Abstract
During an investigation of fungal diversity from freshwater environments in different regions in Jiangxi Province, China, four interesting species were collected. Morphology coupled with combined gene analysis of an ITS, LSU, SSU, and rpb2 DNA sequence data showed that they belong to the family Pleurotheciaceae. Four new species, Pleurotheciella ganzhouensis, Pla. irregularis, Pla. verrucosa, and Pleurothecium jiangxiense are herein described. Pleurotheciella ganzhouensis is characterized by its capsule-shaped conidia and short conidiophores, while Pla. irregularis has amorphous conidiophores and 3-septate conidia. Pleurotheciella verrucosa has cylindrical or verrucolose conidiogenous cells, 1-septate, narrowly fusiform, meniscus or subclavate conidia. Pleurothecium jiangxiense characterized in having conidiogenous cells with dense cylindrical denticles and short conidiophores. Pleurothecium obovoideum was transferred to Neomonodictys based on phylogenetic evidence. All species are compared with other similar species and comprehensive descriptions, micrographs, and phylogenetic data are provided.
Keywords: diversity, four novel species, morphology, phylogeny, Pleurotheciella, Pleurothecium, taxonomy
Introduction
Freshwater fungi refer to fungi that depend on aquatic environments for their entire or partial life cycle (Shearer, 1993; Calabon et al., 2023). They play a vital role in maintaining the balance of the freshwater ecosystem. Freshwater fungi are involved in the nutrient cycling of the ecosystem, they can decompose plant litter and other carbon sources that are difficult to degrade, such as insect bones, fish scales, and some animal hair (Palmer et al., 1997; Yuen et al., 1998; Vijaykrishna et al., 2005; Cai et al., 2006; Hyde et al., 2016). There have been some studies on lignicolous freshwater fungi in Jiangxi Province, China (Chen et al., 2022; Liu et al., 2022; Peng et al., 2022; Zhai et al., 2022; Yan et al., 2023). However, Pleurotheciaceae Réblová species have not been reported in Jiangxi Province so far.
Pleurotheciaceae was introduced by Réblová et al. (2015), and typified by Pleurothecium. Interestingly, most of Pleurotheciaceae species have been recorded from freshwater habitats, and currently eight genera are accepted, such as Adelosphaeria (Réblová et al., 2015), Anapleurothecium (Hernández-Restrepo et al., 2017), Helicoascotaiwania (Dayarathne et al., 2019), Melanotrigonum (Réblová et al., 2015), Neomonodictys (Hyde et al., 2020), Pleurotheciella (Réblová et al., 2012), Pleurothecium (Höhnel, 1919), and Saprodesmium (Dong et al., 2021). Pleurotheciaceae genera are highly varied, both morphologically and phylogenetically. Some are highly diverse with numerous species (e.g., Pleurotheciella and Pleurothecium). The species of Pleurotheciella and Pleurothecium are similar in having hyaline to brown macronematous conidiophores and denticulate conidiogenous cells (Luo et al., 2018; Hyde et al., 2020; Hyde et al., 2023). In addition, almost all of genera are monotypic and need more collections for their expansion (Luo et al., 2018; Réblová et al., 2020). Currently, there are nearly 60 species in Pleurotheciaceae, most of which are asexual morphs recorded from aquatic habitats (Luo et al., 2018).
Pleurotheciella was introduced by Réblová et al. (2012) to accommodate two species, Pla. rivularia and Pla. centenaria, which have nonstromatic peridium, unitunicate asci, persistent paraphyses and hyaline, and 3-septate ascospores. They have dactylaria-like asexual morph characterized by holoblastic, denticulate conidiogenous cells, subhyaline conidiophores and hyaline, septate conidia. Based on morphology and phylogenetic analyses, Réblová et al. (2015) transferred Dactylaria uniseptata Matsushima to Pleurotheciella as Pla. uniseptata. The species subsequently introduced later in Pleurotheciella are mainly asexual morphs. The morphological characteristics of most species in Pleurotheciella are similar to Pleurothecium in terms of conidiophores and denticulate conidiogenous cells, but they usually have conidia with a single septum (Hyde et al., 2018; Luo et al., 2018; Abdel-Aziz et al., 2020; Réblová et al., 2020; Dong et al., 2021; Shi et al., 2021). In this study, we introduce three new species of Pleurotheciella based on morphological characters and analyses of ITS, LSU, SSU, and rpb2 sequence data which are isolated from aquatic habitats in Jiangxi, China.
Pleurothecium is the type genus of Pleurotheciaceae, with P. recurvatum as the type species (Höhnel, 1919). This genus is characterized by distinct brown to light brown conidiophores, polyblastic sympodially extended denticulate conidiogenous cells, and solitary, 3-septate, hyaline or pigmented or bicolored conidia (Tubaki and Matsushima, 1972; Subramanian and Bhat, 1989; Cooper, 2005; Arzanlou et al., 2007; Yueming and Tianyu, 2009; Réblová et al., 2012; Monteiro et al., 2016; Hyde et al., 2017; Luo et al., 2018; Shi et al., 2021; Fryar and Catcheside, 2023; Jayawardena et al., 2023; Hyde et al., 2023). Currently, 16 species are accepted in this genus. In this study, we introduce a new Pleurothecium species based on morphological characters and analyses of ITS, LSU, SSU, and rpb2 sequence data, which is isolated from aqua tic habitats, in Jiangxi province, China.
In this study, we introduce four new species collected from Jiangxi province, China. Detailed descriptions and illustrations of morphological characteristics are provided for the new taxa.
Materials and methods
Samples collection, morphological observation, and isolation
Submerged decaying wood were collected from streams and rivers in Jiangxi province, China, and brought back to the laboratory in sealed plastic bags. The samples were incubated at room temperature (25°C) for 2 weeks in plastic boxes, spraying sterile water for moisturizing during the incubation. The samples were viewed under a Nikon SMZ-1270 microscope (Nikon Corporation, Japan) to observe fungi. Micro-morphological characteristics were observed and captured using a Nikon ECLIPSE Ni-U compound microscope (Nikon Corporation, Japan), equipped with a Nikon DS-Fi3 camera. All measurements were calculated using PhotoRuler Ver. 1.1 software.1 Figures were processed using Adobe Photoshop CS6 Extended version 10.0 software (Adobe Systems, United States) (Zhai et al., 2022).
Pure cultures of fungi were obtained by single spore isolation method described by Chomnunti et al. (2014). The germinated conidia were individually transferred to potato dextrose agar (PDA) and incubated at 25°C for 2 weeks. The fungal cultures were deposited in the Jiangxi Agricultural University Culture Collection (JAUCC) and the herbarium specimens were deposited in the Herbarium of Fungi Jiangxi Agricultural University (HFJAU).
