Summary
Background
Osteoarthritis is a leading cause of disability, and disease-modifying osteoarthritis drugs (DMOADs) could represent a pivotal advancement in treatment. Identifying the potential of antidiabetic medications as DMOADs could impact patient care significantly.
Methods
We designed a comprehensive analysis pipeline involving two-sample Mendelian Randomization (MR) (genetic proxies for antidiabetic drug targets), summary-based MR (SMR) (for mRNA), and colocalisation (for drug-target genes) to assess their causal relationship with 12 osteoarthritis phenotypes. Summary statistics from the largest genome-wide association meta-analysis (GWAS) of osteoarthritis and gene expression data from the eQTLGen consortium were utilised.
Findings
Seven out of eight major types of clinical antidiabetic medications were identified, resulting in fourteen potential drug targets. Sulfonylurea targets ABCC8/KCNJ11 were associated with increased osteoarthritis risk at any site (odds ratio (OR): 2.07, 95% confidence interval (CI): 1.50–2.84, P < 3 × 10−4), while PPARG, influenced by thiazolidinediones (TZDs), was associated with decreased risk of hand (OR: 0.61, 95% CI: 0.48–0.76, P < 3 × 10−4), finger (OR: 0.50, 95% CI: 0.35–0.73, P < 3 × 10−4), and thumb (OR: 0.49, 95% CI: 0.34–0.71, P < 3 × 10−4) osteoarthritis. Metformin and GLP1-RA, targeting GPD1 and GLP1R respectively, were associated with reduced risk of knee and finger osteoarthritis. In the SMR analyses, gene expression of KCNJ11, GANAB, ABCA1, and GSTP1, targeted by antidiabetic drugs, was significantly linked to at least one osteoarthritis phenotype and was replicated across at least two gene expression datasets. Additionally, increased KCNJ11 expression was related to decreased osteoarthritis risk and co-localised with at least one osteoarthritis phenotype.
Interpretation
Our findings suggest a potential therapeutic role for antidiabetic drugs in treating osteoarthritis. The results indicate that certain antidiabetic drug targets may modify disease progression, with implications for developing targeted DMOADs.
Funding
This study was funded by the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant (2022), the Shanghai Municipal Health Commission Health Industry Clinical Research Project (Grant No. 20224Y0139), Beijing Natural Science Foundation (Grant No. 7244458), and the Postdoctoral Fellowship Program (Grade C) of China Postdoctoral Science Foundation (Grant No. GZC20230130).
Keywords: Osteoarthritis, Drug targets, Antidiabetic drugs, Gene expression, Mendelian randomization
Research in context.
Evidence before this study
Previous research has explored the complex interplay between metabolic dysfunctions and osteoarthritis, implicating conditions such as diabetes in the pathophysiology of the disease. Despite extensive research, traditional treatments have not effectively halted osteoarthritis progression, highlighting a substantial gap in effective Disease-Modifying Osteoarthritis Drugs (DMOADs). We conducted a comprehensive literature review using multiple databases, such as PubMed, Web of Science, and Scopus, with no language restrictions and covering publications up to January 2024. Our search was focused on gathering robust evidence from genetic studies, clinical trials, and observational cohorts to inform our understanding of the links between metabolic dysfunction and osteoarthritis. Our search criteria were carefully designed to include relevant studies, assessing their quality and potential biases. This groundwork identified knowledge gaps regarding the use of antidiabetic drugs as potential treatments for osteoarthritis. This gap in knowledge underscored the need for robust genetic studies to clarify these potential relationships, leading directly to the design of our study, which utilises Mendelian Randomization to overcome these limitations.
Added value of this study
Our study uniquely applies employing a comprehensive analysis pipeline involving two-sample Mendelian Randomization, summary-based Mendelian Randomization, and colocalisation analyses to explore the relationship between antidiabetic drug targets and osteoarthritis phenotypes. We found that certain antidiabetic drugs, including sulfonylurea targets like ABCC8/KCNJ11 and the PPARG target of Thiazolidinediones (TZDs), might influence osteoarthritis risk, offering new insights into their potential as disease-modifying drugs.
Implications of all the available evidence
The study opens up new possibilities for osteoarthritis treatment by suggesting antidiabetic drugs as potential disease-modifying osteoarthritis drugs. This has significant implications for clinical practice and future research, particularly in developing targeted treatments and understanding disease mechanisms. Further research is needed to confirm these findings and assess their clinical relevance.
Introduction
Osteoarthritis (commonly abbreviated as OA), the most prevalent form of arthritis, is a whole-joint disease that leads to substantial morbidity due to pain and functional limitations.1 Characterised by progressive cartilage degradation, subchondral bone remodelling, and inflammation, the aetiology of osteoarthritis is multifactorial, involving biomechanical forces and biochemical processes.2 Despite its prevalence, there are currently no approved Disease-Modifying Osteoarthritis Drugs (DMOADs) that effectively halt the progression or reverse the course of the disease.3 Thus, searching for effective DMOADs remains a critical pursuit in osteoarthritis research.4
Recent insights into osteoarthritis pathogenesis have highlighted metabolic dysregulation as a key factor in disease progression, suggesting a strong interplay between metabolic syndrome and osteoarthritis.5 Hyperglycemia and osteoarthritis interact at both local and systemic levels. Locally, oxidative stress and the accumulation of advanced glycation end-products play a role in damaging cartilage.6 Systemically, the build-up of glucose leads to low-grade inflammation, creating a harmful internal environment that can worsen osteoarthritis symptoms.6 This has sparked interest in the potential repurposing of antidiabetic medications as DMOADs, with studies indicating that such drugs may influence pathways relevant to joint health.7 Previous studies have explored the potential of antidiabetic drugs, such as metformin, in influencing osteoarthritis.8,9 Metformin has been observed to modulate inflammatory responses and cartilage matrix homeostasis, while drugs affecting the insulin pathway may influence systemic inflammation and anabolic processes in chondrocytes.10, 11, 12, 13, 14 The recently developed antidiabetic drug class, glucagon-like peptide-1 receptor agonists (GLP1-RA), also demonstrates promising potential as therapeutic treatments for osteoarthritis.15 Given the complexities of osteoarthritis and the multifaceted actions of antidiabetic drugs, understanding the potential of these medications as DMOADs requires robust investigative methods. Mendelian Randomization (MR), by leveraging genetic variants (single nucleotide polymorphisms, SNPs) as instrumental variables (IVs), offers a powerful approach to infer causal relationships from observational data, mitigating confounding and reverse causation that often plague traditional epidemiological studies.16
In this study, we utilise MR to examine the implications of targets of antidiabetic medications on osteoarthritis phenotypes. By using genetic proxies for drug targets, we aim to delineate the potential of these medications to modify disease progression in osteoarthritis. This approach provides insights into the biological mechanisms by which these drugs may serve as DMOADs and underscores the potential for a paradigm shift in osteoarthritis management through the repurposing of existing drugs. The introduction of antidiabetic medications into the realm of osteoarthritis treatment reflects an innovative convergence of metabolic and musculoskeletal research. This study contributes to the ongoing quest to discover effective DMOADs and opens new avenues for therapeutic interventions in osteoarthritis, with the potential to improve the quality of life for millions worldwide.
