Abstract
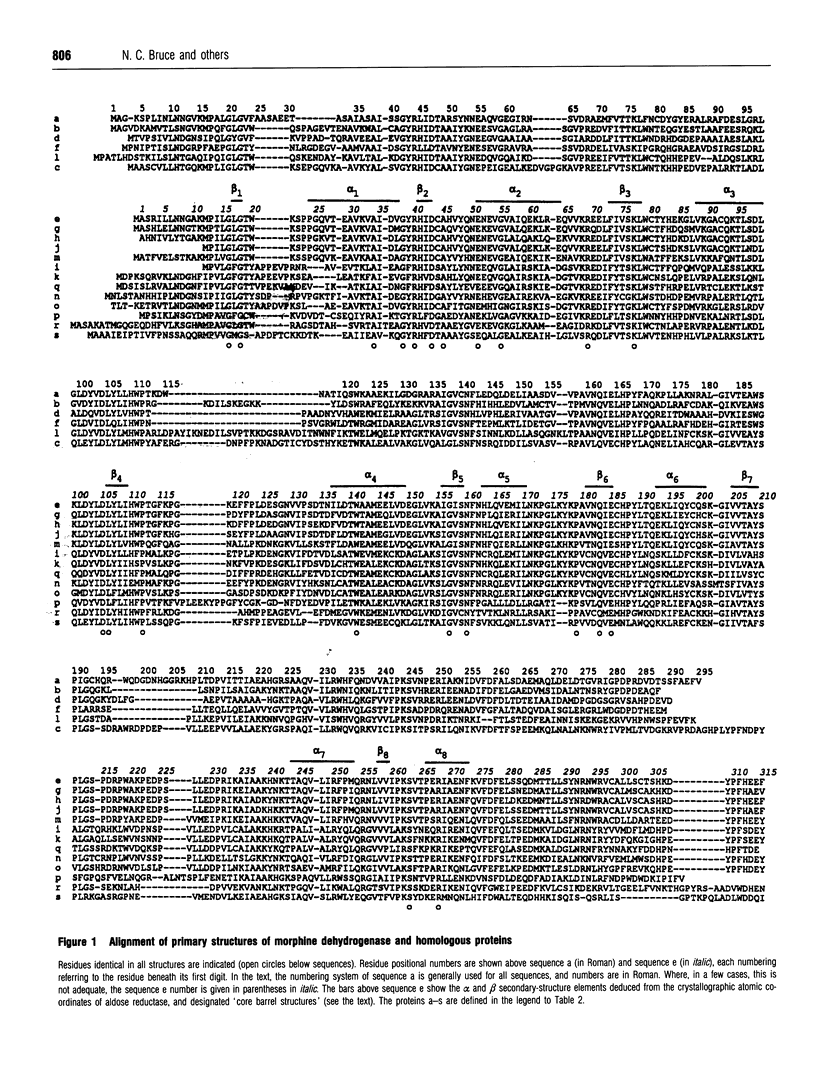
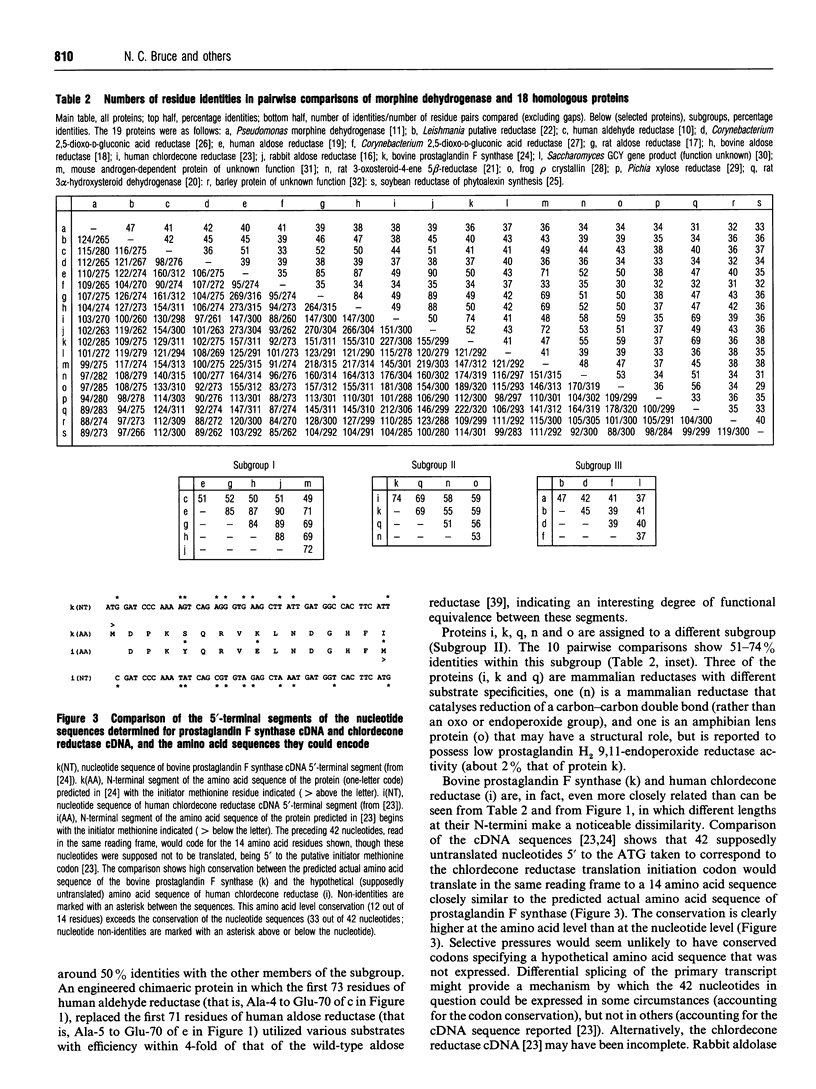
Pseudomonas putida morphine dehydrogenase is shown to be closely homologous to 18 proteins, defining a superfamily within which morphine dehydrogenase particularly resembles two bacterial, 2,5-dioxo-D-gluconic acid reductases, and two eukaryotic proteins of unknown functions. Relationships within the superfamily are extensive and complex. Residue identities between protein pairs range from 29-90%. Three subgroups are proposed. Nevertheless, on the basis of residue conservations/exchanges it is suggested that the nicotinamide coenzyme binding and substrate reduction occur in all the enzymes by broadly analogous mechanisms, among which some probable differences are identified.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Banner D. W., Bloomer A. C., Petsko G. A., Phillips D., Rivers P. S., Wilson I. A. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- Amore R., Kötter P., Küster C., Ciriacy M., Hollenberg C. P. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene. 1991 Dec 20;109(1):89–97. doi: 10.1016/0378-1119(91)90592-y. [DOI] [PubMed] [Google Scholar]
- Anderson S., Marks C. B., Lazarus R., Miller J., Stafford K., Seymour J., Light D., Rastetter W., Estell D. Production of 2-Keto-L-Gulonate, an Intermediate in L-Ascorbate Synthesis, by a Genetically Modified Erwinia herbicola. Science. 1985 Oct 11;230(4722):144–149. doi: 10.1126/science.230.4722.144. [DOI] [PubMed] [Google Scholar]
- Aronson B. D., Somerville R. L., Epperly B. R., Dekker E. E. The primary structure of Escherichia coli L-threonine dehydrogenase. J Biol Chem. 1989 Mar 25;264(9):5226–5232. [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D., Engelhardt K., Roncarati R., Schneider K., Rotter M., Salamini F. An ABA and GA modulated gene expressed in the barley embryo encodes an aldose reductase related protein. EMBO J. 1991 May;10(5):1037–1043. doi: 10.1002/j.1460-2075.1991.tb08042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
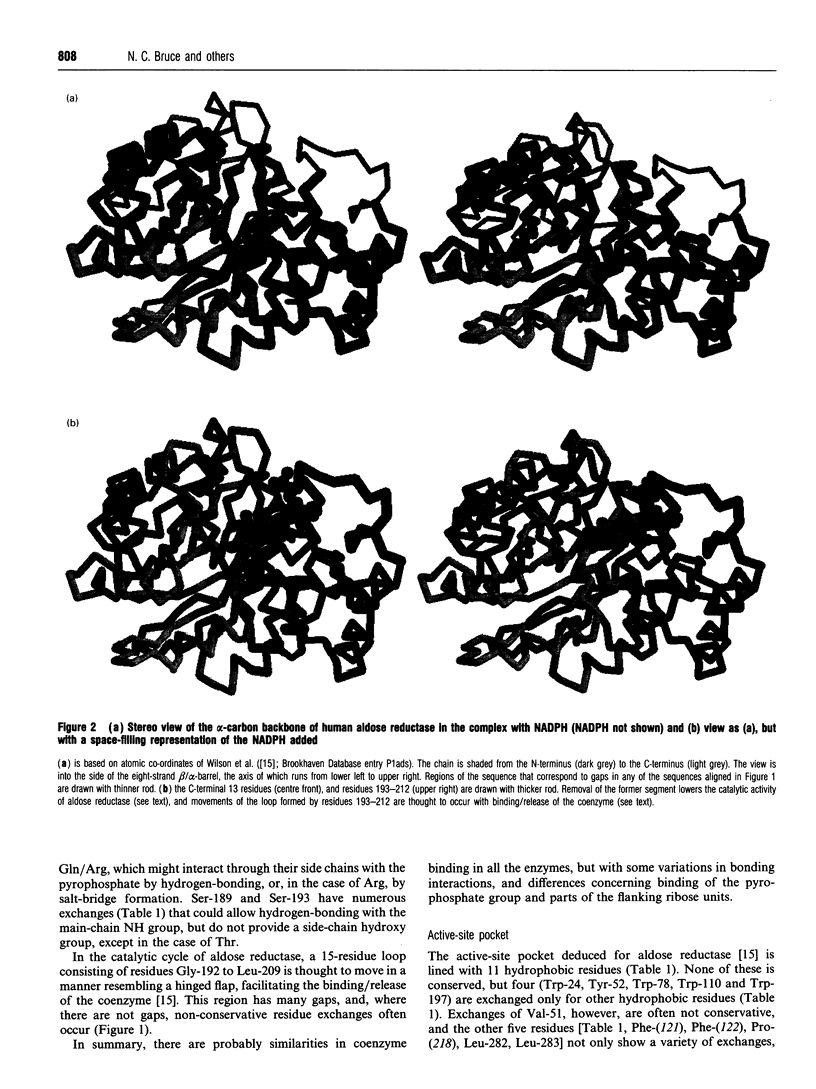