DNA extraction, PCR amplification, and sequencing
DNA was extracted from fresh mycelium on PDA using a modified cetyltrimethy lammonium bromide (CTAB) method (Doyle and Doyle, 1987). Four deoxyribonucleic acid (DNA) barcodes, ITS, LSU, SSU, and rpb2, were selected for polymerase chain reaction (PCR) using the primer pairs ITS1/ITS4 (White et al., 1990), LR0R/LR5 (Hopple and Vilgalys, 1999), NS1/NS4 (White et al., 1990), and fRPB2-5f/fRPB2-7cR (Liu et al., 1999), respectively. Amplification reactions were carried out in a volume of 25 μL, containing 12.5 μL 2 × Taq PCR MasterMix (Qingke, Changsha, China), 1 μL each forward and reverse primer (0.2 μM), 1 μL template DNA (circa 50–100 ng), and 9.5 μL ddH2O. Amplifications (ITS, LSU, and SSU) were conducted under the following conditions: 3 min at 98°C, 35 cycles of 10 s at 98°C, 10 s of annealing at 55°C and extension at 72°C for 10 s, with a final 2-min extension at 72°C. Regions of rpb2 were amplified with initial denaturation of 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 54°C for 90 s, elongation at 72°C for 90 s and the final extension at 72°C for 10 min included for each condition of amplification (Luo et al., 2018). Sequencing reactions were conducted with the corresponding forward and reverse primers commercially by QingKe Biotechnology Co. (Changsha, China). All sequences were edited with Sequencher v.4.14 (GeneCodes Corporation, United States) and have been deposited in the NCBI GenBank database (Table 1).
Table 1.
Sequences used in this study.
Phylogenetic analyses
The combined dataset consists of 39 taxa including our newly generated taxa. Dematipyriforma aquilariae was used as the out-group taxon. Taxa with the highest similarities to our strains were determined with standard nucleotide BLASTn searches in GenBank.2 The other sequences used in the analyses were obtained from the recent publications (Luo et al., 2018; Réblová et al., 2020). Detailed information on fungal strains used in this paper is provided in Table 1.
All obtained sequences were aligned using the online service of MAFFT (Madeira et al., 2019) and refined manually in MEGA v.7.0 (Kumar et al., 2016). Maximum likelihood (ML) analysis was conducted using RAxML 8.0 with GTR-GAMMA model of evolution (Stamatakis, 2014). Non-parametric bootstrap analysis was implemented using 1,000 replicates to estimate ML bootstrap (BS) values. Bayesian Inference analysis was carried out with PhyloSuite_v1.2.2_Win under partitioned models (Ronquist et al., 2012). The best-fit models of nucleotide substitutions were selected according to the Bayesian Information Criterion (BIC) implemented in ModelFinder on PhyloSuite_v1.2.2_Win. The models for ITS(1–599), LSU(600–1,458), SSU(1,459–2,432), and rpb2(2,433–3,322) datasets used for phylogenetic analysis are GTR + F + I + G4 model, SYM + I + G4 model, SYM + I + G4 model, and GTR + F + I + G4 model, respectively. The datasets were run for 10,000,000 generations, with four chains and trees sampled every 1,000th generation. The first 10% trees were discarded as burn-in. The Bayesian consensus tree with posterior probabilities (PP) was visualized with FigTree v.1.4.4 (Rambaut, 2018) and was edited in Adobe Illustrator CS6.
Results
Phylogenetic results
According to the BLAST results, the ITS sequence of Pleurothecium jiangxiense shares 90.3% similarity to Pleurothecium brunius with 51 nucleotide differences (18 gaps). The ITS of Pla. verrucosa shares 93.81% similarity to Pla. Krabiensis with 33 nucleotide differences (four gaps). The ITS sequence of Pla. ganzhouensis shares 95% similarity (25 nucleotide differences, of which 18 are gaps) with that of Pla. rivularia. In addition, Pla. irregularis shares 98.8% similarity (six nucleotide differences) to Pla. centenaria.
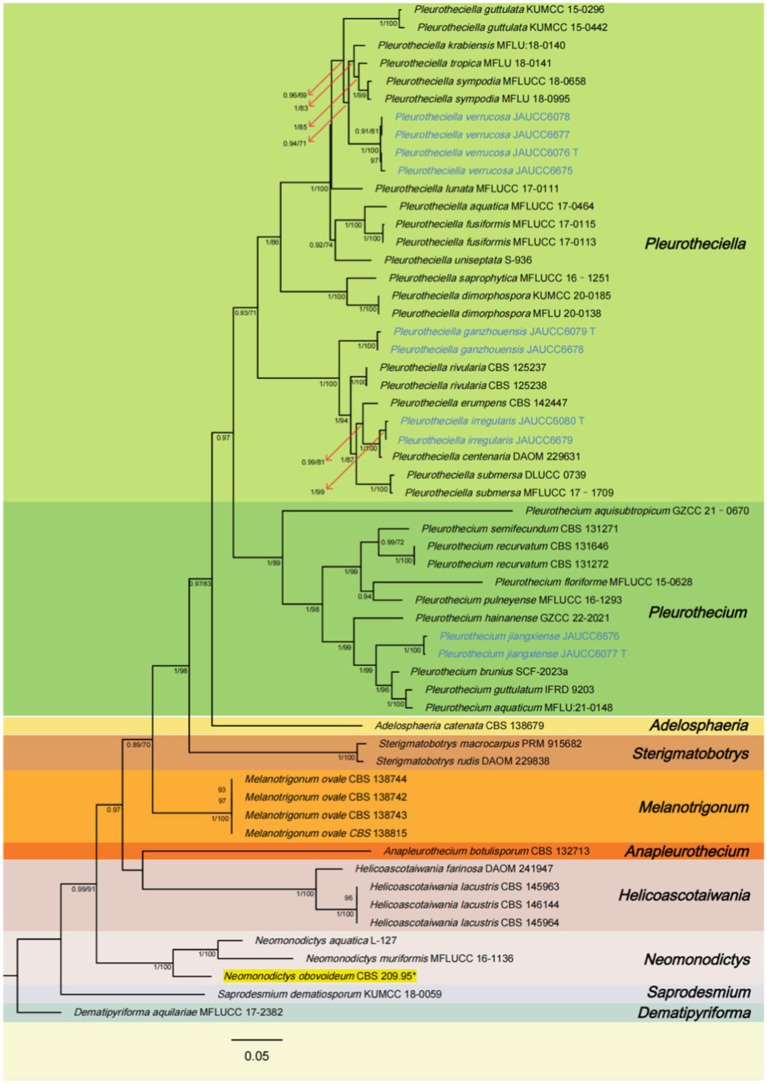
The aligned matrix for the combined analysis, ITS + LSU + SSU + rpb2, comprised 3,322 bp, including 599 bp of ITS, 859 bp of LSU, 974 bp of SSU, and 890 bp of rpb2. The combined ITS, LSU, SSU, and rpb2 dataset consisted of 57 sequences representing 36 species of the Pleurotheciaceae, two sequences representing two species of Sterigmatobotrys, and one sequence representing one species of Dematipyriforma. The topologies of trees generated by ML and BI analyses are congruent. The Bayesian tree with BS and PP is shown in Figure 1.
