Abstract
We present an approach to learning regular spatial transformations between image pairs in the context of medical image registration. Contrary to optimization-based registration techniques and many modern learning-based methods, we do not directly penalize transformation irregularities but instead promote transformation regularity via an inverse consistency penalty. We use a neural network to predict a map between a source and a target image as well as the map when swapping the source and target images. Different from existing approaches, we compose these two resulting maps and regularize deviations of the Jacobian of this composition from the identity matrix. This regularizer – GradICON – results in much better convergence when training registration models compared to promoting inverse consistency of the composition of maps directly while retaining the desirable implicit regularization effects of the latter. We achieve state-of-the-art registration performance on a variety of real-world medical image datasets using a single set of hyperparameters and a single non-dataset-specific training protocol. Code is available at https://github.com/uncbiag/ICON.
1. Introduction
Image registration is a key component in medical image analysis to estimate spatial correspondences between image pairs [14, 53]. Applications include estimating organ motion between treatment fractions in radiation therapy [25, 37], capturing disease progression [64], or allowing for localized analyses in a common coordinate system [19].
Many different registration algorithms have been proposed over the last decades in medical imaging [10, 41, 44, 63, 64] and in computer vision [21, 33]. Contributions have focused on different transformation models (i.e., what types of transformations are considered permissible), similarity measures (i.e., how “good alignment” between image pairs is quantified), and solution strategies (i.e., how transformation parameters are numerically estimated). The respective choices are generally based on application requirements as well as assumptions about image appearance and the expected transformation space. In consequence, while reliable registration algorithms have been developed for transformation models ranging from simple parametric models (e.g., rigid and affine transformations) to significantly more complex nonparametric formulations [41, 44, 63] that allow highly localized control, practical applications of registration typically require many choices and rely on significant parameter tuning to achieve good performance. Recent image registration work has shifted the focus from solutions based on numerical optimization for a specific image pair to learning to predict transformations based on large populations of image pairs via neural networks [10, 15, 17, 34, 35, 56, 57, 68]. However, while numerical optimization is now replaced by training a regression model which can be used to quickly predict transformations at test time, parameter tuning remains a key challenge as loss terms for these two types of approaches are highly related (and frequently the same). Further, one also has additional choices regarding network architectures. Impressive strides have been made in optical flow estimation as witnessed by the excellent performance of recent approaches [34] on Sintel [7]. However, our focus is medical image registration, where smooth and often diffeomorphic transformations are desirable; here, a simple-to-use learning-based registration approach, which can adapt to different types of data, has remained elusive. In particular, nonparametric registration approaches require a balance between image similarity and regularization of the transformation to assure good matching at a high level of spatial regularity, as well as choosing a suitable regularizer. This difficulty is compounded in a multi-scale approach where registrations at multiple scales are used to avoid poor local solutions.
Instead of relying on a complex spatial regularizer, the recent ICON approach [23] uses only inverse consistency to regularize the sought-after transformation map, thereby dramatically reducing the number of hyperparameters to tune. While inverse consistency is not a new concept in image registration and has been explored to obtain transformations that are inverses of each other when swapping the source and the target images [11], ICON [23] has demonstrated that a sufficiently strong inverse consistency penalty, by itself, is sufficient for spatial regularity when used with a registration network. Further, as ICON does not explicitly penalize spatial gradients of the deformation field, it does not require pre-registration (e.g., rigid or affine), unlike many other related works. However, while conceptually attractive, ICON suffers from the following limitations: 1) training convergence is slow, rendering models costly to train; and 2) enforcing approximate inverse consistency strictly enough to prevent folds becomes increasingly difficult at higher spatial resolutions, necessitating a suitable schedule for the inverse consistency penalty, which is not required for GradICON.
Our approach is based on a surprisingly simple, but effective observation: penalizing the Jacobian of the inverse consistency condition instead of inverse consistency directly1 applies zero penalty for inverse consistent transform pairs but 1) yields significantly improved convergence, 2) no longer requires careful scheduling of the inverse consistency penalty, 3) results in spatially regular maps, and 4) improves registration accuracy. These benefits facilitate a unified training protocol with the same network structure, regularization parameter, and training strategy across registration tasks.
Our contributions are as follows:
We develop GradICON (Gradient Inverse CONsistency), a versatile regularizer for learning-based image registration that relies on penalizing the Jacobian of the inverse consistency constraint and results, empirically and theoretically, in spatially well-regularized transformation maps.
We demonstrate state-of-the-art (SOTA) performance of models trained with GradICON on three large medical datasets: a knee magnetic resonance image (MRI) dataset of the Osteoarthritis Initiative (OAI) [46], the Human Connectome Project’s collection of Young Adult brain MRIs (HCP) [60], and a computed tomography (CT) inhale/exhale lung dataset from COPDGene [47].
2. Related work
Nonparametric transformation models & regularization.
There are various ways of modeling a transformation between image pairs. The most straightforward nonparametric approach is via a displacement field [59]. Different regularizers for displacement fields have been proposed [32], but they are typically only appropriate for small displacements [44] and cannot easily guarantee diffeomorphic transformations [2], which is our focus here for medical image registration. Fluid models, which parameterize a transformation by velocity fields instead, can capture large deformations and, given a suitably strong regularizer, result in diffeomorphic transformations. Popular fluid models are based on viscous fluid flow [12, 13], the large deformation diffeomorphic metric mapping (LDDMM) model [5], or its shooting variant [41, 63]. Simpler stationary fluid approaches, such as the stationary velocity field (SVF) approach [1, 61], have also been developed. While diffeomorphic transformations are not always desirable, they are often preferred due to their invertibility, which allows mappings between images to preserve object topologies and prevent foldings that are physically implausible. These models have initially been developed for pair-wise image registration where solutions are determined by numerical optimization, but have since, with minimal modifications, been integrated with neural networks [4, 52, 68]. In a learning-based formulation, the losses are typically the same as for numerical-optimization approaches, but one no longer directly optimizes over the parameters of the chosen transformation model but instead over the parameters of a neural network which, once trained, can quickly predict the transformation model parameters.
Fluid registration models are computationally complex as they require solving a fluid equation (either greedily or via direct numerical integration [42], or via scaling and squaring [1]), but can guarantee diffeomorphic transformations. In contrast, displacement field models are computationally cheaper but make it more difficult to obtain diffeomorphic transformations. Solution regularity can be obtained for displacement field models by adding appropriate constraints on the Jacobian [24]. Alternatively, invertibility can be encouraged by adding inverse consistency losses, either for numerical optimization approaches [11] or in the context of registration networks as is the case for ICON [23]. Similar losses have also been used in computer vision to encourage cycle consistencies [6, 22, 67, 69] though they are, in general, not focused on spatial regularity. Most relevant to our approach, ICON [23] showed that inverse consistency alone is sufficient to approximately obtain diffeomorphic transformations when the displacement field is predicted by a neural network. Our work extends this approach by generalizing the inverse consistency loss to a gradient inverse consistency loss, which results in smooth transformations, faster convergence, and more accurate registration results.
Multi-scale image registration.
Finding good solutions for the optimization problems of image registration is challenging, and one might easily get trapped in an unfavorable local minimum. In particular, this might happen for self-similar images, such as lung vessels, where incorrect vessel alignment might be locally optimal. Further, if there is no overlap between vessels, a similarity measure might effectively be blind to misalignment, which is why it is important for a similarity measure to have a sufficient capture range2.