- Bohren K. M., Bullock B., Wermuth B., Gabbay K. H. The aldo-keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. J Biol Chem. 1989 Jun 5;264(16):9547–9551. [PubMed] [Google Scholar]
- Bohren K. M., Grimshaw C. E., Gabbay K. H. Catalytic effectiveness of human aldose reductase. Critical role of C-terminal domain. J Biol Chem. 1992 Oct 15;267(29):20965–20970. [PubMed] [Google Scholar]
- Bohren K. M., Page J. L., Shankar R., Henry S. P., Gabbay K. H. Expression of human aldose and aldehyde reductases. Site-directed mutagenesis of a critical lysine 262. J Biol Chem. 1991 Dec 15;266(35):24031–24037. [PubMed] [Google Scholar]
- Borhani D. W., Harter T. M., Petrash J. M. The crystal structure of the aldose reductase.NADPH binary complex. J Biol Chem. 1992 Dec 5;267(34):24841–24847. doi: 10.2210/pdb1abn/pdb. [DOI] [PubMed] [Google Scholar]
- Cheng K. C., White P. C., Qin K. N. Molecular cloning and expression of rat liver 3 alpha-hydroxysteroid dehydrogenase. Mol Endocrinol. 1991 Jun;5(6):823–828. doi: 10.1210/mend-5-6-823. [DOI] [PubMed] [Google Scholar]
- Chung S., LaMendola J. Cloning and sequence determination of human placental aldose reductase gene. J Biol Chem. 1989 Sep 5;264(25):14775–14777. [PubMed] [Google Scholar]
- Eklund H. Coenzyme binding in alcohol dehydrogenase. Biochem Soc Trans. 1989 Apr;17(2):293–296. doi: 10.1042/bst0170293. [DOI] [PubMed] [Google Scholar]
- Eklund H., Horjales E., Jörnvall H., Brändén C. I., Jeffery J. Molecular aspects of functional differences between alcohol and sorbitol dehydrogenases. Biochemistry. 1985 Dec 31;24(27):8005–8012. doi: 10.1021/bi00348a025. [DOI] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Flynn T. G., Kubiseski T. J. Chemical modification of an arginine residue in aldose reductase is enhanced by coenzyme binding: further evidence for conformational change during the reaction mechanism. Adv Enzyme Regul. 1993;33:197–206. doi: 10.1016/0065-2571(93)90018-9. [DOI] [PubMed] [Google Scholar]