Methods
Identification and validation of antidiabetic drug targets
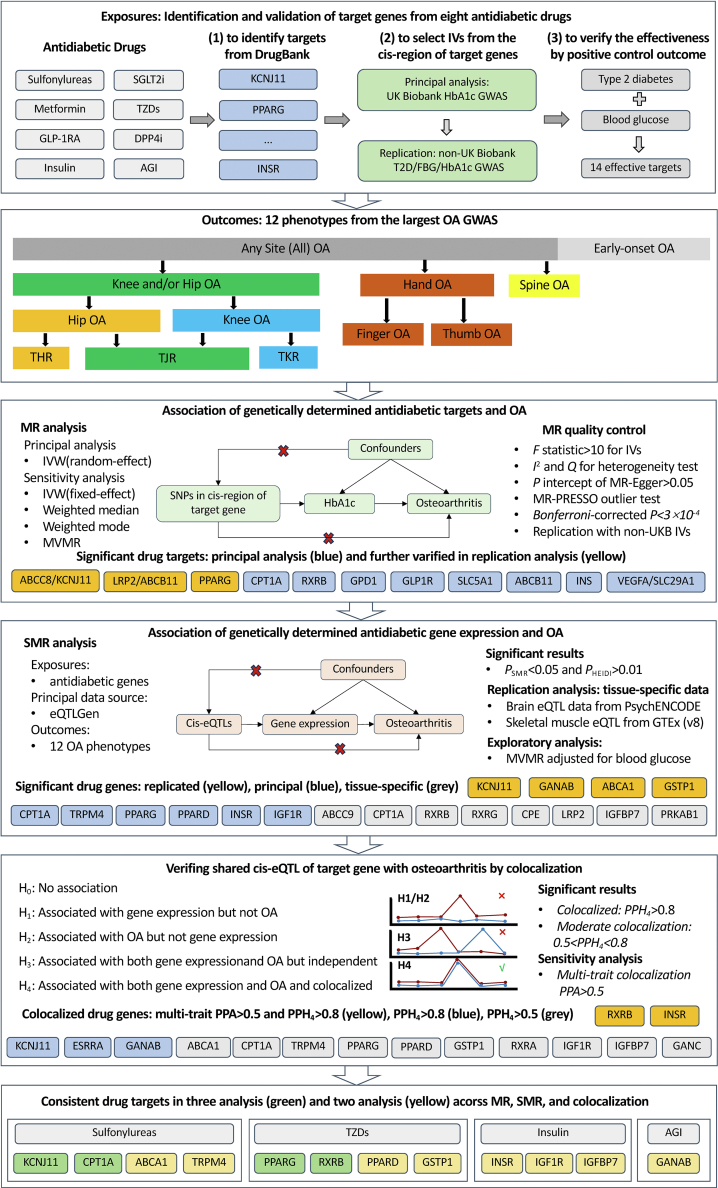
The study's procedural flowchart is depicted in Fig. 1. The identification of antidiabetic drug targets was carried out in a three-step process. Firstly, the gene targets for the existing eight types of clinical antidiabetic drugs were ascertained using the DrugBank (https://go.drugbank.com/) pharmacogenetics database (Table S1 in Supplemental Tables). The specifics of these target genes are catalogued in Table S2. Secondly, we obtained the available IVs in the cis-region (±500 kb) of these targets from a previous genome-wide association study (GWAS) on haemoglobin A1C (HbA1c) in the UK Biobank population,17 using the parameters P < 5 × 10−8 and r2 < 0.2.18 This process allowed us to determine the IVs for the drug targets of seven key antidiabetic drugs: sulfonylureas, metformin, alpha glucosidase inhibitors (AGI), Thiazolidinediones (TZDs), GLP1-RA, insulin and its analogues, and sodium-glucose cotransporter 2 inhibitors (SGLT2i). Despite the relevance of dipeptidyl peptidase-4 inhibitors (DPP4i) as antidiabetic drugs, they were not involved in further analysis due to the lack of available IVs that meet our stringent selection criteria. If two neighbouring target genes shared the same IVs due to being located in the overlapped cis-region, these genes were combined and marked with a slash, as in “ABCC8/KCNJ11”. The IVs for ESRRA were automatically excluded by the default settings of the harmonise_data function in the TwoSampleMR package due to palindromes. Furthermore, we acquired IVs for these gene targets from several non-UK Biobank GWAS databases. Since there is no other GWAS data that includes IVs for all drug targets related to HbA1c, blood glucose levels, or type 2 diabetes mellitus (T2DM), we sourced these IVs from six datasets. These include the HbA1c GWAS from MAGIC19 and the Within Family GWAS consortium,20 the T2DM GWAS from DIAGRAM,21 FinnGen, as well as the fasting glucose data from MAGIC.22 All these datasets predominantly consist of European ancestry samples and are available on the MRC IEU OpenGWAS platform (Table S3).23 Thirdly, four GWAS datasets on blood glucose levels and T2DM were used as positive controls to validate the effectiveness of the identified drug targets (Table S3). The target KCNJ1 was excluded from further analysis as it was not associated with any positive control outcomes (Table S5). The IVs and their corresponding effects for each drug target are presented in Table S6.
Fig. 1.
Flowchart illustrating the systematic approach for identifying the association between antidiabetic drug targets and osteoarthritis phenotypes. This flowchart delineates the Mendelian Randomization framework used to assess the causal impact of antidiabetic drug targets on 12 osteoarthritis phenotypes identified from the largest osteoarthritis genome-wide association studies. If two neighbouring target genes shared the same IVs due to being located in the overlapped cis-region, these genes were combined and marked with a slash, as in “ABCC8/KCNJ11.” Abbreviations: OA, Osteoarthritis; MR, Mendelian randomisation; MVMR, multivariable Mendelian randomisation; SMR, summary-based Mendelian randomisation; IVs, instrumental variables; GWAS, genome-wide association studies; IVW, inverse variance weighted; T2DM, type 2 diabetes mellitus; HbA1c, haemoglobin A1C; FBG, fasting blood glucose; SNPs, single nucleotide polymorphisms; PSMR, P-value for SMR analysis; PHEIDI, P-value for heterogeneity in dependent instruments; eQTLs, expression quantitative trait loci; H0–H4, Hypotheses 0–4; PPA, posterior probability; THR, total hip replacement; TJR, total joint replacement; TKR, total knee replacement.
Determination of study outcomes
We utilised summary statistics from the most comprehensive and current GWAS of osteoarthritis (Genetics of Osteoarthritis (GO) Consortium). The meta-analysis dataset included 826,690 individuals, with 177,517 diagnosed osteoarthritis cases, spanning 13 international cohorts from nine diverse populations.24 This rich dataset provided us with a broad spectrum of osteoarthritis phenotypes to investigate, including knee and/or hip osteoarthritis, knee osteoarthritis, hip osteoarthritis, hand osteoarthritis, finger osteoarthritis, thumb osteoarthritis, spine osteoarthritis, early-onset osteoarthritis, total joint replacements such as total knee replacement (TKR), total hip replacement (THR), total joint replacement (TJR) and all osteoarthritis (or any site osteoarthritis). The overview of the 12 defined osteoarthritis phenotypes and their relationships are shown in Fig. 1. Osteoarthritis may also occur in the shoulder, ankle, foot, toe, and other joints; however, data on these sites are not available or included in this definition of osteoarthritis. The dataset delineates specific relationships between osteoarthritis manifestations, such as the categorisation of TJR—indicating advanced disease—into procedures like TKR for knee osteoarthritis and THR for hip osteoarthritis. It also provides a detailed breakdown of hand osteoarthritis, further dissecting it into finger and thumb osteoarthritis, reflecting the diverse joints that can be affected by osteoarthritis. To ensure the validity of our results, we carefully checked and confirmed the absence of sample overlap between the exposure and outcome datasets. Specifically, in the principal outcome GWAS dataset (GO Consortium), UK Biobank participants (n = 316,467) were also included in the larger cohort comprising 826,690 individuals in the GO Consortium (Table S3), and the percentage of the overlap for each GWAS of osteoarthritis phenotype ranged from about 0.5% to 38%, with all values being less than 50% (detailed in Table S4).