Multi-scale approaches have been proposed for optimization-based registration models [39, 55, 58, 70] to overcome these issues. For these approaches, the loss function is typically first optimized at a coarse resolution, and the image warped via the coarse transformation then serves as the input for the optimization at a finer resolution. This helps to avoid poor local optima, as solutions computed at coarser resolutions effectively increase capture range and focus on large-scale matching first rather than getting distracted by fine local details. Multi-scale approaches have also been used for learning-based registration [16, 18, 29, 36, 45, 52] and generally achieve better results than methods that only consider one scale [4]. These methods all use sub-networks operating at different scales but differ in how the multi-scale strategy is incorporated into the network structure and the training process. A key distinction is if source images are warped as they pass through the different sub-networks [16, 23, 29, 45] or if sub-networks always start from the unwarped, albeit downsampled, source image [18]. The former approach simplifies capturing large deformations as sub-networks only need to refine a transformation rather than capturing it in its entirety. However, these methods compute the similarity measure and the regularizer losses at all scales, which requires balancing the weights of the losses across all scales and for each scale between the similarity measure and the regularizer. Hence, there are many parameters that are difficult to tune. To side-step the tuning issue, it is common to rely on a progressive training protocol to avoid tuning the weights between losses at all scales. We find that our multi-resolution approach trains well when the loss and regularizer are applied only at the highest scale: the coarser components are effectively trained by gradients propagating back through the multi-scale steps.
3. Gradient Inverse Consistency (GradICON)
3.1. Preliminaries
We denote by and the source and the target images in our registration problem. By we denote a transformation map with the intention that . The map is a diffeomorphism if it is differentiable, bijective and its inverse is differentiable as well3. Optimization-based image registration approaches typically solve the optimization problem
| (1) |
where is the similarity measure, is a regularizer, are the transformation parameters, and . In learning-based registration, one does not directly optimize over the transformation parameters of , but instead over the parameters of a neural network that predicts given the source and target images. Such a network is trained over a set of image pairs by solving
| (2) |
with as shorthand for denoting the output of the network given the -th input image pair. By training with and the loss is symmetric in expectation. For ease of notation, we omit the subscripts or in cases where the dependency is clear from the context.
3.2. Regularization
Picking a good regularizer is essential as it implicitly expresses the class of transformations one considers plausible for a registration. Ideally, the space of plausible transformations should be known (e.g., based on physical principles) or learned from the data. As nonparametric image registration (at least for image pairs) is an ill-posed problem [20], regularization is required to obtain reasonable solutions.
Regularizers frequently involve spatial derivatives of various orders to discourage spatial non-smoothness [32]. This typically requires picking a type of differential operator (or, conversely, a smoothing operator) as well as all its associated parameters. Most often, this regularizer is chosen for convenience and not learned from data. Instead of explicitly penalizing spatial non-smoothness, ICON [23] advocates using inverse consistency as a regularizer, which amounts to learning a transformation space from data in the class of (approximately) invertible transforms. When implementing inverse consistency, there is a choice of loss. The ICON approach penalizes the sum-of-squares difference between the identity and the composition of the maps between images and , i.e., the regularizer has the form , where Id denotes the identity transform. In [23], it is shown that this loss has an implicit regularization effect, similar to a sum-of-squares on the gradient of the transformation, i.e., an type of norm. In fact, it turns out that regular invertible maps can be learned without explicitly penalizing spatial gradients. Inspired by this observation, we propose to use the Jacobian of the composition of the maps instead, i.e.,
| (3) |
where the identity matrix, and is the squared Frobenius norm integrated over . As we will see in Sec. 5, this loss equally leads to regular maps by exerting another form of implicit regularization, which we analyze in Sec. 3.3. To understand the implicit regularization of the ICON loss, one makes the modeling choice such that , i.e., the output of the network is inverse consistent up to a white noise term with parameter (artificially introduced to make the discussion clear). This white noise can be used to prove that the resulting maps are regularized via the square of a first-order Sobolev (semi-) norm. Further, [23] empirically showed that an approximate diffeomorphism can be obtained without the white noise when used in the context of learning a neural registration model: if inverse consistency is not exactly enforced, as suggested above, the inconsistency can be modeled by noise, and the observed smoothness is explained by the theoretical result. In our analysis, we follow a conceptually similar idea.
3.3. Analysis
Implicit type regularization.
Since the GradICON loss of Eq. (3) is formulated in terms of the gradient, it is a natural assumption to put the white noise on the Jacobians themselves rather than on the maps, i.e., where is a white noise. This model of randomness is motivated by the stochastic gradient scheme on the global population that drives the parameters of the networks. At the level of maps, we write where . Since integration is a low-pass filter, the noise applies to the low frequencies of . In addition, we expect the low frequencies of the noise to be dampened by the similarity measure between and . Hence, we hypothesize that , which will be used only once in our analysis. This comparison means that our estimates of the gradient and on our grid satisfy this inequality. We assess this hypothesis Appendix A.1. We start by rewriting the GradICON regularizer, by applying the chain rule, as
| (4) |
using and still omitting the integral sign. We now Taylor expand the loss w.r.t. and in particular expand the term from Eq. (4) as
| (5) |
where implies that the approximation
| (6) |
holds as it is only compared with in the expansion. Using the first-order approximation , see Appendix A.1, and simplifying Eq. (4), we obtain
| (7) |
as (and selecting the first-order coefficients in ). Expanding the square then yields
| (8) |
Under the assumption of independence of the white noises and , the contribution of the last term in Eq. (8) vanishes in expectation. We are left with the following loss, at order , now taking expectation,
| (9) |
Note that the expectation is explicit due to the white noise assumption; thus, Eq. (9) can be further simplified to
| (10) |
Eq. (10) amounts to an regularization of the inverse of the Jacobian maps on and which explains why we call it type of regularization, see Appendix A.1; yet, strictly speaking, it is not the standard norm [40].
Comparison with ICON [23].
Interestingly, our analysis shows that GradICON is an type of regularization as for ICON, although we could have expected a second-order regularization from the model. Such higher-order terms appear when taking into account the magnitude of the noise in the expansion. While there are several assumptions that can be formulated differently, such as the form of the noise and the fact that it is white noise for given pairs of images , we believe that Eq. (10) is a plausible explanation of the observed regularity of the maps in practice. Importantly, since this regularization is implicit, GradICON, as well as ICON can learn based on a slightly more informative prior than this regularization which relies on simplifying assumptions.
Why GradICON performs better than ICON.
In practice, we observe that learning a registration model via the GradICON regularizer of Eq. (3) shows a faster convergence than using the ICON regularizer, not only in the toy experiment of Sec. 5.4 but also on real data. While we do not yet have a clear explanation for this behavior, we provide insight based on the following key differences between the two variants.
First, a difference by a translation is not penalized in the GradICON formulation, but implicitly penalized in the similarity measure, assuming images are not periodic. Second, using the Jacobian in Eq. (3) correlates the composition of the map between neighboring voxels. In a discrete (periodic) setting, this can be seen by expanding the squared norm. To shorten notations, define considered as a discrete vector. Then, Eq. (3) is the sum over voxels , which can be rewritten as
| (11) |
where is the (hyper-)parameter for the finite difference estimation of the gradient. This shows that the GradICON regularizer includes more correlation than the ICON regularizer, which would only contain the first factor in Eq. (11). Lastly, it is known [9] that gradient descent on an energy rather than energy is a preconditioning of the gradient flow, emphasizing high-frequencies over low-frequencies. In our case, however, high-frequencies are the dominant cause of folds. Consequently, as GradICON penalizes high frequencies but allows low frequencies of the composition of the maps to deviate from identity, this is beneficial to avoid folds4. Overall, GradICON is more flexible than ICON while retaining the non-folding behavior of the resulting maps.
4. Implementation
To learn a registration model under Eq. (3), we use a neural network that predicts the transformation maps. In particular, we implement a multi-step (i.e., multiple steps within a forward pass), multi-resolution approach trained with a two-stage process, followed by optional instance optimization at test time. We will discuss these parts next.
4.1. Network structure
To succinctly describe our network structure, we hence-forth omit and represent a registration neural network as (or e.g. ). The notation (shorthand for ) represents the output of this network (a transform from ) for input images and . To combine such registration networks into a multistep, multiscale approach, we rely on the following combination operators from [23]:
The downsample operator (Down) is for predicting the warp between two high-resolution images using a network that operates on low-resolution images, and the two-step operator (TS) is for predicting the warp between two images in two steps, first capturing the coarse transform via and then the residual transform via . We use these operators to realize a multi-resolution, multi-step network, see Fig. 2, via
| (12) |
Our atomic (i.e., not composite) registration networks are each represented by a UNet instance5 from [23] taking as input two images and returning a displacement field . These displacement fields are converted to functions since the above operators are defined on networks that return functions from to .
