Abstract
The structural components of the thymus are essential for guiding T cell development, but a thorough spatial view is still absent. Here we develop the TSO-his tool, designed to integrate multimodal data from single-cell and spatial transcriptomics to decipher the intricate structure of human thymus. Specifically, we characterize dynamic changes in cell types and critical markers, identifying ELOVL4 as a mediator of CD4+ T cell positive selection in the cortex. Utilizing the mapping function of TSO-his, we reconstruct thymic spatial architecture at single-cell resolution and recapitulates classical cell types and their essential co-localization for T cell development; additionally, previously unknown co-localization relationships such as that of CD8αα with memory B cells and monocytes are identified. Incorporating VDJ sequencing data, we also delineate distinct intermediate thymocyte states during αβ T cell development. Overall, these insights enhance our understanding of thymic biology and may inform therapeutic interventions targeting T cell-mediated immune responses.
Subject terms: T cells, Lymphocyte differentiation, Systems analysis, miRNA in immune cells
Thymus is important for shaping T cell immunity, but spatial insights at cellular and molecular level are still scarce. Here the author use multi-omics approaches and custom algorithms to reconstruct the spatial organization of human thymi for both confirming known features and unrevealing new components during thymocyte maturation.
Introduction
The thymus is crucial for T cell development, and its lobules consist of cortical (C) and medullary (M) regions that facilitate the movement of developing T lymphocytes1–3. Typically, CD3+CD4-CD8- (double negative or DN) precursor T cells migrate from hematopoietic organs to distal cortical regions, also known as the “sub-capsular zone”4. Following division, these cells migrate to the inner cortical regions and start expressing CD4 and CD8 co-receptors (double positive or DP; CD3+CD4+CD8+). The DP cells that fail to recognize major histocompatibility complex (MHC) molecules presented by cortical thymic epithelial cells (cTEC) are eliminated through a process known as “positive selection”, while the remaining cells pass through the medulla-cortex (MC) boundary to the medullary region with the help of chemokines and their ligands (such as CCL21-CCR7), eventually transforming into CD3+CD4+ or CD3+CD8+ T cells (single positive or SP). The medullary thymic epithelial cells (mTEC) and dendritic cells (DCs) present self-antigens to the developing T cells, and the cells that recognize these self-antigens undergo apoptosis, a process known as “negative selection”. The T cells that are tolerant to the self-antigens are released into the periphery upon maturation5–7. The thymus gradually becomes fibrotic and atrophic with age, leading to a reduction in T-cell output and possible TEC abnormalities, which increases the risk of cancer, infections, and autoimmune diseases8–10. Therefore, gaining an in-depth understanding of the origin, cellular interactions, and localization of thymic T cells is of great significance to the fields of cell therapy and regenerative medicine11.
Single-cell RNA sequencing (scRNA-seq) is a sophisticated technique for identifying previously uncharacterized cell types and constructing cellular atlas of thymus. Studies by Park et al. 8, Zeng et al. 12, and Michelson et al. 7 have used scRNA-seq to gain insights into the developmental continuum from early thymocyte genesis to T lymphocyte maturation. Specifically, Park et al. conducted an in-depth analysis of the thymic cell atlas, uncovering new cell subpopulations, and detailing gene expression profiles across T cell development stages. Zeng et al. focused on early thymic progenitor cells, TECs, and the molecular mechanisms of T cell selection, revealing new genes and pathways. Michelson et al. investigated the later stages of T cell maturation, examining gene expression changes from DP to SP cells, T cell maturation in the medulla, self-tolerance mechanisms, and regulatory networks for T cell differentiation. Despite these advances, scRNA-seq requires tissue dissociation, which results in the loss of spatial information about cells, thereby limiting the understanding of thymic architecture. Spatial transcriptomics (ST) technology addresses this limitation by enabling unbiased mapping of transcripts across tissue sections through spatially encoded oligo-deoxythymidine microarrays13–16, preserving the structural context and cellular relationships, and offering a new perspective for elucidating thymic tissue organization.
To the best of our knowledge, Suo et al. 11 pioneered the use of ST technology with the 10X Visium platform for the thymus, creating a developmental map of the human immune system. They utilized empirical thresholds and image segmentation to determine the cortex, medulla, and cortico-medullary junction of the thymus and employed the Cell2location17 tool to infer thymic cell abundance distribution. While this work provides valuable insights into thymic spatial architecture, the 10X Visium platform aggregates transcriptomic data from 1-10 cells per capture point, limiting spatial resolution to proportional abundance of cell types rather than single-cell resolution. This constraint hampers accurate spatial localization of thymic cells and temporal tracking of single-cell transcriptional changes. Obtaining single-cell resolution of thymic spatial architecture is crucial for understanding thymic cell distribution, interactions, regulatory factors, and T cell development trajectories. Additionally, integrating thymic spatial architecture at single-cell resolution with T cell receptor sequencing (TCR-seq) data can enhance our understanding of the dynamic evolution of TCR chains during the development of αβ T cells18.
In this work, we apply scRNA-seq to generate comprehensive transcriptomic profiles of the human thymus across prenatal, pediatric, adult, and geriatric stages, revealing dynamic changes in thymic cell types throughout development. By integrating scRNA-seq with spatial transcriptomics and developing the TSO-his and TSO-hismap tools, we achieve precise single-cell resolution of thymic cell localization and map their distribution along the cortex-medulla axis. Additionally, TCR sequencing data elucidates distinct intermediate states in αβ T cell development and their spatial characteristics, offering new insights into the role of the thymic microenvironment in T cell development.
Results
Generation of a single-cell atlas of human thymus development
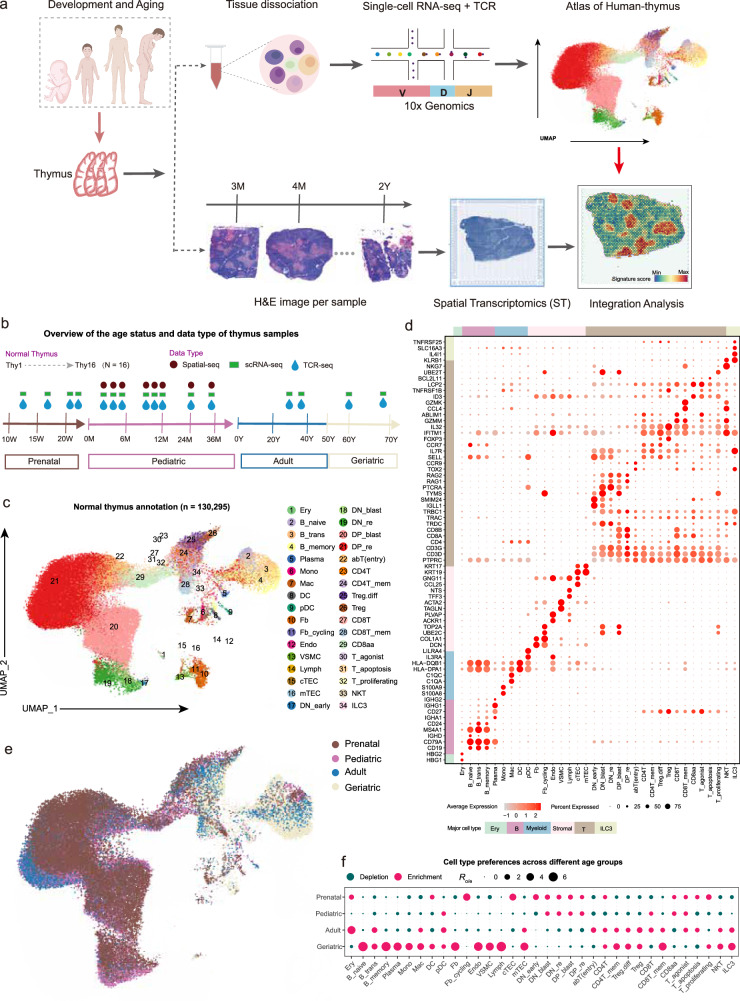
We performed scRNA-seq on 16 thymus samples encompassing the prenatal (4 samples), pediatric (8 samples), adult (2 samples) and geriatric (2 samples) stages (Figs. 1a, b; Supplementary Data 1). After quality control, a total of 130,295 high-quality cells were obtained (Supplementary Figs. 1a, b; see “Methods”). These cells from diverse thymus samples were integrated into a merged object using the Seurat R package (version 4.1.0)19, a comprehensive analysis tool with excellent visualization capabilities (Supplementary Figs. 1c, d). Subsequently, principal component analysis (PCA) and unsupervised clustering were performed (see “Methods”). Utilizing canonical markers, the clustered cells were pre-annotated into six broad lineages, namely erythroid (Ery) cells (HBG1 and HBG2), B cells (CD79A and CD19), plasma cells (IGHG1 and IGHG2), myeloid cells (S100A8, C1QA and IL3RA), stromal cells (ACTA2 and DCN), and T cells (CD3D and CD3E) (Supplementary Fig. 1e). The B cells, myeloid cells, stromal cells, and T cells were further subjected to a second round of clustering, which revealed 34 distinct cell subtypes (Figs. 1c, d; Supplementary Fig. 1e; Supplementary Data 2; see “Methods”).
Fig. 1. Generation of a transcriptional atlas of human thymus development at different age groups.
a Schematic representation of the study design. b Summary of normal thymus samples. Different shapes and colors represent data types, and arrows indicate the transition from prenatal to geriatric stages. c Two-dimensional uniform manifold approximation and projection (UMAP) of single cells collected from normal thymus samples. Annotated cell types are color-coded. d Dot plot showing marker gene expression used for cellular annotation. The size of the dots indicates the proportion of that marker expressed in a particular cell type and the color indicates the average expression level. The annotation bars on the left and top indicate broad cell subsets, with the corresponding color codes. e UMAP colored by age groups, including prenatal, pediatric, adult, and geriatric groups. f Age group preference of each cell type was measured by the ratio (Ro/e)21 of the observed number of cells to the expected number of cells at random (i.e., with no association between subsets and ages, allowing observation of the expected number of cells). Figure 1a created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
The number of UMIs and genes detected for each subset are shown in Supplementary Figs. 1f, g. Additionally, an overview of cell counts from each cell type in the thymus samples was presented in Supplementary Figs. 1h, i (Supplementary Table 1). Within the thymic microenvironment annotations, DN cells (CD4-CD8-), DP cells (CD4+CD8+), and SP cells (CD4+CD8-CD3+ or CD8+CD4-CD3+) are the predominant cell types involved in T cell differentiation. The DN cells included early DN cells (DN_early), DN blast cells (DN_blast), and DN thymocytes undergoing rearrangement (DN_re), and the DP cells consisted of the DP_blast and DP_re subsets. The subpopulations of SP cells include CD4+ T cells, regulatory T cells (Treg), CD8+ T cells, and CD8aa T cells (Figs. 1c, d). Furthermore, B cells are classified into distinct subtypes, including naïve B cells (B_naive), transitional B cells (B_trans), and memory B cells (B_memory). Monocytes (Mono), macrophages (Mac), dendritic cells (DCs), and plasmacytoid DCs (pDC) were the subsets identified in the myeloid population. Stromal cells were further categorized into cTECs, mTECs, fibroblasts (Fb), cycling fibroblast cells (Fb_cycling), vascular smooth muscle cells (VSMC), endothelial cells (Endo), and lymphatic endothelial cells (Lymph) (Figs. 1c, d).
The reliability of cell type annotations forms the basis for downstream analyses. We assessed this by examining two aspects. Firstly, the purity of cell clusters was evaluated using the ROUGE index20 (see “Methods”). With the exception of Plasma, Lymph, and DP_blast, the average index for each subset was above 0.9 (all > 0.85), indicating good cluster purity (Supplementary Fig. 2a). This was further confirmed through pairwise correlations and the purity index defined by Logistic regression model (Supplementary Figs. 2b, c; see “Methods”). Secondly, the consistency of annotations with previous studies was examined. When compared with annotations of the healthy thymus reported by Park et al. 8, a high degree of consistency is observed (Supplementary Fig. 2d). These results indicate that the cell types we identified in the thymus scRNA-seq data are reliable.
To investigate the developmental preferences of different cell types across various age groups, we calculated the ratio of the observed to the expected number of cells (Ro/e)21 to estimate group preference (see “Methods”). As shown in Figs. 1e, f and Supplementary Fig. 2e, immature T cell populations, including DN_early, DN_blast, DN_re, DP_blast, and DP_re, exhibited an antagonistic relationship with age, being most abundant during the prenatal stage (Figs. 1e, f; Supplementary Figs. 2e, f). Conversely, partially mature T cell populations, such as CD4+ T memory (CD4T_mem), CD8+ T memory (CD8T_mem), Treg, differentiating Treg (Treg.diff) and natural killer T cells (NKT), were more prevalent in the adult and geriatric groups. This can be attributed to the age-dependent progressive fibrosis of the thymus, which is characterized by a reduction in thymocytes and an increase in mesenchymal cells like Fb, Endo, VSMC, and Lymph (Fig. 1f; Supplementary Figs. 2e, f)22. CD8aa cells were more abundant in the prenatal and pediatric groups compared to the adult and geriatric groups (Fig. 1f; Supplementary Figs. 2e, f). The abundance of cTECs and mTECs, which play crucial roles in the selection and regulation of T cell development, displayed distinct patterns across age groups. The cTECs were significantly more abundant in the prenatal group, while mTECs were more prevalent in the geriatric group. This finding further supports the notion that epithelial cells and mature T cells synergistically facilitate mutual differentiation23. Thymic DCs that mediate the negative selection of T cells through antigen presentation were enriched during the prenatal and geriatric stages (Fig. 1f; Supplementary Figs. 2e, f). B cell subsets, including B_naive, B_trans, B_memory, and Plasma, predominated in the geriatric thymus group. A previous study24 showed that thymic B cells play a compensatory role in antigen presentation and the elimination of developing thymocytes in the aging thymus, which is crucial for maintaining thymic function and immune homeostasis. In summary, these findings provide valuable insights into the dynamic changes of thymic cell types with age.