Two-sample MR analysis
Two-sample MR method was applied to estimate the causal effect of each antidiabetic drug target on osteoarthritis phenotypes. To distinguish between the effects of blood glucose changes and the effects of antidiabetic drugs on osteoarthritis risk, we used MR to focus on the genetic proxies for drug targets rather than direct measures of blood glucose. This approach helps isolate the specific effects of the drug targets from those of blood glucose levels. According to the causal graph of MR shown in Fig. 1, this method must satisfy three assumptions: (1) the IVs should be associated with the exposure, (2) the IVs should affect the outcome only through the exposure (lower red cross in Fig. 1), and (3) the IVs should not be associated with any confounders (upper red cross in Fig. 1). To meet these assumptions, several approaches were applied. The primary analysis method was the random-effect inverse variance weighted (IVW) MR.25 To ensure the robustness of our findings, we also conducted sensitivity analyses using the fixed-effects IVW, weighted median, and weighted mode approaches. These methods were employed concurrently to address any potential violations of IV assumptions. We compared the odds ratios (ORs) and P-values obtained from all four methods to assess the stability and validity of the findings. Random-effects IVW was used to counteract the potential bias from high heterogeneity among the multiple IVs. Heterogeneity among the IVs was quantified using I2 statistics and Q test. The weighted median method was employed to provide robust estimates even in the presence of some invalid genetic instruments (if the proportion of invalid instruments was less than 50%).26 The weighted mode estimator is deemed reliable when the majority of similar causal effect estimates are derived from valid IVs.27 Findings that were concurrently significant across these methods were considered robust associations. Furthermore, the intercept of MR-Egger regression was used to test for the presence of horizontal pleiotropy.28 The MR analysis was performed using the TwoSampleMR package with the default parameters (https://mrcieu.github.io/TwoSampleMR/).
We also performed the MR-PRESSO analysis to test and correct the presence of specific IV outliers (potential pleiotropy SNPs) as another sensitivity analysis.29 Furthermore, to evaluate the robustness of the MR results, we conducted a multivariable MR (MVMR) analysis (the schematic representation is shown in Figure S1 for Supplemental Figures).30 This analysis also used IVs from the cis-region (cis-window: ±500 kb) of each drug target and adjusted for body mass index, systolic blood pressure, smoking status, and alcohol drinker status, using the same parameters (P < 5 × 10−8, r2 < 0.2). The strength of the IVs was determined using the F-statistic, calculated as , where is the estimate of the SNP-exposure association, and is the standard error of this association.31 In our study, all observed F-statistics comfortably exceeded the threshold of F >10, indicating no weak instrument bias (Table S7). A Bonferroni-corrected P-value threshold of 3 × 10−4 (0.05 divided by the number of tests, which is 14 drug targets times 12 osteoarthritis phenotypes) was set for significance to address multiple testing, with P-values less than 0.05 being deemed suggestive of a statistically significant association. If the IVs for multiple target genes were available for a single antidiabetic drug, the combined IVs were used in the MR analysis to demonstrate the overall effects. The MR estimators in the principal analysis represented a per-standard deviation (SD) decrease in genetically predicted levels of HbA1c when targeting a specific gene by the corresponding drug on the risk of osteoarthritis. Additionally, we scaled these effects to a per-SD decrease in genetically predicted levels of random blood glucose. This process was conducted by the positive control MR analysis of the per-SD HbA1c on per-SD blood glucose for each drug target, which can acquire the corresponding coefficients (β) of their relationship. Then, the estimators from per-SD decreased HbA1c to per-SD decreased blood glucose could be scaled by multiple 1/β (detailed in Table S7).
Gene expression analysis
Gene expression data was sourced from the eQTLGen consortium, which provided us with a substantial sample size (n = 31,684) to identify SNPs associated with the expression of genes targeted by antidiabetic drugs in blood.32 In this analysis, we specifically focused on cis-eQTLs (cis expression quantitative trait locus), located within a one-megabase distance from the target gene, ensuring the relevance of the genetic variation to gene expression changes. Data were normalised and scaled by subtracting from each expression value (abundance of mRNA) the mean of the respective probe and then dividing it by the SD of the respective probe.32 The eQTL data is represented as the effect of each additional allele on per-SD change in gene expression level.
The effects of target genes of all eight antidiabetic drugs (including DPP4i) were evaluated by the summary-based MR (SMR) method to examine the association between antidiabetic drug-related gene expression and osteoarthritis risk.33 The SMR method employs the most significantly associated cis-eQTL SNP as the top IV. The main results are presented as the OR for osteoarthritis per-SD increase in gene expression. The HEIDI (heterogeneity in dependent instruments) test, part of the SMR approach, was conducted to determine if the association observed between gene expression and osteoarthritis was due to a linkage scenario, where an eQTL SNP in linkage disequilibrium with another SNP independently influences the disease outcome, potentially violating one of the MR assumptions. A HEIDI test P-value below 0.01 was considered indicative of an association likely due to linkage rather than the regulation of gene expression.33 We performed the SMR analysis using the default parameters (https://yanglab.westlake.edu.cn/software/smr/#SMR&HEIDIanalysis; P value of cis-eQTL: <5 × 10−8, P value of eQTLs for the HEIDI test: <1.57 × 10−3, window centred around the probe to select cis-eQTLs: 2000 Kb).33 Additionally, serum HbA1c levels, glucose levels, and T2DM were employed as outcomes to confirm the validity of the results' direction.
Furthermore, to evaluate the robustness of the SMR results and determine if the effects of gene expression on osteoarthritis risk were independent of blood glucose levels, we also conducted MVMR analysis (the schematic representation is shown in Figure S1).30 This analysis used IVs from the cis-region and adjusted for blood glucose levels using the same parameters (P < 5 × 10−8, r2 < 0.2, cis-window: ±500 kb). We confirmed that there was no sample overlap between the gene expression and osteoarthritis datasets.
Colocalisation analysis
Colocalisation analysis was conducted to determine if the associations between specific gene expressions and osteoarthritis were attributable to the same causal genetic variant. The analysis was based on a Bayesian model with a posterior probability of five hypotheses (PPH): H0 suggests no association with either trait; H1 indicates an association with the first trait only; H2 with the second trait only; H3 posits that different causal variants are associated with each trait; and H4 implies the same causal variant is associated with both traits.34 The coloc. abf algorithm in R package (http://cran.r-project.org/web/packages/coloc) was used (parameter setting: p1 = 1 × 10−4, p2 = 1 × 10−4, p12 = 1 × 10−3), and we considered a PPH (H4) greater than 0.8 as evidence of strong colocalisation between gene expression targets and osteoarthritis, while a PPH (H4) above 0.5 was indicative of moderate colocalisation (Fig. 1). Considering the classification and similarity of 12 osteoarthritis phenotypes, we also conducted a multiple trait colocalisation analysis for target gene expression across a group of osteoarthritis phenotypes using the moloc method (https://github.com/clagiamba/moloc).35 This method enables the detection of colocalisation across multiple traits at a specific locus. We set the prior variance and prior probability for each trait at 0.1 and 1 × 10−4, respectively. In this analysis, we focused particularly on the results of the posterior probability (PPA), which indicates whether a target gene is co-localised with at least one osteoarthritis phenotype from specific osteoarthritis groups (Fig. 1).
Replication analysis
First, to address the partial overlap between the osteoarthritis GWAS data and the primary HbA1c GWAS used in our two-sample MR analysis, we conducted replication analyses to further validate the robustness of our findings. For these analyses, we selected IVs (in the cis-region (±500 kb) of the targets using the same parameters P < 5 × 10−8 and r2 < 0.2) for target genes/drugs from non-UK Biobank exposure GWAS datasets to ensure there was no potential sample overlap. Second, to expand the scope of gene expression analysis, we also incorporated brain eQTL data from the PsychENCODE project and skeletal muscle eQTL data from the most recent version of the Genotype-Tissue Expression (GTEx) database, both of which are pertinent to the pathophysiology of osteoarthritis.36,37 The PsychENCODE project focuses on understanding the genetic underpinnings of psychiatric disorders by studying brain-specific gene expression, while the GTEx project aims to provide a comprehensive public resource to study tissue-specific gene expression and regulation. SMR analyses were conducted that included all target genes with available cis-eQTLs of antidiabetic drugs.