Figure 2.
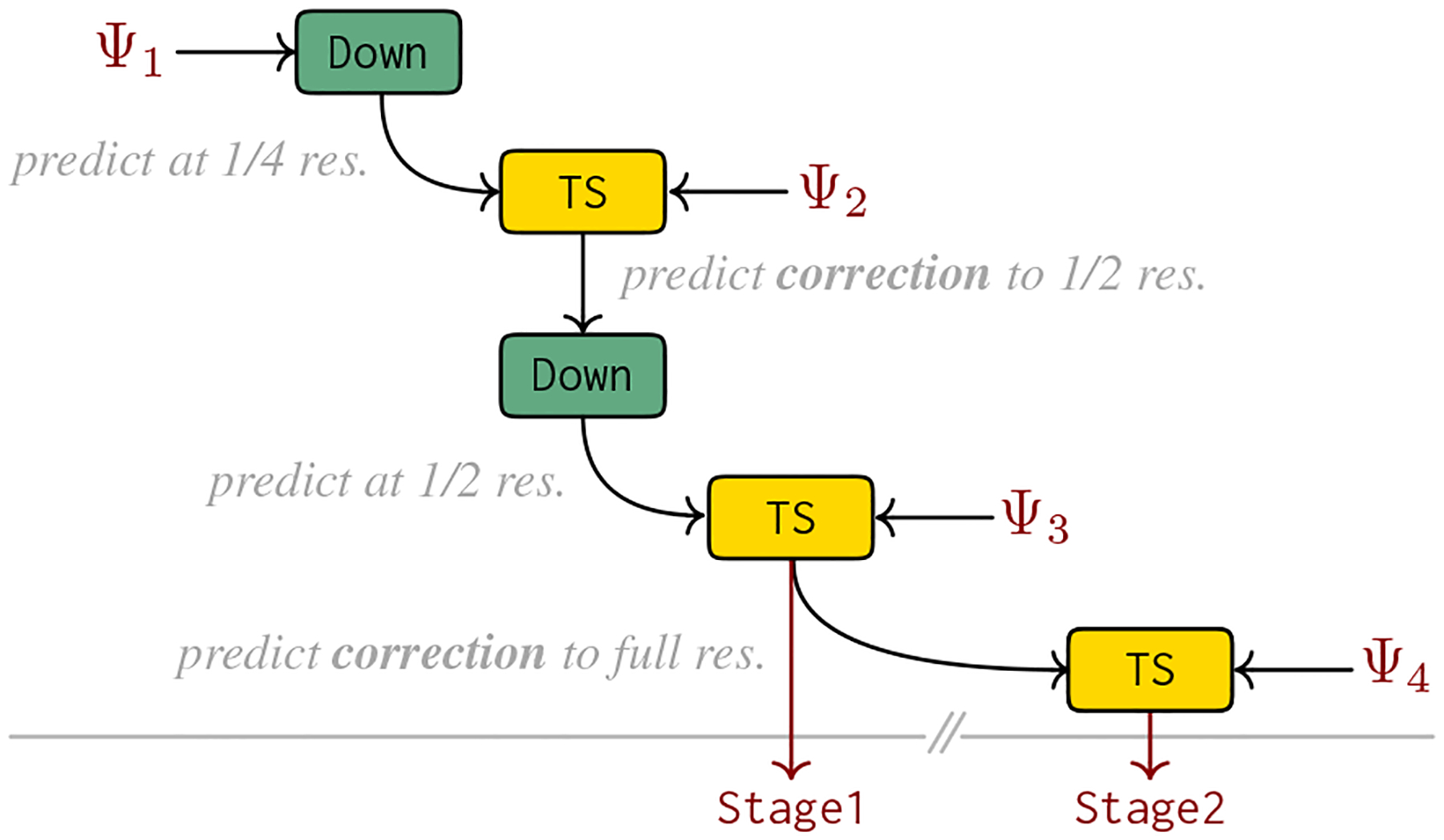
Illustration of the combination steps to create our registration network, see Eq. (12), from the atomic registration networks () via the downsample (Down) and the two-step (TS) operator.
4.2. Training
We define a single training protocol that is applied to train a network on all the registration tasks of Sec. 5. For preprocessing, each image has its intensity clipped and rescaled to [0, 1], with clipping intensities appropriate for the modality and anatomy being registered. For modalities with region of interest (ROI) annotations (brain and lung), all values outside the region of interest are set to zero. For intra-subject registration, we have many fewer pairs to train on6, so we perform augmentation via random flips along axes and small affine warps (see Appendix A.2). In all experiments, the image similarity measure is combined with the GradICON regularizer to yield the overall loss
| (13) |
In our implementation, is computed using one-sided finite differences with is computed by (uniform) random sampling over with number of samples equal to the number of voxels in the image , and we use coordinates where .
Multi-stage training.
We train in two stages. Stage1: we train the multi-resolution network defined in Eq. (12) with the loss from Eq. (13). Stage2: we train with the same loss, jointly optimizing the parameters of Stage1 and .
Instance optimization.
We optionally optimize the loss of Eq. (13) for 50 iterations at test time [66]. This typically improves performance but also increases runtime.
Unless noted otherwise, we use LNCC (local normalized cross-correlation) with a Gaussian kernel (std. of 5 voxels), computed as in [65], and GradICON with balancing parameter . Stage1 and Stage2 are trained using ADAM at a learning rate of 5e-5 for 50,000 iterations each. This protocol remains fixed across all datasets, and any result obtained by exactly this protocol is marked by †; if instance optimization is included, results are marked by ‡.
5. Experiments
Ethics.
We use one 2D synthetic (Triangles and Circles), one real 2D (DRIVE), and four real 3D datasets (OAI, HCP, COPDGene, DirLab). Acquisitions for all real datasets were approved by the respective Institutional Review Boards.
5.1. Datasets
Triangles and Circles.
A synthetic 2D dataset introduced in [23] where images are either triangles or circles. We generate 2000 hollow images with size 128 × 128 which consist of the shape edges and use them as the training set.
DRIVE [54].
This 2D dataset contains 20 retina images and the corresponding vessel segmentation masks. We use the segmentation masks to define a synthetic registration problem. In particular, we take downsampled segmentations as the source (/target) image and warp them with random deformations generated by ElasticDeform7 to obtain the corresponding target (/source) image. We generate 20 pairs per image, leading to 400 pairs at size 292×282 in total.
OAI [46].
We use a subset of 2532 images from the full corpus of MR images of the Osteoarthritis Initiative (OAI8) for training and 301 pairs of images for testing. The images are normalized so that the 1th percentile and the 99th percentile of the intensity are mapped to 0 and 1, respectively. Then we clamp the normalized intensity to be in [0, 1]. We follow the train-test split9 used in [23, 52]. We downsample images from 160×384×384 to 80×192×192
HCP [60].
We use a subset of T1-weighted and brain-extracted images of size 260×311×260 from the Human Connectome Project’s (HCP) young adult dataset to assess inter-patient brain registration performance. We downsample images to 130×155×130 for training but evaluate the average Dice score for 28 manually segmented subcortical brain regions [49, 50]10 at the original resolution. We train on 1076 images, excluding the 44 images we use for testing.
COPDGene [47].
We use a subset of11 999 inspiratory-expiratory lung CT pairs from the COPDGene study12 [47] with provided lung segmentation masks for training. The segmentation masks are computed automatically13. CT images are first resampled to isotropic spacing (2 mm) and cropped/padded evenly along each dimension to obtain a 175×175×175 volume. Hounsfield units are clamped within [−1000, 0] and scaled linearly to [0, 1]. Then, the lung segmentations are applied to the images to extract the lung region of interest (ROI). Among the processed data, we use 899 pairs for training and 100 pairs for validation.
DirLab [8].
This dataset is only used to evaluate a trained network. It contains 10 pairs of inspiration-expiration lung CTs with 300 anatomical landmarks per pair, manually identified by an expert in thoracic imaging. We applied the same preprocessing and lung segmentation as for COPDGene.