Figure 1.
Phylogenetic tree of Pleurotheciaceae, inferred from the combined regions (ITS-LSU-SSU-rpb2) using Bayesian Inference (BI) analysis. The Dematipyriforma aquilariae was used as the outgroup. The lineages with new species were shown in blue font. The lineages with adjusted species were shown in yellow background. PP ≥ 0.90 and BS ≥ 70% were indicated around the branches. Supported clade (PP/BS = 1.00/99) with the lineage consisting of Pleurothecium brunius, Pleurothecium aquaticum, and Pleurothecium guttulatum. Species of Saprodesmium and Neomonodictys have longer genetic distance from other species of Pleurotheciaceae. Sterigmatobotrys groups together with species of Pleurotheciaceae with a strong statistical support.
All species of Pleurotheciaceae form a monophyletic group. Pleurotheciella verrucosa groups together with Pla. krabiensis, Pla. tropica, and Pla. sympodia (PP/BS = 0.94/71), which four collections are from two different regional freshwater habitats with a strong-supported clade (BS/PP = 100/1.00). The strains of Pleurotheciella ganzhouensis form a strong-supported clade (PP/BS = 1.00/100), group together with Pla. rivularia, Pla. erumpens, Pla. irregularis, Pla. centenaria, and Pla. submersa. Collections of Pleurotheciella irregularis form a strong-supported clade (PP/BS = 1.00/99) with Pla. centenaria. Collections of Pleurothecium jiangxiense form a strongly supported clade (PP/BS = 1.00/99) with the lineage consisting of P. brunius, P. aquaticum, and P. guttulatum. Species of Saprodesmium and Neomonodictys have longer genetic distance from other species of Pleurotheciaceae. Sterigmatobotrys groups together with species of Pleurotheciaceae with a strong statistical support.
Taxonomy
Pleurotheciella ganzhouensis W.M. He, D.M. Hu & H.Y. Song, sp. nov.
MycoBank number: MB853181
Typification: Qing Tang Zhen 515 Xiang Dao Xie Cun, Ningdu County, Ganzhou City, Jiangxi Province, China (江西省赣州市宁都县青塘镇515乡道谢村). Longitude: E115.562124° Latitude: N26.404602°, on 30 Jan 2023, W.M. He, (HFJAU 10280, holotype), ex-type culture JAUCC6079. Additional specimen examined: Qing Tang Zhen 515 Xiang Dao Xie Cun, Ningdu County, Ganzhou City, Jiangxi Province, China (江西省赣州市宁都县青塘镇515乡道谢村). Longitude: E115.562124° Latitude: N26.404602°, on 5 Jun 2023, W.M. He, (HFJAU 10413, paratype), ex-paratype culture JAUCC6678. GenBank number: JAUCC6079: ITS = OR853417, LSU = OR853422, SSU = OR853426, rpb2 = PPO78759. JAUCC6678: ITS = PP180192, LSU = PP800214, SSU = PP801261, rpb2 = PP816289.
Etymology: Referring to the collecting place, Ganzhou city, Jiangxi Province, China.
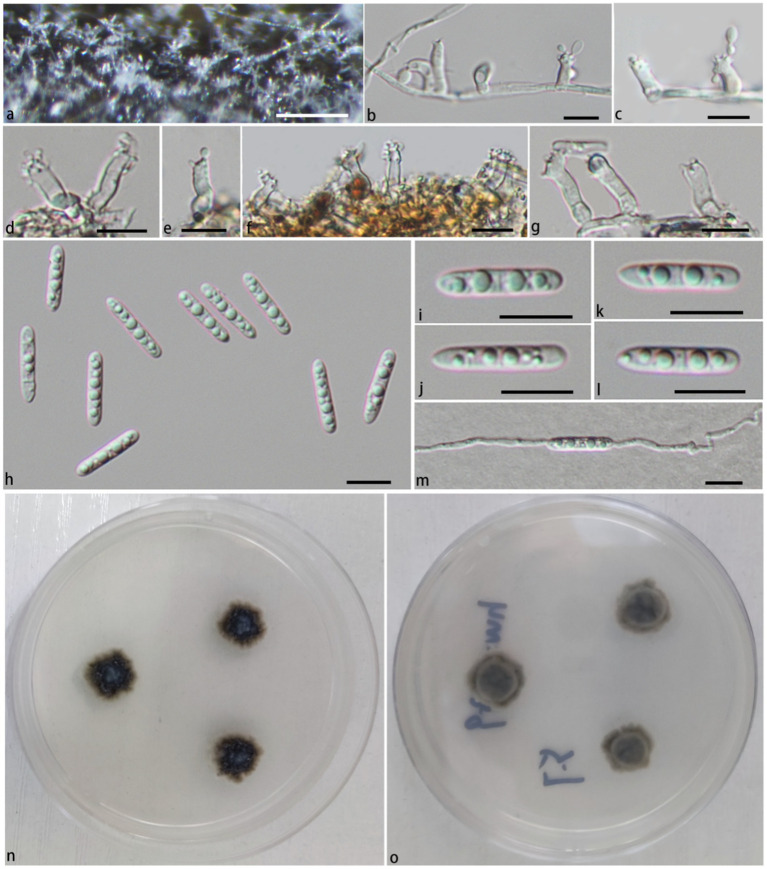
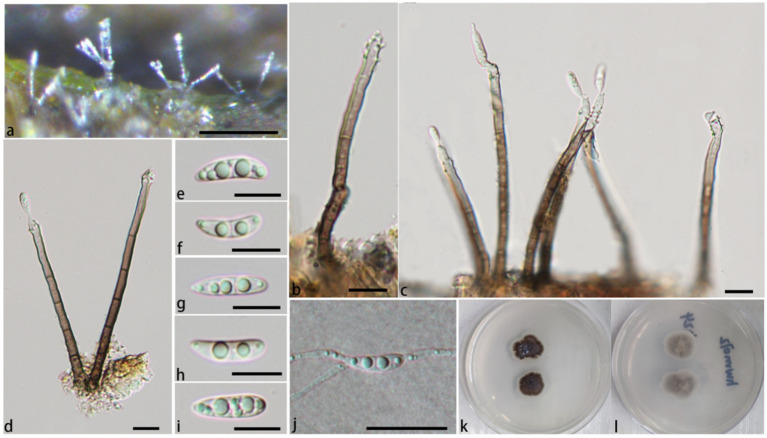
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies grow on the wood surface, white, reflective, clustered, upper part covered with bright white mass of conidia, outward radially distributed. Mycelium immersed, partly superficial, relatively sparse, composed with unbranched, hyaline and 1.4–3.2 μm wide hyphae, with few conidiophores on the superficial part. Conidiophores 9.9–41.9 μm (−x = 16.4 μm, n = 30) long, 2.2–3.7 μm (−x = 2.9 μm, n = 30) wide, macronematous, mononematous, 1-septate or aseptate, smooth, cylindrical, hyaline, erect or slightly curved, the top slightly swollen with denticulate conidiogenous sites. Denticulate. Conidiogenous cells integrated, terminal, polyblastic, cylindrical or verrucous, hyaline, forming conidia sympodially on cylindrical denticles. Conidia 14.4–19.4 (−x = 17.1, n = 30) × 2.5–3.3 (−x = 2.9, n = 30) μm, capsule-shaped, fusiform, cylindrical or subclavate, hyaline, guttulate, straight or slightly arcuate, 1-septate, round and tapering at both ends, one end is usually sharper, smooth-walled.
