. 4a).
An interesting feature arising from applying AutoFocus on two phenotypes was the nesting of enrichment peaks, where the peak of one phenotype was a descendant of a peak of the other. As the NFT and CD phenotypes had a large overlap of significantly associated molecules, their enriched modules tended to occupy similar regions of the tree. Despite this considerable overlap, only 6 internal nodes were identified as enrichment peaks for both phenotypes (Fig. 4a, orange nodes). For all other regions of the hierarchy where both phenotypes had overlapping significant hits, modules enriched for one phenotype contained descendent sub-modules enriched for the other phenotype (Fig. 4b, c). This nesting highlights how different phenotypes within a single disease can manifest at different scales of biological processes, where cognitive decline may be associated with a biological process at a higher level than neurofibrillary tangles pathology, and vice versa.
The ROS/MAP hierarchical structure was strongly affected by the dataset correlation bias as metabolites were condensed within the tree, leading to only 6 of the 170 modules being multi-omic (Fig. 4a, Supplementary Data 2). Within the dense metabolomic region of the ROS/MAP hierarchy, CD had more significant metabolite associations, which resulted in a higher enrichment peak (larger cluster) than the NFT phenotype. This 36-metabolite module was enriched for antioxidants and lipid peroxidation metabolites42,43, indicating that CD interacts with oxidative stress metabolism at a higher biological scale than NFT (Fig. 4b, Supplementary Data 2).
One of the 6 modules identified for both phenotypes was a metabolomic module enriched for amino acids, with 22 out of 29 members being amino acids or their derivatives (Fig. 4b). These include branched-chain amino acids (valine, leucine, and isoleucine) whose dysregulation is a well-known marker of AD pathology, as well as the aromatic amino acids (tyrosine, phenylalanine, and tryptophan) which are substrates for neurotransmitters like serotonin, dopamine, and norepinephrine (Fig. 4c, Supplementary Data 2)44. As this was a module where both phenotypes were enriched at the same level, these processes do not seem to be specific to either phenotype but are a shared feature of both AD traits. Interestingly, this metabolomic module breaks away from the metabolite-dense portion of the hierarchy, and is surrounded by proteins, indicating a closer functional relationship of amino acids to proteomic processes (Fig. 4a).
Of the 6 multi-omic modules was the largest module in the tree, which significantly associated with the NFT phenotype (Fig. 4d). This module contained proteins and metabolites involved in a variety of processes; one sub-module showed multi-omic dysregulation of arginine flux, degradation, and metabolism45–48, one sub-module contained proteins associated with inflammatory mediator TNF-α49–51, while an adjacent sub-module contained glycosylation proteins52. In contrast, the CD phenotype had enrichment peaks for the NFT sub-modules involved in arginine metabolism and inflammation, but not for the region associated with protein glycosylation. This indicates that protein glycosylation has an NFT-specific association, and thus the NFT phenotype is associated at a higher level in the biological hierarchy for this process than the CD phenotype.
Stability of ROS/MAP hierarchy
The bootstrapping procedure applied to the ROS/MAP dataset resulted in a mean cophenetic correlation of 0.62 with a standard deviation of 0.02. These values suggest that the hierarchical structure is less stable for the ROS/MAP dataset compared to the QMDiab dataset. The lower mean cophenetic correlation indicates greater variability in the hierarchical structure when subjected to bootstrapping. While the reason for this remains unclear, insight can be drawn from the differences in the QMDiab and ROS/MAP hierarchical structures. The internal nodes of the QMDiab hierarchy are evenly distributed across the height of the tree. In ROS/MAP, on the other hand, the internal nodes are densest higher up in the tree. This indicates weaker correlations within the data that could lead to a more variable structure when clustering the multi-omics markers.
In summary, overlaying these two phenotypes on the ROS/MAP hierarchy demonstrated the difference in biological manifestation of cognitive decline and tau neurofibrillary tangles in Alzheimer’s disease. These differences highlight phenotype-specific processes, while modules equally enriched for both phenotypes indicate more universal disease processes that may not be attributable to a single phenotype.
Comparison with other clustering methods
To adequately compare the performance of AutoFocus against other methods, we chose existing clustering algorithms that met the following criteria: First, the methods must have been designed to cluster biomolecular features (instead of samples). Second, the methods must have infrastructure for a multi-scale examination of the identified clusters. We identified three statistical methods that met these criteria and are commonly used to identify clusters that show phenotype associations: (1) MoDentify12, a partial correlation network-based method that uses phenotype association to define modules. (2) MEGENA11 (Multiscale Embedded Gene Co-Expression Network Analysis) because of its ability to identify multi-scale clustering structures. (3) WGCNA10 (Weighted Gene Co-Expression Network Analysis) as it is similarly a hierarchical method, in addition to being a widely used tool for clustering omics data.
Structural comparison
Both MoDentify and MEGENA rely on underlying network structures upon which to perform their clustering methods. MoDentify models molecular interactions between biomolecules by transforming omics datasets into a partial correlation network with edges selected using a p-value inclusion criterion12. Similarly, MEGENA models these interactions using a similarity metric (filtered by an FDR p-value cutoff) to construct a Planar Filtered Network (PFN)11. Despite the difference in network derivation methods, the networks of both methods are still largely affected by the correlation bias discussed in Results “Intra- and inter-dataset relationships in the 12-dataset multi-omics QMDiab study”, with intra-dataset edges represented at a much higher rate than inter-dataset edges (Supplementary Fig. 4).
In contrast, WGCNA derives a hierarchical structure using a “topological overlap matrix” (TOM) of a co-expression network10. The dataset correlation bias persisted in the WGCNA hierarchy, as evident by the proteomics dataset being highly segregated in WGCNA’s TOM-based tree (Supplementary Fig. 5a). Taken together, the correlation bias introduced when combining multiple omics datasets will affect the underlying correlation and cluster structure, regardless of the method. The ability to capture the relationships between these datasets then relies on the clustering approach.