Target genes/drugs that passed the replication analysis (highlighted in yellow in Fig. 1) and were verified at least twice in the MR, SMR, and colocalisation analyses were considered key findings. Finally, to ascertain the distribution and specificity of target gene expression across various tissues and cell types, we incorporated single-cell transcriptomic annotations from the Human Protein Atlas (proteinatlas.org), which offers normalised counts of protein-coding transcripts per million reads across 76 cell types derived from 14 distinct healthy tissue types.38
All analyses in this study were performed using R software, version 4.3.1, with the packages TwoSampleMR, coloc, and moloc, and the smr-1.3.1-win software on the Windows platform.
Ethics statement
This study is based on existing publications and public databases; both ethical approval and informed consent have been received by each relevant institutional review committee.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Effective antidiabetic drug targets using blood glucose and T2DM as positive control outcomes
Fourteen viable drug targets were identified that could harmonise with osteoarthritis outcomes across seven antidiabetic drug groups: sulfonylureas (ABCB11, ABCC8/KCNJ11, CPT1A, INS, KCNJ1, KCNJ8/ABCC9, LRP2/ABCB11, and VEGFA/SLC29A1), TZDs (PPARG, RXRB, and VEGFA/SLC29A1), GLP1-RA (GLP1R), insulin and its analogues (LRP2/ABCB11), AGIs (GANC), metformin (GPD1), and SGLT2i (SLC5A1 and SLC5A2) (Table S5). The SNPs and corresponding effects for each drug target are shown in Table S6. This analysis revealed 14 targets significantly associated with blood glucose levels and T2DM, except for KCNJ1 (Table S5).
Effects of genetically determined antidiabetic drug targets on osteoarthritis by MR
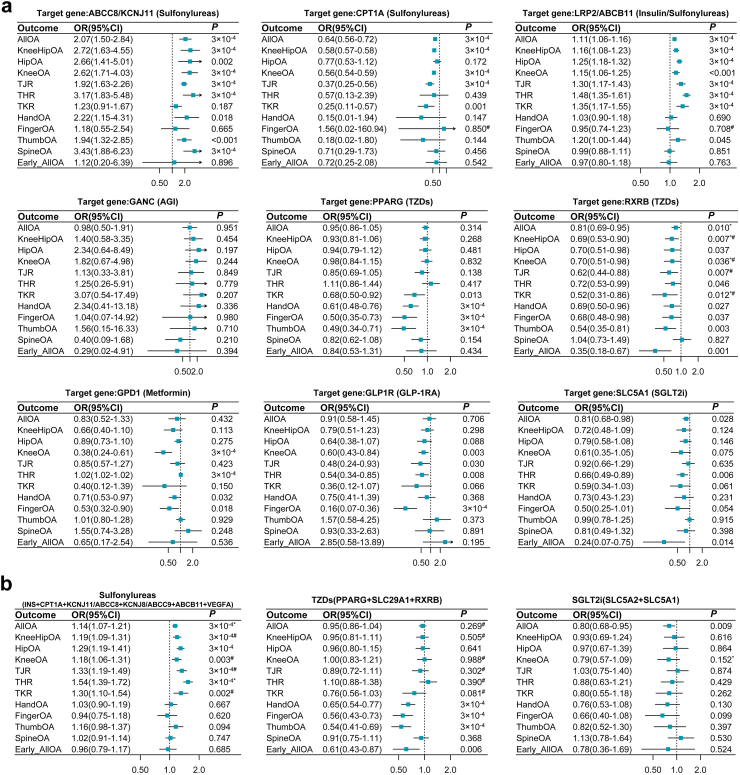
Under the principal analysis of IVW (random-effect), the sensitivity analyses (fixed-effect IVW, weighted median, and weighted mode method), and the quality control of MR (F statistic >10 for IVs; Q test for heterogeneity <50%; P intercept of MR-Egger >0.05; Bonferroni-corrected P < 3 × 10−4), there were 11 significant drug targets, except GANC, KCNJ8/ABCC9, and SLC5A2 (Fig. 2a, Table S6, Figure S2), associated with at least one osteoarthritis phenotype in the MR analyses. Representative targets for each drug, such as ABCC8/KCNJ11 for sulfonylureas, LRP2/ABCB11 for insulin and sulfonylureas, PPARG for TZDs, GPD1 for metformin, and GLP1R for GLP1-RA, surpassed the multiple testing threshold (P < 3 × 10−4) (Fig. 2a).
Fig. 2.
Forest plots of the effects of the five antidiabetic drug targets on 12 osteoarthritis phenotypes. All results were derived from the random-effects inverse variance weighted Mendelian Randomization (MR). The effects (ORs) were adjusted to reflect a per-SD decrease in genetically predicted levels of HbA1c when targeting the specific gene with the corresponding drug on the risk of osteoarthritis. Each subtitle identifies the target gene and its associated drug class. Panel a) displays results for individual drug targets, and Panel b) presents results for combined multiple drug targets within a specific drug class. Additional results for other single drug targets are presented in Figure S2. ∗denotes the P < 0.05 for the test of intercept by the MR Egger method. #denotes the P < 0.05 of the Q test for heterogeneity. Abbreviations: OA, Osteoarthritis; THR, total hip replacement; TJR, total joint replacement; TKR, total knee replacement; OR, odds ratio; CI: confidence interval.
In the random-effect IVW MR analysis (Fig. 2a), for sulfonylureas targeting ABCC8/KCNJ11, a decrease in HbA1c by this target was significantly linked to an increased risk of osteoarthritis at any site (OR: 2.07, 95% CI: 1.50–2.84, P < 3 × 10−4), knee and/or hip osteoarthritis (OR: 2.72, 95% CI: 1.63–4.55, P < 3 × 10−4), knee osteoarthritis (OR: 2.62, 95% CI: 1.71–4.03, P < 3 × 10−4), TJR (OR: 1.92, 95% CI: 1.63–2.26, P < 3 × 10−4), THR (OR: 3.17, 95% CI: 1.83–5.48, P < 3 × 10−4), and spine osteoarthritis (OR: 3.43, 95% CI: 1.88–6.23, P < 3 × 10−4). However, sulfonylureas targeting CPT1A demonstrated varied effects, showing an association with a decreased risk of osteoarthritis at any site, knee and/or hip osteoarthritis, knee osteoarthritis, and TJR. The target LRP2 of insulin, which shares IVs with the sulfonylureas target ABCB11, also exhibited consistent effects similar to those of the sulfonylureas target ABCC8/KCNJ11. In contrast, TZDs targeting PPARG were associated with a reduced risk of hand (OR: 0.61, 95% CI: 0.48–0.76, P < 3 × 10−4), finger (OR: 0.50, 95% CI: 0.35–0.73, P < 3 × 10−4), and thumb (OR: 0.49, 95% CI: 0.34–0.71, P < 3 × 10−4) osteoarthritis, suggesting a protective effect. Metformin and GLP1-RA, which target GPD1 and GLP1R, respectively, were associated with decreased knee and finger osteoarthritis risk. Additionally, the target RXRB of TZDs and SLC5A1 of SGLT2i were both associated with a reduced risk of any site osteoarthritis, THR, and early-onset osteoarthritis, although these findings did not achieve statistical significance after multiple testing corrections (Fig. 2a).
Three drugs with multiple available target genes, including sulfonylureas, TZDs, and SGLT2i, were also significantly associated with at least one osteoarthritis phenotype (Fig. 2b). The combined targets of sulfonylureas were significantly associated with an increased risk of any site osteoarthritis, knee and/or hip osteoarthritis, hip osteoarthritis, TJR, and THR (all P < 3 × 10−4). Targeting PPARG, SLC29A1, and RXRB of TZDs demonstrated protective effects on hand, finger, and thumb osteoarthritis (all P < 3 × 10−4). SGLT2i was also negatively associated with any site osteoarthritis, although this association did not pass the multiple correction threshold.