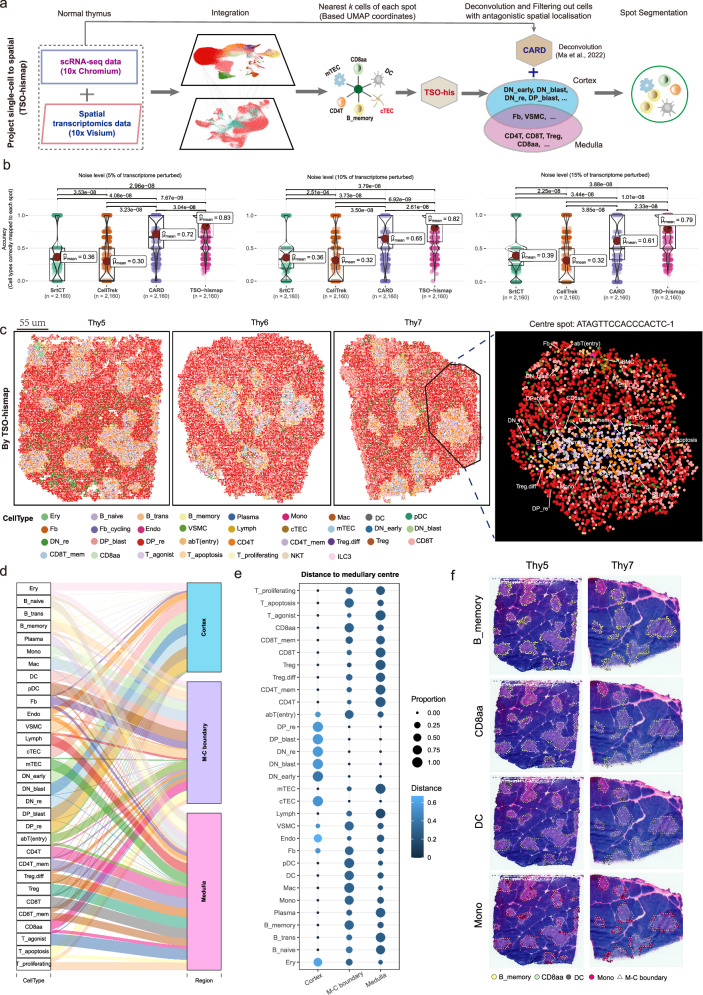
Revealing thymic tissue organization with TSO-his
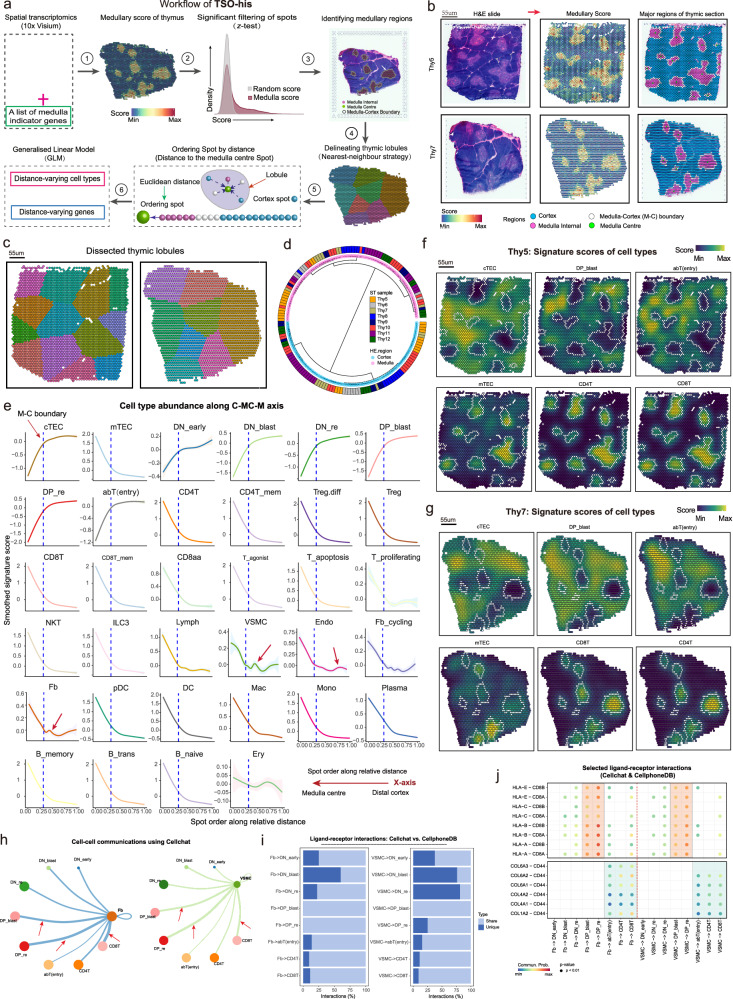
ST sequencing was performed on eight pediatric thymus samples (3 months to 3 years of age) based on the 10X Genomics Visium Spatial Gene Expression platform (Figs. 1a, b; Supplementary Figs. 3a–c; Supplementary Data 1; see “Methods”). With the 10X Visium platform, tissue organization can be determined based on microdomains (spots) measuring 55 µm in diameter, and a center-to-center distance of 100 µm. The scRNA-seq data and ST bulk gene expression data of matched thymus samples were significantly correlated (R > 0.89; Supplementary Fig. 3d). Typically, T cell development and maturation occur along the C-MC-M axis within the thymus lobules (C: cortical regions; M: medullary regions; MC: Medulla-Cortex boundary). Thus, accurate identification of these regions can provide insights into the possible roles of different cell types during T cell development. To this end, we developed the TSO-his (Transcriptomic Segmentation of Histological Structure) algorithm for segmenting the histological structures in thymic H&E sections. The signature (also called medullary score) scores of 12 medullary marker genes obtained from previous studies were initially computed in the thymic ST spots (Supplementary Table 2). Subsequently, cortical spots, medullary spots and M-C boundary spots were determined using z-test framework, and medullary centers were also defined. Finally, utilizing a nearest neighbor strategy, the lobules were accurately segmented by assigning spots to the nearest centers (Fig. 2a; see “Methods”). We applied TSO-his on eight ST slices of thymic tissue and found that the scores associated with medullary marker genes were prominently emphasized in the medullary region (Fig. 2b), which is consistent with previous reports18,25,26. Furthermore, TSO-his accurately detected distinctive anatomical features such as the cortical region, medullary region, M-C boundaries and lobules, which closely resembled the structures observed in H&E sections (Fig. 2b, c, Supplementary Figs. 3e, f). By clustering the structural regions in ST slices using the Pearson correlation of aggregated expression profiles, we found that the medullary and cortical regions in lobules from eight samples could be clearly distinguished (Fig. 2d). Notably, the ratio of the number of spots in the medullary and cortical regions was approximately 1:3 (Supplementary Fig. 3g). Taken together, TSO-his has the ability to recognize both the medullary and cortical regions of the thymus section, as well as to segment its lobules.
Fig. 2. Identification of critical regions in thymic spatial transcriptomics (ST) slices, and depiction of changes in cell type with spatial distance.
a Algorithm for identifying cortical and medullary regions of thymus and segmenting thymic lobules based on spatial transcriptome data (TSO-his). Details are provided in “Methods” section. b Critical tissue regions of Thy5 (top) and Thy7 (bottom) thymus sections as determined by TSO-his analysis. Left: Hematoxylin and Eosin (H&E) slices (control); Middle: medullary scores of spatial spots; Right: cortex, medulla, and Medulla-Cortex (M-C) boundaries. Medullary center spots in thymus sections are marked in green. Segmentation results for other ST slices can be found in Supplementary Figs. 3e, f (n = 8 biological replicates). c Dissection of thymus ST slices into lobules based on the nearest-neighbor strategy using TSO-his. Left: Thy5; Right: Thy7. d Hierarchical clustering of identified cortical and medullary regions in lobules of eight thymus slices, with similarity measured by Pearson correlation of averaged expression profiles between regions. e Spatial-distance applied to eight thymus ST slices, illustrating the distribution of signature gene scores for 34 cell types from the distal cortex to medullary center. The lines, representing the relationship between spatial distance and signature scores of cell types, were smoothed using the generalized linear models, and the shaded areas around these lines denote the 95% confidence intervals for the fitted values. f, g Spatial spot scores of Thy 5 (f) and Thy7 (g) samples based on signature genes of cell types and smoothed using the “wkde” method in the Nebulosa R package70. Enriched populations in cortical regions (cTECs, DP_blast, and abT(entry)) and medullary regions (mTEC, CD4+ T and CD8+ T) are shown. h Network plots showing cell-cell communications derived from CellChat. (left) Fb vs. T cells and (right) VSMC vs. T cells. The thickness of the edges indicates the strength of the interaction. Nodes are color-coded according to the cell types. (i) Comparison of shared and unique significant ligand-receptor interactions (%) between CellChat31 and CellPhoneDB32 tools. (left) Fb vs. T cells, (right) VSMC vs. T cells. (j) Dot plot showing the selected significant ligand-receptor pairs. (left) Fb vs. T cells and (right) VSMC vs. T cells. The color of the dots reflects communication probabilities and dot size represents computed p-values. Empty space indicates that the communication probability is zero. P-values were computed from one-sided permutation tests. cTEC: Cortical thymic epithelial cells, mTEC: Medullary thymic epithelial cells, VSMC: Vascular Smooth Muscle Cell, Fb: Fibroblasts, DP_blast: Double positive blast cells, DP_re: Double positive rearrangement cells. Source data are provided as a Source Data file.
Vertical distribution of thymic cell types along with C-MC-M axis
Using the TSO-his tool, we generated a vertical distribution atlas of thymic cells along the C-MC-M axis, to identify the cell types that potentially regulate T-cell development. A generalized linear model was integrated into TSO-his to fit the signature scores of 34 cell types to spatial distances on the basis of eight ST slices (see “Methods”). As shown in Fig. 2e, the abundance of cTECs and mTECs showed opposite trends when transitioning from the distal cortical region to the medullary center. The cTECs predominated in the cortical region and declined rapidly at the M-C boundary, while mTEC numbers increased significantly in the medullary regions. These findings are consistent with the results shown in Fig. 2f and Fig. 2g. The immature states of T cells, namely DN_early, DN_blast, DP_blast, DP_re, and abT (entry) subsets were enriched in the cortical region and declined along the C-MC-M axis. In contrast, the mature CD4+ T, CD4 T_mem, CD8+ T, CD8+ T_mem, CD8aa, and Treg cells predominantly occupied the medullary region and exhibited a sharp increase at the M-C boundary (Figs. 2e–g and Supplementary Fig. 4a), in agreement with previous studies27,28.
Other immune cells, including B_naive, B_trans, B_memory, Plasma, pDC, DC, Mac, and Mono, were primarily localized in the medullary region (Figs. 2e–g; Supplementary Fig. 4a). Stromal cells, including Endo, VSMC, Fb, and Lymph, were predominantly detected in the medullary region and to a lesser extent in the cortical region (Figs. 2e–g; Supplementary Fig. 4a). Notably, we observed a protrusion phenomenon of Endo cells in the distal cortical region. Previous studies29,30 have shown that an increase in their abundance enhances vascular permeability and facilitates the recruitment of T cell precursors to the distal cortical area (Fig. 2e). The Fb cells and VSMCs were aggregated cells exhibited a sudden accumulation near the M-C boundary, suggesting their potential role in the positive selection process of T cells (Fig. 2e). To validate this hypothesis, we investigated the interactions between Fb, VSMC, and thymic T cells using the CellChat (version 1.1.3)31 tool (see “Methods”). As shown in Fig. 2h, both Fb and VSMCs interacted strongly with DP_blast, DP_re, and CD8+ T cells (Fig. 2h). To enhance the credibility of these interaction strengths, we performed proportional gradient subsampling of Fb (also for VSMC), along with thymic cells, revealing stable and consistent strengths between Fb (VSMC) and thymic cells (Supplementary Figs. 4b, c). Specifically, significant ligand-receptor pairs identified by CellChat between Fb (or VSMC) and DP and SP cells, revealing a substantial overlap (almost greater than 80%) with CellphoneDB32 (Fig. 2i; see “Methods”). Extracting the significant ligand-receptor pairs identified by both CellChat and CellphoneDB further demonstrated that Fb and VSMC in the cortical region engage with CD8 receptors on DP cells by presenting MHC class I molecules, thereby mediating T cell recognition and selection, in line with our initial hypothesis (Fig. 2j). Moreover, Fb and VSMCs interact with SP T cells in the medullary region through collagen-related genes and their ligand CD44, potentially facilitating the localization and migration of mature T cells into the bloodstream. However, it is challenging to determine the relationship between Ery and spatial distance due to contamination in H&E sections (Fig. 2e; Supplementary Figs. 4d, e).
Overall, these results provide important insights into the dynamics of cell types along the C-MC-M spatial axis within the thymic microenvironment. We also show that thymic Fb and VSMCs interact with DP cells via MHC class I molecules, potentially facilitating T cell recognition and selection.
Identification of significant distance-varying genes along the C-MC-M axis
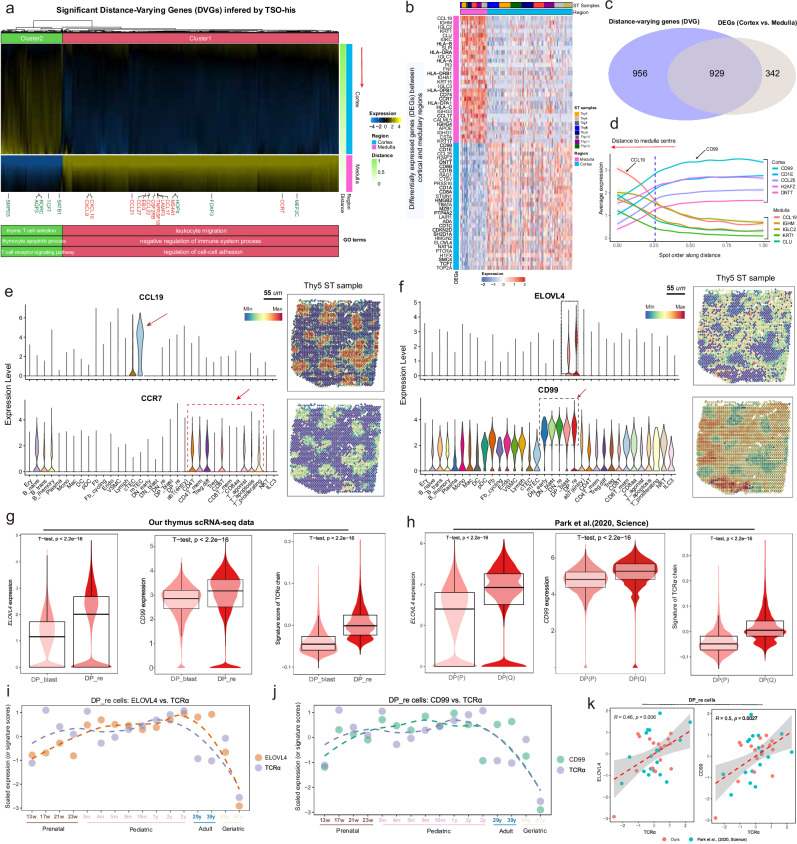
The gene expression changes across anatomical regions of the thymus are crucial for T cell development at different stages. To identify the novel regulatory relationship between gene expression and T cell development, we used the TSO-his tool to screen for significant distance-varying genes (DVGs) along the C-MC-M axis (see “Methods”). In combination with eight ST samples, we identified 1885 DVGs (pooled p ≤ 1%), including medulla markers (marked in red) and classical stage markers of T-cell development (marked in green)8,33. Hierarchical clustering of these genes revealed enrichment of specific pathways in different regions (Fig. 3a; Supplementary Data 3; see “Methods”). The cortical regions were enriched for thymic T cell selection, thymocyte apoptotic process and T cell receptor signaling pathway, while medulla regions were involved in processes such as negative regulation of immune system process, leukocyte migration and regulation of cell-cell adhesion (Fig. 3a). We explored the relationship between the DVGs and the differentially expressed genes (DEGs) in medullary and cortical regions and detected considerable overlap (Figs. 3b, c; Supplementary Data 4). TSO-his was used to determine the top five most prominently expressed genes in the cortical and medullary regions (Fig. 3d). CCL19 was the most significantly upregulated gene in the medulla and specifically expressed in mTECs, while its ligand CCR7 was expressed at high levels in mature T cells, confirming that CCL19-CCR7 synergistically guide the migration of mature T cells from the thymus to the peripheral blood (Fig. 3e and Supplementary Figs. 5a–c)1. Intriguingly, genes enriched in the cortical region exhibited notable relevance to thymocyte differentiation and/or T cell function (Supplementary Data 4). For instance, Recombination Activating Gene 1 (RAG1) emerged as a pivotal contributor, crucially involved in V(D)J recombination and essential for the survival and positive selection of DP thymocytes34. Similarly, Terminal Deoxynucleotidyl Transferase (DNTT), indispensable for random nucleotide insertion during T-cell receptor rearrangement35, was identified. Key roles in the development of a major subset of Natural Killer T (NKT) cells were attributed to CD1 family proteins, specifically CD1C and CD1D36. Additionally, Stathmin 1 (STMN1), a modulator of microtubule dynamics, surfaced as crucial in the maturation, activation, and functional orchestration of T cells37. Interestingly, certain transcripts retained elusive connections to the aforementioned processes, including Elongation of very long chain fatty acids 4 (ELOVL4), linked to macular degeneration diseases, and CD99, also known as MIC2 or E2 antigen, a cell surface glycoprotein, stood out among these transcripts. The expression of ELOVL4 and CD99 was prominently elevated within the cortical region, particularly at the DP stages (Fig. 3f). Importantly, the expression of ELOVL4 and CD99 were significantly upregulated in DP_re compared to DP_blast, in line with the expression pattern of genes associated with the TCRα chain (Figs. 3g, h). Furthermore, we investigated how the expression of ELOVL4 and CD99 relates to that of the TCRα chain across various age groups. Remarkably, the expression patterns of these two genes closely paralleled the fluctuations observed in the TCRα chain, particularly among geriatric individuals, where a notable decline was observed simultaneously (Figs. 3i, j). This decline may be attributed to the degeneration and shrinkage of the aging thymus, leading to diminished functionality. Additionally, correlation analysis provided quantitative evidence of a strong positive relationship between the expression of ELOVL4 and CD99 and the TCRα chain (Fig. 3k).
Fig. 3. Exploring spatial gene expression patterns and functional consequences in thymic T cell development and differentiation.
a Heat map showing the smoothed expression of genes that exhibited significant variance with distance in pediatric spatial transcriptomics (ST) slices. The red arrow indicates the direction of gene expression change from the distal cortex to the medullary center (Supplementary Data 3). Two broad gene clusters (Cluster1 and Cluster2) were assigned by cutting the hierarchical clustering tree, and the biological processes (BP) terms enriched in each cluster are selectively shown. The medulla indicator genes (Supplementary Table 2) closely linked to T-cell differentiation status8,33 are highlighted. b Heat map of marker genes, only the top 30 over-expressed genes are shown for the cortical and medullary regions, respectively. Spots are ordered by ST samples. c Overlap between genes that vary significantly with distance (DVGs) and the differentially expressed genes (DEGs) between medullary and cortical regions. d Spatial distribution of the top ten DEGs in eight thymus ST slices; five were up-regulated in the medullary region and the remainder in the cortical region. The lines, representing the relationship between spatial distance and gene expressions, were smoothed using the natural spline regression model, and the shaded areas around these lines denote the 95% confidence intervals for the fitted values. e, f Expression levels (left) and spatial localization (right) of (e) CCL19, CCR7 and (f) ELOVL4 and CD99 in thymic cell types. g, h Violin plots combined with box plots showing the distribution of ELOVL4 (left) and CD99 (middle) expression, and T-cell receptor α (TCRα)-related gene scores (right) in DP_blast and DP_re cell types in our thymic single-cell dataset. Median value, interquartile range (IQR) as bounds of the box and whiskers that extends from the box to upper/lower quartile ± IQR × 1.5. P-values were obtained from the two-sided t-test. Our thymus data (DP_blast=15,812 cells; DP_re=52,320 cells; (g) Park et al.’s data, (DP(P) = 12,057 cells; DP(Q) = 11,219 cells; (h). i, j Scatter plot showing the expression of the selected gene and the signature score of TCRα changing with age in the DP_re stage. Each data point represents an individual sample, with the dashed line indicating the loess fit. The signature score of TCRα is inferred by its associated TRAV* genes. (i) ELOVL4 vs. TCRα; (j) CD99 vs. TCRα. k Scatter plot showing the associations between the expression of CD99 (left) and ELOVL4 (right) with the signature score of TCRα separately. The shaded areas indicate the 95% confidence interval from the linear regression models, respectively. “R” represents the Pearson correlation coefficient, and the p-value is obtained from the two-sided t-test. DP_blast: Double positive blast cells, DP_re: Double positive rearrangement cells. Source data are provided as a Source Data file.