5.2. Ablation study
In Table 1, we investigate 1) which UNet structure should be used and 2) how multi-resolution, multi-stage training, instance optimization, and data augmentation affect the registration results. To this end, we train on COPDGene and evaluate on DirLab using the same similarity measure, regularizer weight , and number of iterations. We report the mean target registration error (mTRE) on landmarks (in mm) and the percentage of voxels with negative Jacobian . First, we observe that the UNet from [23] performs better than the UNet from [4]. Hence, in all experiments, we adopt the former as our backbone. Notably, this model has considerably more parameters than the variant from [4] (≈ 17M vs. 300k, see Appendix A.6), but uses less V-RAM as it concentrates parameters in the heavily downsampled layers of the architecture. Second, we find that the multi-resolution approach, including Stage2 training, clearly improves performance, reducing the mTRE from 5.176mm to 3.478mm (1 vs. 3 res.) and further down to 3.153mm (with Stage2). In this setting, there is also a noticeable benefit of (affine) data augmentation, with the mTRE dropping to 1.258mm. This is not unexpected, though, as we only have 899 pairs of training images for this (inter-patient) registration task. While this is considered a large corpus for medical imaging standards, it is small from the perspective of training a large neural network. Finally, we highlight that adding instance optimization yields the overall best results, but the benefits are less noticeable in combination with augmentation.
Table 1.
Ablation results for training on COPDGene and evaluating on Dirlab. We assess the effect of the backbone network, the number of resolutions (Res.), including Stage2 training (2nd), instance optimization (Opt.), and affine augmentation (Aug.).
5.3. Comparison to other regularizers
We study different regularizers in terms of the trade-off between transform regularity and image similarity on the training set when varying . We use Stage1 from Eq. (12) for all experiments, setting the regularizer to either Bending Energy , Diffusion , ICON, or GradICON. Specifically, we pick for each regularizer such that is kept at roughly the same level and train multiple networks with for the same number of iterations. Fig. 3 shows results on Triangles and Circles and DRIVE. We observe that all the regularizers lose a certain level of accuracy when increasing and GradICON in general has the least sacrifice in similarity. This is presumably because of the possible magnitude of deformation each regularizer allows. More results can be found in Appendix A.3.
Figure 3.
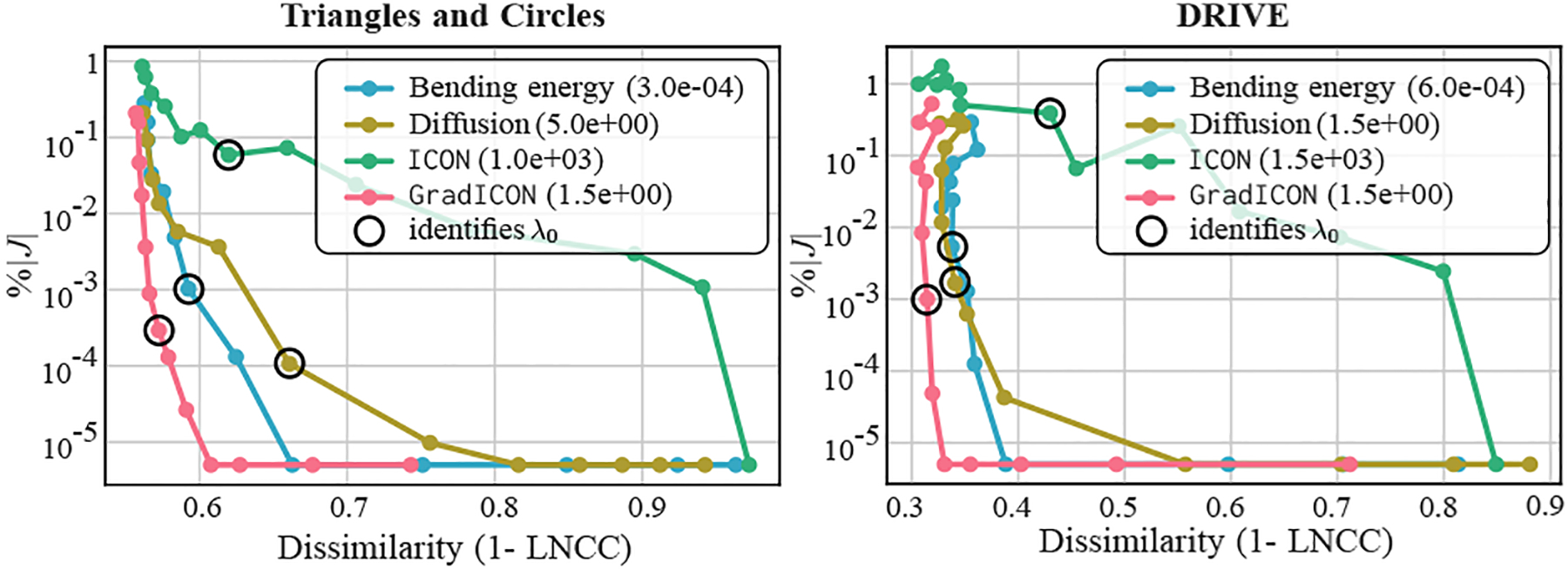
GradICON vs. other regularization techniques.
5.4. Empirical convergence analysis
To demonstrate improved convergence when training models with our GradICON regularizer vs. models trained with the ICON regularizer of [23], we assess the corresponding loss curves under the same network architecture, in particular, the network described in Sec. 4. We are specifically interested in (training) convergence behavior when both models produce a similar level of map regularity. To this end, we choose to approximately achieve the same similarity loss under both regularizers and plot the corresponding curves for , see Fig. 4, for Triangles and Circles and DRIVE.
Figure 4.
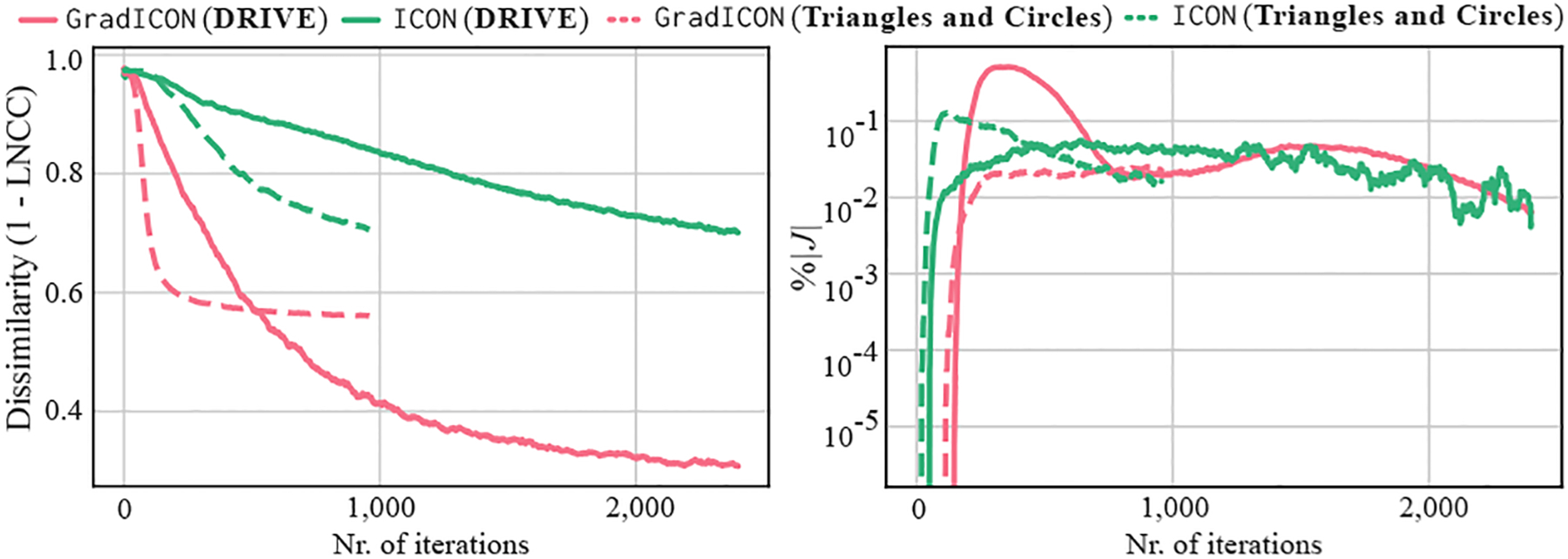
Comparison of the convergence speed (left), visualized as 1-LNCC (i.e., dissimilarity), for ICON and GradICON when is set to produce a similar level of map regularity (right).