The MR results scaled by random blood glucose levels are presented in Table S7. The results from different MR methods in the sensitivity analysis were largely consistent with the principal findings, as shown in Table S7 and Figures S3–S15. In MVMR analysis, the effects of representative targets, including ABCC8/KCNJ11 for sulfonylureas, PPARG and PPARD for TZDs, GPD1 for metformin, and GLP1R for GLP1-RA et al. were further verified (Figure S16). Furthermore, only a few target–outcome pairs in the MR analysis detected outlier SNPs by the MR-PRESSO global test; however, the outlier-corrected results also support the above findings, suggesting limited bias from horizontal pleiotropy (Table S8). In the replication analysis using non-UK Biobank IVs, the results for targets ABCC8/KCNJ11, PPARG, and LRP2/ABCB11 were further replicated at least once, avoiding bias from potential sample overlap (Tables S9, Figures S17 and S18).
Antidiabetic drugs target gene expression and osteoarthritis
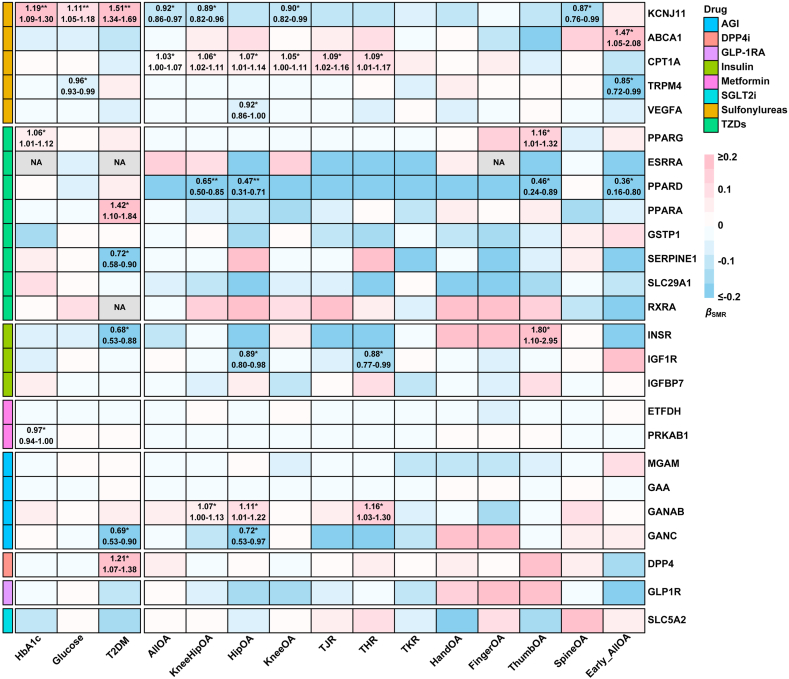
In the primary SMR analyses, gene expression of KCNJ11, CPT1A, PPARD, IGF1R, and GANAB in eQTLGen, targeted by antidiabetic drugs, was significantly linked to at least one osteoarthritis phenotype (Fig. 3, Table S10). Specifically, an increase in KCNJ11 expression within the blood by per-SD was inversely associated with the presence of osteoarthritis across several phenotypes, including osteoarthritis at any site (OR: 0.92, 95% CI: 0.86–0.97, P = 0.005), knee and/or hip osteoarthritis (OR: 0.89, 95% CI: 0.82–0.96, P = 0.005), knee osteoarthritis (OR: 0.90, 95% CI: 0.82–0.99, P = 0.027), and spine osteoarthritis (OR: 0.87, 95% CI: 0.76–0.99, P = 0.036) (Fig. 3 and 4 and Table S10). The gene expression of GANAB, targeted by AGI, was associated with an increased risk of osteoarthritis across various phenotypes, including knee and/or hip osteoarthritis (OR: 1.07, 95% CI: 1.00–1.13, P = 0.048), hip osteoarthritis (OR: 1.11, 95% CI: 1.01–1.22, P = 0.030), and THR (OR: 1.16, 95% CI: 1.03, 1.30, P = 0.014) (Fig. 3 and 4 and Table S10). Additionally, the expression of the PPARD gene, targeted by TZDs, exhibited a protective effect against various osteoarthritis phenotypes (Fig. 3, Table S10).
Fig. 3.
Gene expression analysis of antidiabetic drug targets and osteoarthritis. The heatmap illustrates the association between the expression of drug target genes in the blood (from eQTLGen) and 12 osteoarthritis phenotypes, with serum HbA1c, glucose levels, and T2DM as outcomes to confirm the direction of the results. A red region indicates a positive association, a blue region indicates a negative association, and the ORs (95% CIs) are displayed if the results are significant and pass the HEIDI test. Asterisks denote the level of significance (∗P < 0.05, ∗∗P < 0.05/25 for multiple corrections). Abbreviations: OA, Osteoarthritis; SMR, summary-based Mendelian randomisation; HbA1c, Hemoglobin A1C; T2DM, Type 2 Diabetes Mellitus; THR, Total Hip Replacement; TKR, Total Knee Replacement; AGI, alpha glucosidase inhibitors; DPP4i, dipeptidyl peptidase-4 inhibitors; GLP-1RA, glucagon-like peptide-1 receptor agonists; SGLT2i, sodium-glucose cotransporter 2 inhibitors; TZDs, Thiazolidinediones.
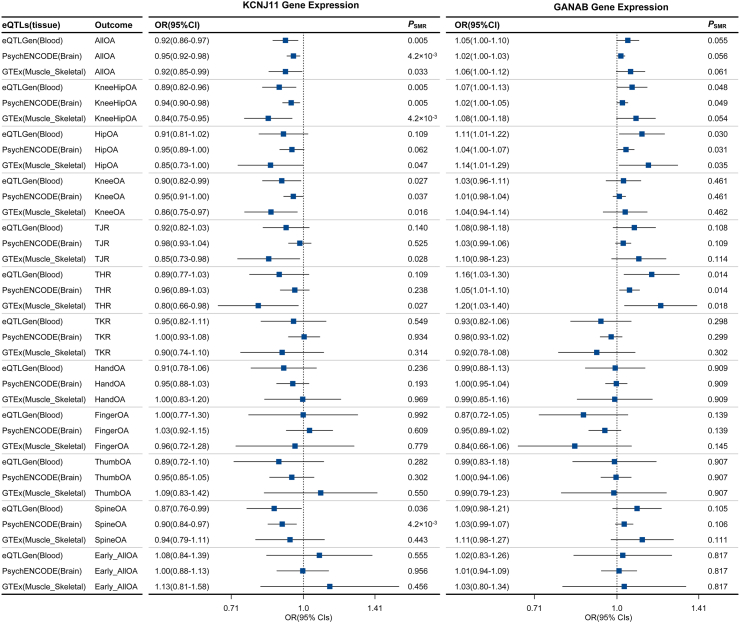
Fig. 4.
Results of the SMR analysis for the associations of KCNJ11 and GANAB gene expression in the blood (sourced from eQTLGen), brain (from PsychENCODE), and skeletal muscle (from GTEx) with 12 osteoarthritis phenotypes. The forest plot visualises the effect sizes, with the vertical dashed line representing an odds ratio (OR) of 1. Values to the left indicate a protective effect, and values to the right suggest a risk effect. The PSMR values indicate the significance levels, with lower values providing stronger evidence against the null hypothesis of no association. A P-value of <4.2 × 10−3 indicates a statistically significant level, adjusted for multiple testing across 12 osteoarthritis phenotypes. All results passed the HEIDI test of the SMR method. For detailed eQTL results, please refer to Table S10. Abbreviations: OA, Osteoarthritis; SMR, summary-based Mendelian randomisation; eQTLs, expression quantitative trait loci; HEIDI, heterogeneity in dependent instruments; OR, odds ratio; CI, confidence interval.