- Fujii Y., Watanabe K., Hayashi H., Urade Y., Kuramitsu S., Kagamiyama H., Hayaishi O. Purification and characterization of rho-crystallin from Japanese common bullfrog lens. J Biol Chem. 1990 Jun 15;265(17):9914–9923. [PubMed] [Google Scholar]
- Garcia-Perez A., Martin B., Murphy H. R., Uchida S., Murer H., Cowley B. D., Jr, Handler J. S., Burg M. B. Molecular cloning of cDNA coding for kidney aldose reductase. Regulation of specific mRNA accumulation by NaCl-mediated osmotic stress. J Biol Chem. 1989 Oct 5;264(28):16815–16821. [PubMed] [Google Scholar]
- Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley J. F., Payton M. A., van de Pol H., Hardy K. G. Conversion of Glucose to 2-Keto-l-Gulonate, an Intermediate in l-Ascorbate Synthesis, by a Recombinant Strain of Erwinia citreus. Appl Environ Microbiol. 1988 Jul;54(7):1770–1775. doi: 10.1128/aem.54.7.1770-1775.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horjales E., Brändén C. I. Docking of cyclohexanol-derivatives into the active site of liver alcohol dehydrogenase. Using computer graphics and energy minimization. J Biol Chem. 1985 Dec 15;260(29):15445–15451. [PubMed] [Google Scholar]
- Jeffery J., Jörnvall H. Sorbitol dehydrogenase. Adv Enzymol Relat Areas Mol Biol. 1988;61:47–106. doi: 10.1002/9780470123072.ch2. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Persson B., Jeffery J. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem. 1987 Sep 1;167(2):195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Persson M., Jeffery J. Alcohol and polyol dehydrogenases are both divided into two protein types, and structural properties cross-relate the different enzyme activities within each type. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4226–4230. doi: 10.1073/pnas.78.7.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Koivusalo M., Baumann M., Uotila L. Evidence for the identity of glutathione-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEBS Lett. 1989 Oct 23;257(1):105–109. doi: 10.1016/0014-5793(89)81797-1. [DOI] [PubMed] [Google Scholar]
- Nishimura C., Wistow G., Carper D. Rat lens aldose reductase: characterization of its primary structure, mRNA, and gene complexity. Prog Clin Biol Res. 1989;290:211–220. [PubMed] [Google Scholar]