In the phylogenetic tree (Figure 1), Pla. ganzhouensis formed an independent lineage sister to a clade containing Pla. rivularia, Pla. erumpens, Pla. centenaria, Pla. amorphous, and Pla. submersa (PP/BS = 1.00/100). Morphologically, compared with other species of Pleurotheciella, Pla. ganzhouensis have unique capsule-shaped conidia and shorter conidiophores (9.9–41.9 × 2.2–3.7 μm), and with few hyaline conidiophores on superficial part of mycelium, which can be clearly distinguished from other species of Pleurotheciella (Hyde et al., 2018; Luo et al., 2018; Abdel-Aziz et al., 2020; Réblová et al., 2020; Dong et al., 2021; Shi et al., 2021). In addition, combined with its short conidiophores, it is similar to the species of Neta, but its 1-septate, slender and capsule-shaped conidia can be clearly distinguished from Neta (Figure 2).
Figure 2.
Pleurotheciella ganzhouensis (HFJAU 10280, holotype). (A) Colonies on submerged wood. (B–G) Conidiophores and conidiogenous cells. (H–L) Conidia. (M) A germinating conidium. (N,O) Cultures on PDA medium (N from above, O from below). Scale bars: (A)= 100 μm. (B–M) = 10 μm.
Pleurotheciella irregularis W.M. He, D.M. Hu & H.Y. Song, sp. nov.
MycoBank number: MB853182
Material examined: Shenlong Tan, Taiping town, Xinjian District, Nanchang City, Jiangxi Province, China (江西省南昌市新建区太平镇神龙潭). Longitude: 115.697°E Latitude: 28.766°N, on 19 Mar 2023, W.M. He, (HFJAU 10281, holotype), ex-type culture JAUCC6080. Additional specimen examined: Shenlong Tan, Taiping town, Xinjian District, Nanchang City, Jiangxi Province, China (江西省南昌市新建区太平镇神龙潭). Longitude: 115.697°E Latitude: 28.766°N, on 7 Oct 2023, W.M. He, (HFJAU 10414, paratype), ex-paratype culture JAUCC6679. GenBank number: JAUCC6080: ITS = OR853418, LSU = OR853423, SSU = PP801258, rpb2 = PP816286. JAUCC6679: ITS = PP180193, SSU = PP801262.
Etymology: Refers to the irregular shape of conidiophores.
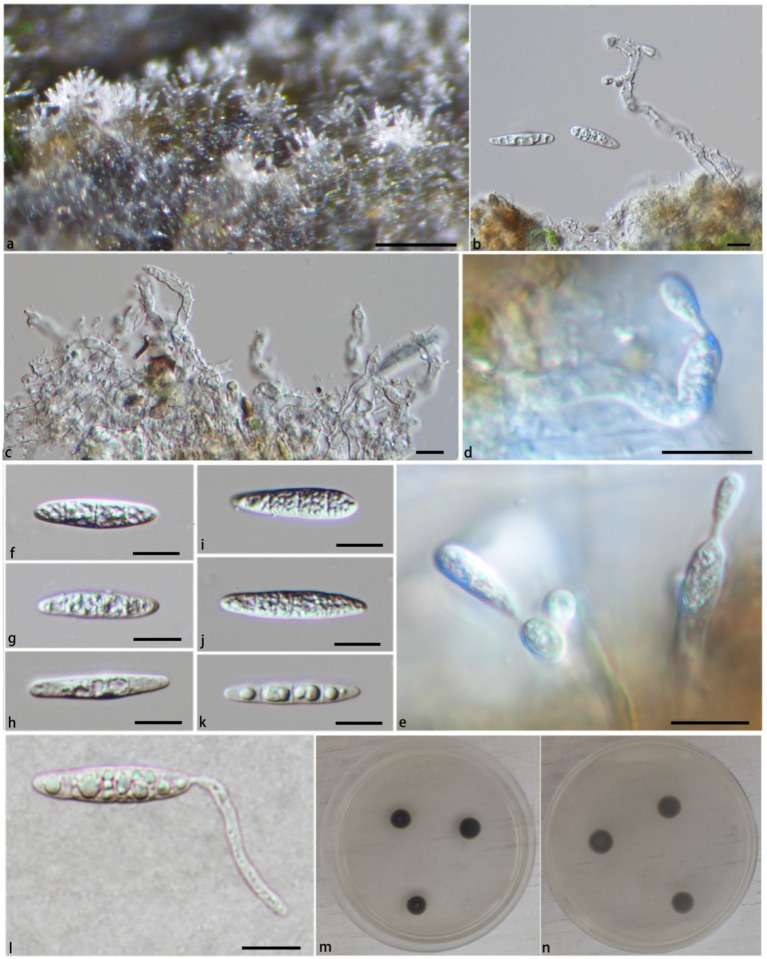
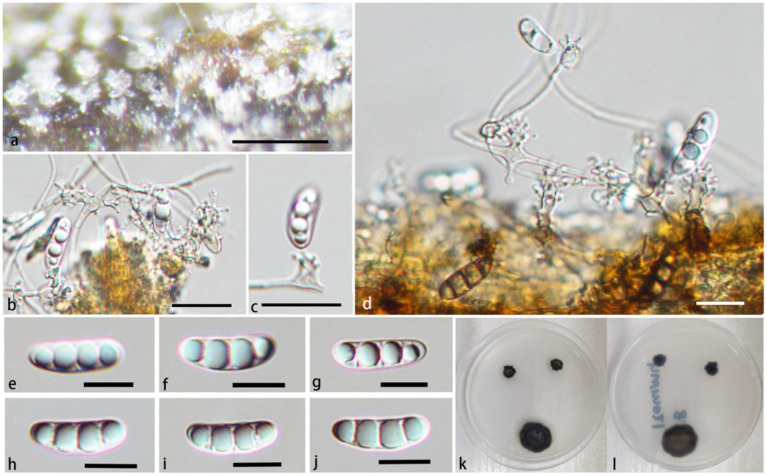
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies grow on the submerged wood surface, white, reflective, clustered, radiant growth from the bottom, upper part covered with bright white mass of conidia. Mycelium immersed, relatively dense, brown, composed with 2.3–3.5 μm wide, unbranched hyphae. Conidiophores 50–110 μm long, 2.6–3.5 μm wide, macronematous, mononematous, mostly curved, hyaline, aseptate, verrucous, irregular, cylindrical mostly tapering toward the apex, some with a terminal node of denticles, with verruca in the middle and upper part. Conidiogenous cells integrated, terminal, polyblastic, cylindrical or verrucous, hyaline forming conidia sympodially on cylindrical denticles. Conidia 24.2–33.9 μm (−x = 29.4 μm, n = 30) × 4.2–6.4 μm (−x = 5.4 μm, n = 30), narrowly fusiform, subclavate, hyaline, guttulate, straight or slightly arcuate, 1-3-septate, slightly constricted at the septum, pointed at one end the other round an wider in the middle, smooth-walled.