Cluster comparison
The three methods compared to AutoFocus have vastly different designs for identifying clusters.
MoDentify identifies clusters by selecting seed nodes in their network and performing a greedy neighborhood search, integrating each new visited node into a cluster, and testing for significant association with phenotype based on a conglomerative measure12. Because of this stepwise network expansion design, MoDentify does not identify nested structures. Due to these discrepancies between the approaches, the resulting clusters from MoDentify and AutoFocus are substantially different (Fig. 4a). Of note, MoDentify was considerably computationally expensive on the QMDiab dataset, taking 10 h over 4 2.7 GHz Quad-Core Intel Core i7 CPUs.
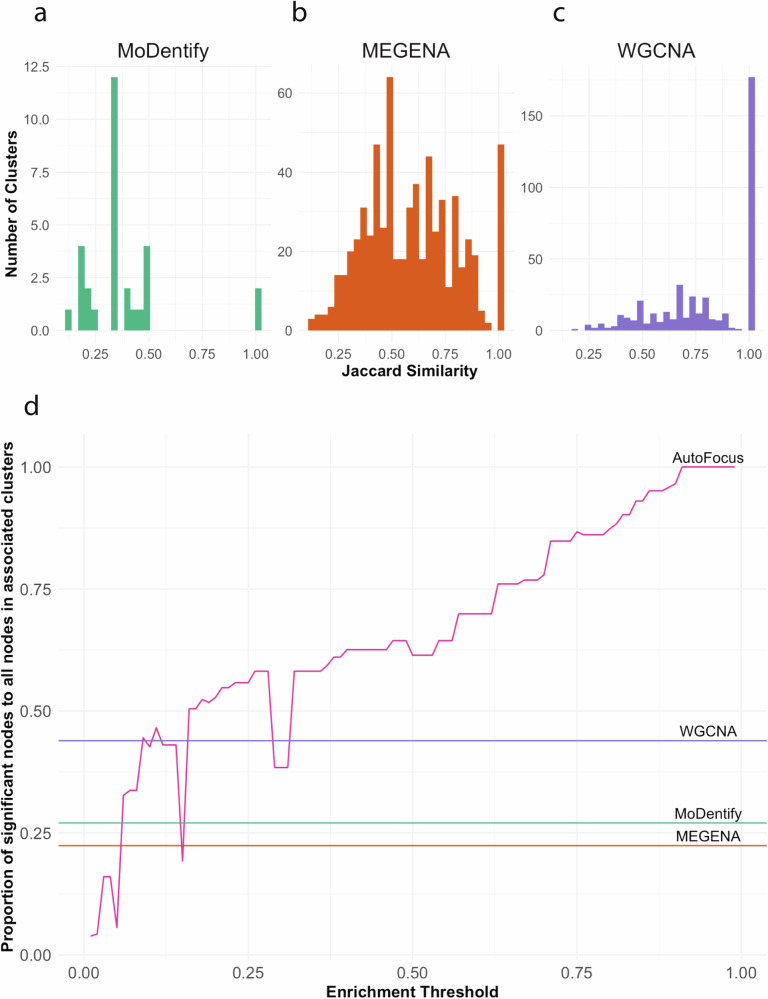
MEGENA initially splits its PFN into clusters based on Newman’s modularity measure, then iteratively splits those clusters into subclusters based on a compactness evaluation11. Consequently, the MEGENA method produces nested clusters, similar to AutoFocus. This leads to a larger similarity between MEGENA and AutoFocus clusters, with two-thirds of MEGENA’s clusters with a Jaccard similarity score of over 0.5 with at least one cluster in AutoFocus’s hierarchy (Fig. 5b). However, MEGENA lacks the functionality to organize sub-clusters within their parent clusters, leaving the multi-scale relationships between clusters to manual inspection. AutoFocus allows the user to analyze these relationships in the R Shiny app, which visualizes nested clusters within their parent clusters and contextualizes their relationships.
Fig. 5. Comparison of AutoFocus with MEGENA, WGCNA, and MoDentify.
Highest Jaccard similarities of clusters from MoDentify (a), MEGENA (b), and WGCNA (c) with clusters from AutoFocus hierarchy. d Proportion of significantly associated molecules in associated clusters found by the four methods. AutoFocus’ proportion increases with a more stringent threshold, surpassing the proportion in WGCNA, MoDentify and MEGENA comfortably after a threshold of 0.35.
WGCNA produced the most similar results to AutoFocus, likely due to the use of hierarchical clustering in both methods10. This similarity is evident as 40% of WGCNA clusters have an exact match in the AutoFocus tree, and 90% have a Jaccard similarity of at least 0.5 (Fig. 5C). However, each analyte is placed into a single cluster and the resulting clusters have no overlap. This leads to WGCNA results being a single scale per cluster, with no regard for potential sub- or super-processes. In fact, 17 of the 26 WGCNA clusters that significantly associated with type 2 diabetes had their highest Jaccard similarity score to sub-clusters found within the largest AutoFocus cluster described in Fig. 3. AutoFocus was not only able to identify the processes found within WGCNA clusters but was also able to contextualize them into the broader system in which they participate.
Further, the AutoFocus framework can add this multi-scale context to a WGCNA hierarchy. The method’s framework is applicable to any input tree structure, including the hierarchical structure derived from the “topological overlap matrix” (TOM) of a co-expression network used in WGCNA10. The QMDiab dataset was rerun using WGCNA’s TOM-based hierarchical structure, the results and a comparison to the original analysis of which can be seen in Supplementary Fig. 5.
Beyond the clustering approaches, all three methods use aggregated metrics of cluster members to associate clusters with disease, rather than looking at the individual molecules within. As such, the interpretability of individual node association to disease is lost, and the association signal of a single member can be dispersed into large clusters that are otherwise full of noise. The use of enrichment as an association metric, alongside the added functionality of ‘piggy-backers’ in the hierarchy leads AutoFocus to have the most signal-dense clusters among the four methods, even at moderately low thresholds (Fig. 5d).