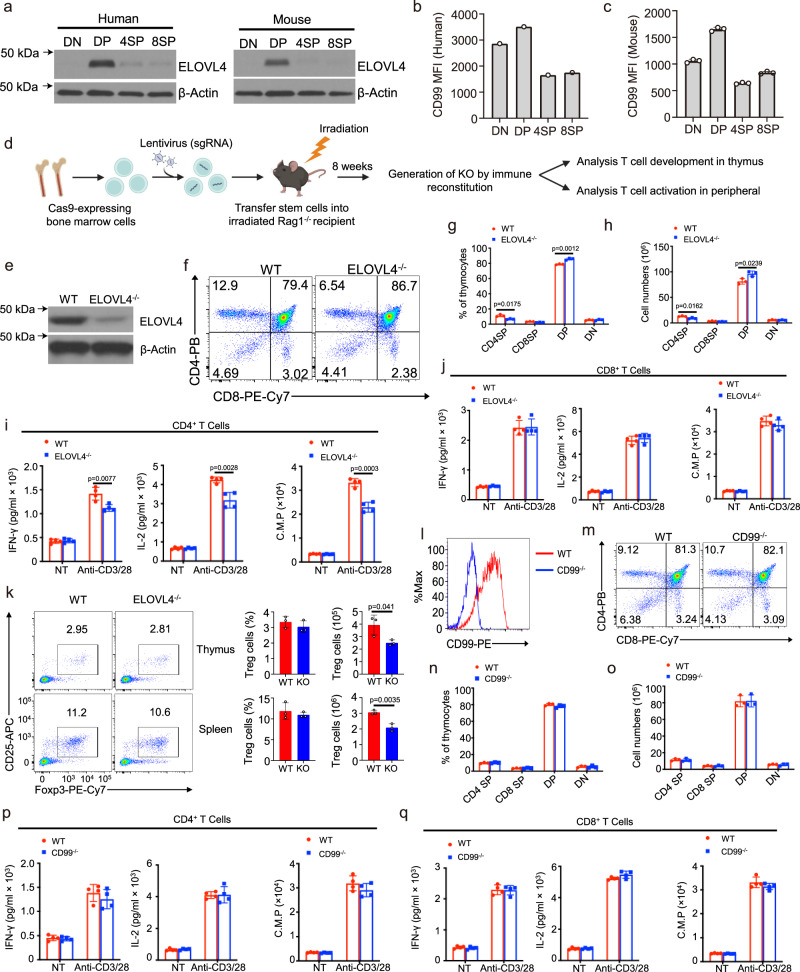
Given the involvement of the DP_re stage in TCRα chain rearrangement and T cell positive selection, we postulate that ELOVL4 and CD99 play pivotal roles in these processes, particularly in TCRα chain development. To ascertain the contribution of these genes, we isolated thymocytes from both mice and humans at various developmental stages and examined the protein levels of ELOVL4 and CD99. Our analysis revealed an enrichment of ELOVL4 and CD99 expression at the DP_re stage, aligning with their corresponding RNA expression profiles (Figs. 4a–c). To further validate the functions of ELOVL4 and CD99, we employed the CHimeric IMmune Editing approach38, utilizing a CRISPR-Cas9 bone marrow delivery system for rapid gene function evaluation in this context (Fig. 4d; Supplementary Figs. 6a, b; see “Methods”). Chimeric mice exhibited a notable decrease in the frequency of thymic CD4+ T cells derived from ELOVL4-KO bone marrow, a phenomenon not observed with CD99-KO bone marrow compared to control counterparts (Figs. 4e–h, l–o). Intriguingly, the absence of ELOVL4 resulted in decreased production of cytokines, including interleukin 2 (IL-2) and interferon-γ (IFN-γ), coupled with reduced proliferation upon TCR + CD28 stimulation in naïve CD4+ T cells, as evidenced by ELISA and 3H-thymidine incorporation assays (Figs. 4i, j). Conversely, CD99 deficiency had no discernible impact on this process (Figs. 4p, q). Furthermore, upon analysis of the Treg population, we found no significant differences between the wild-type (WT) and ELOVL4-KO groups in the percentages of Tregs. However, the reduced Treg numbers observed in the ELOVL4-KO group may be attributed to the loss of total CD4+ T cells (Fig. 4k). These findings establish ELOVL4 as a novel regulator governing the maturation and activation of CD4+ T cells. The significance of these compelling results underscores the effectiveness of this analytical approach in rapidly identifying crucial genes that orchestrate intricate immunological processes.
Fig. 4. Using CRISPR/Cas9-mediated knockout to investigate cortical region-enriched genes in thymus development.
a Immunoblot analysis of the ELOVL4 proteins using mouse and human thymocytes at different stages as indicated. b, c Flow cytometric analysis of CD99 expression using median fluorescence intensity (MFI) on gated thymocytes as indicated. Summary graphs are presented as mean ± SD. P values were determined by an unpaired two-tailed Student’s t test. d Schematic of Lentiviral CRISPR/Cas9-Mediated knockout using bone marrow chimeric mice. e Immunoblot analysis of ELOVL4 proteins using naïve T cells from CRISPR/Cas9-Mediated ELOVL4 knockout and control mice. f–h Flow cytometric analysis of thymocyte subpopulations showing a representative FACS plot (left) and summary graph (right) of total thymocyte numbers and frequency of indicated subpopulations (n = 3 biological replicates). i, j ELISA analyzes of IFN-γ and IL-2 expression using supernatants collected from naïve CD4+ T cells and naïve CD8+ T cells purified from splenocytes of CRISPR/Cas9-Mediated ELOVL4 knockout and control mice (n = 4 biological replicates), stimulated for 48 hours with plate-bound anti-CD3 plus anti-CD28 antibodies. T-cell proliferation assays were measured after 40 hours by pulse-labeling the stimulated T cells with [3H] thymidine for 8 hours. k Flow cytometric analysis of Treg cells (CD4+CD25+Foxp3+) showing a representative FACS plot (left) and summary graph (right) of absolute numbers and frequency (n = 3 biological replicates). (l) Flow cytometric analysis of the surface expression of CD99 on splenocyte T cells within the indicated group. m–o Flow cytometric analysis of thymocyte subpopulations showing a representative FACS plot (left) and summary graph (right) of total thymocyte numbers and frequency of indicated subpopulations. p, q ELISA analyzes of interferon-gamma (IFN-γ) and IL-2 expression using supernatants collected from naïve CD4+ T cells (CD4+CD44loCD62Lhi) and naïve CD8+ T cells (CD8+CD44loCD62Lhi) purified from splenocytes of CRISPR/Cas9-Mediated CD99 knockout and control mice (n = 4 biological replicates), stimulated for 48 hours with plate-bound anti-CD3 plus anti-CD28 antibodies. T-cell proliferation assays were measured after 40 h by pulse-labeling the stimulated T cells with [3H] thymidine for 8 hours. 8-week-old female mice were used for experiment. Figure 4d created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. ELISA: Enzyme-Linked immunosorbent assay, FACS: Fluorescence-Activated cell sorting. Source data are provided as a Source Data file.
Optimizing TSO-his for thymic cortex and medulla segmentation with distance-varying genes
Since the identification of medullary and cortical regions by TSO-his mainly relies on the hypothesis testing framework using multiple ST slices (Fig. 2a; step 2), our aim was to explore the feasibility of efficiently and rapidly identifying regions in ST slices, even when using a single ST slice. To this end, we utilized a combination of feature selection based on both significantly DVGs and DEGs, and the Extreme Gradient Boosting (XGBoost)39 model to develop a classifier capable of replacing step 2 in the TSO-his method (see “Methods”; Figs. 2a, 3c; Supplementary Fig. 7a). The results demonstrated a good performance, with an overall accuracy of 95.8% and an area under the curve (AUC) of 0.986 for the training set, as well as an accuracy of 95.1% and an AUC of 0.977 for the testing set (Supplementary Figs. 7b, c). Additionally, when the predictions from the XGBoost classifier were projected onto the three ST slices, we observed a strong concordance with the regions identified using the hypothesis testing framework employed in TSO-his (Supplementary Fig. 7d). Moreover, we analyzed the contributions of features in the classifier, noting that the CC-chemokine family of genes was the most prominent (Supplementary Fig. 7e). To further validate the performance of our trained XGBoost classifier, we used the thymic ST slices of Suo et al. 11 as an independent testing set, and demonstrated that the identified medullary and cortical regions aligned well with the corresponding H&E images, as well as the regions highlighted by medullary scores (Supplementary Fig. 7f), thus confirming reliable performance and successful integration into the TSO-his methodology.
Spatial architecture of the thymus at single-cell resolution
Given the limited resolution of ST technology, the spots represent only the average gene expression of multiple cells. To this end, we developed the TSO-hismap tool to accurately map thymic single cells to ST coordinates, and determine the physical proximity between different cell types within the spots during T cell development. It can reconstruct a spatial atlas of the thymus at single-cell resolution by integrating scRNA-seq and ST data through TSO-his and the spatial deconvolution tool CARD40 (Fig. 5a; Supplementary Figs. 8a, b; see “Methods”). To assess the performance of TSO-hismap, we conducted simulations of thymus ST data by incorporating known cell type locations at various noise levels (see “Methods”). By mapping single cells to these simulated ST data, TSO-hismap consistently demonstrated superior accuracy compared to Seurat19 Coordinate Transfer (SrtCT), CellTrek41, and CARD (Fig. 5b). Furthermore, we projected thymic single cells onto eight real ST sections using TSO-hismap and the other tools (Fig. 5c; Supplementary Figs. 8c–e and 9a). While CellTrek and SrtCT showed limited ability to differentiate cell types within the cortex and medulla regions (Supplementary Fig. 8e), both TSO-hismap and CARD demonstrated significant differences in the cellular composition of the medullary and cortical regions that align with the thymic anatomical structures (Fig. 5c and Supplementary Fig. 9a). To further compare the performance of TSO-hismap and CARD, we visualized the relative proportions of each cell type in the cortex and medulla regions using a Sankey diagram (Fig. 5d; Supplementary Fig. 9b). Most thymic cell types displayed nearly identical distributions (Fig. 5d; Supplementary Figs. 9b, c). However, TSO-hismap accurately projected cTEC cells into the cortex region, whereas CARD mapped a significant proportion of cTEC cells to the medulla region (Supplementary Fig. 9d). TSO-hismap placed B_memory cells at the boundary of the medulla region, which is consistent with a previous report42, while CARD primarily mapped them to the cortex region (Supplementary Fig. 9d). These findings indicate that the results obtained with TSO-hismap aligned more consistently with the localization studies of thymic cell types compared to CARD. In addition, we compared the relative abundance of cell types observed in the scRNA-seq data with that obtained after projection to spatial coordinates, and found that TSO-hismap exhibited the highest correlation (R = 0.99; Supplementary Fig. 9e, f). These findings demonstrate that TSO-hismap can accurately reconstruct the spatial distribution of different thymic cell types at single-cell resolution.
Fig. 5. Projection of single cells from the normal thymus to spatial coordinates.
a The strategy of TSO-hismap for projecting single cells onto spatial transcriptomics (ST) slices. Details of the full algorithm can be found in the “Methods” section. b Boxplots showing the performance of SrtCT, CellTrek, CARD, and TSO-hismap. Various noise levels (5%, 10%, and 15%) were introduced to evaluate the accuracy of assigning individual cells to the correct spot in simulated ST datasets. Median value, interquartile range (IQR) as bounds of the box and whiskers that extends from the box to upper/lower quartile ± IQR × 1.5. P-values were obtained using the two-sided t-tests. c Projection of thymic single cells to ST coordinates using TSO-hismap. Examples include Thy5, Thy6 and Thy7. Each dot represents a single cell from the normal thymus, with cell types color-coded. An enlarged lobule with a medullary center of ATAGTTCCACCCACTC-1 is shown. d Sankey plot showing the proportion of single cells of each cell type projected onto the medullary, cortical, and Medulla-Cortex (M-C) boundary regions of ST slices using TSO-hismap. The thickness of the lines indicates the proportion of one cell type projecting to that region. e Bubble plot showing a panoramic view of thymocytes in the cortical, medullary, and M-C boundary regions inferred by TSO-hismap, based on eight pediatric ST sections. The size of the dots indicates the proportion of that cell type in a particular region, and the color shade represents the average distance of that cell type from the medullary centers. f Single-cell resolved distribution of B_memory, CD8aa, DC, and Mono in ST slices (left: Thy5, right: Thy7). Each dot represents an individual cell. B_memory: Memory B cells, DC: Dendritic cells, Mono: Monocytes. The cell type icons in Fig. 5a were created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
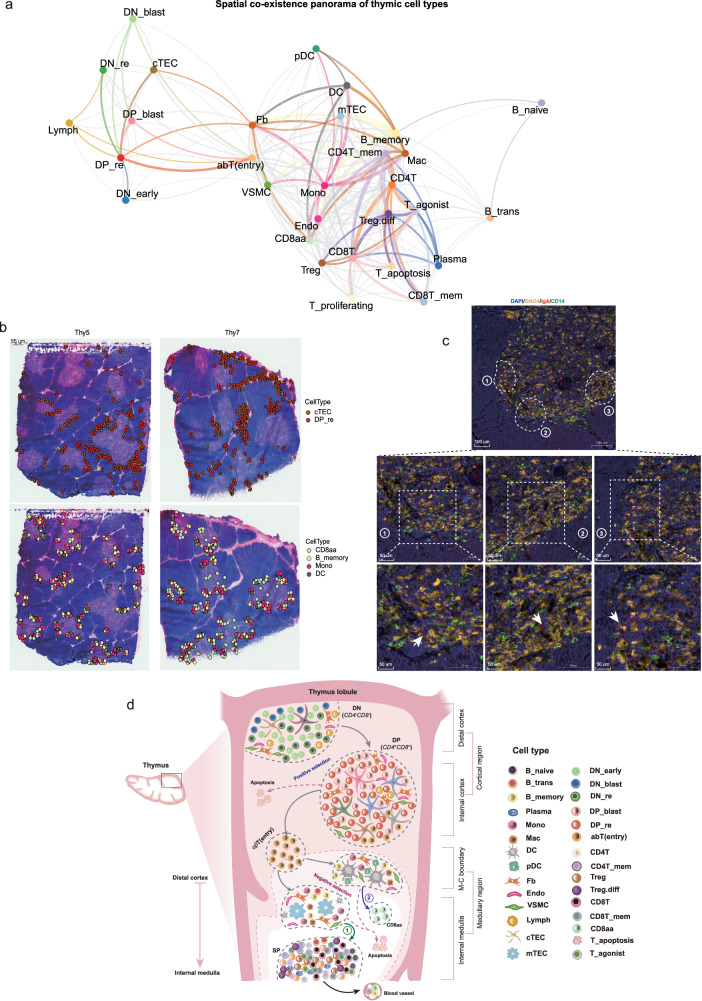
Based on the spatial atlas reconstructed by TSO-hismap, we first quantified the relative distances of different cell types from the medullary center (Fig. 5e). Interestingly, CD8aa, B_memory, DC, pDC, Mono, and Mac exhibited a higher abundance at the boundary with the cortex (Figs. 5e, f). Furthermore, we assessed the proximity of different cell types by segmenting the spots at a single-cell resolution using TSO-hismap. The J-index (see “Methods”) was introduced to measure the strength of co-localization based on segmented spots and illustrate the co-localization patterns. cTECs were highly concentrated at the proximal cortico-medullary junction and around blood vessels, and strongly co-localized with certain DP_re cells (Figs. 6a, b; J-index = 0.959; Supplementary Data 5). We also observed that Fb cells were enriched in the cortical regions surrounding blood vessels and co-localized with cTECs (Fig. 6a; Supplementary Fig. 9g; J-index = 0.613; Supplementary Data 5). Previous studies have suggested a role for Fb in TEC maintenance and T cell development, although the underlying mechanism remains unknown43,44. This finding provides potential evidence of their involvement in T cell differentiation. In the medullary region, we observed strong co-localization of mTECs with CD4+ T and CD8+ T cells (Supplementary Fig. 9g; J-indexes = 0.768 and 0.759). Furthermore, CD8aa cells (Supplementary Figs. 9h–j) were enriched near the M-C boundary and co-localized strongly with DC cells (J-index = 0.821) but weakly with mTECs (J-index = 0.254). This suggests that DCs may mediate the negative selection of CD8aa cells (Fig. 6a, b), which is consistent with previous studies8,11. Moreover, CD8aa cells also showed significant co-localization with B_memory cells (Fig. 6b; J-index = 0.759) and, as well as with Mono cells. These co-existences were further confirmed by H&E staining (Figs. 6b, c; J-index = 0.807; CD8aa: GNG4; B_memory: IgA; Mono: CD14). However, the nature of the interaction between these cells and CD8aa cells remains unclear and warrants further exploration. To summarize, the spatial distribution and coexistence of various cell types in the thymus provide a panoramic view of T cell development (Fig. 6d).