Notes: In the phylogenetic tree (Figure 1), Pla. irregularis form a strongly supported clade (PP/BS = 1.00/100) with the strains of Pla. centenaria. Morphologically, Pla. irregularis is unique in its amorphous conidiophores. In addition, our collection can be distinguished from Pla. centenaria by larger conidia (24.2–33.9 × 4.2–6.4 μm vs. 18.0–22.5 × 4.0–5.5 μm) and longer conidiophores (50–110 × 2.6–3.5 μm vs. 12.0–35.0 × 3.0–4.5 μm) (Réblová et al., 2012) (Figure 3).
Figure 3.
Pleurotheciella irregularis (HFJAU 10281, holotype). (A) Colonies on submerged wood. (B,C) Conidiophores and conidia. (D,E) Conidiophores and conidiogenous cells bearing conidia. (F–K) Conidia. (L) A germinating conidium (M from above, N from below). (M,N) Culture on PDA. Scale bars: (A) = 100 μm; (B–L) = 10 μm.
Pleurotheciella verrucosa W.M. He, D.M. Hu & H.Y. Song, sp. nov.
MycoBank number: MB853180
Typification: Tang long, 824 County highway, Wanan County, Jian City, Jiangxi Province (江西省吉安市万安县824县道塘咙). Longitude: E115.029429° Latitude: N26.224633°, on 30 Jun 2022, W.M. He (HFJAU 10277, holotype), ex-type culture JAUCC6076, ex-paratype culture JAUCC6675. Additional specimen examined: Tang long, 824 County highway, Wanan County, Jian City, Jiangxi Province (江西省吉安市万安县824县道塘咙). Longitude: E115.029429° Latitude: N26.224633°, on 3 October 2022, W.M. He (HFJAU 10410, paratype), ex-paratype culture JAUCC6675. Qing Tang Zhen 515 Xiang Dao Xie Cun, Ningdu County, Ganzhou City, Jiangxi Province, China (江西省赣州市宁都县青塘镇515乡道谢村). Longitude: E115.562124° Latitude: N26.404602°, on 30 Jan 2023, W.M. He, (HFJAU 10279, paratype), ex-paratype culture JAUCC6078. Qing Tang Zhen 515 Xiang Dao Xie Cun, Ningdu County, Ganzhou City, Jiangxi Province, China (江西省赣州市宁都县青塘镇515乡道谢村). Longitude: E115.562124° Latitude: N26.404602°, on 5 Jun 2023, W.M. He, (HFJAU 10412, paratype), ex-paratype culture JAUCC6677. GenBank number: JAUCC6076: ITS = OR853414, LSU = OR853419, SSU = OR853424, rpb2 = PPO78756. JAUCC6675: ITS = PP800189, LSU = PP800211, SSU = PP801259, rpb2 = PP816287. JAUCC6078: ITS = OR853416, LSU = OR853421, SSU = PP801257, rpb2 = PPO78758. JAUCC6677: ITS = PP800191, LSU = PP800213.
Etymology: Referring to apical conidiophores with verrucose conidiogenous warts.
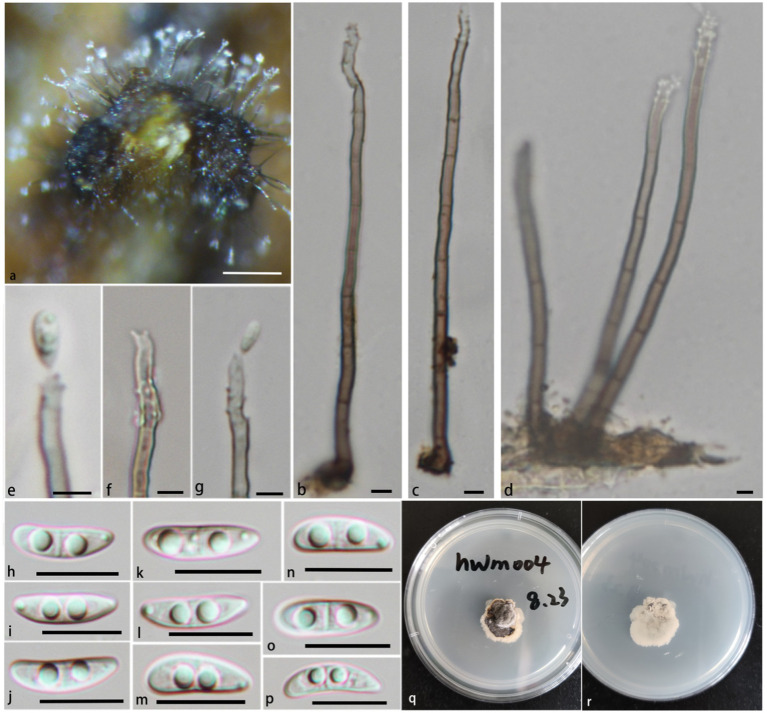
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies grow on the surface of wood, light brown, reflective, clustered, most upright, upper part covered with bright white mass of conidia. Mycelium 1.2–2.2 μm wide, immersed, relatively sparse, light brown, with few or no branches. Conidiophores 51.3–131.8 μm (−x = 89.4 μm, n = 60) long, 1.9–3.4 μm (−x = 2.62 μm, n = 60) wide, macronematous, mononematous, septate, smooth, cylindrical, dark brown at the base, becoming paler toward the apex, erect or slightly curved, bottom slightly swollen, Conidiogenous cells integrated, terminal, polyblastic, cylindrical or verrucolose, pale brown to hyaline, forming conidia sympodially on cylindrical denticles or wart. Conidia 10.2–16.9 μm (−x = 14.1 μm, n = 60) × 2.3–4.3 μm (−x = 3.4 μm, n = 60), narrowly fusiform, meniscus or subclavate, hyaline, guttulate, straight or arcuate, uniseptate, pointed at one end, the other round and wide in the middle smooth-walled.