Significantly, KCNJ11, GANAB, ABCA1, and GSTP1 were replicated across at least two gene expression datasets, including eQTLGen (blood), PsychENCODE (brain), and GTEx (skeletal muscle) (Figures S19 and S20). KCNJ11 and GANAB were consistently replicated across all three datasets (Fig. 4). Analysis of antidiabetic drug target gene expression in different tissues (brain and skeletal muscle) revealed that KCNJ11 expression in the brain was inversely associated with osteoarthritis at various sites, including any site osteoarthritis (OR: 0.95, 95% CI: 0.92–0.98, P = 4.2 × 10−3), knee and/or hip osteoarthritis (OR: 0.94, 95% CI: 0.90–0.98, P = 0.005), knee osteoarthritis (OR: 0.95, 95% CI: 0.91–1.00, P = 0.037), and spine osteoarthritis (OR: 0.90, 95% CI: 0.84–0.97, P = 4.2 × 10−3). Similarly, KCNJ11 expression in skeletal muscle was associated with a decreased likelihood of osteoarthritis at any site (OR: 0.92, 95% CI: 0.85–0.99, P = 0.033), knee and/or hip osteoarthritis (OR: 0.84, 95% CI: 0.75–0.95, P = 4.2 × 10−3), knee osteoarthritis (OR: 0.86, 95% CI: 0.75–0.97, P = 0.016), hip osteoarthritis (OR: 0.85, 95% CI: 0.73–1.00, P = 0.047), TJR (OR: 0.85, 95% CI: 0.73–0.98, P = 0.028), and THR (OR: 0.80, 95% CI: 0.66–0.98, P = 0.027) (Fig. 4). Moreover, the effects of GANAB expression in blood on knee and/or hip osteoarthritis, hip osteoarthritis, and THR were also replicated by gene expression in either brain or skeletal muscle (Fig. 4). Furthermore, the findings from the principal SMR analysis were corroborated by the MVMR analysis adjusted for blood glucose (Figure S21), suggesting that the effects of this gene expression on osteoarthritis were independent of blood glucose levels.
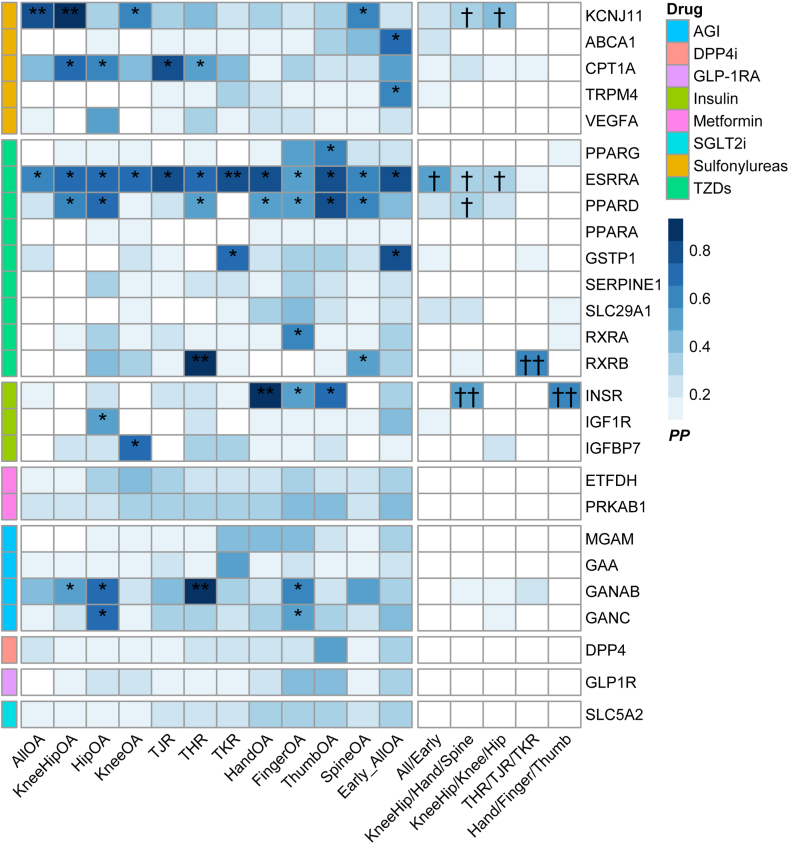
Colocalisation of the putative proteins with osteoarthritis
The colocalisation analysis presented consistent results of these genes with osteoarthritis (Fig. 5, Table S11 and S12). Among them, the KCNJ11, ESRRA, RXRB, INSR, and GANAB were co-localised with at least one osteoarthritis phenotype (PPH4 >0.8) (Fig. 3). Meanwhile, the ABCA1, CPT1A, TRMP4, PPARG, PPARD, GSTP1, RXRA, IGF1R, IGFBP7, and GANC were moderately co-localized with osteoarthritis (PPH4 >0.5). Specifically, KCNJ11 presented a consistent association with osteoarthritis at any site, as well as knee and/or hip osteoarthritis in both SMR and colocalisation analysis. In the multi-trait colocalisation analysis, RXRB and INSR demonstrated moderate colocalisation with at least one trait within specific osteoarthritis phenotype groups (PPA >0.5). Additionally, KCNJ11, ESRRA, and PPARD showed suggestive colocalisation (PPA >0.3), although they did not meet the stricter criteria. The regional association plot displaying results with PPH4 >0.8 is presented in Figure S22.
Fig. 5.
Colocalisation results of antidiabetic drug gene expression and osteoarthritis. The heatmap depicts the two-trait colocalisation analysis of putative targets with 12 osteoarthritis phenotypes (left) and the multiple-trait colocalisation analysis with specific osteoarthritis phenotype groups (right), where darker blue shading and asterisks indicate stronger evidence of colocalisation. ∗∗indicates PPH4 >0.8 in coloc analysis. ∗indicates 0.5 <PPH4 <0.8 in coloc analysis. ††indicates PPA >0.5 in moloc analysis. †indicates PPA >0.3 in moloc analysis. Abbreviations: OA, Osteoarthritis; THR, Total Hip Replacement; TKR, Total Knee Replacement; AGI, alpha glucosidase inhibitors; DPP4i, dipeptidyl peptidase-4 inhibitors; GLP-1RA, glucagon-like peptide-1 receptor agonists; SGLT2i, sodium-glucose cotransporter 2 inhibitors; TZDs, Thiazolidinediones.
Single-cell RNA annotations
The single-cell RNA annotations for tissue cluster specificity revealed that the KCNJ11 gene was expressed predominantly in skeletal muscle and skeletal myocytes (Table S13). Similarly, the ABCC9, PPARG, ESRRA, RXRA, RXRG, and GPD1 genes also exhibited high tissue specificity in skeletal muscle or adipose tissue and demonstrated specific cell type specificity in cells such as skeletal myocytes and adipocytes.
Discussion
In this study, we sought to elucidate the role of antidiabetic drug targets in the risk of osteoarthritis using MR. The significant associations found between certain drug targets and osteoarthritis outcomes suggest that metabolic dysregulation plays a role in osteoarthritis pathogenesis and that antidiabetic medications may have therapeutic potential as DMOADs. The distinct associations between sulfonylurea targets like ABCC8/KCNJ11 and the PPARG target of TZDs suggest potential pathways through which these drugs may influence osteoarthritis progression. Our findings extend beyond the scope of previous research, which predominantly assessed the progression of preexisting osteoarthritis, offering new insights into the potential for antidiabetic drugs to also affect the onset of osteoarthritis.