Overall, we see that GradICON converges significantly faster than ICON. We hypothesize that this is due to the fact that maps produced by GradICON contain larger motions than maps produced by ICON. I.e., for the same level of regularity, ICON more strongly limits deformations, effectively slowing down and resulting in less accurate image alignments.
5.5. Inter-patient registration
We evaluate inter-patient registration performance of our model with GradICON regularization on OAI and HCP. We report (as in Sec. 5.2) and the mean DICE score between warped and target image for the segmentations of the femoral and tibial cartilage (OAI), and of a set of 28 subcortical brain regions (HCP). Both measures, averaged over the evaluation data, are listed in Table 2. In particular, we compare GradICON to the methods reported in [52] and [23] on OAI, and compare to ANTs SyN [3] and SynthMorph [31]14 (sm-shapes/brains) on HCP. Segmentations are not used during training and allow quantifying if the network yields semantically meaningful registrations. Table 2 shows that on the OAI dataset GradICON can significantly outperform the state-of-the-art with minimal parameter tuning and, just as ICON, without the need for affine pre-alignment (i.e., a step needed by many registration methods on OAI). On HCP, GradICON performs better than the standard non-learning approach (SyN) and matches SynthMorph while not requiring affine pre-alignment and producing much less folds.
Table 2.
Results on OAI, HCP and DirLab. † and ‡ indicate results obtained using our standard training protocol (Sec. 4.2), w/o (†) and w/ (‡) instance optimization (Opt.). Only when GradICON is trained with MSE do we set . Results marked with * are obtained using the official source code; otherwise, values are taken from the literature (see A.5). Top and bottom table parts denote non-learning and learning-based methods, resp. For DirLab, results are shown in the common inspiration→expiration direction. A: affine pre-registration, BE: bending energy, MI: mutual information, TB: Tukey’s biweight, DV: displacement vector of sparse key points, TV: total variation, Curv: curvature regularizer, VCC: volume change control, NGF: normalized gradient flow, TVD: sum of squared tissue volume difference, VMD: sum of squared vesselness measure difference, Diff: diffusion, VF: velocity field, SVF: stationary VF, DVF: displacement vector field. PLOSL50: 50 iterations of instance optimization with PLOSL.
| Method | Trans. | DICE ↑ | %|J| ↓ | ||
|---|---|---|---|---|---|
| OAI | |||||
| Initial | 7.6 | ||||
| Demons [62] | A,DVF | Gaussian | MSE | 63.5 | 0.0006 |
| SyN [3] | A,VF | Gaussian | LNCC | 65.7 | 0.0000 |
| NiftyReg [43] | A,B-Spline | BE | NMI | 59.7 | 0.0000 |
| NiftyReg [43] | A,B-Spline | BE | LNCC | 67.9 | 0.0068 |
| vSVF-opt [52] | A,vSVF | m-Gauss | LNCC | 67.4 | 0.0000 |
| VM [4] | SVF | Diff. | MSE | 46.1 | 0.0028 |
| VM [4] | A,SVF | Diff. | MSE | 66.1 | 0.0013 |
| AVSM [52] | A,vSVF | m-Gauss | LNCC | 68.4 | 0.0005 |
| ICON* [23] | DVF | ICON | MSE | 65.1 | 0.0040 |
| Ours (MSE, λ=0.2) | DVF | GradICON | MSE | 69.5 | 0.0000 |
| Ours (MSE, λ=0.2, Opt.) | DVF | GradICON | MSE | 70.5 | 0.0001 |
| Ours (std. protocol) | DVF | GradICON | LNCC | 70.1† | 0.0261 |
| DVF | GradICON | LNCC | 71.2‡ | 0.0042 | |
| HCP | |||||
| Initial | 53.4 | ||||
| FreeSurfer-Affine* [48] | A | — | TB | 62.1 | 0.0000 |
| SyN* [3] | A,VF | Gaussian | MI | 75.8 | 0.0000 |
| sm-shapes* [31] | A,SVF | Diff. | DICE | 79.8 | 0.2981 |
| sm-brains* [31] | A,SVF | Diff. | DICE | 78.4 | 0.0364 |
| Ours (std. protocol) | DVF | GradICON | LNCC | 78.7† | 0.0012 |
| DVF | GradICON | LNCC | 80.5‡ | 0.0004 | |
| DirLab | |||||
| Method | Trans. | mTRE ↓ (in mm) |
%|J| ↓ | ||
| Initial | 23.36 | ||||
| SyN [3] | A,VF | Gaussian | LNCC | 1.79 | — |
| Elastix [38] | A,B-Spline | BE | MSE | 1.32 | — |
| NiftyReg [43] | A,B-Spline | BE | MI | 2.19 | — |
| PTVReg [65] | DVF | TV | LNCC | 0.96 | — |
| RRN [28] | DVF | TV | LNCC | 0.83 | — |
| VM* [4] | A,SVF | Diff. | NCC | 9.88 | 0.0000 |
| LapIRN* [45] | SVF | Diff. | NCC | 2.92 | 0.0000 |
| LapIRN* [45] | DVF | Diff. | NCC DICE |
4.24 | 0.0105 |
| Hering et al. [30] | DVF | Curv+VCC | +KP+NGF | 2.00 | 0.0600 |
| GraphRegNet [26] | DV | — | MSE | 1.34 | — |
| PLOSL [66] | DVF | Diff. | TVD+VMD | 3.84 | 0.0000 |
| PLOSL50 [66] | DVF | Diff. | TVD+VMD | 1.53 | 0.0000 |
| ICON* [23] | DVF | ICON | LNCC | 7.04 | 0.3792 |
| Ours (std. protocol) | DVF | GradICON | LNCC | 1.26† | 0.0003 |
| DVF | GradICON | LNCC | 0.96‡ | 0.0002 | |
5.6. Intra-patient registration
We demonstrate the ability of our model with GradICON regularization to predict large deformations between lung exhale (source)/inhale (target) pairs (within patient) from COPDGene. This dataset is challenging as deformations are complex and large. Motion is primarily visually represented by the deformation of lung vessels, which form a complex tree-like structure that creates capture-range and local minima challenges for registration. As in our ablation study (Sec. 5.2), we report and the mTRE (in mm) for manually annotated lung vessel landmarks [8] averaged over all 10 DirLab image pairs. We assess GradICON against traditional optimization-based methods and state-of-the-art (SOTA) learning-based methods. Table 2 shows that our approach with instance optimization (0.96mm) yields a mTRE very close to the SOTA techniques and exceeds the SOTA for learning-based approaches of any sort. Further, our performance (1.26mm) in a single forward pass is the best of any one-forward-pass neural method.
6. Conclusion
We introduced and theoretically analyzed GradICON, a new regularizer to train deep image registration networks. In contrast to ICON [23], GradICON penalizes the Jacobian of the inverse consistency constraint. This has profound effects: we obtain dramatically faster training convergence, higher registration accuracy, do not require scale-dependent regularizer tuning, and retain desirable implicit regularization effects resulting in approximately diffeomorphic transformations. Remarkably, this allows us to train registration networks using GradICON regularization with one standard training protocol for a range of different registration tasks. In fact, using this standard training protocol, we match or outperform state-of-the-art registration methods on three challenging and diverse 3D datasets. This uniformly good performance without the need for dataset-specific tuning takes the pain out of training deep 3D registration networks and makes our approach highly practical.
Limitations and future work.
We only explored first-order derivatives of the inverse-consistency constraint and intensity-based registration. This might have limited registration performance. Studying higher-order derivatives, more powerful image similarity measures (e.g., based on deep features), as well as extensions to piecewise diffeomorphic transformations would be interesting future work.