Notes: In the phylogenetic tree (Figure 1), Pla. verrucosa groups together with a clade containing Pla. krabiensis, Pla. tropica, and Pla. sympodia with moderate statistical support (PP/BS = 0.94/71). Morphologically, Pla. verrucosa is easily distinguished from Pla. krabiensis by its shorter and finer conidiophores (51.3–131.8 × 1.9–3.4 μm vs. 240–390 × 3.3–4.8 μm) and smaller conidia (10.2–16.9 × 2.3–4.3 μm vs. 19–25 × 4.5–6 μm) (Hyde et al., 2018). Pleurotheciella tropica differs from Pla. verrucosa in having longer and wider conidiophores (100–250 × 4–4.8 μm vs. 51.3–131.8 × 1.9–3.4 μm) and bigger conidia (16–21 × 5.5–7 μm vs. 10.2–16.9 × 2.3–4.3 μm) (Hyde et al., 2018). Pleurotheciella sympodia differs from Pleurotheciella verrucosa in having longer conidiophores (135–355 × 1.5–3.5 μm vs. 51.3–131.8 × 1.9–3.4 μm) and larger conidia (22.5–29 × 4.5–6.5 μm vs. 10.2–16.9 × 2.3–4.3 μm) (Shi et al., 2021) (Figures 4, 5).
Figure 4.
Pleurotheciella verrucosa (HFJAU 10277, holotype). (A) Colonies on submerged wood. (B–G) Conidia and conidiogenous cells. (H–P) Conidia. (Q,R) Cultures on PDA medium (Q from above, R from below). Scale bars: (A,B) = 100 μm. (B–G) = 5 μm. (H–P) = 10 μm.
Figure 5.
Pleurotheciella verrucosa (HFJAU 10279). (A) Colonies on submerged wood. (B–D) Conidiophores and conidiogenous cells. (E–I) Conidia. (J) A germinating conidium. (K,L) Cultures on PDA medium (K from above, L from below). Scale bars: (A) = 100 μm. (B–J) = 10 μm.
Pleurothecium jiangxiense W.M. He, D.M. Hu & H.Y. Song, sp. nov.
MycoBank number: MB853179
Typification: Shunfeng village, Wanan country, Jian city, Jiangxi province, China (江西省吉安市万安县顺峰乡). Longitude: E115.015900° Latitude: N26.198737°, on 30 Jun 2022, W.M. He, (HFJAU 10278, holotype), ex-type culture JAUCC6077. Additional specimen examined: Shunfeng village, Wanan country, Jian city, Jiangxi province, China (江西省吉安市万安县顺峰乡). Longitude: E115.015900° Latitude: N26.198737°, on 3 Oct 2022, W.M. He, (HFJAU 10411, paratype), ex-paratype culture JAUCC6676. GenBank number: JAUCC6077: ITS = OR853415, LSU = OR853420, SSU = OR853425, rpb2 = PPO78757. JAUCC6676: ITS = PP800190, LSU = PP800212, SSU = PP801260, rpb2 = PP816288.
Etymology: Referring to the collecting place, Jiangxi Province, China.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies on the substratum superficial, effuse, solitary, shiny, white to hyaline. Mycelium 1–1.5 μm wide, composed of partly immersed, partly superficial, light brown to hyaline, septate, unbranched hyphae, superficial part differentiating into conidiogenous cells, with some conidia. Conidiophores 6.2–17.8 μm (−x = 11.8 μm, n = 15) long, 2.2–5.3 μm (−x = 3.4 μm, n = 15) wide, mononematous, cylindrical, unbranched, aseptate, straight or slightly curved, light brown to hyaline, rough-walled. Conidiogenous cells 2.6–8.4 μm long, 2.6–4 μm diam, integrated, holoblastic, polyblastic, ellipsoidal or cylindrical, hyaline to light brown, distributed at the middle and apex of the conidiophores, conidiogenous loci denticulate, denticles discrete, determinate, 1.8–4.2 × 0.4–0.6 μm. Conidia 15.1–20.5 μm (−x = 17.5 μm, n = 30) × 4.3–6.2 μm (−x = 5.3 μm, n = 30), acrogenous, holoblastic, grow on denticles, hyaline, 3-septate, guttulate, slightly curved, subclavate, cylindrical, ellipsoidal, rounded at both ends, conidial secession schizolytic, smooth-walled.
Notes: In the phylogenetic analysis (Figure 1), P. jiangxiense clades with P. aquaticum, P. brunius, and P. guttulatum with strong statistical support (PP/BS = 1.00/99), then clades with P. hainanense with strong statistical support (PP/BS = 1.00/99).
Morphologically, the conidiophores of P. jiangxiense are significantly short (6.2–17.8 × 2.2–5.3 μm), which are significantly different from other species of the genus. P. jiangxiense have obvious superficial hyphae between conidiophores, which can differentiate into conidiogenous cells and with some conidia, this phenomenon has not been observed in other species of the genus (Tubaki and Matsushima, 1972; Subramanian and Bhat, 1989; Cooper, 2005; Arzanlou et al., 2007; Yueming and Tianyu, 2009; Réblová et al., 2012; Monteiro et al., 2016; Hyde et al., 2017; Luo et al., 2018; Shi et al., 2021; Fryar and Catcheside, 2023; Jayawardena et al., 2023; Hyde et al., 2023). Compare with P. brunius, P. aquaticum and P. guttulatum, conidia of P. jiangxiense have more rounded bottom, and not constricted at the septa. The size of the conidia of P. jiangxiense is shorter than P. aquaticum (15.1–20.5 × 4.3–6.2 μm vs. 19–21 × 4.5–5.5 μm), P. guttulatum (15.1–20.5 × 4.3–6.2 μm vs. 22–28 × 5–6 μm), and P. yunnanensis (15.1–20.5 × 4.3–6.2 μm vs. 17–25.6 × 2.8–9 μm), almost similar to P. brunius (15.1–20.5 × 4.3–6.2 μm vs. 16–19 × 5–6 μm) (Luo et al., 2018; Shi et al., 2021; Fryar and Catcheside, 2023; Chun-Sheng et al., 2023).
In addition, P. jiangxiense have similar conidiophores and conidiogenous cells to genus Neta, but its denticles are denser than Neta. Pleurothecium jiangxiense have 3-septate conidia, while species of Neta often have 1-septate conidia (Figure 6).
Figure 6.
Pleurothecium jiangxiense (HFJAU 10278, holotype). (A) Colonies on submerged wood. (B–D) Conidiophores and conidiogenous cells. (E–J) Conidia. (K,L) Cultures on PDA medium (K from above; L from below). Scale bars: (A) = 100 μm. (B–J) = 12.5 μm.