The causal association of antidiabetic drug targets with osteoarthritis phenotypes, as identified through MR analyses in our study, suggests that sulfonylurea targets (ABCC8/KCNJ11) may increase osteoarthritis risk. Supporting these findings, SMR analysis indicates that lower expression of the KCNJ11 gene in blood and skeletal muscle was associated with increased osteoarthritis risk, in line with the inhibitory effects of sulfonylureas on this gene expression (drug target). This observation is in line with the known pharmacological actions of sulfonylureas, which involve the inhibition of KCNJ11 gene expression (as detailed in Table S2). Sulfonylureas operate primarily by blocking the KCNJ11 (Kir6.2) subunit of ATP-sensitive potassium channels.39 Under normal physiological conditions, this subunit may protect against osteoarthritis. However, this protective mechanism is likely compromised by sulfonylureas, consequently heightening the risk of developing the disease. In our further exploratory analysis, single-cell RNA data indicated that KCNJ11 gene expression was primarily observed in skeletal muscle and myocytes. MicroRNAs (miRNAs) in circulation analyses also revealed that KCNJ11-targeted miRNAs were downregulated in the serum of patients with osteoarthritis.40 To the best of our knowledge, the association between sulfonylureas and osteoarthritis has been minimally explored in observational studies, with the primary focus of this medication being on T2DM. A recent publication reported that genetic variation in the target (KCNJ11) of sulfonylurea was associated with a rheumatoid arthritis risk (OR: 1.25), although this finding did not achieve statistical significance.41
The subunits of ATP-sensitive K+ channels (K(ATP) channels) encoded by KCNJ11 and ABCC8, were expressed by the human chondrocytes at the mRNA and protein levels.42,43 In chondrocytes, K(ATP) channels are believed to be crucial for maintaining cartilage metabolism and modulating intracellular ATP concentrations.44 They may act indirectly as part of the glucose-sensing mechanism in chondrocytes, influencing the cell's capacity to adjust to varying extracellular glucose concentrations by regulating the availability of glucose transporters, affecting the cells' overall glucose transport capacity.42,44 This means K(ATP) channels are important in the regulation of cartilage metabolism and intracellular ATP sensing.
The protective effects observed with PPARG-targeting drugs on hand, finger, and thumb osteoarthritis suggest that modulation of this pathway could benefit osteoarthritis in smaller joints. This finding is particularly interesting as it may indicate a differential impact of antidiabetic drugs on osteoarthritis based on the affected joints. Studies found that PPARG maintains articular cartilage homeostasis, in part, by regulating the mTOR pathway, and epigenetic PPARG suppression plays a key role in osteoarthritis development.45,46 While the SMR analyses did not yield consistent findings for the PPARG gene expression in our study, the exploratory analyses revealed that the expression of the PPARD gene, which is also a target of TZDs, showed a protective effect against osteoarthritis phenotypes. Significantly, the drug repurposing potential highlighted by the GO consortium identified ABCC8, PPARD, and PPARG as druggable targets, albeit they are presently being explored for other clinical indications.24
While the SMR and colocalisation analyses did not verify our findings due to data limitations, our results indicated that metformin and GLP1-RA, targeting GPD1 and GLP1R respectively, were associated with reduced risk of knee and finger osteoarthritis. This aligns with existing research, which consistently supports metformin's beneficial effects on chondroprotection, immunomodulation, and pain reduction in knee osteoarthritis.10 Similarly, current studies suggest that GLP1 analogues could serve as potential DMOADs due to their anti-inflammatory, immunoregulatory, and differentiation-enhancing properties.15 Additionally, SGLT2i (targeted by SLC5A2 and SLC5A1) were negatively associated with osteoarthritis at any site, though this association did not meet the threshold for statistical significance after multiple corrections in this study. Recognised primarily as a new class of glucose-lowering drugs, emerging research has increasingly highlighted the broader roles of these inhibitors beyond blood sugar reduction, including cardiorenal protective effects and benefits related to bone health.47,48
Our study has limitations. The causal inferences made by MR rely on the assumptions that the genetic variants used as instruments are associated with the drug target, that they affect osteoarthritis risk only through that target, and that they are not related to any confounders of the drug–osteoarthritis relationship. While we have taken steps to ensure the robustness of our instruments, potential pleiotropy and other biases inherent to MR analyses cannot be entirely ruled out. Our study's reliance on HbA1c as a biomarker in the MR analyses may still be confounded by red blood cell traits, affecting the accuracy of our causal inferences, though the IVs are selected from the cis-region of each antidiabetic target gene rather than from broader genomic areas that may involve the red blood cell related genes. Most of the IVs and drug targets were obtained from individuals of European ancestry, while our outcome data encompassed 13 international cohorts, as detailed in Table S3. Although European ancestry is predominant, this could introduce some bias into the MR results. This limitation should be considered when interpreting our findings. Since current drug-target MR typically depends on SNPs within the cis-regions of specific genes rather than on GWAS data of different traits, adjusting multiple targets with each other using MVMR is challenging. Despite these issues, we successfully implemented MVMR using IVs in the cis-region, highlighting the need for future developments in statistical analysis methods to address these complexities adequately.49 Furthermore, the observational nature of GWAS data used in MR limits the ability to establish definitive causal relationships. Thus, our findings should be validated by pharmacoepidemiology studies or randomised controlled trials. Moreover, the differential impact on various osteoarthritis phenotypes highlights the need for a nuanced understanding of the mechanisms by which these drugs may exert their effects. Our study also acknowledges the potential selection bias inherent in MR analysis, given that participants are selected based on the presence of both genetically determined genes and outcomes.
Our study observed that sulfonylureas, which target KCNJ11, are associated with an increased risk of osteoarthritis. Conversely, TZDs that target PPARG, have shown a protective effect against osteoarthritis. These findings suggest that the impact of antidiabetic drugs on osteoarthritis risk is more complex than previously thought and extends beyond their glucose-lowering properties. The potential therapeutic strategy emerging from our study involves exploring drugs that counteract the adverse effects on osteoarthritis risk posed by these targets. However, such an approach must be approached cautiously, considering the delicate balance between alleviating osteoarthritis symptoms and not exacerbating or inducing diabetes.
In conclusion, our study supports the repurposing of antidiabetic drugs or target genes for osteoarthritis treatment and underscores the need for a precision medicine approach, considering the heterogeneity of osteoarthritis and the specific actions of different drugs. Future research should focus on targeted clinical trials, investigating drug effects in distinct osteoarthritis phenotypes, and exploring the molecular pathways mediating these effects to fully realise the potential of antidiabetic drugs as DMOADs.
Contributors
KF, SS, DJH and CZ conceptualised and designed the study. SS contributed to the study design and was responsible for the acquisition of data. XJ, YZ, VD, QC, CB, YQZ, YG, CZ, and DJH contributed to the interpretation of data and provided critical revisions of the manuscript for important intellectual content. KF and SS conducted the statistical analyses and drafted the manuscript. All authors have participated in the revision of the draft manuscript, and have approved the final version to be published. KF and SS had directly accessed and verified the underlying data reported in the manuscript.
Data sharing statement
Data related to the identification and validation of antidiabetic drug targets, derived from the DrugBank pharmacogenetics database and various GWAS sources, can be accessed and downloaded from original studies. These include instrumental variables from the MRC IEU OpenGWAS platform and detailed osteoarthritis phenotype data. Restrictions may apply based on ethical approvals and data protection regulations.
Declaration of interests
Professor David J. Hunter provides consulting advice to Merck Serono, Pfizer, Lilly, TLCBio, and Novartis, outside the submitted work.
Acknowledgements
We express our sincere thanks to all participants and researchers who contributed data and summary statistics vital to this analysis. Your invaluable contributions have significantly advanced this research. This study was funded by the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant (2022), the Shanghai Municipal Health Commission Health Industry Clinical Research Project (Grant No. 20224Y0139), Beijing Natural Science Foundation (Grant No. 7244458), and the Postdoctoral Fellowship Program (Grade C) of China Postdoctoral Science Foundation (Grant No. GZC20230130).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105285.
Contributor Information
Changqing Zhang, Email: zhangcq@sjtu.edu.cn.
Youshui Gao, Email: gaoyoushui@sjtu.edu.cn.