In summary, compared to MoDentify, MEGENA, and WGCNA, AutoFocus provides clusters that are denser in signal and more interpretable at the node level, alongside a visualization tool that is able to contextualize the scale of clusters and how they interact.
Discussion
The AutoFocus method provides a novel computational approach for identifying disease-perturbed, multi-omic modules of biomolecules at various resolutions of biological hierarchy. By testing for enrichment at each internal node in a hierarchical tree, AutoFocus allows relationships between molecules across all platforms, fluids, and omics to be analyzed in the context of phenotypic perturbations. The identified modules are better able to model multi-omic, multi-fluidic, and multi-dataset biological interactions as compared to clustering methods which rely on modules defined at a fixed level and explored as standalone processes. The hierarchical framework allows for the exploration of one or more phenotypes at fine granularity or at a larger, zoomed-out scale. Furthermore, the method’s implementation in an interactive application makes navigation of the complex biological structure, and the modules within, easy and intuitive.
We applied AutoFocus to two multi-omic datasets, QMDiab and ROS/MAP. For both datasets, AutoFocus was able to find a multitude of disease-associated modules at various levels of correlation. For the type 2 diabetes (T2D) phenotype in QMDiab, AutoFocus was able to detect multi-omic modules enriched for known T2D associated processes, such as energy metabolism pathways and bone degradation, distinguishing them as separate but related processes. We were able to integrate the TOM-based hierarchical structure of the WGCNA method into AutoFocus to identify discrepancies in the module results stemming from the underlying hierarchy. Applying AutoFocus to the ROS/MAP Alzheimer’s disease dataset with multiple phenotypes, we were able to distinguish the different scales at which two different pathophenotypes associated with dysregulated processes within a single disease. Without the hierarchical perspective and tool allowing us to explore multiple levels within our dataset, neither of these findings would have been possible.
A core limitation of the field of biomolecular clustering across multi-platform and multi-omic datasets is the dataset correlation bias in which intra-dataset correlations are systematically higher than inter-dataset correlations. In the context of AutoFocus, this bias affects the hierarchical structure between the molecules, and therefore the modules identified by the algorithm will be more likely to contain relationships within one dataset than cross-dataset interactions. Notably however, this bias will affect any method that uses statistical similarity measures between molecules. By testing clusters at all levels of the hierarchy rather than cutting clusters into disparate groups that potentially sever ties between datasets, the AutoFocus design increases the likelihood of identifying multi-omic modules if they exist.
In addition to the bias, the AutoFocus tool is limited by the lack of a gold standard against which to compare the resulting modules. Experimental evidence for interactions of molecules from high throughput experiments are far from comprehensive, especially between omics layers, and therefore there is no ground truth against which modules can be compared. However, similar to the correlation bias mentioned above, all clustering methods run into this problem. AutoFocus should then be seen as a tool for exploring existing associations between molecules and disease within the context of the whole biological system measured by high-throughput experiments.
In conclusion, AutoFocus is a new approach to detect modules in complex, multi-omics data at any scale of association. It allows for multiple phenotype comparison and comes with an interactive Shiny app for result exploration. Our results show that AutoFocus is effective at identifying interactions between biological systems and disease perturbations and can distinguish molecular modules affected by different phenotypes in complex disease.
Methods
Datasets
The QMDiab study was conducted at the Dermatology Department of Hamad Medical Corporation (HMC) in Doha, Qatar. The study population was predominantly of Arab, South Asian, and Filipino descent, with participants falling between the ages of 23 and 71. Data was collected between February and June of 2012; collection and sampling methods have been previously described elsewhere34. The study was approved by the Institutional Review Boards of HMC and Weill Cornell Medicine-Qatar (WCM-Q). Written informed consent was obtained from all participants. For the analysis described in this paper, samples came from 388 distinct subjects (192 females, 196 males; 195 diabetic, 193 non-diabetic).
The Religious Order Study (ROS) and Rush Memory and Aging Project (MAP) are two studies conducted by the Rush Alzheimer’s Disease Center. ROS started recruiting individuals from religious communities across the United States in 1994, and MAP started recruiting individuals from a wide range of backgrounds and socio-economic statuses from Northeastern Illinois in 1997. Data collection and sampling methods have been previously described elsewhere41. For this study, data from post-mortem tissue of 500 distinct subjects was included (352 females, 148 males; 220 with Alzheimer’s Disease, 119 with mild cognitive impairment, 153 with no cognitive impairment, 8 with other forms of dementia). Both cohorts were approved by an institutional review board of Rush University Medical center. All participants provided informed consent, an Anatomic Gift Act, and a repository consent to allow their data and biospecimens to be shared.
All ethical regulations relevant to human research participants were followed.
Multi-omic measurements
QMDiab
Plasma metabolomic profiling was performed by running plasma samples through 5 separate platforms: 1) The Metabolon Inc. HD2 platform, which uses non-targeted ultrahigh-performance liquid chromatography (UHPLC) and gas chromatography (GC) separation coupled with mass spectrometry (MS/MS)53. This yielded 466 measured metabolites. 2) The Metabolon UHPLC-MS/MS and GC-MS/MS HD4 platform (843 metabolites). 3) The Metabolon LipidyzerTM platform, which resolved fatty acid side chains (1133 lipids)54. 4) The Biocrates Life Sciences AG AbsoluteIDQTM p150 metabolomics kit, which used targeted flow injection analysis tandem mass spectrometry (FIA-MS/MS) from (161 molecules)55. 5) The targeted Nuclear Magnetic Resonance (NMR) platform of Nightingale Ltd. (224 metabolites)56.