Discussion
Pleurotheciaceae members are mainly distributed in the tropics and subtropics, with a few in central and southern Europe, such as P. semifecundum, P. recurvatum, Pla. erumpens, and Pla. rivularia (Réblová et al., 2012; Réblová et al., 2020). Most species of Pleurotheciella and Pleurothecium have been reported in aquatic habitat from Yunnan province, China, and Thailand (Luo et al., 2018). Inaddition, two species, Pla. nilotica and Pla. obovoideum, have been reported in Japan and and Egypt, respectively, (Arzanlou et al., 2007; Abdel-Aziz et al., 2020). Currently there are no reports of Pleurotheciella or Pleurothecium from Jiangxi Province, China.
Ten strains including four new species introduced in this paper are all from the freshwater environment in Jiangxi Province. Pleurotheciaceae species observed mostly from submerged decaying woods (Luo et al., 2018; Réblová et al., 2020). The 10 strains examined in this study were all recovered from decaying wood within a freshwater ecosystem. These strains were observed and isolated from the wood over a period of time. Upon observing a substantial collection of specimens, no sexual reproductive patterns have been identified. Consequently, it appears that species within the genera Pleurotheciella and Pleurothecium may predominantly exist in an asexual form within their natural habitat.
According to phylogenetic analyses, species of Saprodesmium and Neomonodictys have longer genetic distance from other species of Pleurotheciaceae. This observation underscores the need for additional research and discovery to enhance the taxonomic classification within the Pleurotheciaceae. Sterigmatobotrys has not been definitively identified as a genus within the Pleurotheciaceae family, but Sterigmatobotrys groups together with species of Pleurotheciaceae with a strong statistical support (Figure 1). Previously, P. obovoideum was reported with a separate phylogenetic tree not fully integrated into other species sequences of Pleurotheciaceae, which have some problems in its taxonomic position (Arzanlou et al., 2007). According to phylogenetic analysis, we adjusted the taxonomic position of Pleurothecium obovoideum to Neomonodictys obovoideum by its strong clade support (PP/BS = 1.00/100) clades with Neomonodictys aquatica and Neomonodictys muriformis.
Previously, all asexual morphs of Pleurotheciella collected from natural environment have macronematous conidiophores, which distinguished from Dactylaria species (Luo et al., 2018). Previous studies have shown that Pleurotheciella species have macronematous conidiophores in natural environment, our specimens further confirm this phenomenon. At present, there are still five Pleurothecium species without molecular data, other species of Pleurothecium form a well-supported monophyletic clade in the Pleurotheciaceae. Morphologically, Pleurothecium species are similar to Neta in conidiogenous cell and conidiophores, but Pleurothecium have denser denticles and more obvious cylindrical conidiogenous site and 3-septate conidia, which significantly distinguished from Neta.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funds for research were provided by the National Natural Science Foundation of China (NSFC 32060014 and NSFC 32070023), the earmarked fund for Jiangxi Agriculture Research System (2024), and the Forestry Science and Technology Innovation Project of Jiangxi Forestry Bureau of China [Innovation Special (2022) No. 9].
Footnotes
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
W-MH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. J-BZ: Writing – review & editing. Z-JZ: Writing – review & editing. DT: Writing – review & editing. C-YC: Writing – review & editing. J-PZ: Writing – review & editing. M-HC: Writing – review & editing. H-JH: Writing – review & editing. HY: Writing – review & editing. YG: Writing – review & editing. D-MH: Writing – review & editing. H-YS: Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1452499/full#supplementary-material
References
- Abdel-Aziz F. A., Bahkali A. H., Elgorban A. M., Abdel-Wahab M. A. (2020). Pleurotheciella nilotica sp. nov. (Pleurotheciales, Ascomycota) from freshwater habitats in Egypt. Nova Hedwigia 110, 91–98. doi: 10.1127/nova_hedwigia/2020/0570 [DOI] [Google Scholar]
- Arzanlou M., Groenewald J. Z., Gams W., Braun U., Shin H. D., Crous P. W. (2007). Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 58, 57–93. doi: 10.3114/sim.2007.58.03, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmee S., Wanasinghe D. N., Calabon M. S., Huanraluek N., Chandrasiri S. K. U., Jones G. E. B., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Ji K., Hyde K. D. (2006). Variation between freshwater and terrestrial fungal communities on decaying bamboo culms. Antonie Van Leeuwenhoek 89, 293–301. doi: 10.1007/s10482-005-9030-1, PMID: [DOI] [PubMed] [Google Scholar]
- Calabon M., Hyde K. D., Jones E., Bao D. F., Bhunjun C. S., Phukhamsakda C., et al. (2023). Freshwater fungal biology. Mycosphere 14, 195–413. doi: 10.5943/mycosphere/14/1/4 [DOI] [Google Scholar]
- Chen J., Hu D., Song H., Zhai Z., Lai L., Lin K. (2022). Menisporopsis aquatica sp. nov. (Sordariomycetes, Chaetosphaeriales, Chaetosphaeriaceae), from freshwater habitat in China. Biodivers. Data J. 10:e91008. doi: 10.3897/BDJ.10.e91008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomnunti P., Hongsanan S., Aguirre-Hudson B., Tian Q., Peršoh D., Dhami M. K., et al. (2014). The sooty moulds. Fungal Divers. 66, 1–36. doi: 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Chun-Sheng L., You-Peng W., Xu Z., Yan L., Xiang-Chun S., Jian M., et al. (2023). Additions to hyphomycetes from Yungui plateau, China with three new species (Ascomycota, Sordariomycetes). Biodivers. Data J. 11:e101629. doi: 10.3897/BDJ.11.e101629, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A. (2005). New Zealand hyphomycete fungi: additional records, new species, and notes on interesting collections. N. Z. J. Bot. 43, 323–349. doi: 10.1080/0028825X.2005.9512957 [DOI] [Google Scholar]
- Dayarathne M. C., Maharachchikumbura S. S., Jones E. B., Dong W., Devadatha B., Yang J., et al. (2019). Phylogenetic revision of Savoryellaceae and evidence for its ranking as a subclass. Front. Microbiol. 10:840. doi: 10.3389/fmicb.2019.00840, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Jeewon R., Hyde K. D., Yang E., Zhang H., Yu X., et al. (2021). Five novel taxa from freshwater habitats and new taxonomic insights of Pleurotheciales and Savoryellomycetidae. J. Fungi. 7:711. doi: 10.3390/jof7090711, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–13. doi: 10.1016/0031-9422(80)85004-7 [DOI] [Google Scholar]
- Ertz D., Heuchert B., Braun U., Freebury C. E., Common R. S., Diederich P. (2016). Contribution to the phylogeny and taxonomy of the genus Taeniolella, with a focus on lichenicolous taxa. Fungal Biol. 120, 1416–1447. doi: 10.1016/j.funbio.2016.05.008, PMID: [DOI] [PubMed] [Google Scholar]