Appendix A. Supplementary data
References
- 1.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 2.Mobasheri A., Batt M. An update on the pathophysiology of osteoarthritis. Ann Phys Rehabil Med. 2016;59(5-6):333–339. doi: 10.1016/j.rehab.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Oo W.M., Hunter D.J. Repurposed and investigational disease-modifying drugs in osteoarthritis (DMOADs) Ther Adv Musculoskelet Dis. 2022;14 doi: 10.1177/1759720X221090297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oo W.M., Little C., Duong V., Hunter D.J. The development of disease-modifying therapies for osteoarthritis (DMOADs): the evidence to date. Drug Des Devel Ther. 2021;15:2921–2945. doi: 10.2147/DDDT.S295224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mobasheri A., Rayman M.P., Gualillo O., Sellam J., van der Kraan P., Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2017;13(5):302–311. doi: 10.1038/nrrheum.2017.50. [DOI] [PubMed] [Google Scholar]
- 6.Zhuo Q., Yang W., Chen J., Wang Y. Metabolic syndrome meets osteoarthritis. Nat Rev Rheumatol. 2012;8(12):729–737. doi: 10.1038/nrrheum.2012.135. [DOI] [PubMed] [Google Scholar]
- 7.Richardson S., Neama G., Phillips T., et al. Molecular characterization and partial cDNA cloning of facilitative glucose transporters expressed in human articular chondrocytes; stimulation of 2-deoxyglucose uptake by IGF-I and elevated MMP-2 secretion by glucose deprivation. Osteoarthritis Cartilage. 2003;11(2):92–101. doi: 10.1053/joca.2002.0858. [DOI] [PubMed] [Google Scholar]
- 8.Song P., Hwang J.S., Park H.C., et al. Therapeutic applications of type 2 diabetes mellitus drug metformin in patients with osteoarthritis. Pharmaceuticals (Basel) 2021;14(2):152. doi: 10.3390/ph14020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirinsky I.V., Shirinsky V.S. Effects of medication-treated diabetes on incidence and progression of knee osteoarthritis: a longitudinal analysis of the osteoarthritis Initiative data. Rheumatol Int. 2017;37(6):983–991. doi: 10.1007/s00296-017-3676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim Y.Z., Wang Y., Estee M., et al. Metformin as a potential disease-modifying drug in osteoarthritis: a systematic review of pre-clinical and human studies. Osteoarthritis Cartilage. 2022;30(11):1434–1442. doi: 10.1016/j.joca.2022.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Baker M.C., Sheth K., Liu Y., Lu D., Lu R., Robinson W.H. Development of osteoarthritis in adults with type 2 diabetes treated with metformin vs a sulfonylurea. JAMA Netw Open. 2023;6(3) doi: 10.1001/jamanetworkopen.2023.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Zhang B., Liu W.X., et al. Metformin limits osteoarthritis development and progression through activation of AMPK signalling. Ann Rheum Dis. 2020;79(5):635–645. doi: 10.1136/annrheumdis-2019-216713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Ding X., Terkeltaub R., et al. Exploration of metformin as novel therapy for osteoarthritis: preventing cartilage degeneration and reducing pain behavior. Arthritis Res Ther. 2020;22(1):34. doi: 10.1186/s13075-020-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro M., López de Figueroa P., Blanco F.J., Mendes A.F., Caramés B. Insulin decreases autophagy and leads to cartilage degradation. Osteoarthritis Cartilage. 2016;24(4):731–739. doi: 10.1016/j.joca.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Meurot C., Jacques C., Martin C., et al. Targeting the GLP-1/GLP-1R axis to treat osteoarthritis: a new opportunity? J Orthop Translat. 2022;32:121–129. doi: 10.1016/j.jot.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanderson E., Glymour M.M., Holmes M.V., et al. Mendelian randomization. Nat Rev Methods Primers. 2022;2:6. doi: 10.1038/s43586-021-00092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbatchou J., Barnard L., Backman J., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53(7):1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 18.Yarmolinsky J., Bouras E., Constantinescu A., et al. Genetically proxied glucose-lowering drug target perturbation and risk of cancer: a Mendelian randomisation analysis. Diabetologia. 2023;66(8):1481–1500. doi: 10.1007/s00125-023-05925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N., Sanna S., Wheeler E., et al. Common variants at 10 genomic loci influence hemoglobin A₁(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe L.J., Nivard M.G., Morris T.T., et al. Within-sibship genome-wide association analyses decrease bias in estimates of direct genetic effects. Nat Genet. 2022;54(5):581–592. doi: 10.1038/s41588-022-01062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahajan A., Go M.J., Zhang W., et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott R.A., Lagou V., Welch R.P., et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsworth B., Lyon M., Alexander T., et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 2020 doi: 10.1101/2020.08.10.244293. [DOI] [Google Scholar]
- 24.Boer C.G., Hatzikotoulas K., Southam L., et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184(18):4784–4818.e17. doi: 10.1016/j.cell.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S., Thompson S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S., Thompson S.G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J., Del Greco M.F., Minelli C., Davey Smith G., Sheehan N.A., Thompson J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45(6):1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Võsa U., Claringbould A., Westra H.J., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z., Zhang F., Hu H., et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 34.Foley C.N., Staley J.R., Breen P.G., et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12(1):764. doi: 10.1038/s41467-020-20885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giambartolomei C., Zhenli Liu J., Zhang W., et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34(15):2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandal M.J., Zhang P., Hadjimichael E., et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420) doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlsson M., Zhang C., Méar L., et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7(31) doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letourneau L.R., Greeley S.A.W. Precision medicine: long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations. Curr Diab Rep. 2019;19(8):52. doi: 10.1007/s11892-019-1175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ntoumou E., Tzetis M., Braoudaki M., et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9:127. doi: 10.1186/s13148-017-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin C., Diaz-Gallo L.M., Tang B., et al. Repurposing antidiabetic drugs for rheumatoid arthritis: results from a two-sample Mendelian randomization study. Eur J Epidemiol. 2023;38(7):809–819. doi: 10.1007/s10654-023-01000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mobasheri A., Gent T.C., Nash A.I., Womack M.D., Moskaluk C.A., Barrett-Jolley R. Evidence for functional ATP-sensitive (K(ATP)) potassium channels in human and equine articular chondrocytes. Osteoarthritis Cartilage. 2007;15(1):1–8. doi: 10.1016/j.joca.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Mobasheri A., Lewis R., Ferreira-Mendes A., Rufino A., Dart C., Barrett-Jolley R. Potassium channels in articular chondrocytes. Channels (Austin) 2012;6(6):416–425. doi: 10.4161/chan.22340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rufino A.T., Rosa S.C., Judas F., Mobasheri A., Lopes M.C., Mendes A.F. Expression and function of K(ATP) channels in normal and osteoarthritic human chondrocytes: possible role in glucose sensing. J Cell Biochem. 2013;114(8):1879–1889. doi: 10.1002/jcb.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasheghani F., Zhang Y., Li Y.H., et al. PPARγ deficiency results in severe, accelerated osteoarthritis associated with aberrant mTOR signalling in the articular cartilage. Ann Rheum Dis. 2015;74(3):569–578. doi: 10.1136/annrheumdis-2014-205743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X., Chen F., Lu K., Wei A., Jiang Q., Cao W. PPARγ preservation via promoter demethylation alleviates osteoarthritis in mice. Ann Rheum Dis. 2019;78(10):1420–1429. doi: 10.1136/annrheumdis-2018-214940. [DOI] [PubMed] [Google Scholar]
- 47.Ni L., Yuan C., Chen G., Zhang C., Wu X. SGLT2i: beyond the glucose-lowering effect. Cardiovasc Diabetol. 2020;19(1):98. doi: 10.1186/s12933-020-01071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko H.Y., Bea S., Jeong H.E., et al. Sodium-glucose cotransporter 2 inhibitors vs incretin-based drugs and risk of fractures for type 2 diabetes. JAMA Netw Open. 2023;6(9) doi: 10.1001/jamanetworkopen.2023.35797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes M.V., Richardson T.G., Ference B.A., Davies N.M., Davey Smith G. Integrating genomics with biomarkers and therapeutic targets to invigorate cardiovascular drug development. Nat Rev Cardiol. 2021;18(6):435–453. doi: 10.1038/s41569-020-00493-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.