Urine metabolomic profiling was performed through non-targeted ultrahigh-performance liquid chromatography and gas chromatography separation, coupled with mass spectrometry on the Metabolon Inc. HD2 platform (695 metabolites) and the targeted proton Nuclear Magnetic Resonance (1H NMR) platform of Chenomx, Inc. (32 metabolites)57.
Saliva metabolomic profiling was performed through non-targeted ultrahigh-performance liquid chromatography and gas chromatography separation, coupled with mass spectrometry on the Metabolon Inc. platform (251 metabolites).
Glycomics profiling was performed on 3 separate platforms; 356 plasma samples were sent to Genos, Ltd. (Zagreb, Croatia) for the analysis of total plasma N-glycosylation using ultra-performance liquid chromatography (UPLC) and IgG Fc N-glycosylation using liquid chromatography mass spectrometry lycol-profiling58 (39 and 60 measured glycans, respectively). IgA glycomics measurements were collected at Leiden University Medical Center using UPLC coupled to a quadrupole-TOF-MS, resulting in 90 measured IgA molecules as previously described59,60.
Plasma proteomics profiling was performed on 356 samples at the WCM-Q proteomics core, using the SOMAscan assay (version 1.1) protocols and instrumentation provided and certified by SomaLogic Inc. (Boulder, CO)61 (1141 proteins).
ROS/MAP
For 500 of the brain tissue samples of the ROS/MAP cohort, brain metabolomic profiling was performed through non-targeted ultrahigh-performance liquid chromatography and gas chromatography separation, coupled with mass spectrometry on the Metabolon Inc. platform (667 metabolites)41. Brain proteomic profiles were collected on 265 ROS/MAP samples using tandem mass tag (TMT)-MS and downloaded from the AMP-AD Knowledge Portal (https://adknowledgeportal.synapse.org, 7526 proteins), details of data collection and processing have been previously described62.
Data preprocessing
QMDiab
For each dataset, samples with more than 20% missing molecules and molecules with more than 10% missing samples were removed. Molecular abundance levels were then probabilistic quotient normalized to correct for sample-wise variation63 and log-transformed. In the case of IgA glycomics, proportions (rather than raw counts) of glycans for each protein class were reported. Quotient normalization was applied protein-wise to convert data back to a log-normal distribution, and then log-transformed all together. Data was then scaled, and all outliers with abundance levels above , with n representing the number of samples, were set to missing. Missing values were imputed using a k-nearest neighbors (k-nn) imputation method64. All data preprocessing was performed with the maplet R package65.
ROS/MAP
The ROS/MAP preprocessing steps have been outlined in Batra et al.41 and Johnson et al.62. Briefly, metabolites with over 25% missing values were filtered out, samples were quotient-normalized and subsequently log-transformed. Outlier samples were removed using the local outlier factor method and abundance level outliers were set to NA. Missing values were imputed with a k-nn algorithm. Proteins were log2-transformed and corrected for batch effects using ‘median polish’ approach. Missing values and outliers were treated with same approach as the metabolomics data. Duplicated proteins with same Uniprot IDs were averaged.
Besides the IgA glycomics platform, all other platforms in both the QMDiab and ROS/MAP datasets measured abundance data of their respective omics. As such, the distributions of each of these omics platforms was log-normal and thus the log-transformation step performed for both datasets resulted in normal distributions for each dataset. After protein-wise quotient normalization on the IgA dataset as described above, this platform also resulted in normally distributed abundance measurements.
After pre-processing the individual dataset, the data matrices were concatenated into a final data matrix. For the QMDiab dataset, this final data matrix consisted of 388 samples and 5135 analytes, and for the ROS/MAP dataset, this final data matrix consisted of 500 samples and 8193 analytes.
AutoFocus method
Hierarchical clustering
Once all datasets were preprocessed and concatenated (Fig. 1a), the data matrix was hierarchically clustered. The distance metric between two analytes was derived as one minus the absolute value of their Pearson correlation coefficient value such that the stronger the correlation (either positive or negative), the closer the analytes were in the hierarchy. This distance matrix was transformed into a hierarchical structure using the average-linkage method, which has been shown to maximize the cophenetic correlation between a hierarchical structure and its correlation-based distance matrix as compared to other common linkage methods66 (Fig. 1c). On the hierarchical tree, “leaf” nodes represented biomolecules. Internal nodes represented the root of all their leaf descendants; therefore, a cluster was defined at each internal node. Each internal node also had a right and left child, which could be either a leaf or another internal node.
Univariate analysis
All measured molecules were associated with a phenotype of interest, p, using a linear model with added confounding terms to correct for applicable covariates (e.g., age, sex, BMI). P-values from this linear model were used to determine molecule significance after adjustment for multiple hypothesis testing.
Enrichment “peak” calculation
To find phenotype association enrichment among clusters of the hierarchical tree, the internal nodes of the hierarchy were scanned from top to bottom. At each internal node, the set of leaves descending from that internal node was considered; if the proportion of these leaves that were significantly associated with the phenotype of interest surpassed a user-defined enrichment threshold, this internal node was labeled as an enrichment “peak”. Once a cluster was found at which this enrichment point was met, the scanning stopped for its descendants as we reached the highest level at which disease signal was detected at the desired enrichment threshold. The AutoFocus R package includes a threshold analysis module to assess the impact of the enrichment threshold on the resulting clusters. Details of this analysis can be found in Supplementary Fig. 6.
This process sometimes resulted in “piggy-backers”, defined as peaks that reached the enrichment only due to one child reaching the enrichment threshold, and the joining of the two children diluted the signal (reduced the fraction of significant molecules in the cluster). Once all peaks had been identified in the hierarchy, each peak was assessed for the individual contributions from either child. A peak whose signal could be attributed to a single child was removed (details in Supplementary Fig. 7), the child node that met enrichment was labeled as a peak instead, and the other child that did not meet the threshold continued to be scanned. This iterative process continued until all piggy-backers were removed (Supplementary Fig. 7).