Fig. 6. Spatial co-localization and characterization of thymic cell types.
a Atlas of thymic cell types coexisting in spatial transcriptomics (ST) slices. Each vertex represents a cell type and the thickness of the edges indicates the strength of the J-index (see “Methods”). Edges with J-index values below 0.3 are not shown, those with values less than 0.6 are gray, and those above 0.6 are highlighted by the color code of the specific cell type. In addition, Ery was not considered due to contamination in ST slices. b In combination with the ST images (Thy5 and Thy7 as examples), the spatial co-localization of the cell types is visualized. cTEC vs. DP_re (top) and CD8aa vs. B_memory vs. Mono vs. DC (bottom). Each dot represents a cell projected onto spatial coordinates by TSO-hismap, with cell types color-coded. c Immunofluorescence staining showing the expression patterns of GNG4 (CD8aa), IgA (B_memory), and CD14 (Mono) in the thymus (5 months old). Scale bars: 100 um (middle panel) and 50 um (bottom panel; magnified slices) (n = 3 technical replicates). d Schematic representation of the spatial co-localization of thymic cell types at different stages of human T cell development, from the distal cortex to the inner medulla. Ery: Erythrocytes, B_memory: Memory B cells, cTEC: Cortical thymic epithelial cells, Mono: Monocytes, DC: Dendritic cells, DP_re: Double positive rearrangement cells. The thymus, lobule, and cell type icons in Fig. 6d were created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
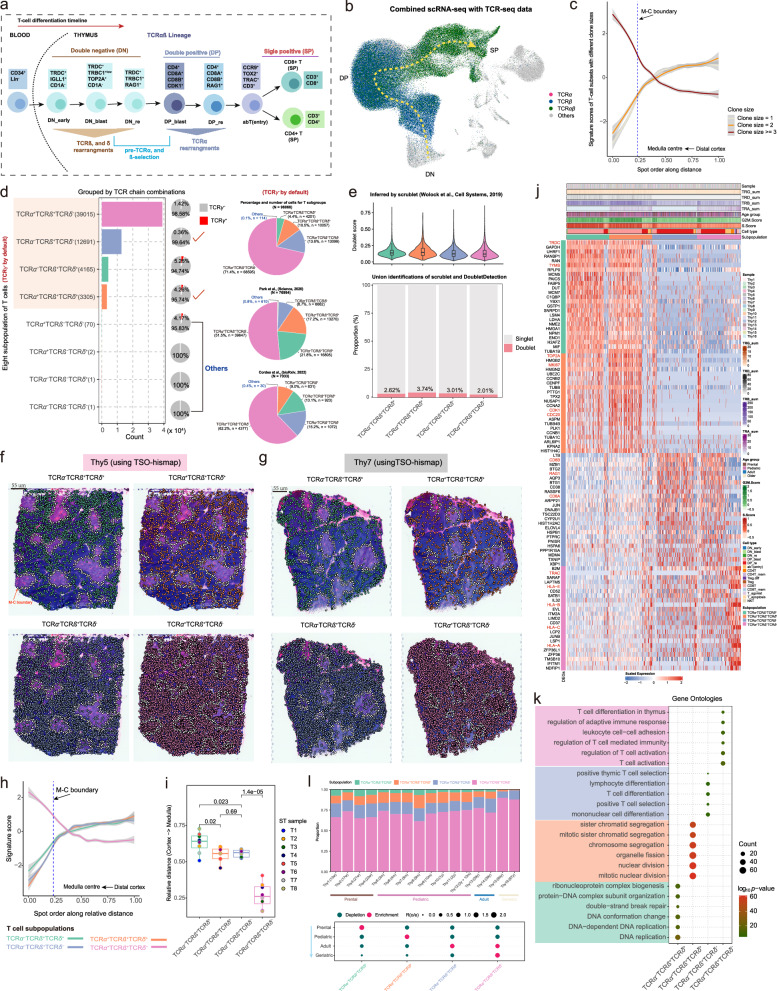
Revealing multiple enigmatic intermediate subpopulations in αβ T cell development through TCR signaling
We were able to reconstruct the developmental route of T cells by mapping out cell types at different stages using conventional marker genes (Fig. 6d and Fig. 7a). Since TCR chain recombination is a critical aspect of T cell development and maturation, we generated and analyzed αβ-VDJ sequencing data from 16 normal thymus samples (Fig. 1a and Fig. 7b; Supplementary Fig. 10a). The diversity of TCR clones was highest in the pediatric group and lowest in the geriatric group (Supplementary Fig. 10b; see “Methods”). By combining thymic TCR-seq, scRNA-seq, and ST-seq data, we observed that T cells with a large clonal size (≥3) were usually enriched in the medullary regions, while T cells with a clone size of 1 were primarily in the cortical region (Figs. 7b, c; Supplementary Figs. 10c, d). Furthermore, leveraging scTCR-seq data, T cells lacking β chains were filtered out (Fig. 7b). The remaining αβT cells were stratified into distinct T cell subpopulations based on the expression of α, β, δ, and γ chain-associated genes (see “Methods”). Notably, four TCRγ- T-cell subpopulations predominated: TCRα-TCRβ+TCRδ+, TCRα+TCRβ+TCRδ+, TCRα-TCRβ+TCRδ-, and TCRα+TCRβ+TCRδ- (Fig. 7d). Conversely, other T-cell subpopulations, particularly those expressing γ chain related genes (i.e, TCRγ + ), were excluded due to their limited representation (Fig. 7d; see “Methods”). Analysis of single-cell data from Park et al. 8 and Cordes et. al.33 confirmed the existence of these four T-cell subpopulations (Fig. 7d). Classical theory outlines two distinct developmental routes for T cells at the DN stage: successful beta rearrangement leads to αβT cells, while successful delta rearrangement results in γδT cells. In our investigation focusing on αβT cells, both TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ subpopulations contain δ chain information. This raises the question of whether these subsets harbor an abundance of double cells. To address this, we employed Scrublet45 and DoubletDetection46 tools for identifying doublets (Fig. 7e; Supplementary Figs. 10e–g; see “Methods”). Our analysis revealed consistently low doublet scores across all subpopulations, with fewer than 4% of cells detected as doublets (Fig. 7e). These findings suggest the consistent presence of four T-cell subpopulations during αβT cell development.
Fig. 7. Spatio-temporal dynamics of four T-cell subpopulations.
a Schematic representation of human T cell development. b Uniform Manifold Approximation and Projection (UMAP) plot showing the cells with rearranged T-cell receptor α (TCRα) (purple), TCRβ (dark blue), or fully rearranged TCRαβ (dark green). c TSO-his applied to eight thymus spatial transcriptomics (ST) slides showing the variation in clone size with spatial distance. Lines smoothed using generalized linear models, with shaded areas representing 95% confidence intervals. The blue dashed line represents the Medulla-Cortex (M-C) boundary. T cells with larger clone sizes are more enriched in the medullary region. d T-cell subpopulations generated using four TCR chains (see “Methods”). Due to significant under-representation (almost below 5%) of the TCRγ+ subpopulation, only the TCRγ- T-cell subpopulations are presented. The bar plot showing counts for TCRα-TCRβ+TCRδ+, TCRα+TCRβ+TCRδ+, TCRα-TCRβ+TCRδ- and TCRα+TCRβ+TCRδ- subpopulations. The pie charts on the right show the relative abundance of these subpopulations in the datasets of this study (top), Park et al. 8 (middle) and Cordes et al. 33 (bottom); “Others” include the TCRα+TCRβ-TCRδ-, TCRα-TCRβ-TCRδ-, TCRα+TCRβ-TCRδ+ and TCRα-TCRβ-TCRδ+ populations. e Violin combined with box plot showing the distribution of doublet scores inferred by Scrublet45 for four T-cell subpopulations. Median value, interquartile range (IQR) as bounds of the box and whiskers that extend from the box to upper/lower quartile ± IQR × 1.5 (top). Stacked bar plot showing the proportions of doublet and singlet cells inferred by Scrublet and DoubletDetection46 within each T cell subpopulation (bottom). Subpopulations: TCRα-TCRβ+TCRδ+ (n = 4,165 cells), TCRα+TCRβ+TCRδ+ (n = 3305 cells), TCRα-TCRβ+TCRδ- (n = 12,691 cells), and TCRα+TCRβ+TCRδ- (n = 39,015 cells). f, g Projection of the four T cell subpopulations onto ST slices using TSO-hismap for Thy5 (f) and Thy7 (g). Each dot represents a T cell and the white dotted line indicates the M-C boundary. h Spatial-distance applied to eight thymus ST slides, showing the trend of signature scores for the four T-cell subpopulations from the distal cortex to medullary center spots. Lines smoothed using generalized linear models, with shaded areas representing 95% confidence intervals. The blue dashed line represents the M-C boundary. i Box plot showing the distances of the four T-cell subpopulations relative to the medullary center. Each dot indicates an ST sample. Median value, interquartile range (IQR) as bounds of the box and whiskers that extend from the box to upper/lower quartile ± IQR × 1.5. The two-sided t-test was used for statistical analysis. j Heatmap showing the top 25 significantly upregulated genes in each T cell subpopulation (FDR ≤ 0.01 and logFC ≥ 0.25). Representative genes in each subpopulation are marked in red. T cells within each subgroup are arranged in chronological order of differentiation. k Dot plot showing the six most significantly enriched biological processes terms (by log10(p-value)) for each subgroup established in Fig. 7j. P-values were obtained using Fisher’s exact test and adjusted by the Benjamini-Hochberg method. (l) Top panel: Stacked bar plot showing the relative proportions of the four T cell subpopulations in each thymus sample, marked by color codes. Bottom panel: Age group preference of T cell subpopulation measured by Ro/e21. TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ are enriched in the younger group, while TCRα-TCRβ+TCRδ- and TCRα+TCRβ+TCRδ- are more prevalent in the geriatric group. Source data are provided as a Source Data file.
Next, the spatial localization of the four aforementioned subpopulations was traced using TSO-hismap, indicating that TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ cells were predominantly distributed in cortical regions, away from the medullary center. TCRα-TCRβ+TCRδ- and TCRα+TCRβ+TCRδ- cells, especially the latter, were enriched in the medulla (Fig. 7f-h; Supplementary Figs. 10h, i; see “Methods”). By measuring the Euclidean distance from representative regions of T-cell subpopulations to the center of the medulla, the TCRα-TCRβ+TCRδ+ population was the most distant, while TCRα+TCRβ+TCRδ+ and TCRα-TCRβ+TCRδ- were almost equidistant, and TCRα+TCRβ+TCRδ- was closest to the medullary center (Fig. 7i; see “Methods”). Furthermore, to investigate the biological functions of the four T-cell subpopulations, we screened for the DEGs (Fig. 7j; Supplementary Fig. 10j). The proliferation-related genes TYMS, TOP2A and MKI67 were significantly overexpressed in the TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ subpopulations, and are involved in DNA replication and chromosome sister monomer division. TRDC, a marker gene of the early stage of T cell differentiation, was also highly expressed in both subpopulations (Figs. 7j, k; Supplementary Data 6). CD8B and RAG1 were highly expressed in the TCRα-TCRβ+TCRδ- subgroup, and as markers of DP rearrangement, are enriched during the positive selection of T cells. TRAC and MHC class I antigens were significantly overexpressed in the TCRα+TCRβ+TCRδ- population, suggesting a close association between the latter and mature T cells (Figs. 7j, k; Supplementary Data 6).
On the age axis, we examined the dynamic changes in abundance of the four T-cell subpopulations across age groups. Notably, TCRα-TCRβ+TCRδ+ cells displayed a significant negative correlation with age, being more abundant in the prenatal group (Fig. 7l; Supplementary Data Fig. 11a). Conversely, TCRα+TCRβ+TCRδ+ and TCRα-TCRβ+TCRδ- were enriched in the Pediatric and Adult stages, respectively (Fig. 7l). Moreover, TCRα+TCRβ+TCRδ- exhibited a positive correlation with age and was predominantly found in the geriatric group (Fig. 7l; Supplementary Fig. 11a). These findings underscore the dynamic alterations in the composition of thymic T-cell subpopulations throughout the aging process.
Overall, these four T-cell subpopulations within αβT cells exhibit distinct spatial distributions, unique gene expression patterns, and dynamic changes in abundance across various age groups, offering valuable insights into their biological processes and developmental traits.
αβ T cell maturation is a continuous differentiation process across multiple intermediate states
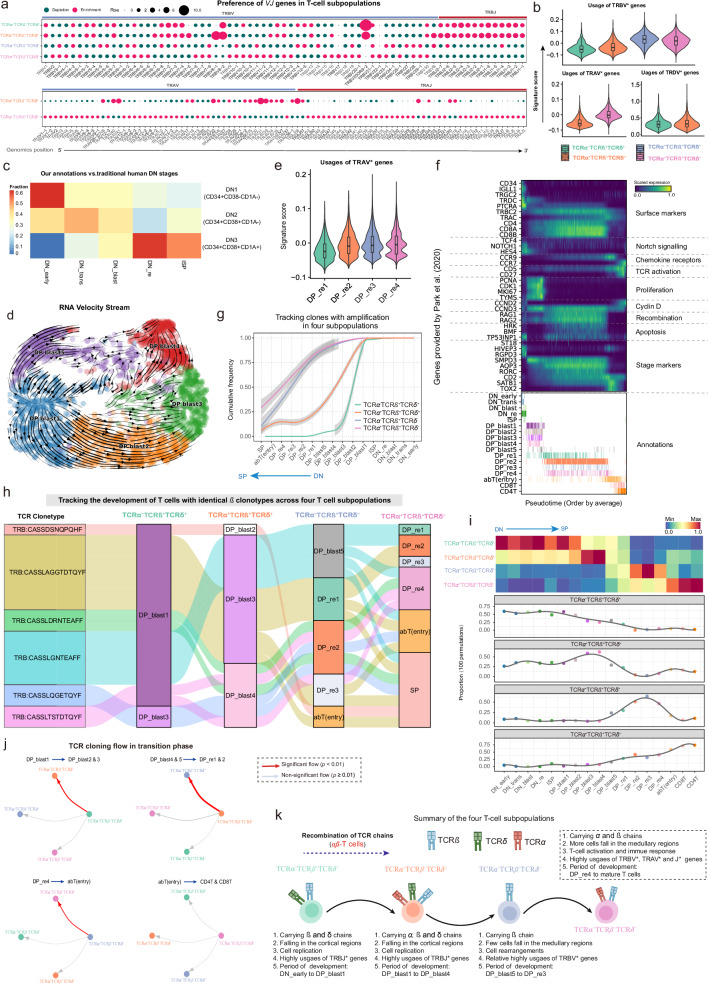
Given the close relationship between T-cell development and the TCR repertoire as well as TCR diversity, we observed pronounced biases in VJ gene usage among the four T-cell subpopulations for TCRα and TCRβ chains. The TRBV* gene exhibited prominence in both TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ subpopulations, whereas TRBJ* was prevalent in the TCRα-TCRβ+TCRδ- and TCRα+TCRβ+TCRδ- subpopulations (Figs. 8a, b). Additionally, TRAV* and TRAJ* displayed a distinct preference in both TCRα+TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ- subpopulations, with TRAJ* apparently enriched in TCRα+TCRβ+TCRδ- cells (Fig. 8a). While certain TRAV* genes, such as TRAV40, TRAV41, and TRAV31, displayed a preference for TCRα+TCRβ+TCRδ+ subpopulations, the overall enrichment of TRAV* genes were observed in TCRα+TCRβ+TCRδ- cells (Figs. 8a, b). Notably, the signature score of TRDV* appeared comparable between the TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ subpopulations, indicating that these δ chain genes are used with similar frequency. Classical theory suggests that mature TCR chains typically manifest a broader spectrum of V gene segment utilization. However, during the initial stages of T cell development, a preference emerges for the selection of J gene segments. Consequently, the TCRα-TCRβ+TCRδ- and TCRα+TCRβ+TCRδ- subpopulations exhibit comparatively more mature β chains than the TCRα-TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ+ subpopulations. In terms of α chains, the TCRα+TCRβ+TCRδ- subpopulation demonstrates a heightened degree of maturity compared to the TCRα+TCRβ+TCRδ+ subpopulation.