Supplementary Material
Figure 1.
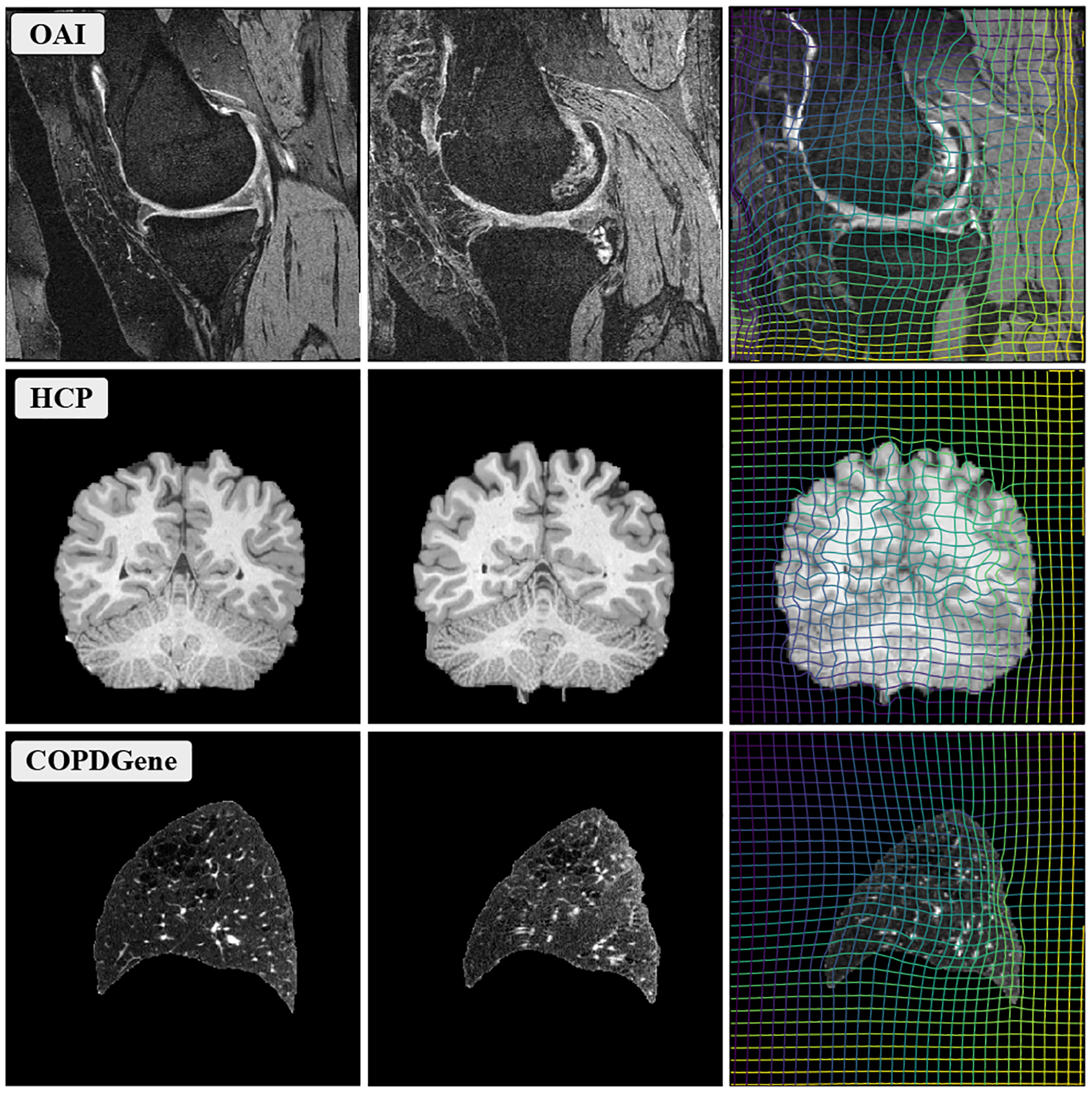
Example source (left), target (middle) and warped source (right) images obtained with our method, trained with a single protocol, using the proposed GradICON regularizer.
Acknowledgements
This work was supported by NIH grants 1R01AR072013, 1R01HL149877, 1R01EB028283, RF1MH126732, R41MH118845 R01MH112748, R01NS125307 and 5R21LM013670. The work expresses the views of the authors, not of NIH. Roland Kwitt was supported in part by the Austrian Science Fund (FWF): project FWF P31799-N38 and the Land Salzburg (WISS 2025) under project numbers 20102- F1901166-KZP and 20204-WISS/225/197-2019. The knee imaging data were obtained from the controlled access datasets distributed from the Osteoarthritis Initiative (OAI), a data repository housed within the NIMH Data Archive. OAI is a collaborative informatics system created by NIMH and NIAMS to provide a worldwide resource for biomarker identification, scientific investigation and OA drug development. Dataset identifier: NIMH Data Archive Collection ID: 2343. The brain imaging data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The lung imaging data was provided by the COPDGene study.
Footnotes
i.e., penalizing deviations from instead of deviations from .
Note that keypoint approaches [26] and approaches based on optimal transport [51] can overcome some of these issues. However, in this work, we focus on the registration of images with grid-based displacement fields.
Basically, we are interested in properties of , as this is the region that can affect the image similarity, but since many maps (e.g., translations) carry points outside of must be defined at those points for to be defined on all of . In practice, this is achieved for a displacement field by .
Indeed, the neighborhood of identity of invertible maps is much larger for small-frequency perturbations than for high-frequency perturbations; what matters for invertibility is the norm of the gradient.
For reference, networks. tallUnet2 in the ICON source code.
For intra-subject reg., instead of .
Available at https://github.com/uncbiag/ICON
The dataset contains 1000 pairs, but 1 pair is also in the DirLab challenge set, and so was excluded from training.
Using SynthMorph networks trained on HCP aging data, which differs slightly from the HCP Young Adults data we use; see [27] for a comparison.