- Oechsner U., Magdolen V., Bandlow W. A nuclear yeast gene (GCY) encodes a polypeptide with high homology to a vertebrate eye lens protein. FEBS Lett. 1988 Sep 26;238(1):123–128. doi: 10.1016/0014-5793(88)80240-0. [DOI] [PubMed] [Google Scholar]
- Onishi Y., Noshiro M., Shimosato T., Okuda K. Molecular cloning and sequence analysis of cDNA encoding delta 4-3-ketosteroid 5 beta-reductase of rat liver. FEBS Lett. 1991 Jun 3;283(2):215–218. doi: 10.1016/0014-5793(91)80591-p. [DOI] [PubMed] [Google Scholar]
- Pailhoux E. A., Martinez A., Veyssiere G. M., Jean C. G. Androgen-dependent protein from mouse vas deferens. cDNA cloning and protein homology with the aldo-keto reductase superfamily. J Biol Chem. 1990 Nov 15;265(32):19932–19936. [PubMed] [Google Scholar]
- Pawlowski J. E., Huizinga M., Penning T. M. Cloning and sequencing of the cDNA for rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase. J Biol Chem. 1991 May 15;266(14):8820–8825. [PubMed] [Google Scholar]
- Penning T. M., Abrams W. R., Pawlowski J. E. Affinity labeling of 3 alpha-hydroxysteroid dehydrogenase with 3 alpha-bromoacetoxyandrosterone and 11 alpha-bromoacetoxyprogesterone. Isolation and sequence of active site peptides containing reactive cysteines; sequence confirmation using nucleotide sequence from a cDNA clone. J Biol Chem. 1991 May 15;266(14):8826–8834. [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991 Sep 1;200(2):537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Petrash J. M., Harter T. M., Devine C. S., Olins P. O., Bhatnagar A., Liu S., Srivastava S. K. Involvement of cysteine residues in catalysis and inhibition of human aldose reductase. Site-directed mutagenesis of Cys-80, -298, and -303. J Biol Chem. 1992 Dec 5;267(34):24833–24840. [PubMed] [Google Scholar]
- Samaras N., Spithill T. W. The developmentally regulated P100/11E gene of Leishmania major shows homology to a superfamily of reductase genes. J Biol Chem. 1989 Mar 5;264(7):4251–4254. [PubMed] [Google Scholar]