Fig. 8. Characterizing the temporal sequence of differentiation dynamics in four T-cell subpopulations.
a Bubble plots showing the preference of each TRB[VJ]* gene segment present in the four T-cell subpopulations (top) and TRA[VJ]* in the TCRα+TCRβ+TCRδ+ and TCRα+TCRβ+TCRδ- subpopulations (bottom). Gene segments are positioned according to their genomic location. b Violin plots showing the distribution of signature scores across the four T-cell subpopulations, as inferred by TCRα, TCRβ, and TCRγ V gene segments. Median value, interquartile range (IQR) as bounds of the box and whiskers that extends from the box to upper/lower quartile ± IQR × 1.5. Subpopulations: TCRα-TCRβ+TCRδ+ (n = 4165 cells), TCRα+TCRβ+TCRδ+ (n = 3305 cells), TCRα-TCRβ+TCRδ- (n = 12,691 cells), and TCRα+TCRβ+TCRδ- (n = 39,015 cells). c Heatmap showing the association between the DN subsets defined by our clustering strategy and the traditional human DN stages. The color gradient reflects the relative proportions of our annotated DN subsets across the DN1-DN3 developmental stages. d RNA velocity stream from DP_blast1 to DP_blast5, with cell subsets marked with color codes. e Violin plot showing the distribution of signature scores across DP_re1 to DP_re4, as inferred by TCRα V gene segments. Median value, interquartile range (IQR) as bounds of the box and whiskers that extends from the box to upper/lower quartile ± IQR × 1.5. Cell subsets: DP_re1 (n = 3865), DP_re2 (n = 18,337), DP_re3 (n = 2152), and DP_re4 (n = 3010). f Heatmap showing representative genes provided by Park et al.8 across T cell differentiation pseudotime. Top panel: The x-axis represents pseudo-temporal ordering. Gene expression levels across the pseudotime axis are maximum-normalized and smoothened, and grouped by their functional categories and expression patterns. Bottom panel: Cell type annotation of cells are aligned along the pseudotime axis. g Tracking of T cells with identical TCRβ chains in the four T-cell subpopulations from DN_early to SP (single positive) development. Cumulative distribution curves showing the number of T cells at different stages (see “Methods”). Lines smoothed using generalized additive models (GAM), with shaded areas representing 95% confidence intervals. h Sankey plot tracking the differentiation routes for cells sharing identical TCRβ clonotypes across T-cell subpopulations. i Heatmap (top) and line plot (bottom) showing the relative abundance of the four T-cell subpopulations at each stage from DN_early to SP development. Abundance was averaged for each cell type after 100 down-samplings for the four T-cell subpopulations respectively. j Significance of the transition of each T-cell subpopulation to the other three subpopulations at specific differentiation stages. P-values obtained by one-sided hypergeometric tests, with p ≤ 0.01 indicating a significant transition to a specific subpopulation (see “Methods”). k Schematic diagram of the developmental timing of the four T-cell subpopulations. The solid line indicates a transitional relationship between two subpopulations and the dotted line indicates the absence of such a relationship. The thickness of the solid line represents the strength of significance obtained in Fig. 8j. DN: Double negative, DP: Double positive, SP: Single positive, DP_blast: DP blast cells, DP_re: DP rearrangement cells, TCR: T-cell receptor. The TCR icons in Fig. 8k were created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a Source Data file.
To ascertain the timing and transition points of differentiation for the TCRα+TCRβ+TCRδ+ and TCRα-TCRβ+TCRδ- populations, the T-cells in the DN and DP stages were re-clustered (Supplementary Figs. 11b, c). Refined subgroups were identified within the DN stage, namely DN_early, DN_trans, DN_blast, DN_re, and ISP (Supplementary Figs. 11c, d). Typically, the developing human DN compartment is categorized into the CD34+CD38-CD1A- (DN1) stage, representing the most immature thymocyte subset, followed by the CD34+CD38+CD1A- (DN2) and CD34+CD38+CD1A+ (DN3) stages. Our comparison revealed that DN_early is predominantly found in the DN1 stage, DN_trans, DN_blast primarily in the DN2 stage, while the DN3 stage is closely linked with DN_re and ISP (Fig. 8c). Similarly, distinct clusters were observed within the DP_blast stage, namely DP_blast1 (UNG), DP_blast2 (TYMS), DP_blast3 (TOP2A and CDK1), DP_blast4 (CDC20), and DP_blast5 (RAG1), based on RNA velocity, pseudo-time, and cell-cycle scores (Fig. 8d; Supplementary Fig. 11c, e-i; see “Methods”). Additionally, the DP_re cells were re-clustered into DP_re1 (ACTG1 and GAPDH), DP_re2 (STMN1 and MZB1), DP_re3 (MT1X and CDR1), and DP_re4 (PTPRC and HSPH1) subsets based on RNA velocity, particularly the signature scores of TRAV* genes (the higher the score, the more mature it indicates). (Fig. 8e; Supplementary Figs. 11j, k). To gain a more systematic understanding of the differentiation pattern of αβT cells, pseudo-time analysis was performed on all T cells, which consistently demonstrated an ordered arrangement of T cells based on known marker genes and transcription factors (Fig. 8f and Supplementary Fig. 11l)8.
Based on refined annotations of T cells, cells exhibiting TCR amplification were isolated from all four T-cell subpopulations, and their respective cumulative distributions were computed across various stages of T cell differentiation (see “Methods”). As shown in Fig. 8g, the TCRα-TCRβ+TCRδ+ cells accumulated significantly in the DN and DP_blast stages, suggesting their early differentiation (Fig. 8g). Similarly, the TCRα+TCRβ+TCRδ+ subpopulation ranked second in accumulation, succeeded by TCRα-TCRβ+TCRδ-. As the TCRα+TCRβ+TCRδ- cells accumulated predominantly at the SP stage, indicative of a closer proximity to mature T-cell phenotypes, thus representing a later stage of differentiation (Fig. 8g). Prompted by these observations, we pose a fundamental question: Do the identified four T-cell subpopulations accurately delineate continuous developmental stages within T cell maturation? To address this pivotal inquiry, we meticulously curated cells exhibiting identical β chains (CDR3 sequences) from the four T-cell subpopulations. Subsequently, we diligently tracked the fate of these clonal cells, with the sorting of T-cell subpopulations aligning with the chronological sequence outlined in Fig. 8g. Our findings unveil a distinct and coherent developmental trajectory of these cells, progressing from the nascent TCRα-TCRβ+TCRδ+ subpopulation to the TCRα+TCRβ+TCRδ- subpopulation (Fig. 8h). Notably, exemplified by TRB:CASSDSNQPQHF, this progression encompasses transitions from DP_blast1 (TCRα-TCRβ+TCRδ+), DP_blast2 (TCRα+TCRβ+TCRδ+), abT (entry) (TCRα-TCRβ+TCRδ-) to the SP state (TCRα+TCRβ+TCRδ-).
To further determine the transition points of the four T-cell subpopulations, the relative proportions across cell types were examined. As shown in Fig. 8i, the TCRα-TCRβ+TCRδ+ cells were highly enriched from the DN_early to DP_blast1 stages, while TCRα+TCRβ+TCRδ+ cells were more abundant from the DP_blast2 to DP_blast4 stages. Likewise, the TCRα-TCRβ+TCRδ- cells were mainly involved in the differentiation of DP_re1 and DP_re4 stages. The stage from abT(entry) to mature T-cells was predominantly composed of the TCRα+TCRβ+TCRδ- cells (Fig. 8i). In addition, there was a significant expansion of TCRα-TCRβ+TCRδ+ towards TCRα+TCRβ+TCRδ+ during differentiation from the DP_blast1 to DP_blast2 and DP_blast3 stages (Fig. 8j; see “Methods”). TCRα+TCRβ+TCRδ+ significantly transitioned towards the TCRα-TCRβ+TCRδ- subpopulation from the DP_blast4/5 to DP_re1/re2 stages (Fig. 8j). From the DP_re4 to abT(entry) stage, TCRα-TCRβ+TCRδ- underwent significant expansion towards TCRα+TCRβ+TCRδ-, with no significant flow of TCRα+TCRβ+TCRδ- to other subpopulations (Fig. 8j). These results collectively indicate that DP_blast1, DP_blast4/5 and DP_re4 represent the transition points of the four T-cell subpopulations, and the formation and maturation of TCR chains constitute a continuous rather than a transient differentiation process.
Based on these findings, we propose a novel perspective on the developmental trajectory of αβ T cells. Specifically, during the early stages of T cell development, both the segment genes of the β and δ chains are concurrently active (TCRα-TCRβ+TCRδ+). This finding aligns with prior investigations suggest that the expression levels of δ segment genes in early T cells may influence the cellular fate (α/β or γ/δ)47. Moreover, concomitant with the expression of α segment genes, a triple-positive intermediate T cell state emerges (TCRα+TCRβ+TCRδ+). Notably, during this stage, the α chain exhibits discernible immature characteristics, speculation of its potential pivotal regulatory role in facilitating the β-selection process of T cells. As T cells progress into the DP rearrangement stage, the expression of δ chain is wholly suppressed. Intriguingly, the segment genes of immature α chain are also subject to suppression, resulting in the formation of TCRα-TCRβ+TCRδ- cells. However, it is imperative to underscore that the underlying regulatory mechanism governing the suppressed expression of α segment genes remains elusive. Ultimately, to form functional TCR complexes, the TCRαβ complex is formed by integrating with the β chain following α rearrangement during the DP stage, culminating in the development of SP T cells (TCRα+TCRβ+TCRδ-). Additionally, we summarize the developmental timing and characteristics of the four T-cell subpopulations in Fig. 8k to depict αβ T cell development.
Discussion
The proper development of T cells is crucial for maintaining a healthy adaptive immune system6,28,48. Although numerous studies utilizing scRNA-seq have uncovered the developmental mechanisms of thymic T cells, the spatial distribution of the cells at single-cell resolution within the thymic microenvironment, and the inter-cellular interactions remain to be elucidated8,11,49–51. Additionally, previous studies have primarily focused on thymus development in fetal or early pediatric stages, with limited information available regarding the thymus in elderly individuals8,11,33,52. In this study, we mapped thymocyte development at single-cell resolution from the fetal to geriatric stages. Our findings revealed a higher enrichment of immature-state T cells in the prenatal and pediatric thymus, while mature-state T cells predominated in the adult and geriatric groups. This suggests that the microenvironment of the prenatal and pediatric thymus is more conducive for T-cell development, whereas the geriatric thymus tends to undergo fibrosis and atrophy, leading to the development of autoimmune diseases53.
The spatial localization of cell types in the thymic microenvironment is closely associated with their function54. To investigate the spatial distribution of thymic cell types, we developed the TSO-his tool based on ST data from eight pediatric donors. TSO-his effectively identified the critical structural regions and lobular segmentation in ST slices. By applying this tool, we comprehensively examined the relationship between cell types and their spatial distance, and inferred the spatial location of these cells. We found that Fb and VSMCs interact with DP cells via MHC class I molecules, potentially facilitating T-cell recognition and selection. Furthermore, we identified significant distance-varying genes and characterized their expression patterns and biological functions. While projecting single cells into ST slices can effectively characterize the spatial cell atlas of the thymus, accurate localization of 34 cell types proved to be challenging. The widely used tool SrtCT55, as well as the recently published CellTrek41, showed unsatisfactory results. To address this issue, we developed the thymus-specific TSO-hismap by integrating histological structures determined by TSO-his and the spatial deconvolution tool CARD. TSO-hismap allowed accurate projection of thymic cell types in ST slices, which aligned with the anatomical structure of the thymus. Based on the spatial architecture of the thymus at single-cell resolution obtained by TSO-hismap, we systematically characterized the spatial localization and interactions among cell types in the thymic microenvironment, and identified several novel co-localizations, including CD8aa with B_memory and Mono. These findings provide important insights into T-cell development in the thymus.
Four distinct subpopulations within the αβT cell lineage have been identified, each characterized by unique patterns of TCR receptors, providing valuable insights into the intricate stages of T cell development within the thymus. By leveraging TCR sequencing data, scRNA-seq, and spatial transcriptomics, we have systematically delineated the spatial distribution of these four T-cell subpopulations within the thymic architecture. Early precursors (TCRα-TCRβ+TCRδ+, TCRα+TCRβ+TCRδ+) primarily inhabit the cortex, whereas more mature T cells (TCRα-TCRβ+TCRδ-, TCRα+TCRβ+TCRδ-) are predominantly located in the medulla, highlighting the nuanced interplay between diverse thymic microenvironments and T cell maturation. Moreover, dynamic alterations in the TCR repertoire during T cell maturation were observed, with discernible biases in the utilization of VJ genes across different subpopulations, elucidating the intricate mechanisms governing TCR repertoire selection. Meticulous tracking of TCR clones has further facilitated the delineation of the developmental timeline and critical transition points among these four T-cell subpopulations. Significantly, as T cells transition from the TCRα+TCRβ+TCRδ+ to the TCRα-TCRβ+TCRδ- stage, the suppressed expression of α chain-related genes may result from regulatory constraints imposed by other cellular proteins or pathways during TCR chain folding and assembly into functional complexes within the cell.
Particularly, the expression of MHC class II genes was suppressed in cTECs, leading to impaired positive selection of CD4+ T cells. The discovery of ELOVL4’s regulatory role in CD4+ T cell maturation and its correlation with the population of CD4+ T cells provides valuable insights into both fundamental immunological processes and the clinical implications of its dysfunction. n-3 VLCPUFAs (very long-chain polyunsaturated fatty acids) have been identified as specific components of membrane microdomains crucial for cellular signaling processes during T lymphocyte activation56. Consequently, the downregulation of ELOVL4 may potentially inhibit the synthesis of n-3 VLCPUFAs, thereby opposing the formation of the microdomains required for CD4+ T cell maturation and activation.
In summary, our study delivers an in-depth investigation of thymocyte development, spatial organization at single-cell resolution, and the dynamic changes in TCR signaling associated with T-cell maturation. It offers valuable insights and data resources for both fundamental immunological mechanisms and potential clinical applications
Methods
All studies comply with all relevant ethical regulations. All research protocols involving human samples were subjected to review by the Ethics Committee of Nanjing Drum Tower Hospital, and received approval for the study protocols as described in detail below.
Thymic tissue acquisition
The study adhered to the principles delineated in the Declaration of Helsinki, and ethical clearance was secured from the Research Ethics Committee of Nanjing Drum Tower Hospital (ID 2020-015-01). 16 individuals were recruited between January 2020 and November 2022, encompassing healthy thymus subjects spanning various life stages (prenatal, pediatric, adult, and elderly). Fetal specimens were sourced from miscarriages or stillbirths, while pediatric samples were obtained from patients with congenital heart disease undergoing surgical intervention, necessitating the removal of obstructing thymic tissue. Specimens from adult and elderly subjects were procured posthumously from organ donors. The patients included males and females, as the information on sex and gender was not relevant in our study. The clinical characteristics of the patients were shown in Supplementary Data 1. All patients provided written informed consent for sample collection and data analyzes prior to operation.
Detailed demographic characteristics of the study population were succinctly presented in Supplementary Data 1.
Tissue dissociation and preparation of single-cell suspensions
To initiate tissue dissociation, samples were rinsed in ice-cold 1×PBS at 4°C. They were then transferred to a sterile RNase-free culture dish for processing. Using surgical scissors, the tissues were finely cut into small fragments, approximately 0.5 mm² in size. Unwanted tissues such as blood stains, fatty layers, and connective tissues were eliminated during the washing process with 1×PBS. The tissue samples were then incubated in a constant-temperature water bath set at 37 °C, utilizing the Miltenyi MACS Human Tumor Dissociation kit (130-095-929). Typically, the incubation period ceased once the digestive solution turned turbid and the tissue mass dissolved. Following this, the cell suspension was filtered using a 40 µm cell strainer (Corning, 352340).
Upon filtration, the cell suspension underwent centrifugation at 400rcf at 4 °C for 5 minutes. The supernatant was discarded, and 10 mL of 1X pre-diluted Red Blood Cell Lysis Solution (10x) (Miltenyi,130-094-183) was added to the tube, ensuring even distribution. After incubating at 4°C for 5–10 minutes, centrifugation was promptly performed at 400rcf at 4°C for 5 minutes. The supernatant was then removed, and the cells were washed two to three times with pre-cooled 1X PBS. To assess cell viability, trypan blue staining was conducted using the TC20 automated cell counter (Bio-Rad, 1450102). Based on the results, adjustments were made to achieve a target concentration of 700–1200 cells/µL with a cell viability exceeding 90%. Once the desired cell concentration and viability were attained, the cells were kept on ice, ready for the 10X Genomics single-cell immune profiling chip on-board experiment to commence within 30 minutes.
Library preparation for 10X genomics single-cell 5’ gene expression and V(D)J sequencing
The scRNA-seq and V(D)J libraries were generated using the 10X Genomics Chromium Controller instrument, Chromium Single Cell 5’ Library & Gel Bead Kit, and V(D)J Enrichment Kit according to the manufacturer’s instructions. Briefly, single-cell suspensions of thymus tissues (>90% viability) were loaded onto the controller to generate gel bead-in-emulsions for individual cells. The mRNA was reverse transcribed and sample indexed to obtain barcoded cDNA, which was purified using DynaBeads and amplified by PCR. To construct the 5’ gene expression library, the amplified barcoded cDNA was fragmented, end repaired, A-tailed, sample indexed, and subjected to double-size selection (average size 450 bp). For the V(D)J library, V(D)J sequences of human T cells were enriched from the amplified cDNA, followed by fragmentation, end repairing, A-tailing, sample indexing, and double-size selection (average size 600 bp). DNA was quantified using the Qubit dsDNA HS assay kit (Thermo, Q32851), and the fragment size distribution of the libraries was determined using the Agilent 2100 BioAnalyzer High Sensitivity DNA kit (Agilent Technologies, 5067-4626). Subsequently, the pooled libraries were sequenced on the Illumina high-output sequencing platform, and both RNA-seq and V(D)J libraries had 150 bp paired-end reads.