Cluster driver analysis
Once enrichment peaks were identified, an additional analysis was performed on molecules within the biological cluster descending from each peak to identify potential drivers of the disease signal. While a significant univariate association indicated a biological link between a molecule and a phenotype, this effect could have been indirect, meaning the association was relayed through an intermediate variable that was directly associated with the phenotype. Therefore, a driver analysis was performed to identify which molecules had a direct effect.
To this end, the data matrix consisting of abundance data from the molecules descending from the enriched peak was combined with the phenotype vector and all covariates and used to build a mixed graphical model using the mgm package in R67. Graphical models use conditional dependency estimates between molecules, covariates, and disease diagnosis to extract direct correlations and to exclude indirect effects through confounding. Mixed graphical models in particular are capable of generating the conditional independence structure of many underlying distributions, including Gaussian, Poisson, and categorical68.
For our application, molecules and/or covariates were labeled as drivers of a disease phenotype if they shared an edge with the disease phenotype in the resulting MGM graph, as they shared a direct correlation with the phenotype.
AutoFocus code and interactive tool
The AutoFocus method is accompanied by an interface developed using the Shiny app environment69 under R version 4.2.2. The code for the app is freely available as a GitHub at https://github.com/krumsieklab/autofocus. The user has the option to choose between Pearson and Spearman correlation methods for distance calculation, and between Bonferroni or FDR for p-value adjustment methods.
Runtime performance and complexity
Running the AutoFocus method can be parallelized over multiple CPUs to reduce computation time for large datasets. Generating the results of the ROS/MAP dataset with 8193 molecules and two phenotypes took 4.5 h on a single 2.7 GHz Quad-Core Intel Core i7 CPU, with most of this time spent creating the MGMs for each cluster. If MGM calculation is omitted from the analysis, this runtime reduces drastically to just over 6 min on the same architecture.
The computational complexity of AutoFocus (without MGM calculation) is dominated by the pairwise correlation operation used to determine the distance between molecules. The time complexity of AutoFocus on features is ). The space complexity of the algorithm is dominated by the intermediate matrix used to store pairwise distances between molecules. Therefore, the algorithm’s space complexity is .
Stability of hierarchical clustering
The modules returned by the AutoFocus algorithm rely heavily on the underlying hierarchical structure. To test the stability of this structure for both the QMDiab and ROS/MAP datasets, the following bootstrapping-based procedure was performed: First, bootstrapped samples were generated by randomly sampling with replacement from the original datasets, maintaining the original sample sizes (n = 388 for QMDiab, n = 500 for ROS/MAP). Second, a hierarchical tree was calculated from this bootstrapped data using the method outlined in Section “AutoFocus Method”. Finally, the cophenetic correlation was calculated between the bootstrapped hierarchical tree and the hierarchy derived from the full dataset. Cophenetic correlation calculates the similarity between two hierarchies by correlating the heights at which each pair of nodes is merged into clusters in each tree70. This bootstrapping procedure was repeated 100 times to create a distribution of cophenetic correlations. From this distribution, the mean and standard deviation of the cophenetic correlations were calculated. A high mean cophenetic correlation with low variance would indicate a stable hierarchical structure.
Comparison to other clustering methods
The MoDentify method12 was run on the QMDiab dataset using the generateNetwork and identifyModules functions in the publicly available R package (https://github.com/krumsieklab/MoDentify). The partial correlation network on which to run this analysis was created using Bonferroni p-value adjustment with a p-value cutoff of 0.01. Overlapping modules were merged for the comparison analysis.
The MEGENA method11 was run on the QMDiab dataset using a standard pipeline from the MEGENA R package (https://github.com/songw01/MEGENA). To allow for the widest range of cluster sizes, the min.size and max.size parameters were set to 1 and the total number of analytes in the QMDiab dataset, respectively.
The WGCNA method10 was run on the QMDiab dataset using a standard pipeline from the WGCNA R package ((https://CRAN.R-project.org/package=WGCNA) with the minModuleSize parameter set to 1. For both WGCNA and MEGENA, clusters were associated with the type 2 diabetes phenotype through correlation of the cluster’s eigenanalyte (the clusters first principal component) with the disease phenotype. P-values were then Bonferroni corrected.
Jaccard similarities71 (cluster member intersection divided by cluster member union) were then calculated between the resulting clusters of the three comparison methods and those created by the AutoFocus hierarchy’s internal nodes, both phenotype-associated and not. The highest Jaccard similarities for each cluster for the three methods were kept for downstream analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was supported by Biomedical Research Programme funds at Weill Cornell Medicine in Qatar, a program funded by the Qatar Foundation. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The results published here are in whole or in part based on data obtained from the AD Knowledge Portal. Study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Additional phenotypic data can be requested at www.radc.rush.edu. ROSMAP metabolomics data is provided by the Alzheimer’s Disease Metabolomics Consortium (ADMC). The investigators within the ADMC, not listed specifically in this publication’s author’s list, provided data along with its pre-processing and prepared it for analysis, but did not participate in analysis or writing of this manuscript. A complete listing of ADMC investigators can be found at: https://sites.duke.edu/adnimetab/team/. The Metabolon datasets were generated at Metabolon and pre-processed by the ADMC. J.K. and R.B. are supported by the National Institute of Aging of the National Institutes of Health under awards U19AG063744, R01AG069901-01 and Alzheimer’s association award AARFD-22-974775. ROS/MAP data collection was supported through funding by NIA grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNAseq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation, ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNAseq), R01AG48015 (monocyte RNAseq) RF1AG57473 (single nucleus RNAseq), U01AG32984 (genomic and whole exome sequencing), U01AG46152 (ROSMAP AMP-AD, targeted proteomics), U01AG46161(TMT proteomics), U01AG61356 (whole genome sequencing, targeted proteomics, ROSMAP AMP-AD), the Illinois Department of Public Health (ROSMAP), and the Translational Genomics Research Institute (genomic). ROSMAP metabolomics data is funded wholly or in part by the following grants and supplements thereto: NIA R01AG046171, RF1AG051550, RF1AG057452, R01AG059093, RF1AG058942, U01AG061359, U19AG063744 and FNIH: #DAOU16AMPA awarded to Dr. Kaddurah-Daouk at Duke University in partnership with many academic institutions.