- Fryar S., Catcheside D. (2023). Freshwater ascomycetes from southern Australia: Melanascomaceae fam. nov., Melanascoma panespora gen. et. sp. nov., and Pleurothecium brunius sp. nov. Fungal System. Evol. 11, 85–93. doi: 10.3114/fuse.2023.11.07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M., Gené J., Castañeda-Ruiz R. F., Mena-Portales J., Crous P. W., Guarro J. (2017). Phylogeny of saprobic microfungi from southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhnel F. (1919). Fünfte vorlaüfige Mitteilungen mykologischer Ergebnisse (Nr. 399–500). Ber. Deut. Bot. Ges. 37, 153–161. doi: 10.1111/j.1438-8677.1919.tb07337.x [DOI] [Google Scholar]
- Hopple JS Jr., Vilgalys R. (1999). Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol. Phylogenet. Evol. 13, 1–19. doi: 10.1006/mpev.1999.0634, PMID: [DOI] [PubMed] [Google Scholar]
- Huang S., Bao D., Shen H., Su H., Luo Z. (2022). Neomonodictys aquatica sp. nov. (Pleurotheciaceae) from a plateau lake in Yunnan Province, China. Biodivers. Data J. 10:e76842. doi: 10.3897/bdj.10.e76842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde K. D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K. W., et al. (2018). Mycosphere notes 169-224. Mycosphere 9, 271–430. doi: 10.5943/MYCOSPHERE/9/2/8 [DOI] [Google Scholar]
- Hyde K. D., Fryar S. C., Tian Q., Tian Q., Tian Q., Bahkali A. H., et al. (2016). Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/J.FUNECO.2015.07.002 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Abreu V. P., Bazzicalupo A., Thilini Chethana K. W., Clericuzio M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Ma J., Yang H., Zhang J., Du T., et al. (2023). Mycosphere notes 387–412 – novel species of fungal taxa from around the world. Mycosphere 14, 663–744. doi: 10.5943/mycosphere/14/1/8 [DOI] [Google Scholar]
- Hyde K. D. Y., Phookamsak R., Jeewon R., Bhat D., Jones E., Liu N. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Jayawardena R. S., Hyde K. D., Wang S., Sun Y. R., Suwannarach N. (2023). Fungal diversity notes 1512–1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-022-00513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). Mega7: Molecular evolutionary genetic analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 33, 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A026092, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Y., Xu G., Yan X., Chen M., Gao Y., Hu H., et al. (2022). Phaeoisarialaianensis (Pleurotheciales, Pleurotheciaceae), a new species from freshwater habitats in China. Biodivers. Data J. 10:e94088. doi: 10.3897/BDJ.10.e94088, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Hyde K. D., Bhat D. J., Jeewon R., Maharachchikumbura S. S., Bao D., et al. (2018). Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 17, 511–530. doi: 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. S., Gusmao L. F., Castañeda-Ruiz R. F. (2016). Pleurothecium bicoloratum & Sporidesmiopsis pluriseptata spp. nov. from Brazil. Mycotaxon 131, 145–152. doi: 10.5248/131.145 [DOI] [Google Scholar]
- Palmer M. E., Covich A. P., Finlay B. J., Gilbert J., Hyde K. D., Johnson R. K., et al. (1997). Biodiversity and ecosystem processes in freshwater sediments. Ambio 26, 571–577. [Google Scholar]
- Peng S., Liu Y., Huang J., Li X., Yan X., Song H., et al. (2022). Aquapteridospora jiangxiensis, a new aquatic hyphomycetous fungus from a freshwater habitat in China. Arch. Microbiol. 204:378. doi: 10.1007/s00203-022-02942-6, PMID: [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2018) FigTree v1.4.4: Tree figure drawing tool. Available at: https://github.com/rambaut/figtree/releases (Accessed August, 2023).
- Réblová M., Hernández-Restrepo M., Fournier J., Nekvindová J. (2020). New insights into the systematics of Bactrodesmium and its allies and introducing new genera, species and morphological patterns in the Pleurotheciales and Savoryellales (Sordariomycetes). Stud. Mycol. 95, 415–466. doi: 10.1016/j.simyco.2020.02.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M., Seifert K. A., Fournier J., Štěpánek V. (2012). Phylogenetic classification of Pleurothecium and Pleurotheciella gen. Nov. and its dactylaria-like anamorph (Sordariomycetes) based on nuclear ribosomal and protein-coding genes. Mycologia 104, 1299–1314. doi: 10.3852/12-035, PMID: [DOI] [PubMed] [Google Scholar]
- Réblová M., Seifert K. A., Fournier J., Štěpánek V. (2015). Newly recognised lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia 37, 57–81. doi: 10.3767/003158516X689819, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer C. A. (1993). The freshwater Ascomycetes. Nova Hedwigia 56, 1–33. [Google Scholar]
- Shi L., Yang H., Hyde K. D., Wijayawardene N. N., Wang G., Yu X., et al. (2021). Freshwater Sordariomycetes: new species and new records in Pleurotheciaceae, Pleurotheciales. Phytotaxa 518, 143–166. doi: 10.11646/phytotaxa.518.2.4 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C. V., Bhat D. J. (1989). Hyphomycetes from South India I. Some new taxa. Kavaka 15, 41–74. [Google Scholar]
- Sun L., Li H., Sun X., Guo L. (2017). Dematipyriforma aquilaria gen. Et sp. nov., a new Hyphomycetous taxon from Aquilaria crassna. Cryptogam. Mycol. 38, 341–351. doi: 10.7872/crym/v38.iss3.2017.341 [DOI] [Google Scholar]
- Tubaki K., Matsushima T. (1972). Microfungi of the Solomon Islands and Papua-New Guinea. Mycologia 64:1208. doi: 10.2307/3758096 [DOI] [Google Scholar]
- Vijaykrishna D., Jeewon R., Hyde K. D. (2005). Fusoidispora aquatica: a new freshwater ascomycete from Hong Kong based on morphology and phylogeny inferred from rDNA gene sequences. Sydowia 57, 267–280. [Google Scholar]
- White T. J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 38, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yan X., Huang J., Song H., Gao Y., Hu H., Zhai Z., et al. (2023). A new species of Dictyochaeta (Sordariomycetes, Chaetosphaeriales, Chaetosphaeriaceae) from freshwater habitats in China. Biodivers. Data J. 11:e97439. doi: 10.3897/bdj.11.e97439, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueming W., Tianyu Z. (2009). New species of Phialosporostilbe and Pleurothecium from soil. Mycotaxon 110, 1–4. doi: 10.5248/110.1 [DOI] [Google Scholar]
- Yuen T. K., Hyde K. D., Hodgkiss I. J. (1998). Physiological growth parameters and enzyme production in tropical freshwater fungi. Mater. Und Organism. 32, 1–16. [Google Scholar]
- Zhai Z., Yan J., Li W., Gao Y., Hu H., Zhou J., et al. (2022). Three novel species of Distoseptispora (Distoseptisporaceae) isolated from bamboo in Jiangxi Province, China. Myco Keys 88, 35–54. doi: 10.3897/mycokeys.88.79346, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.