10X library sequencing
The scRNA libraries were sequenced using the Illumina Novaseq platform, ensuring a minimum sequencing depth of 50,000 reads per cell. The sequencing setup included 150 bp read 1, 8 bp i7 index, and 150 bp read 2. Similarly, the single-cell TCR libraries were sequenced on the same platform, with a minimum sequencing depth of 5000 reads per cell, and using the same sequencing setup. For scRNA-seq analysis, 180,039 cells were sequenced, resulting in 7,169,639,936 reads. Each sample had 358,481,997 reads, and each cell had 39,823 reads on average. In addition, an average of 20,816 genes were detected per sample. For scTCR-seq, 147,694 cells were sequenced, yielding 835,189,411 reads. On average, each sample had 41,759,471 reads, and each cell had 5,655 reads. Detailed statistics on the mapping of individual cells are shown in Supplementary Data 1.
10X Visium experiment and spatial transcriptome sequencing
Sample preparation and tissue optimization
Fresh tissues were frozen and embedded in an optical cutting tissue (OCT) compound using liquid nitrogen. The RNA quality of the OCT embedded block was evaluated using Agilent 2100. Only tissues with an RNA integrity number (RIN) greater than 7 were selected for the Visium spatial gene expression experiments. To optimize the tissue, the Visium Spatial Tissue Optimization Slide & Reagent kit (10X Genomics, PN-1000193) was utilized following the guidelines provided in the Visium Spatial Tissue Optimization User Guide (CG000238, 10X Genomics). The optimization process involved placing tissue sections onto 7 Capture Areas on a Visium Tissue Optimization slide. These sections were fixed, stained, and permeabilized for different durations. During permeabilization, the released mRNA molecules bound to oligonucleotides present on the Capture Areas. Subsequently, fluorescent cDNA was synthesized on the slide and subjected to imaging. The optimal permeabilization time was determined by maximizing the fluorescence signal while minimizing signal diffusion. If the signal remained the same at two different time points, the longer permeabilization time was considered the optimal choice.
Staining and imaging
Cryosections were cut to a thickness of 10 μm and then mounted onto the GEX arrays. These sections were placed on a Thermocycler Adapter with the active surface facing upwards and incubated at 37 °C for 1 minute. Subsequently, they were fixed for 6 minutes using methyl alcohol at −20°C. Following the fixation, the sections were stained with H&E (Eosin, Dako CS701, Hematoxylin Dako S3309, bluing buffer CS702). The brightfield images were captured using a Leica Aperio Versa8 whole-slide scanner at a resolution of 20×.
cDNA library preparation for sequencing
The Visium spatial gene expression analysis was conducted using the Visium spatial gene expression slide and Reagent Kit (10X Genomics, PN-1000184). To create leakproof wells for reagent addition, the Slide Cassette was utilized for each well. Subsequently, 70 μl of Permeabilization enzyme was added and incubated at 37°C for 30 minutes. Afterward, each well was washed with 100 μl of SSC buffer, followed by the addition of 75 μl of reverse transcription Master Mix for cDNA synthesis. Upon completion of the first-strand synthesis, the RT Master Mix was removed from the wells. A solution of 75 μl of 0.08 M KOH was added and incubated at room temperature for 5 minutes. Then, the KOH was removed from the wells, and they were washed with 100 μl of EB buffer. Subsequently, 75 μl of Second Strand Mix was added to each well for second-strand synthesis. For cDNA amplification, a S1000TM Touch Thermal Cycler (Bio Rad) was employed. Following the manufacturer’s instructions, the Visium spatial libraries were constructed using the Visium spatial Library construction kit (10X Genomics, PN-1000184). Finally, the libraries were sequenced using an Illumina NovaSeq 6000 sequencer with a sequencing depth of at least 50,000 reads per spot using a pair-end 150 bp (PE150) reading strategy.
Antibodies and plasmid
The functional-grade antibodies utilized in this study included CD3 (Cat: 14-0031-82, clone 145-2C11,1:1000) and CD28 (Cat: 16-0281-82, clone 37.51, 1:1000), both sourced from eBioscience. Fluorescence-labeled antibodies for CD3 (Cat: 11-0032-82, 17A2,1:200), CD4 (Cat: MCD0428, RM4-5, 1:200), CD8 (Cat: A15385, 53-6.7, 1:200), CD25 (Cat: 17-0251-82, PC61.5, 1:200), FOXP3 (Cat: 25-5773-82, FJK-16s, 1:200), CD44 (Cat: 17-0441-82, IM7, 1:200), CD62L (Cat: 12-0621-82, MEL-14, 1:200) and CD99 (Cat: 12-0997-42, 3B2/TA8, 1:200) were from eBioscience. Mouse CD99 PE-conjugated Antibody were from R&D (Cat: FAB3905P,1:200). The antibody targeting mouse ELOVL4 (Cat: 55023-1-AP,clone 55023-1-AP, 1:1000) was procured from Proteintech. Anti-Actin (Cat: MAB1501, C-4, 1:10,000) was from Sigma. psPAX2, pMD2.G and lentiviral sgRNA vector were co-transfected into HEK293T cells to produce lentiviruses.
Cell line, mice and BM adoptive transfer
HEK293T cells were originally purchased from ATCC (catalog no. CRL-3216). Rag1-KO (C57BL/6, Strain NO. T004753) mice were purchased from Gempharmatech Co., Ltd. ROSA26-Cas9 mice (C57BL/6, Cat. NO. NM-KI-00120) were purchased from Shanghai Model Organisms Center, Inc. Cas9-expressing bone marrow cells isolated from ROSA26-Cas9 mice were spin transduced with lentiviral constructs on a Retronectin-coated plate and adoptively transferred into irradiated (950 rad on an X-Rad irradiator) Rag1-KO mice. After 8weeks, the chimeric mice were euthanized using CO2 asphyxiation followed by cervical dislocation for analysis of thymus T cell development and peripheral T cell activation. All animal experiments were conducted in accordance with the guidelines set forth by the institutional and national committees. Explicit permission to perform animal experiments was granted by the Institutional Animal Care and Use Committee (IACUC) at Nanjing Medical University, under the protocol number 2308051. The animals were housed in a specific pathogen-free (SPF) facility. For each experiment, sex-matched and age-matched (6–8 weeks old) mice were used, with experimental and control animals co-housed to ensure consistent environmental conditions. The facility maintains strict protocols to prevent pathogen contamination and to ensure the well-being of the animals. All sgRNA sequences and their oligos are listed in Supplementary Table 5.
Naïve T cells isolation and stimulation
Total T cells were isolated from the spleen utilizing a pan T cell isolation kit (Miltenyi Biotec), in accordance with established protocols. Enriched T cell populations were subsequently subjected to flow cytometric cell sorting based on distinctive cell surface markers. Specifically, the sorting strategy involved selecting cells displaying the CD4+CD44loCD62Lhi for naïve CD4+ T cells, and the CD8+CD44loCD62Lhi for naïve CD8+ T cells. The obtained naïve CD4+ and CD8+ T cells were subjected to stimulation through the utilization of plate-bound anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml) antibodies (eBioscience).
Flow Cytometry (FACS)
Thymic, splenic, and lymph node single-cell suspensions were prepared by gently homogenizing the tissues using a tissue homogenizer. Cells were washed in PBS and resuspended in FACS buffer (PBS containing 2% FBS and 0.1% sodium azide). The cells were then incubated with Fc-block (anti-mouse CD16/CD32) for 10 minutes at 4°C to block nonspecific binding. Subsequently, cells were stained with fluorochrome-conjugated antibodies against surface markers for 30 minutes at 4°C in the dark. Data acquisition was performed on a BD FACSCelesta flow cytometer, and data were analyzed using FlowJo (v10).
Western Blot (WB)
Protein extracts were prepared from thymocytes or naïve T cells using RIPA buffer containing protease and phosphatase inhibitors (Pierce). Protein concentrations were determined using the BCA assay (Pierce). Equal amounts of protein (30 μg) were separated by SDS-PAGE on a 12% polyacrylamide gel and transferred to PVDF membranes. Membranes were blocked with 5% non-fat dry milk in TBS-T (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature, followed by overnight incubation at 4 °C with primary antibodies. After washing, membranes were incubated with HRP-conjugated secondary antibodies for 1 hour at room temperature. Bands were visualized using ECL substrate (Thermo Scientific) and imaged with Bio-Rad ChemiDoc.
Enzyme-Linked Immunosorbent Assay (ELISA)
Serum or cell culture supernatants were collected and stored at −80°C until analysis. ELISA kits for IL-2 and IFN-γ were purchased from Biolegend and assays were performed according to the manufacturer’s instructions. Briefly, samples and standards were added to 96-well plates pre-coated with capture antibodies. After incubation and washing, detection antibodies were added, followed by HRP-conjugated secondary antibodies. The plates were developed using TMB substrate, and the reaction was stopped with 2 N H2SO4. Absorbance was measured at 450 nm using a microplate reader (Multiskan FC, Thermo Scientific). Concentrations were calculated based on the standard curve.
Gating strategy
For immune cells, lymphocytes were gated based on FSC-A and SSC-A. Singlet cells were gated according to the pattern of FSC-H vs FSC-A. Detailed gating strategies are shown in Supplementary Figs. 6a, b.
Multiplex immunofluorescence assays
Multiplex staining was performed using the Opal 4- Color Manual IHC Kit (NEL810001KT) with the anti-GNG4 (1:500; 13780-1-AP, Protein Tech), anti-IgA (1:200; ab124716, Abcam), and anti-CD14 (undiluted, GT229807, Gene Tech) antibodies. The staining was visualized with fluorescein AF-690 (1:75), AF-520 (1:75) and AF-570 (1:50), and the nuclei were counterstained with 4’, 6- diamidino- 2- phenylindole (1:3,000). All sections were covered with Vectashield Hardset 895 mounting media, and scanned using the Vectra slide scanner (PerkinElmer).
Single-cell RNA sequencing (scRNA-seq) data processing, integration, and dimensionality reduction
The raw sequencing data of normal thymus samples were processed using the CellRanger pipeline (version 4.0.0, 10X Genomics) and aligned to the GRCH38 v93 genome assembly with default parameters. The resulting UMI count matrices by cell barcode were loaded into the Seurat R package (version 4.1.0)19. Cells with less than 200 or more than 5000 expressed genes, more than 15% mitochondrial counts, more than 50% ribosomal counts, or less than 500, or more than 20,000 UMI counts were discarded (Supplementary Fig. 1a). Doublet cells were detected using DoubletFinder57 (version 2.0.3) and filtered out (Supplementary Fig. 1b). A total of 130,295 cells were obtained after quality control. For each sample, the raw UMI counts were log-normalized using a scale of 10,000, and the top 2000 highly variable genes were determined using the FindVariableGenes function with the “vst” selection method. To remove donor effects, canonical correlation analysis (CCA) was performed using the FindIntegrationAnchors and IntegrateData functions. Following integration, all samples were merged into a single Seurat object with the “integrated” assay. The ScaleData function was then used to regress out the effects of UMI count, percentage of mitochondrial and ribosomal genes, and the S and G2/M scores (obtained using the CellCycleScoring function). For dimensionality reduction, principal component analysis (PCA) was conducted on the 2000 variable genes, and 50 principal components (PCs) were retained for subsequent analysis. The first 30 principal components were used for uniform manifold approximation and projection (UMAP), with a minimum distance of 0.3 and 30 neighbors (Supplementary Fig. 1e).
Cell type annotation and visualization for normal thymus
The cell types in the thymus were identified by unsupervised clustering and comparison of the differentially expressed genes (DEGs) with known marker genes from literature (Fig. 1d; Supplementary Fig. 1e). To annotate the broad cell categories, the cells were initially clustered based on the first 30 PCs using the Shared Nearest Neighbor (SNN) algorithm with the FindNeighbors and FindClusters function, and parameters k = 30 and resolution = 0.8. The DEGs were screened using the FindAllMarkers function in Seurat, with min.pct = 0.25 and logfc.threshold = 0.25. Six major cell types, including Ery (HBG1 and HBG2), B cells (CD79A and CD19), plasma cells (IGHG1 and IGHG2), myeloid cells (S100A8, C1QA and IL3RA), stromal cells (ACTA2 and DCN) and T cells (CD3D and CD3E)8,58, were identified by comparing the DEGs of each cluster to the canonical marker genes. The B cells, myeloid cells, stromal cells, and T cells were subjected to a second round of clustering. Briefly, the first 30 PCs were re-extracted from the subset of the “integrated” assay, and unsupervised clustering was conducted with a resolution range of 0.3–1.5, followed by differential expression analysis based on the “RNA” assay. Each sub-cluster was required to have a minimum of 20 significantly highly expressed genes (FDR ≤ 0.01 and logFC ≥ 0.25; Wilcoxon test) compared to other cells.
B cells were re-clustered at a resolution of 0.3, and the B_naive (CD19, CD79A, IGHD), B_trans (MS4A1, CD24), and B_memory (IGHA1, CD27) subtypes were identified. Myeloid cells were re-clustered into four subtypes at a resolution of 0.5, namely Mono (S100A8, S100A9), Mac (C1QA, C1QC), DC (HLA-DPA1, HLA-DQB1), and pDC (IL3RA, LILRA4). Stromal cells were divided into five subtypes at a resolution of 0.3, including Fb (DCN, COL1A1), Fb_cycling (DCN, COL1A1, UBE2C, TOP2A), VSMC (TAGLN, ACTA2), Endo (ACKR1, PLVAP), Lymph (TFF3, NTS), and TEC (KRT17, KRT9). TEC cells were further isolated and re-clustered into two groups, namely cTEC (CCL25, GNG11) and mTEC (CCL19, KRT19). The DN (CD4-CD8-), DP (CD4+CD8+), abT (entry) and SP (CD3+CD4+CD8- or CD3+CD4-CD8+) subtypes of T cells were identified after clustering at a resolution of 0.5 and comparing with the thymus data of Park et al. 8 and Cordes et al. 33. The DN cells were further divided into the DN_early (IGLL1, SMIM24, TRDC), DN_blast (TOP2A, TYMS), and DN_re (PTCRA, RAG1, RAG2) sub-clusters at a resolution of 0.5. The DP cells were classified into DP_blast (CDK1, TOP2A) and DP_re (RAG1, RAG2) subtypes at a resolution of 0.3. The SP cells were annotated as CD4+ T (CD4, SELL), CD4+ T_mem (IL7R, CCR7), Treg.diff (FOXP3, IFITM1), Treg (FOXP3, IL32), CD8+ T (CD8A, CD8B, GZMM, ABLIM1), CD8+ T_mem (CCL4, GZMK), CD8aa (CD8A, CD27, ID3), agonist T cells (T_agonist) (TNFRSF1B, LCP2), apoptosis T cells (T_apoptosis) (BCL2L11), proliferating T cells (T_proliferating) (TYMS, UBE2T), NKT (NKG7, KLRB1), and innate lymphoid cell type 3 (ILC3) (IL4I1, SLC16A3, TNFRSF25) at a high resolution (1.5). Overall, the thymus cells were classified into 34 clusters within six major cell lineages (Figs. 1c, d; Supplementary Fig. 1e). The signature marker genes for each cluster are listed in Supplementary Data 2.
To visualize the refined annotations more efficiently, we employed the FindAllMarkers function in Seurat was used to identify the top 30 significantly upregulated genes for each cell type, and the subset of these genes was exported to Scanpy (version 1.8.1)59 python package. Using gene expression as the response variable, and the cell type, percentage of mitochondria and ribosomes, and cell-cycle scores as covariates, an L2-regularized linear model was fitted and the residual matrix containing biological information was retained8. The PCs were extracted using the scanpy.pp.pca function with default parameters, and the batches were aligned using the scanpy.api.pp.bbknn function to achieve a high-resolution and batch-mixed manifold. The UMAP coordinates of cells were generated using the scanpy.tl.umap function in Scanpy and exported as a new embedding in the merged normal thymus Seurat object for visualization (Fig. 1c).
Assessment of the purity of single-cell populations
ROGUE Index
The ROGUE index20 (version 1.0) ranging from zero to one was used to assess the purity of single cell populations (Supplementary Fig. 2a). One represents complete purity and zero represents the most heterogeneous state of a population.
Calculation of purity index by logistic regression model
The purity index was defined using a logistic regression model (Supplementary Fig. 2c). Briefly, 50% of the cells from each cell type were down-sampled to generate the training set, then a logistic regression model was trained using the sklearn.LogisticRegression function from the sklearn60 Python package (version 1.3.2) with the parameters penalty = “l2” and C = 0.2. The features were genes and the labels were cell types. The remaining 50% of the single cell dataset was used to test the model. These processes were repeated 100 times, and the purity index was calculated using the following equation:
| 1 |
where Cj indicates the number of cells of cell type j in the test set, represents the number of cells accurately predicted to be of type j in the ith iteration, Purityj indicates the purity index of cell type j. The purity index ranges from 0 to1, and larger value indicates higher purity and less heterogeneity.
Differential expression analysis
The differentially expressed genes (DEGs) between the subsets were screened using FindAllMarkers function (Wilcoxon rank-sum test, min.pct = 0.25, and logfc.threshold = 0.25) in Seurat, with adjusted p-value ≤ 0.01 (corrected using the Bonferroni method) as the threshold.