Author contributions
K.S. and A.H. provided the QMDiab data. R.K.-D. provided the ROS/MAP data which was preprocessed by R.B. A.S. and J.K conceived of and designed the research study. A.S. preprocessed the QMDiab dataset, wrote the main functionality of the AutoFocus algorithm and shiny application, analyzed all data and results. K.C. formalized all code, added key shiny functionality, and made the AutoFocus R package. A.S. and J.K. wrote the manuscript. All authors gave final approval to publish.
Peer review
Peer review information
Communications Biology thanks Jie Tan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Yuedong Yang and Tobias Goris. [A peer review file is available.]
Data availability
The preprocessed, concatenated QMDiab dataset used in this paper can be found at 10.6084/m9.figshare.23934933.v1. The ROS/MAP data used in this paper can be obtained from two sources: (1) Metabolomics and proteomics data for the ROS/MAP cohort are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses, and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data is available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.org/DataAccess/Instructions). For access to content described in this manuscript see: 10.7303/syn26401311. (2) The full complement of clinical and demographic data for the ROS/MAP cohort are available via the Rush AD Center Resource Sharing Hub and can be requested at https://www.radc.rush.edu.
Code availability
Codes used in this study are available at the GitHub repository https://github.com/krumsieklab/autofocus and 10.5281/zenodo.1313843572.
Competing interests
The authors declare the following competing interests: J.K. holds equity in Chymia LLC, owns intellectual property in PsyProtix, serves as an advisor for celeste, and is a co-founder of iollo. R.K-D. in an inventor on a series of patents on use of metabolomics for the diagnosis and treatment of CNS diseases and holds equity in Metabolon Inc., Chymia LLC and PsyProtix.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06724-2.
References
- 1.Palsson, B. & Zengler, K. The challenges of integrating multi-omic data sets. Nat. Chem. Biol.6, 10.1101/gr.107540.110 (2010). [DOI] [PubMed]
- 2.Halama, A. et al. A roadmap to the molecular human linking multiomics with population traits and diabetes subtypes. Nat. Commun.15, 7111 (2024). [DOI] [PMC free article] [PubMed]
- 3.Bartel, J. et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet.11, 10.1371/journal.pgen.1005274 (2015). [DOI] [PMC free article] [PubMed]
- 4.Ritchie, M. D., Holzinger, E. R., Li, R., Pendergrass, S. A. & Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet.16, 85–97 (2015). 10.1038/nrg3868 [DOI] [PubMed] [Google Scholar]
- 5.Kopczynski, D. et al. Multi-OMICS: a critical technical perspective on integrative lipidomics approaches. Biochim. Biophys. Acta Mol. Cell Biol. Lipids1862, 808–811 (2017). 10.1016/j.bbalip.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 6.Hampel, H. et al. Omics sciences for systems biology in Alzheimer’s disease: State-of-the-art of the evidence. Pitié Ageing Res. Rev.69, 101346 (2021). 10.1016/j.arr.2021.101346 [DOI] [PubMed] [Google Scholar]
- 7.Joshi, A., Rienks, M., Theofilatos, K. & Mayr, M. Systems biology in cardiovascular disease: a multiomics approach. Nat. Rev. Cardiol.18, 313 (2021). 10.1038/s41569-020-00477-1 [DOI] [PubMed] [Google Scholar]
- 8.Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol.18, 83 (2017). 10.1186/s13059-017-1215-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen, H. K. et al. A computational framework to integrate high-throughput ‘-omics’ datasets for the identification of potential mechanistic links. Nat. Protoc.13, 2781–2800 (2018). 10.1038/s41596-018-0064-z [DOI] [PubMed] [Google Scholar]
- 10.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma.9, 559 (2008). 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song, W.-M. & Zhang, B. Multiscale Embedded Gene Co-expression Network Analysis. PLoS Comput. Biol.11, e1004574 (2015). 10.1371/journal.pcbi.1004574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Do, K. T., Rasp, D. J. N.-P., Kastenmüller, G., Suhre, K. & Krumsiek, J. MoDentify : phenotype-driven module identification in metabolomics networks at different resolutions. Bioinformatics35, 532–534 (2019). 10.1093/bioinformatics/bty650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wörheide, M. A., Krumsiek, J., Kastenmüller, G. & Arnold, M. Multi-omics integration in biomedical research – A metabolomics-centric review. Anal. Chim. Acta1141, 144–162 (2021). 10.1016/j.aca.2020.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra, K., Carvunis, A.-R., Ramesh, S. K. & Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet.14, 10.1038/nrg3552 (2013). [DOI] [PMC free article] [PubMed]
- 15.Spirin, V. & Mirny, L. A. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA100, 12123–12128 (2003). 10.1073/pnas.2032324100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blatti, C. et al. Knowledge-guided analysis of ‘omics’ data using the KnowEnG cloud platform. PLoS Biol.18, e3000583 (2020). 10.1371/journal.pbio.3000583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reshetova, P., Smilde, A. K., van Kampen, A. H. C. & Westerhuis, J. A. Use of prior knowledge for the analysis of high-throughput transcriptomics and metabolomics data. BMC Syst. Biol.8, S2 (2014). 10.1186/1752-0509-8-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntyre, L. M. et al. GAIT-GM integrative cross-omics analyses reveal cholinergic defects in a C. elegans model of Parkinson’s disease. Sci. Rep.12, 3268 (2022). [DOI] [PMC free article] [PubMed]