References
- [1].Arsigny Vincent, Commowick Olivier, Ayache Nicholas, and Pennec Xavier. A fast and log-Euclidean polyaffine framework for locally linear registration. J. Math. Imaging Vis, 33(2):222–238, 2009. 2 [Google Scholar]
- [2].Ashburner John. A fast diffeomorphic image registration algorithm. Neuroimage, 38(1):95–113, 2007. 2 [DOI] [PubMed] [Google Scholar]
- [3].Avants Brian B, Epstein Charles L, Grossman Murray, and Gee James C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. MedIA, 12(1):26–41, 2008. 7, 8, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Balakrishnan Guha, Zhao Amy, Sabuncu Mert R, Guttag John, and Dalca Adrian V. VoxelMorph: a learning framework for deformable medical image registration. TMI, 38(8):1788–1800, 2019. 2, 3, 6, 8, 21, 22 [DOI] [PubMed] [Google Scholar]
- [5].Beg M Faisal, Miller Michael I, Trouvé Alain, and Younes Laurent. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. IJCV, 61(2):139–157, 2005. 2 [Google Scholar]
- [6].Bian Zhangxing, Jabri Allan, Efros Alexei A, and Owens Andrew. Learning pixel trajectories with multiscale contrastive random walks. In CVPR, 2022. 3 [Google Scholar]
- [7].Butler Daniel J, Wulff Jonas, Stanley Garrett B, and Black Michael J. A naturalistic open source movie for optical flow evaluation. In ECCV, 2012. 2 [Google Scholar]
- [8].Castillo Richard, Castillo Edward, Fuentes David, Ahmad Moiz, Wood Abbie M, Ludwig Michelle S, and Guerrero Thomas. A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive. Phys. Med. Biol, 58(9):2861, 2013. 6, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Charpiat Guillaume, Maurel Pierre, Pons Jean-Philippe, Keriven Renaud, and Faugeras Olivier. Generalized gradients: Priors on minimization flows. IJCV, 73:325–344, 07 2007. 5 [Google Scholar]
- [10].Chen Xiang, Diaz-Pinto Andres, Ravikumar Nishant, and Frangi Alejandro F. Deep learning in medical image registration. Prog. biomed. eng, 3(1):012003, 2021. 1 [Google Scholar]
- [11].Christensen Gary E and Johnson Hans J. Consistent image registration. TMI, 20(7):568–582, 2001. 2 [DOI] [PubMed] [Google Scholar]
- [12].Christensen Gary E, Rabbitt Richard D, and Miller Michael I. 3D brain mapping using a deformable neuroanatomy. Phys. Med. Biol, 39(3):609, 1994. 2 [DOI] [PubMed] [Google Scholar]
- [13].Christensen Gary E, Rabbitt Richard D, and Miller Michael I. Deformable templates using large deformation kinematics. TMI, 5(10):1435–1447, 1996. 2 [DOI] [PubMed] [Google Scholar]
- [14].William R Crum Thomas Hartkens, and Hill DLG. Nonrigid image registration: theory and practice. Brit. J. Radiol, 77(suppl_2):S140–S153, 2004. 1 [DOI] [PubMed] [Google Scholar]
- [15].Dalca Adrian V, Balakrishnan Guha, Guttag John, and Sabuncu Mert R. Unsupervised learning for fast probabilistic diffeomorphic registration. In MICCAI, 2018. 1 [DOI] [PubMed] [Google Scholar]
- [16].De Vos Bob D, Berendsen Floris F, Viergever Max A, Hessam Sokooti, Staring Marius, and Išgum Ivana. A deep learning framework for unsupervised affine and deformable image registration. MedIA, 52:128–143, 2019. 3 [DOI] [PubMed] [Google Scholar]
- [17].Dosovitskiy Alexey, Fischer Philipp, Ilg Eddy, Hausser Philip, Hazirbas Caner, Golkov Vladimir, Van Der Smagt Patrick, Cremers Daniel, and Brox Thomas. Flownet: Learning optical flow with convolutional networks. In CVPR, 2015. 1 [Google Scholar]
- [18].Eppenhof Koen AJ, Lafarge Maxime W, Veta Mitko, and Pluim Josien PW. Progressively trained convolutional neural networks for deformable image registration. TMI, 39(5):1594–1604, 2019. 3 [DOI] [PubMed] [Google Scholar]
- [19].Evans, Janke Andrew L, Collins D Louis, and Baillet Sylvain. Brain templates and atlases. Neuroimage, 62(2):911–922, 2012. 1 [DOI] [PubMed] [Google Scholar]
- [20].Fischer Bernd and Modersitzki Jan. Ill-posed medicine—an introduction to image registration. Inverse problems, 24(3):034008, 2008. 3 [Google Scholar]
- [21].Fortun Denis, Bouthemy Patrick, and Kervrann Charles. Optical flow modeling and computation: A survey. Comput. Vis. Image Underst, 134:1–21, 2015. 1 [Google Scholar]
- [22].Godard Clément, Aodha Oisin Mac, and Brostow Gabriel J. Unsupervised monocular depth estimation with left-right consistency. In CVPR, 2017. 3 [Google Scholar]
- [23].Greer Hastings, Kwitt Roland, Vialard François-Xavier, and Niethammer Marc. ICON: Learning regular maps through inverse consistency. In ICCV, 2021. 2, 3, 4, 5, 6, 7, 8, 15, 16, 21, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Haber Eldad and Modersitzki Jan. Image registration with guaranteed displacement regularity. IJCV, 71(3):361–372, 2007. 2 [Google Scholar]
- [25].Han Xu, Hong Jun, Reyngold Marsha, Crane Christopher, Cuaron John, Hajj Carla, Mann Justin, Zinovoy Melissa, Greer Hastings, Yorke Ellen, Mageras Gig, and Niethammer Marc. Deep-learning-based image registration and automatic segmentation of organs-at-risk in cone-beam CT scans from high-dose radiation treatment of pancreatic cancer. J. Med. Phys, 48(6):3084–3095, 2021. 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hansen Lasse and Heinrich Mattias P. GraphRegNet: Deep graph regularisation networks on sparse keypoints for dense registration of 3D lung CTs. TMI, 40(9):2246–2257, 2021. 3, 8, 21 [DOI] [PubMed] [Google Scholar]
- [27].Harms Michael P, Somerville Leah H, Ances Beau M, Andersson Jesper, Barch Deanna M, Bastiani Matteo, Bookheimer Susan Y, Brown Timothy B, Buckner Randy L, Burgess Gregory C, Coalson Timothy S, Chappell Michael A, Dapretto Mirella, Douaud Gwenaëlle, Fischl Bruce, Glasser Matthew F, Greve Douglas N, Hodge Cynthia, Jamison Keith W, Jbabdi Saad, Kandala Sridhar, Li Xiufeng, Mair Ross W, Mangia Silvia, Marcus Daniel, Mascali Daniele, Moeller Steen, Nichols Thomas E, Robinson Emma C, Salat David H, Smith Stephen M, Sotiropoulos Stamatios N, Terpstra Melissa, Thomas Kathleen M, Tisdall M Dylan, Ugurbil Kamil, van der Kouwe Andre, Woods Roger P., Zöllei Lilla, Van Essen David C., and Yacoub Essa. Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects. NeuroImage, 183:972–984, 2018. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He Xinzi, Guo Jia, Zhang Xuzhe, Bi Hanwen, Gerard Sarah, Kaczka David, Motahari Amin, Hoffman Eric, Reinhardt Joseph, R Graham Barr Elsa Angelini, and Laine Andrew. Recursive refinement network for deformable lung registration between exhale and inhale CT scans. arXiv preprint arXiv:2106.07608, 2021. 8, 21 [Google Scholar]
- [29].Hering Alessa, van Ginneken Bram, and Heldmann Stefan. mlvirnet: Multilevel variational image registration network. In MICCAI, 2019. 3 [Google Scholar]
- [30].Hering Alessa, Stephanie Häger Jan Moltz, Lessmann Nikolas, Heldmann Stefan, and van Ginneken Bram. CNN-based lung CT registration with multiple anatomical constraints. MedIA, 72:102139, 2021. 8, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hoffmann Malte, Billot Benjamin, Greve Douglas N, Iglesias Juan Eugenio, Fischl Bruce, and Dalca Adrian V. Synthmorph: learning contrast-invariant registration without acquired images. TMI, 41(3):543–558, 2022. 7, 8, 15, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Holden Mark. A review of geometric transformations for nonrigid body registration. TMI, 27(1):111–128, 2007. 2, 3 [DOI] [PubMed] [Google Scholar]
- [33].Horn Berthold KP and Schunck Brian G. Determining optical flow. Artif. Intell, 17(1–3):185–203, 1981. 1 [Google Scholar]
- [34].Huang Zhaoyang, Shi Xiaoyu, Zhang Chao, Wang Qiang, Cheung Ka Chun, Qin Hongwei, Dai Jifeng, and Li Hongsheng. Flowformer: A transformer architecture for optical flow. arXiv preprint arXiv: 2203.16194, 2022. 1, 2 [Google Scholar]
- [35].Ilg Eddy, Mayer Nikolaus, Saikia Tonmoy, Keuper Margret, Dosovitskiy Alexey, and Brox Thomas. Flownet 2.0: Evolution of optical flow estimation with deep networks. In CVPR, 2017. 1 [Google Scholar]