Group preferences analysis
To measure the preference of each cluster across different groups, the observed and expected (as inferred by the Chi-square test) number of cells in each cluster were compared according to a previously established formula21 (Figs. 1f, 7l, 8a; Supplementary Fig. 2e,f). A cluster was considered to be enriched in a specific group status if the ratio of observed to expected (Ro/e) number of cells was greater than 1.
| 2 |
Here, “Observed” represents the actual cell counts of subsets within different age groups, while “Expected” indicates the frequencies anticipated if there were no association between subsets and age groups. These expected frequencies were computed by multiplying the respective row and column totals of each subset and then dividing by the total number of cells.
Spatial transcriptomics (ST) data processing
Raw sequencing data were processed using the Spaceranger pipeline (version 2.0.0) from 10X Genomics with the GRCh38 v93 genome assembly as the reference, and resulting UMI count spot matrices were sorted by cell barcode. Raw UMI count spot matrices, along with the corresponding images, spot-image coordinates, and scale factors, were loaded into the Seurat R package (version 4.1.0)19. Only the spots that overlaid tissue sections were retained for further analysis.
For each ST sample, the raw UMI counts were log-normalized using the ScaleData function with a scale factor of 10,000. The number of spots detected in these ST samples ranged from 2138 to 2838, with a mean of approximately 2605 (Supplementary Fig. 3a). On average, each spot contained approximately 2933 genes, with an approximate UMI count of 8612 (Supplementary Figs. 3b, c).
TSO-his algorithm
The TSO-his algorithm was developed to determine the medullary-cortical regions in ST slices, delineate lobules, and measure changes in gene expression or cell abundance with spatial distance (Fig. 2a). The detailed steps were as follows:
(1) Scoring ST spots: The ST spots were scored by combining medulla-indicator genes, including EBI3, CCL17, CCR7, CSF2RB, CCL21, CCL22, TNFRSF18, CCL27, CXCL10, CXCL9, MS4A1, and LAMP3 (Supplementary Table 2)6,28,48, using the AddModuleScore function from the Seurat package. The higher scoring spots were more likely to fall in the medullary region, while the lower scoring spots were associated with the cortical regions.
(2) Screening high scoring spots: In the ST slices, regions with high brightness occupy a relatively small area, and most spots do not deviate significantly, allowing the medullary scores to fit a normal distribution. Briefly, kernel density was used to find the center of the normal distribution, and the scores under the center point were used to fit the normal distribution. The p-value of the medullary score for each spot was obtained using the fitted normal distribution (z-test), and corrected by Benjamin and Hochberg method61. Spots with adjusted p-values ≤ 1e-5 were considered significantly highlighted, and denoted as S.
(3) Determining medullary and cortical regions: The Euclidean distance between spots in the set S was calculated based on their image coordinates. A spot was considered an outlier and removed if its distance to its two nearest neighbors exceeded the mean of the distances plus three times the standard deviation (as inferred from the entire slices; denoted as ε). This process resulted in an updated set, S’. To determine candidate medullary regions, M, a distance matrix was initially constructed from the spots in S′. Each spot in S’ was then assigned to the i-th medullary region Mi if it met the following condition:
| 3 |
Therefore, if M contains m medullary regions, the following condition are satisfied:
| 4 |
Based on the determined medullary set M, candidate medullary regions containing fewer than eight spots were excluded to reduce false positives. Spots not classified as part of the medullary regions were considered cortical regions. To delineate the medulla-cortex (M-C) boundaries, a subset of spots from the cortical region was initially extracted if at least one of their six nearest neighbors was within the medullary region. Spots within the medullary regions were then used to define the M-C boundary if at least one of their six nearest neighbors was part of the previously extracted cortical subset.
(4) Segmenting lobules: Lobules, the fundamental structural units of thymic T-cell development, encompass both medullary and cortical regions. To achieve precise segmentation of these lobules, a nearest neighbor strategy was employed. Specifically, for each ST slice, the medullary center of each medullary region was defined as the spot closest to the arithmetic mean of all coordinates within that region. Subsequently, spots were allocated to the nearest medullary central spot based on Euclidean distances, thereby delineating clusters that represent the thymic lobules.
(5) Measuring dynamic change in cell type signature score: To measure the dynamic change in cell type signature score with spatial distance, generalized linear Gaussian models with four degrees of freedom were fitted. Notably, the signature score was derived using the AddModuleScore function from Seurat, incorporating the top 20 highly expressed genes for the corresponding cell type as features. The distance between the spot of each thymus lobule and its medullary center was normalized to 0–1 scale by the maximum value, and the spots across all ST slices were organized in descending order.
(6) Identifying significant distance-varying genes (DVGs): To pinpoint significant DVGs in ST slices, generalized linear Gaussian models with four degrees of freedom were fitted for each gene and distance within each slice, and then the dependence of gene expression on spot distance was tested. P values corresponding to each natural spline were corrected using the Benjamin-Hochberg method in each slice. Corrected p values for each gene from eight ST slices were then pooled together using Stouffer’s Z-score method. Genes with pooled p-value ≤ 0.01 were considered to be significantly co-varying with spatial distance (i.e., DVGs; Fig. 3a; Supplementary Data 3).
Cell-cell communication analysis
Cellchat
The CellChat (version 1.1.3)31 R package was used to evaluate the interaction between thymic cell subsets based on known ligand-receptor pairs. The normalized counts of merged thymus samples were loaded into CellChat, and the data were pre-processed using the identifyOverExpressedGenes, identifyOverExpressedInteractions, and projectData functions based on the “CellChatDB.human” database. The core functions of CellChat, namely computeCommunProb, computeCommunProbPathway and aggregationNet, were applied in a sequential manner to infer significant ligand-receptor pairs. The resulting significant interaction pairs were visually represented using the netVisual_bubble function in CellChat (Fig. 2j).
To assess the robustness of the inferred interactions predicted by CellChat, a thorough evaluation utilizing a down-sampling strategy was employed. The procedure was outlined as follows: Down-sampling was performed for each cell type based on given cell proportions. Starting with a 100% initial sampling ratio, the ratio was iteratively decreased by 5% until reaching 50% of the original cell type count. Subsequently, inference of interactions using CellChat was carried out for each down-sampled dataset (Supplementary Fig. 4b). Finally, the measurement of interaction strength consistency was conducted using the Coefficient of Variation (CV) (Supplementary Fig. 4c).
CellPhoneDB
CellPhoneDB32 (version v5.0), a repository of ligand-receptor interactions, was employed to identify enriched interactions between various cell types in single-cell transcriptomics data. To manage computational load and ensure fair representation, the dataset was down-sampled by randomly selecting 1000 cells from each cell type. Analysis was conducted using default parameters with the “cellphonedb method statistical_analysis” command (Fig. 2i). To enhance the reliability of significant ligand-receptor interactions, only those identified by both CellChat and CellphoneDB were considered for selection (Fig. 2j).
XGBoost model for predicting cortical and medullary spots
Given that TSO-his requires multiple ST slices to establish a reliable background signal, an improved strategy based on the XGBoost model was used to predict cortical and medullary spots within a single ST slice (Supplementary Fig. 7a). Briefly, the genes at the intersection of significant distance-varying genes (DVGs) and differentially expressed genes (DEGs) were selected as candidate features. ST slices from Thy9 to Thy12 were used as the training set, while Thy5 to Thy8 were used as the test set. Medullary and cortical labels corresponding to each spot in the training set were provided by TSO-his. The features with weak importance were filtered out using the wrapper_feat_select function in the FeatureSelection R package (version 1.0.0), with the parameters objective = ‘reg:linear’ and max_depth = 5. Only the top 50 genes with the highest importance, ranked by the “Cover” index in descending order, were selected as confidence features (Supplementary Table 3). The XGBoost model was trained using the xgb.cv function in the xgboost package (version 1.7.5.1), employing five-fold cross-validation with the parameters objective = ‘binary:logistic’ and max_depth = 10. Model performance was evaluated based on the area under the curve (AUC) and the accuracy of the test set. In addition, the thymic ST data from So et al. 11 were used as an independent validation set to further assess the reliability of the model (Supplementary Fig. 7f). Notably, A structured description for the XGBoost model based on the DOME62 (Data, Optimization, Model, Evaluation) framework, as detailed in Supplementary Table 4. This standardized approach aims to establish a unified method to enhance clarity in understanding models.
Spatial mapping of single-cells
TSO-hismap method
The TSO-hismap method was developed to project thymic cells into spatial transcriptomics (ST) slices (Fig. 5a). The steps were detailed as follows: (1) To determine the ST regions wherein thymic cell types reside, the scRNA-seq was integrated with the ST data using the Canonical Correlation Analysis (CCA) method from the Seurat package (Supplementary Fig. 8a), followed by data scaling and dimensionality reduction. Cells and spots were clustered based on the first 30 PCs using the shared nearest neighbor (SNN) algorithm implemented as FindNeighbors with parameters “k = 30”, and the five nearest neighbor spots of each single cell were obtained using the Seurat:::NNHelper function. Based on the cortical regions, medullary regions, and medulla-cortex (M-C) boundary of the ST slices depicted by TSO-his, the relative proportion f of nearest-neighbor spots of each cell type in the three regions was counted. If f region < 0.05, the cell type was considered not to be in that region. (2) The createCARDObject function from the R package CARD (version 1.0)40 was used to create objects for ST and single cell data, with parameters “minCountGene = 100” and “minCountSpot = 5”, and the CARD_deconvolution function was employed to deconvolute the ST spots with default parameters (Supplementary Fig. 8b). (3) The single cells were mapped to spatial locations using the CARD_SCMapping function of the CARD package with the parameter “numCell = 20”. Since this method initially does not consider the anatomical structure of the thymus, the CARD_SCMapping function was modified to incorporate the spatial location information of thymic cell types obtained in step (1). Specifically, if neighboring cells of a spot include cell types that were not enriched in the region where the spot was located, these cells were excluded from the spot’s assignment.
CellTrek analysis
CellTrek utilizes ST data to train a multivariate random forest model, and predict spatial coordinates by leveraging dimension reduction features shared with scRNA-seq data41. The traint function in the CellTrek R package (version 0.0.94) was used to obtain the co-embedding of ST and scRNA-seq data, and the celltrek function with default parameters was applied to project single cells onto the ST coordinates (Supplementary Fig. 8e).
Seurat coordinate transfer (SrtCT) analysis
SrtCT utilizes the data transfer approach to assign single-cell labels to ST spots. Specifically, the FindTransferAnchors function from Seurat was initially used, with ST data as the query and scRNA-seq data as the reference, then the TransferData function was applied to transfer the single-cell labels (i.e., cell type) to ST spots using default parameters (Supplementary Fig. 8e).
Generation of synthetic spatial transcriptomics (ST) datasets
To assess the accuracy and robustness of TSO-hismap, synthetic ST datasets with predefined cell type compositions and spatial coordinates of cells were generated. Specifically, Thy5 ST data served as a spatial template, and a subset of 500 cells from each cell type in the thymus scRNA-seq data was randomly selected to form the training set. The FindVariableFeatures function from Seurat was employed to identify the top 2,000 highly variable genes (HVGs) within the training dataset. These HVGs were then used to match each spot to the ten closest cells in the training set based on the Pearson correlation coefficient of gene expression. For each spot, synthetic expression was calculated as the sum of UMI counts for each gene across the ten closest cells, with cell type compositions and cell coordinates documented accordingly. Importantly, before HVGs screening, perturbations were introduced by shuffling gene expressions among different cells. Simulated ST datasets with varying noise levels were generated using perturbation proportions of 5%, 10%, and 15%.
Subsequently, the remaining thymus single-cell datasets were used as test sets and mapped to the simulated ST datasets using SrtCT, CellTrek, CARD, and TSO-hismap, respectively. The mapping accuracy was assessed using the following formula:
| 5 |
represents the set of cell types mapped to spot i, while Si represents the set of actual cell types in the simulated data for spot i. The term represents the count of common cell types between and Si, while represents the count of cell types in the union of and Si.
Spatial co-localization analysis of cell types
To explore the spatial co-localization panorama of thymic cell types, the single cells were projected onto ST slices using TSO-hismap, where each spot was split into 20 single cells and the J-index was employed to measure the strength of spatial co-localization between cell types.
| 6 |
|Si| and |Sj| denote the number of spots containing cell types i,and j respectively. indicates the number of spots containing both cell type i and j. Jij represents the co-localization index of cell types i and j, ranging from zero to one. An index of one suggests complete co-localization of the cell types in ST slices, while an index of zero indicates lack of co-localization. The spatial co-localization network of thymic cell types was visualized using the textplot_network function in quanteda R package (version 2.1.2), where lines with a J-index less than 0.3 were removed (Fig. 6a; Supplementary Data 5). Notably, Ery cells were excluded due to their contamination in ST slices (Supplementary Figs. 4d, e).
T-cell receptor (TCR) sequencing analysis
αβTCR VDJ sequence analysis
The single-cell αβTCR sequencing data was mapped using the CellRanger pipeline (version 4.0.0, 10X Genomics) with the “vdj” mode to the GRCH38 v93 genome assembly. The VDJ sequence information for each cell was extracted from the output file “filtered_contig_annotations.csv”, and subsequently added to the “meta.data” slot of the merged thymus Seurat object, based on the cell barcodes from the same donor.
Diversity of TCRs in different age groups
To assess T-cell receptor (TCR) diversity across different age groups, each sample was down-sampled to a standardized count of 1,000 cells, ensuring a consistent sample size and reducing any potential biases arising from discrepancies in T cell counts among the samples. This process was repeated 100 times. For each iteration, Shannon’s entropy was used to estimate TCR diversity as per the formula below:
| 7 |
The p(xi) represents the frequency of a given TCR clone among all T cells with TCR identified in the i-th iteration. Finally, box plots were used to visualize the differences in the TCR diversity index H across age groups (Supplementary Fig. 10b).
Projection of T cells with different clone sizes to spatial transcriptomics (ST) slices
To examine the distribution of T cells in ST slices, they were categorized into three groups on the basis of the TCR clone size: clone size = 1, clone size = 2, and clone size ≥ 3. To this end, the FindAllMarkers function from Seurat was used to identify the top 20 genes with the highest expression in each group. Subsequently, the AddModuleScore function was utilized to assign scores to the ST spots based on these genes. Finally, the spot scores were visualized using the SpatialFeaturePlot function (Supplementary Fig. 10d).
Delineation of T-cell subpopulations for αβT cells
To determine the temporal dynamics of TCR chain formation, the cells were classified on the basis of the different combinations of TCRα, TCRß, TCRδ and TCRγ chains. The TCRα+ or TCRß+ cells harbored the specific CDR3 sequence associated with that particular chain. In the TCRδ+ or TCRγ+ cells, the cumulative UMIs of TRD[V | J | C] or TRG[V | J | C] genes exceeded 1. Nevertheless, due to the negligible proportion of TCRγ+ T-cell subpopulations in the αβTCR-seq data, these entities were disregarded for analytical purposes. Among the TCRγ- T cells, TCRα-TCRß+TCRδ+, TCRα+TCRß+TCRδ+, TCRα-TCRß+TCRδ- and TCRα+TCRß+TCRδ- were the predominant subpopulations of αβT cells (Fig. 7d). These subpopulations were externally validated using thymic single-cell data from Park et al. and Cordes et al. 8,33. Notably, to avoid the influence of doublets on T-cell subpopulations, Scrublet45 (version 0.2.2) and DoubletDetection46 (version 3.0) tools were used, as per the official guidelines, to estimate doublet scores for each subpopulation and identify potential doublet cells, with all parameters set to default values.
Spatial distance assessment for T-cell subpopulations
To measure the relative distances of the four T-cell subpopulations from the medullary centers, spots in ST slices were scored using the AddModuleScore function from Seurat, based on the top 20 highly expressed markers for each subpopulation. Spots with signature scores exceeding the 75th percentile in each ST slice were then selected for each T-cell subpopulation. The relative distances of these selected spots to the nearest medullary center were calculated, characterizing the proximity of each T-cell subpopulation to the medullary centers (Fig. 7i).
Gene ontology (GO) enrichment analysis of T-cell subpopulations
The top 100 highly expressed genes of each T cell subpopulation were subjected to GO analysis. The gene identifiers were mapped using the bitr function from the clusterProfiler package (version 4.2.0)63 in conjunction with the annotation package “org.Hs.eg.db”. The enrichGO function of the clusterProfiler package was used to obtain the significantly enriched GO terms for each subpopulation with the Benjamini-Hochberg correction method. The top five or six significantly enriched terms (adjusted p ≤ 0.01) in each group were visualized using the ggplot2 package (version 3.3.5) (Fig. 7j, k).
Trajectory analysis
Monocle2 analysis
Monocle2 (version 2.22.0)64 was utilized to conduct pseudotime trajectory analyzes for the DP_blast cell populations. Specifically, the newCellDataSet function was used to create a “cell_data_set” object, and cells were arranged on based on differentially expressed genes (DEGs) identified by differentialGeneTest with q-value ≤ 0.01. Dimensionality reduction was performed using the reduceDimension function with the “DDRTree” algorithm. The minimum spanning tree of the cells was visualized with the plot_cell_trajectory function (Supplementary Fig. 11f).