- 19.Dugourd, A. et al. Causal integration of multi-omics data with prior knowledge to generate mechanistic hypotheses. Mol. Syst. Biol.17, e9730–e9730 (2021). 10.15252/msb.20209730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalise, P., Koestler, D. C., Bimali, M., Yu, Q. & Fridley, B. L. Integrative clustering methods for high-dimensional molecular data. Transl. Cancer Res.3, 202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng, C. et al. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform17, 628–641 (2016). 10.1093/bib/bbv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauvel, C., Novoloaca, A., Veyre, P., Reynier, F. & Becker, J. Evaluation of integrative clustering methods for the analysis of multi-omics data. Brief. Bioinform21, 541–552 (2020). 10.1093/bib/bbz015 [DOI] [PubMed] [Google Scholar]
- 23.Krumsiek, J., Suhre, K., Illig, T., Adamski, J. & Theis, F. J. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol.5, 21 (2011). 10.1186/1752-0509-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noor, E., Eden, E., Milo, R. & Alon, U. Central Carbon Metabolism as a Minimal Biochemical Walk between Precursors for Biomass and Energy. Mol. Cell39, 809–820 (2010). 10.1016/j.molcel.2010.08.031 [DOI] [PubMed] [Google Scholar]
- 25.Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. From molecular to modular cell biology. Nature402, C47–C52 (1999). 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- 26.Barabási, A.-L. & Oltvai, Z. N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet.5, 101–113 (2004). 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 27.Do, K. T. et al. Phenotype-driven identification of modules in a hierarchical map of multifluid metabolic correlations. NPJ Syst. Biol. Appl.3, 1–12 (2017). 10.1038/s41540-017-0029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martignetti, L., Calzone, L., Bonnet, E., Barillot, E. & Zinovyev, A. ROMA: Representation and quantification of module activity from target expression data. Front. Genet.7, 18 (2016). 10.3389/fgene.2016.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y. et al. A gene module identification algorithm and its applications to identify gene modules and key genes of hepatocellular carcinoma. Sci. Rep.11, 5517 (2021). 10.1038/s41598-021-84837-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y.-A., Cho, D.-Y., Dao, P. & Przytycka, T. M. MEMCover: integrated analysis of mutual exclusivity and functional network reveals dysregulated pathways across multiple cancer types. Bioinformatics31, i284–i292 (2015). 10.1093/bioinformatics/btv247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett, D. A. et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimer’s Dis.64, S161–S189 (2018). 10.3233/JAD-179939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benedetti, E. et al. A strategy to incorporate prior knowledge into correlation network cutoff selection. Nat. Commun.11, 5153 (2020). 10.1038/s41467-020-18675-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mardinoglu, A. et al. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell Metab.26, 281–283 (2017). 10.1016/j.cmet.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 34.Mook-Kanamori, D. O. et al. 1,5-Anhydroglucitol in Saliva Is a Noninvasive Marker of Short-Term Glycemic Control. J. Clin. Endocrinol. Metab.99, E479–E483 (2014). 10.1210/jc.2013-3596 [DOI] [PubMed] [Google Scholar]
- 35.Puttanna, A. & Padinjakara, R. N. K. Diabetic ketoacidosis in type 2 diabetes mellitus. Practical Diabetes31, 155–158 (2014). 10.1002/pdi.1852 [DOI] [Google Scholar]
- 36.Skenteris, N. T. et al. Osteomodulin attenuates smooth muscle cell osteogenic transition in vascular calcification. Clin. Transl Med.12, 10.1002/ctm2.682 (2012). [DOI] [PMC free article] [PubMed]
- 37.Haug, A. T. et al. Gene expression changes in cancellous bone of type 2 diabetics: A biomolecular basis for diabetic bone disease. Langenbecks Arch. Surg.399, 639–647 (2014). 10.1007/s00423-014-1188-4 [DOI] [PubMed] [Google Scholar]
- 38.Yue, R., Shen, B. & Morrison, S. J. Clec11a/osteolectin is an osteogenic growth factor that promotes the maintenance of the adult skeleton. Elife5, 27 (2016). 10.7554/eLife.18782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hušek, P., Švagera, Z., Všianský, F., Franeková, J. & Šimek, P. Prolyl-hydroxyproline dipeptide in non-hydrolyzed morning urine and its value in postmenopausal osteoporosis. Clin. Chem. Lab. Med.46, 1391–1397 (2008). 10.1515/CCLM.2008.259 [DOI] [PubMed] [Google Scholar]
- 40.Picke, A. K., Campbell, G., Napoli, N., Hofbauer, L. C. & Rauner, M. Update on the impact of type 2 diabetes mellitus on bone metabolism and material properties. Endocr. Connect8, R55 (2019). 10.1530/EC-18-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batra, R. et al. The landscape of metabolic brain alterations in Alzheimer’s disease. Alzheimers Dement.19, 980–998 (2023). 10.1002/alz.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biringer, R. G. The Role of Eicosanoids in Alzheimer’s Disease. Int. J. Environ. Res. Public Health16, 10.3390/IJERPH16142560 (2019). [DOI] [PMC free article] [PubMed]
- 43.Gulliksson, M. et al. Expression of 15-lipoxygenase type-1 in human mast cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids1771, 1156–1165 (2007). 10.1016/j.bbalip.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 44.Griffin, J. W. D. & Bradshaw, P. C. Amino Acid Catabolism in Alzheimer’s Disease Brain: Friend or Foe? Oxid. Med Cell Longev.2017, 5472792 (2017). 10.1155/2017/5472792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braissant, O., Gotoh, T., Loup, M., Mori, M. & Bachmann, C. Differential expression of the cationic amino acid transporter 2(B) in the adult rat brain. Mol. Brain Res.91, 189–195 (2001). 10.1016/S0169-328X(01)00113-9 [DOI] [PubMed] [Google Scholar]