- Schade S. Z., Early S. L., Williams T. R., Kézdy F. J., Heinrikson R. L., Grimshaw C. E., Doughty C. C. Sequence analysis of bovine lens aldose reductase. J Biol Chem. 1990 Mar 5;265(7):3628–3635. [PubMed] [Google Scholar]
- Stolz A., Rahimi-Kiani M., Ameis D., Chan E., Ronk M., Shively J. E. Molecular structure of rat hepatic 3 alpha-hydroxysteroid dehydrogenase. A member of the oxidoreductase gene family. J Biol Chem. 1991 Aug 15;266(23):15253–15257. [PubMed] [Google Scholar]
- Watanabe K., Fujii Y., Nakayama K., Ohkubo H., Kuramitsu S., Kagamiyama H., Nakanishi S., Hayaishi O. Structural similarity of bovine lung prostaglandin F synthase to lens epsilon-crystallin of the European common frog. Proc Natl Acad Sci U S A. 1988 Jan;85(1):11–15. doi: 10.1073/pnas.85.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle R., Schröder G., Schiltz E., Grisebach H., Schröder J. Induced plant responses to pathogen attack. Analysis and heterologous expression of the key enzyme in the biosynthesis of phytoalexins in soybean (Glycine max L. Merr. cv. Harosoy 63). Eur J Biochem. 1991 Mar 14;196(2):423–430. doi: 10.1111/j.1432-1033.1991.tb15833.x. [DOI] [PubMed] [Google Scholar]
- Willey D. L., Caswell D. A., Lowe C. R., Bruce N. C. Nucleotide sequence and over-expression of morphine dehydrogenase, a plasmid-encoded gene from Pseudomonas putida M10. Biochem J. 1993 Mar 1;290(Pt 2):539–544. doi: 10.1042/bj2900539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanns M., Hyde C. C., Davies D. R., Kirschner K., Jansonius J. N. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991 Sep 24;30(38):9161–9169. doi: 10.1021/bi00102a006. [DOI] [PubMed] [Google Scholar]
- Wilson D. K., Bohren K. M., Gabbay K. H., Quiocho F. A. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992 Jul 3;257(5066):81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- Winters C. J., Molowa D. T., Guzelian P. S. Isolation and characterization of cloned cDNAs encoding human liver chlordecone reductase. Biochemistry. 1990 Jan 30;29(4):1080–1087. doi: 10.1021/bi00456a034. [DOI] [PubMed] [Google Scholar]