Diffusion map analysis
T cells with TCR clones from the normal thymus were used for diffusion map analysis. Differentially expressed genes (DEGs) (adjusted p-value ≤ 0.01, and logFC ≥ 0.25) for each T cell subset were identified using the FindAllMarkers function in Seurat. These DEGs were then used to recalculate the PCs, which were input into SCANPY65 for diffusion map analysis. The analysis involved sequential application of the scanpy.pp.neighbors and scanpy.tl.diffmap functions, with a neighborhood graph computed using 50 neighbors and the first 30 PCs. A randomly selected DN_early cell was designated as the root for the analysis. To visualize the results, the cells were binned according to pseudotime ordering, and the expression of specific markers, as reported by Park et al. 8, was depicted using a heat map (Fig. 8f).
RNA velocity-based cell fate tracing
For the RNA velocity analysis, the scvelo Python package (version 0.3.2)66 was employed to recount spliced and unspliced reads from pre-aligned BAM files of scRNA-seq data. Following this, RNA velocity values for each gene of each cell were calculated, and the resulting RNA velocity vectors were embedded into low-dimensional space using the scvelo Python pipeline. Subsequently, the developmental trajectories of DP_blast and DP_re cells were inferred by embedding RNA velocity vectors into the UMAP space (Fig. 8e; Supplementary Fig. 11k).
Cell cycle analysis of DP_blast cell subtypes
To determine the cell cycle phase of the subtypes (i.e., DP_blast1 to DP_blast5) in the DP_blast, a list of genes associated with the cell cycle (G1/S, S, G2/M, M, and M/G1 phases) was obtained from a previous study67. The AddModuleScore function in Seurat was then used to calculate scores for each cell at different cycle phases. Finally, the cycle scores in DP_blast subtypes were visualized using the ComplexHeatmap package (version 2.13.1)68,69 from Bioconductor (Supplementary Fig. 11g, h).
Order of differentiation of T-cell subpopulations
To determine the order of differentiation among the four T-cell subpopulations, cells with identical CDR3 sequences of TCRβ chains across the four T-cell types were selected. From DN_early to SP (single positive) development, the cell numbers in each subpopulation were quantified at different stages using the previously selected cells. The cumulative distribution curves were plotted and normalized to 1 based on the total number of cells in each subgroup (Fig. 8g).
Tracing the significant flow of TCRβ clonotypes
To determine the transition points between the T-cell subpopulations, the TCRβ clonotypes that were amplified in adjacent cell types at specific differentiation time points were tracked (Fig. 8j). Specifically, if x unique clonotypic expansions were present in cell type Ci of T-cell subpopulation A to cell type Ci+1 of cell subpopulation B, m unique clonotypes overlapped in A and B, n unique clonotypes overlapped with A in all other subpopulations, and k clonotypes were common to A and B in cell types Ci and Ci+1. The significant p-value for the flow of cell type Ci in T-cell subpopulation A to cell type Ci+1 in subpopulation B was estimated using a hypergeometric test.
| 8 |
Statistical analysis
Statistical analyzes were conducted using R (version 4.0.2). The correlation between cell subsets or paired samples (SC vs. ST) were determined by Pearson’s method. The t-test, hypergeometric test, Fisher’s exact test, permutation test, and Wilcoxon test were used to determine statistical significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by grants from Youth Fund of the National Natural Science Foundation of China (3230080508 to S.P.J.), the Nanjing Medical University Launches Research Initiation Fund for Talent Recruitment (NMUR20230007 to Y.C.L.), Jiangsu Funding Program for Excellent Postdoctoral Talent (2022ZB699 to H.M.L.), National Natural Science Foundation of China (82173378 to H.L.L.), National High-Level Hospital Clinical Research Funding (2023-NHLHCRF-DJMS-06 to H.L.L.). Additionally, Figs. 1a, 4d, 5a, 6d, and 8k were created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Author contributions
S.P.J., B.C.S., Q.X., and L.H.L. conceived the project and designed experiments. S.P.J., B.C.S., and L.H.L. recruited patients. Y.C.L., C.P., and G.M. collected all tissue samples and carried out experiments. H.M.L. and Q.X. performed bioinformatics data analyses. H.M.L. wrote and revised the manuscript with input from all authors. H.L.L., L.C., Y.J.L, H.Z., Z.X., and L.Y.S. provided useful suggestions. All authors contributed to the discussion and critically reviewed and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw sequence data generated in this study have been deposited in the Genome Sequence Archive (GSA-Human) and are publicly accessible via the following links: GSA-Human: HRA007984 (scRNA-seq) at https://ngdc.cncb.ac.cn/gsa-human/s/C87MlGbL, HRA007980 (Spatial transcriptomics data) at https://ngdc.cncb.ac.cn/gsa-human/s/eOQ2yy34, and HRA007988 (scTCR-seq) at https://ngdc.cncb.ac.cn/gsa-human/s/38O9Q5t8. The processed data (Seurat object) corresponding to these datasets can be accessed from Zenodo [10.5281/zenodo.13207776]. Additionally, the publicly available datasets reused in this study include Park et al. 8 thymus scRNA-seq and scTCR-seq datasets, which can be found in the Zenodo repository (10.5281/zenodo.3572422). Cordes et al. 33 human thymus scRNA-seq data coupled with scTCR-seq data are accessible via GEO: GSE195812. Three thymus ST samples are accessible from the study by Suo et al. 11 (https://developmental.cellatlas.io/fetal-immune). The remaining data are available within the article, Supplementary Information or Source Data file Source data are provided with this paper.
Code availability
All custom code used in this work are available at Github: https://github.com/lihuamei/Thymus. And can also be accessed on Zenodo via 10.5281/zenodo.12803343.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanchuan Li, Huamei Li.
Contributor Information
Lihong Liu, Email: llh-hong@outlook.com.
Qing Xiong, Email: qingx@tcrximmune.cn.
Beicheng Sun, Email: sunbc@nju.edu.cn.
Shiping Jiao, Email: jiaoshp@tcrximmune.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51767-y.
References
- 1.Ueno, T. et al. Role for CCR7 ligands in the emigration of newly generated T lymphocytes from the neonatal thymus. Immunity16, 205–218 (2002). 10.1016/S1074-7613(02)00267-4 [DOI] [PubMed] [Google Scholar]
- 2.Fontaine-Perus, J., Calman, F., Kaplan, C. & Le Douarin, N. Seeding of the 10-day mouse embryo thymic rudiment by lymphocyte precursors in vitro. J. Immunol.126, 2310–2316 (1981). 10.4049/jimmunol.126.6.2310 [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson, B., Owen, J. & Jenkinson, E. Factors regulating stem cell recruitment to the fetal thymus. J. Immunol.162, 3873–3881 (1999). 10.4049/jimmunol.162.7.3873 [DOI] [PubMed] [Google Scholar]
- 4.Halkias, J., Melichar, H. J., Taylor, K. T. & Robey, E. A. Tracking migration during human T cell development. Cell. Mol. life Sci.71, 3101–3117 (2014). 10.1007/s00018-014-1607-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar, B. V., Connors, T. J. & Farber, D. L. Human T cell development, localization, and function throughout life. Immunity48, 202–213 (2018). 10.1016/j.immuni.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luan, R., Liang, Z., Zhang, Q., Sun, L. & Zhao, Y. Molecular regulatory networks of thymic epithelial cell differentiation. Differentiation107, 42–49 (2019). 10.1016/j.diff.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 7.Michelson, D. A., Hase, K., Kaisho, T., Benoist, C. & Mathis, D. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell185, 2542–2558. e2518 (2022). 10.1016/j.cell.2022.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park, J. E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science367, eaay3224 (2020). [DOI] [PMC free article] [PubMed]
- 9.Palmer, S., Albergante, L., Blackburn, C. C. & Newman, T. Thymic involution and rising disease incidence with age. Proc. Natl Acad. Sci.115, 1883–1888 (2018). 10.1073/pnas.1714478115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch, H. E. et al. Thymic involution and immune reconstitution. Trends Immunol.30, 366–373 (2009). 10.1016/j.it.2009.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suo, C. et al. Mapping the developing human immune system across organs. Science376, eabo0510 (2022). [DOI] [PMC free article] [PubMed]
- 12.Zeng, Y. et al. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity51, 930–948. e936 (2019). 10.1016/j.immuni.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 13.Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol.38, 333–342 (2020). 10.1038/s41587-019-0392-8 [DOI] [PubMed] [Google Scholar]
- 14.Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science353, 78–82 (2016). 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 15.Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell182, 497–514. e422 (2020). 10.1016/j.cell.2020.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asp, M. et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell179, 1647–1660. e1619 (2019). 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- 17.Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol.40, 661–671 (2022). 10.1038/s41587-021-01139-4 [DOI] [PubMed] [Google Scholar]
- 18.Takahama, Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol.6, 127–135 (2006). 10.1038/nri1781 [DOI] [PubMed] [Google Scholar]
- 19.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902 e1821 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, B. et al. An entropy-based metric for assessing the purity of single cell populations. Nat. Commun.11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature564, 268–272 (2018). 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- 22.Rezzani, R., Nardo, L., Favero, G., Peroni, M. & Rodella, L. F. Thymus and aging: morphological, radiological, and functional overview. Age36, 313–351 (2014). 10.1007/s11357-013-9564-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes, N., Sergé, A., Ferrier, P. & Irla, M. Thymic crosstalk coordinates medulla organization and T-cell tolerance induction. Front. Immunol.6, 365 (2015). 10.3389/fimmu.2015.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perera, J. & Huang, H. The development and function of thymic B cells. Cell. Mol. life Sci.72, 2657–2663 (2015). 10.1007/s00018-015-1895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spits, H. Development of αβ T cells in the human thymus. Nat. Rev. Immunol.2, 760–772 (2002). 10.1038/nri913 [DOI] [PubMed] [Google Scholar]
- 26.Muro, R., Takayanagi, H. & Nitta, T. T cell receptor signaling for γδT cell development. Inflamm. Regeneration39, 6 (2019). 10.1186/s41232-019-0095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. et al. Development of double-positive thymocytes at single-cell resolution. Genome Med.13, 1–18 (2021). 10.1186/s13073-021-00861-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, L., Kyewski, B., Allen, P. M. & Hogquist, K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol.14, 377–391 (2014). 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg, E. V. T cell lineage commitment: identity and renunciation. J. Immunol.186, 6649–6655 (2011). 10.4049/jimmunol.1003703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, J. & Zúñiga-Pflücker, J. C. A 2020 view of thymus stromal cells in T cell development. J. Immunol.206, 249–256 (2021). 10.4049/jimmunol.2000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1–20 (2021). 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc.15, 1484–1506 (2020). 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 33.Cordes, M. et al. Single-cell immune profiling reveals novel thymus-seeding populations, T cell commitment, and multi-lineage development in the human thymus. Sci. Immunol.7, eade0182 (2022). [DOI] [PubMed]
- 34.Yannoutsos, N. et al. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med.194, 471–480 (2001). 10.1084/jem.194.4.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilfillan, S., Dierich, A., Lemeur, M., Benoist, C. & Mathis, D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science261, 1175–1178 (1993). 10.1126/science.8356452 [DOI] [PubMed] [Google Scholar]
- 36.Gálvez, N. M. et al. Type I natural killer T cells as key regulators of the immune response to infectious diseases. Clinical Microbiol. Rev.34, e00232–20 (2021). [DOI] [PMC free article] [PubMed]
- 37.Filbert, E. L., Le Borgne, M., Lin, J., Heuser, J. E. & Shaw, A. S. Stathmin regulates microtubule dynamics and microtubule organizing center polarization in activated T cells. J. Immunol.188, 5421–5427 (2012). 10.4049/jimmunol.1200242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaFleur, M. W. et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun.10, 1668 (2019). 10.1038/s41467-019-09656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, T. et al. Xgboost: extreme gradient boosting. R. package version 0. 4-21, 1–4 (2015). [Google Scholar]
- 40.Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol.40, 1349–1359 (2022). [DOI] [PMC free article] [PubMed]
- 41.Wei, R. et al. Spatial charting of single-cell transcriptomes in tissues. Nat. Biotechnol.40, 1190–1199 (2022). [DOI] [PMC free article] [PubMed]
- 42.Castañeda, J. et al. The multifaceted roles of B cells in the thymus: from immune tolerance to autoimmunity. Front. Immunol.12, 766698 (2021). [DOI] [PMC free article] [PubMed]
- 43.Nitta, T., Ota, A., Iguchi, T., Muro, R. & Takayanagi, H. The fibroblast: an emerging key player in thymic T cell selection. Immunological Rev.302, 68–85 (2021). 10.1111/imr.12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray, D. H. et al. A unique thymic fibroblast population revealed by the monoclonal antibody MTS-15. J. Immunol.178, 4956–4965 (2007). 10.4049/jimmunol.178.8.4956 [DOI] [PubMed] [Google Scholar]
- 45.Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst.8, 281–291. e289 (2019). 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xi, N. M. & Li, J. J. Protocol for executing and benchmarking eight computational doublet-detection methods in single-cell RNA sequencing data analysis. STAR Protoc.2, 100699 (2021). 10.1016/j.xpro.2021.100699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu, Y. et al. γδ T cells: origin and fate, subsets, diseases and immunotherapy. Signal Transduct. Target. Ther.8, 434 (2023). 10.1038/s41392-023-01653-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadouri, N., Nevo, S., Goldfarb, Y. & Abramson, J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol. 20, 239–253 (2020). [DOI] [PubMed]
- 49.Kernfeld, E. M. et al. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and Developmental maturation. Immunity48, 1258–1270 e1256 (2018). 10.1016/j.immuni.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng, Y. et al. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity51, 930–948 e936 (2019). 10.1016/j.immuni.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 51.Baran-Gale, J. et al. Ageing compromises mouse thymus function and remodels epithelial cell differentiation. Elife9, e56221 (2020). [DOI] [PMC free article] [PubMed]
- 52.Bautista, J. L. et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun.12, 1–15 (2021). 10.1038/s41467-021-21346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shilo, K. et al. Diffuse thymic fibrosis: histologic pattern of injury or distinct entity? Am. J. surgical Pathol.34, 211–215 (2010). 10.1097/PAS.0b013e3181c91301 [DOI] [PubMed] [Google Scholar]
- 54.Fawkner-Corbett, D. et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell184, 810–826. e823 (2021). 10.1016/j.cell.2020.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587. e3529 (2021). 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwabuchi, K., Nakayama, H., Iwahara, C. & Takamori, K. Significance of glycosphingolipid fatty acid chain length on membrane microdomain-mediated signal transduction. FEBS Lett.584, 1642–1652 (2010). 10.1016/j.febslet.2009.10.043 [DOI] [PubMed] [Google Scholar]
- 57.McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. Doubletfinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst.8, 329–337. e324 (2019). 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren, X. et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell184, 1895–1913. e1819 (2021). 10.1016/j.cell.2021.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 1–5 (2018). 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 61.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B (Methodol.)57, 289–300 (1995). 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 62.Walsh, I. et al. DOME: recommendations for supervised machine learning validation in biology. Nat. methods18, 1122–1127 (2021). 10.1038/s41592-021-01205-4 [DOI] [PubMed] [Google Scholar]
- 63.Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a J. Integr. Biol.16, 284–287 (2012). 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. methods14, 979–982 (2017). 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf, F. A., Angerer, P. & Theis, F. J. J. G. B. SCANPY: large-scale single-cell gene expression data analysis. 19, 15 (2018). [DOI] [PMC free article] [PubMed]
- 66.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell161, 1202–1214 (2015). 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics32, 2847–2849 (2016). 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 69.Gu, Z. Complex heatmap visualization. iMeta1, e43 (2022). 10.1002/imt2.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alquicira-Hernandez, J. & Powell, J. E. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics37, 2485–2487 (2021). 10.1093/bioinformatics/btab003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw sequence data generated in this study have been deposited in the Genome Sequence Archive (GSA-Human) and are publicly accessible via the following links: GSA-Human: HRA007984 (scRNA-seq) at https://ngdc.cncb.ac.cn/gsa-human/s/C87MlGbL, HRA007980 (Spatial transcriptomics data) at https://ngdc.cncb.ac.cn/gsa-human/s/eOQ2yy34, and HRA007988 (scTCR-seq) at https://ngdc.cncb.ac.cn/gsa-human/s/38O9Q5t8. The processed data (Seurat object) corresponding to these datasets can be accessed from Zenodo [10.5281/zenodo.13207776]. Additionally, the publicly available datasets reused in this study include Park et al. 8 thymus scRNA-seq and scTCR-seq datasets, which can be found in the Zenodo repository (10.5281/zenodo.3572422). Cordes et al. 33 human thymus scRNA-seq data coupled with scTCR-seq data are accessible via GEO: GSE195812. Three thymus ST samples are accessible from the study by Suo et al. 11 (https://developmental.cellatlas.io/fetal-immune). The remaining data are available within the article, Supplementary Information or Source Data file Source data are provided with this paper.
All custom code used in this work are available at Github: https://github.com/lihuamei/Thymus. And can also be accessed on Zenodo via 10.5281/zenodo.12803343.