- [36].Jiang Zhuoran, Yin Fang-Fang, Ge Yun, and Ren Lei. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys. Med. Biol, 65(1):015011, 2020. 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kessler Marc L. Image registration and data fusion in radiation therapy. Brit. J. Radiol, 79(S1):S99–S108, 2006. 1 [DOI] [PubMed] [Google Scholar]
- [38].Klein Stefan, Staring Marius, Murphy Keelin, Viergever Max A, and Pluim Josien PW. Elastix: a toolbox for intensity-based medical image registration. TMI, 29(1):196–205, 2009. 8, 21 [DOI] [PubMed] [Google Scholar]
- [39].Maes Frederik, Vandermeulen Dirk, and Suetens Paul. Comparative evaluation of multiresolution optimization strategies for multimodality image registration by maximization of mutual information. MedIA, 3(4):373–386, 1999. 3 [DOI] [PubMed] [Google Scholar]
- [40].Mang Andreas and Biros George. Constrained H1-regularization schemes for diffeomorphic image registration. SIAM J. Imaging Sci, 9:1154–1194, 08 2016. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Miller Michael I, Trouvé Alain, and Younes Laurent. Geodesic shooting for computational anatomy. J. Math. Imaging Vis, 24(2):209–228, 2006. 1, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Miller Michael I and Younes Laurent. Group actions, homeomorphisms, and matching: A general framework. IJCV, 41(1):61–84, 2001. 2 [Google Scholar]
- [43].Modat Marc, Ridgway Gerard R, Taylor Zeike A, Lehmann Manja, Barnes Josephine, Hawkes David J, Fox Nick C, and Ourselin Sébastien. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed, 98(3):278–284, 2010. 8, 21 [DOI] [PubMed] [Google Scholar]
- [44].Modersitzki Jan. Numerical methods for image registration. OUP Oxford, 2003. 1, 2 [Google Scholar]
- [45].Mok Tony CW and Chung Albert. Large deformation diffeomorphic image registration with Laplacian pyramid networks. In MICCAI, 2020. 3, 8, 21 [Google Scholar]
- [46].Nevitt Michael C, Felson David T, and Lester Gayle. The osteoarthritis initiative. Protocol for the cohort study, 1, 2006. 2, 6 [Google Scholar]
- [47].Regan Elizabeth A, Hokanson John E, Murphy James R, Make Barry, Lynch David A, Beaty Terri H, Curran-Everett Douglas, Silverman Edwin K, and Crapo James D. Genetic epidemiology of COPD (COPDGene) study design. COPD: J. Chronic Obstr. Pulm. Dis, 7(1):32–43, 2011. 2, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Reuter Martin, Rosas H Diana, and Fischl Bruce. Highly accurate inverse consistent registration: a robust approach. Neuroimage, 53(4):1181–1196, 2010. 8, 15, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rushmore R. Jarrett, Sutherland Kyle, Carrington Holly, Chen Justine, Halle Michael, Lasso Andras, Papadimitriou George, Prunier Nick, Rizzoni Elizabeth, Vessey Brynn, Wilson-Braun Peter, Rathi Yogesh, Kubicki Marek, Bouix Sylvain, Yeterian Edward, and Makris Nikos. Anatomically curated segmentation of human subcortical structures in high resolution magnetic resonance imaging: An open science approach. Front. Neuroanat, 16, 2022.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rushmore R. Jarrett, Sutherland Kyle, Carrington Holly, Chen Justine, Halle Michael, Lasso Andras, Papadimitriou George, Prunier, Rizzoni Elizabeth, Vessey Brynn, Wilson-Braun Peter, Rathi Yogesh, Kubicki Marek, Bouix Sylvain, Yeterian Edward, and Makris Nikos. HOA-2/SubcorticalParcellations: release-50-subjects-1.1.0. Sept. 2022. 6
- [51].Shen Zhengyang, Feydy Jean, Liu Peirong, Curiale Ariel H, San Jose Estepar Ruben, San Jose Estepar Raul, and Niethammer Marc. Accurate point cloud registration with robust optimal transport. In NeurIPS, 2021. 3 [Google Scholar]
- [52].Shen Zhengyang, Han Xu, Xu Zhenlin, and Niethammer Marc. Networks for joint affine and non-parametric image registration. In CVPR, 2019. 2, 3, 6, 7, 8, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sotiras Aristeidis, Davatzikos Christos, and Paragios Nikos. Deformable medical image registration: A survey. TMI, 32(7):1153–1190, 2013. 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Staal Joes, Abràmoff Michael D, Niemeijer Meindert, Viergever Max A, and Van Ginneken Bram. Ridge-based vessel segmentation in color images of the retina. TMI, 23(4):501–509, 2004. 6 [DOI] [PubMed] [Google Scholar]
- [55].Studholme Colin, Hill Derek LG, and Hawkes David J. Multiresolution voxel similarity measures for MR-PET registration. In IPMI, 1995. 3 [DOI] [PubMed] [Google Scholar]
- [56].Sun Deqing, Yang Xiaodong, Liu Ming-Yu, and Kautz Jan. Pwc-net: CNNs for optical flow using pyramid, warping, and cost volume. In CVPR, 2018. 1 [Google Scholar]
- [57].Teed Zachary and Deng Jia. Raft: Recurrent all-pairs field transforms for optical flow. In ECCV, 2020. 1 [Google Scholar]
- [58].Thévenaz Philippe and Unser Michael. Optimization of mutual information for multiresolution image registration. TMI, 9(12):2083–2099, 2000. 3 [DOI] [PubMed] [Google Scholar]
- [59].Thirion Jean-Philippe. Image matching as a diffusion process: an analogy with Maxwell’s demons. MedIA, 2(3):243–260, 1998. 2 [DOI] [PubMed] [Google Scholar]
- [60].Van Essen David C, Ugurbil Kamil, Auerbach Edward, Barch Deanna, Behrens Timothy EJ, Bucholz Richard, Chang Acer, Chen Liyong, Corbetta Maurizio, Curtiss Sandra W, Penna Stefania Della, Feinberg David, Glasser Matthew F, Harel Noam, Heath Andrew C, Larson-Prior Linda, Marcus Daniel, Michalareas Georgios, Moeller Steen, Oostenfeld Robert, Peterson Steve E, Prior Fred, Schlaggar Brad L, Smith Stephen M, Snyder Abraham Z, Xu Junqian, Yacoub Essa, and WU-Minn HCP Consortium. The human connectome project: a data acquisition perspective. Neuroimage, 62(4):2222–2231, 2012. 2, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vercauteren Tom, Pennec Xavier, Perchant Aymeric, and Ayache Nicholas. Symmetric log-domain diffeomorphic registration: A Demons-based approach. In MICCAI, 2008. 2 [DOI] [PubMed] [Google Scholar]
- [62].Vercauteren Tom, Pennec Xavier, Perchant Aymeric, and Ayache Nicholas. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage, 45(1):61–72, 2009. 8, 21 [DOI] [PubMed] [Google Scholar]
- [63].Vialard François-Xavier, Risser Laurent, Rueckert Daniel, and Cotter Colin J. Diffeomorphic 3D image registration via geodesic shooting using an efficient adjoint calculation. IJCV, 97(2):229–241, 2012. 1, 2 [Google Scholar]
- [64].Viergever Max A, Maintz JB Antoine, Klein Stefan, Murphy Keelin, Staring Marius, and Pluim Josien PW. A survey of medical image registration. MedIA, 33:140–144, 2016. 1 [DOI] [PubMed] [Google Scholar]
- [65].Vishnevskiy Valery, Gass Tobias, Szekely Gabor, Tanner Christine, and Goksel Orcun. Isotropic total variation regularization of displacements in parametric image registration. TMI, 36(2):385–395, 2017. 6, 8, 21 [DOI] [PubMed] [Google Scholar]
- [66].Wang Di, Pan Yue, Durumeric Oguz C, Reinhardt Joseph M, Hoffman Eric A, Schroeder Joyce D, and Christensen Gary E. PLOSL: Population learning followed by one shot learning pulmonary image registration using tissue volume preserving and vesselness constraints. MedIA, 79:102434, 2022. 6, 8, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wang Xiaolong, Jabri Allan, and Efros Alexei A.. Learning correspondence from the cycle-consistency of time. In CVPR, 2019. 3 [Google Scholar]
- [68].Yang Xiao, Kwitt Roland, Styner Martin, and Niethammer Marc. Quicksilver: Fast predictive image registration–a deep learning approach. NeuroImage, 158:378–396, 2017. 1, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yin Zhichao and Shi Jianping. GeoNet: Unsupervised learning of dense depth, optical flow and camera pose. In CVPR, 2018. 3 [Google Scholar]
- [70].Zhao Shengyu, Dong Yue, Chang Eric I, Xu Yan, et al. Recursive cascaded networks for unsupervised medical image registration. In CVPR, 2019. 3 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