- 46.Yin, Y. et al. Arginase 2 Deficiency Promotes Neuroinflammation and Pain Behaviors Following Nerve Injury in Mice. J. Clin. Med.9, 10.3390/JCM9020305 (2020). [DOI] [PMC free article] [PubMed]
- 47.Morland, C. & Nordengen, K. N-Acetyl-Aspartyl-Glutamate in Brain Health and Disease. Int. J. Mol. Sci.23, 1268 (2022). 10.3390/ijms23031268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, G. & Morris, S. M. Arginine metabolism : nitric oxide and beyond. Biochem. J.336, 1–17 (1998). 10.1042/bj3360001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai, H., Wang, L., Li, L., Huang, Z. & Ye, L. Metallothionein 1: A New Spotlight on Inflammatory Diseases. Front. Immunol.12, 4604 (2021). 10.3389/fimmu.2021.739918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrasco, J., Hernandez, J., Bluethmann, H. & Hidalgo, J. Interleukin-6 and tumor necrosis factor-α type 1 receptor deficient mice reveal a role of IL-6 and TNF-α on brain metallothionein-I and -III regulation. Mol. Brain Res.57, 221–234 (1998). 10.1016/S0169-328X(98)00087-4 [DOI] [PubMed] [Google Scholar]
- 51.Bellezza, I. et al. A Novel Role for Tm7sf2 Gene in Regulating TNFα Expression. PLoS One8, 10.1371/JOURNAL.PONE.0068017 (2013). [DOI] [PMC free article] [PubMed]
- 52.Oka, T. et al. Genetic Analysis of the Subunit Organization and Function of the Conserved Oligomeric Golgi (COG) Complex: studies of COG5- an COG7-deficient mammalian cells. J. Biol. Chem.280, 32736–32745 (2005). 10.1074/jbc.M505558200 [DOI] [PubMed] [Google Scholar]
- 53.Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M. & Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/ electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem.81, 6656–6667 (2009). 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- 54.Quell, J. D. et al. Characterization of Bulk Phosphatidylcholine Compositions in Human Plasma Using Side-Chain Resolving Lipidomics. Metabolites9, 109 (2019). 10.3390/metabo9060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Römisch-Margl, W. et al. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics, 10.1007/s11306-011-0293-4 (2012).
- 56.Würtz, P. et al. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am. J. Epidemiol.186, 1084–1096 (2017). 10.1093/aje/kwx016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mall, R., Berti-Equille, L. & Bensmail, H. Metabolomic Data Profiling for Diabetes Research in Qatar; Metabolomic Data Profiling for Diabetes Research in Qatar, 10.1109/DEXA.2016.12 (2016).
- 58.Zaghlool, S. B. et al. Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation. Hum. Mol. Genet27, 1106–1121 (2018). 10.1093/hmg/ddy006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dotz, V. et al. O- and N-Glycosylation of Serum Immunoglobulin A is Associated with IgA Nephropathy and Glomerular Function. J. Am. Soc. Nephrol.32, 2455–2465 (2021). 10.1681/ASN.2020081208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Momčilović, A. et al. Simultaneous Immunoglobulin A and G Glycopeptide Profiling for High-Throughput Applications. Anal. Chem.92, 4518–4526 (2020). 10.1021/acs.analchem.9b05722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suhre, K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun.8, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med.10.1038/s41591-020-0815-6 (2020). [DOI] [PMC free article] [PubMed]
- 63.Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem.78, 4281–4290 (2006). 10.1021/ac051632c [DOI] [PubMed] [Google Scholar]
- 64.Do, K. T. et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics14, 10.1007/s11306-018-1420-2 (2018). [DOI] [PMC free article] [PubMed]
- 65.Chetnik, K. et al. maplet : an extensible R toolbox for modular and reproducible metabolomics pipelines. Bioinformatics38, 1168–1170 (2022). 10.1093/bioinformatics/btab741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saraçli, S., Doǧan, N. & Doǧan, I. Comparison of hierarchical cluster analysis methods by cophenetic correlation. J. Inequal. Appl2013, 203 (2013). 10.1186/1029-242X-2013-203 [DOI] [Google Scholar]
- 67.Haslbeck, J. M. B. & Waldorp, L. J. mgm: Estimating Time-Varying Mixed Graphical Models in High-Dimensional Data. J. Stat. Softw.93, 10.18637/jss.v093.i08 (2015).
- 68.Wainwright, M. J. & Jordan, M. I. Graphical Models, Exponential Families, and Variational Inference. Found. Trends R Mach. Learn.1, 1–305 (2008). [Google Scholar]
- 69.Chang, W. et al. shiny: Web Application Framework for R, 10.32614/CRAN.package.shiny (2021).
- 70.Sokal, R. et al. The comparison of dendrograms by objective methods. Taxon11, 33–40 (1962). 10.2307/1217208 [DOI] [Google Scholar]
- 71.Tang, M. et al. Evaluating single-cell cluster stability using the Jaccard similarity index. Bioinformatics37, 2212–2214 (2021). 10.1093/bioinformatics/btaa956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schweickart, A. et al. krumsieklab/autofocus: Final version for publication, 10.5281/zenodo.13138435 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The preprocessed, concatenated QMDiab dataset used in this paper can be found at 10.6084/m9.figshare.23934933.v1. The ROS/MAP data used in this paper can be obtained from two sources: (1) Metabolomics and proteomics data for the ROS/MAP cohort are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses, and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data is available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.org/DataAccess/Instructions). For access to content described in this manuscript see: 10.7303/syn26401311. (2) The full complement of clinical and demographic data for the ROS/MAP cohort are available via the Rush AD Center Resource Sharing Hub and can be requested at https://www.radc.rush.edu.
Codes used in this study are available at the GitHub repository https://github.com/krumsieklab/autofocus and 10.5281/zenodo.1313843572.