Abstract
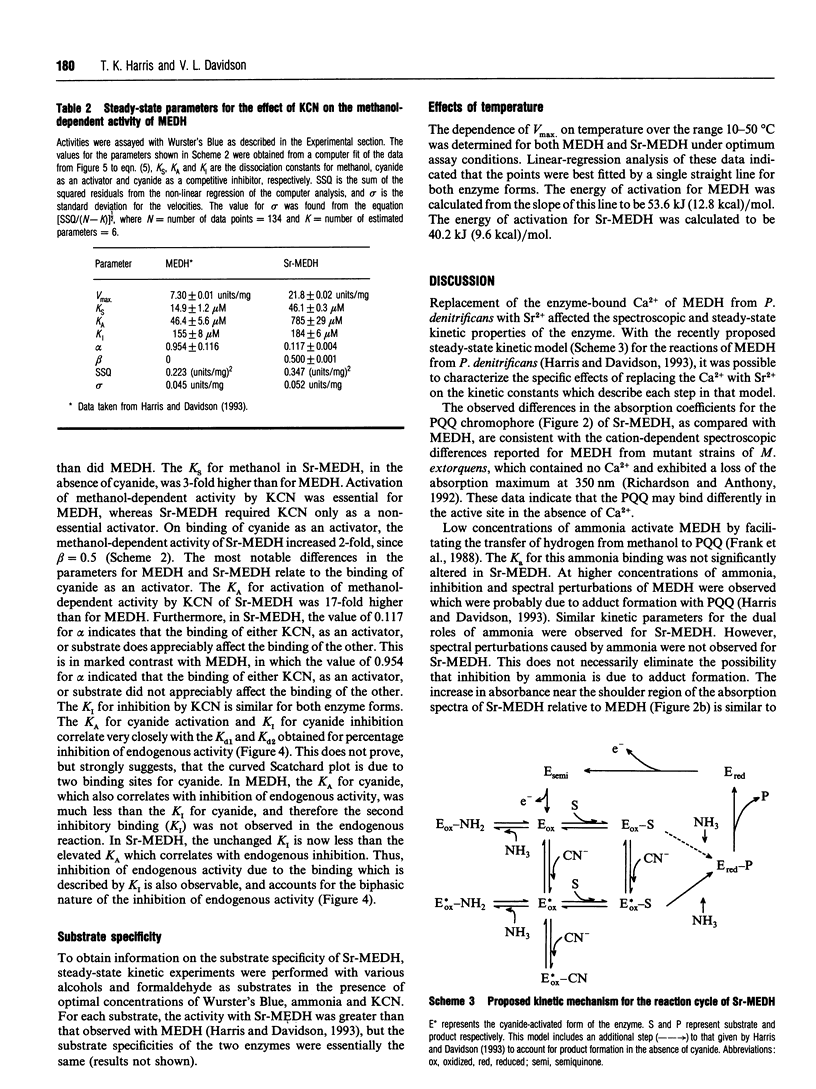
Methanol dehydrogenase (MEDH) possesses tightly bound Ca2+ in addition to its pyrroloquinoline quinone (PQQ) prosthetic group. Ca2+ was replaced with Sr2+ by growing the host bacterium, Paracoccus denitrificans, in media in which Ca2+ was replaced with Sr2+. MEDH, which was purified from these cells (Sr-MEDH), exhibited an increased absorption coefficient for the PQQ chromophore, and displayed certain kinetic properties which were different from those of native MEDH. Native MEDH exhibits an endogenous activity which is not stimulated by substrate and which is inhibited by cyanide. Sr-MEDH exhibited lower endogenous activity which was stimulated by substrate, and was much less sensitive to inhibition by cyanide. The Vmax. for the methanol-dependent activity of Sr-MEDH was 3-fold greater than that of the native enzyme, and the Ks for methanol was altered. Cyanide also acts as an obligatory activator and competitive inhibitor of methanol-dependent activity in native MEDH from P. denitrificans [Harris and Davidson (1993) Biochemistry 32, 4362-4368]. Sr-MEDH exhibited a similar K1 for cyanide inhibition of methanol-dependent activity, but the KA for cyanide activation of this activity was 17-fold greater than that for the native enzyme. The activation energy of Sr-MEDH was 13.4 kJ (3.2 kcal)/mol lower than that of the native enzyme. These data confirm and significantly extend the conclusions from genetic [Richardson and Anthony (1992) Biochem. J. 287, 709-715] and crystallographic [White, Boyd, Mathews, Xia, Dai, Zhang and Davidson (1993) Biochemistry 32, 12955-12958] studies that suggest an apparently unique role for Ca2+ in MEDH compared with other Ca(2+)-dependent proteins and enzymes.
Full text
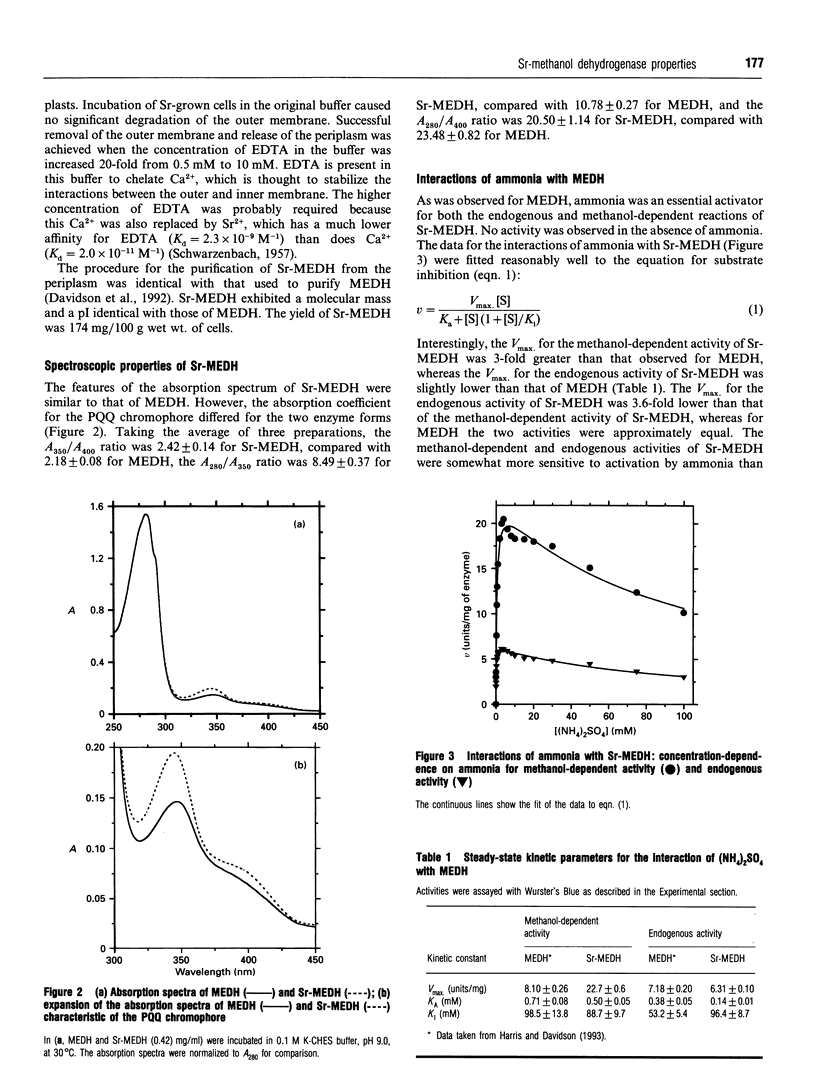
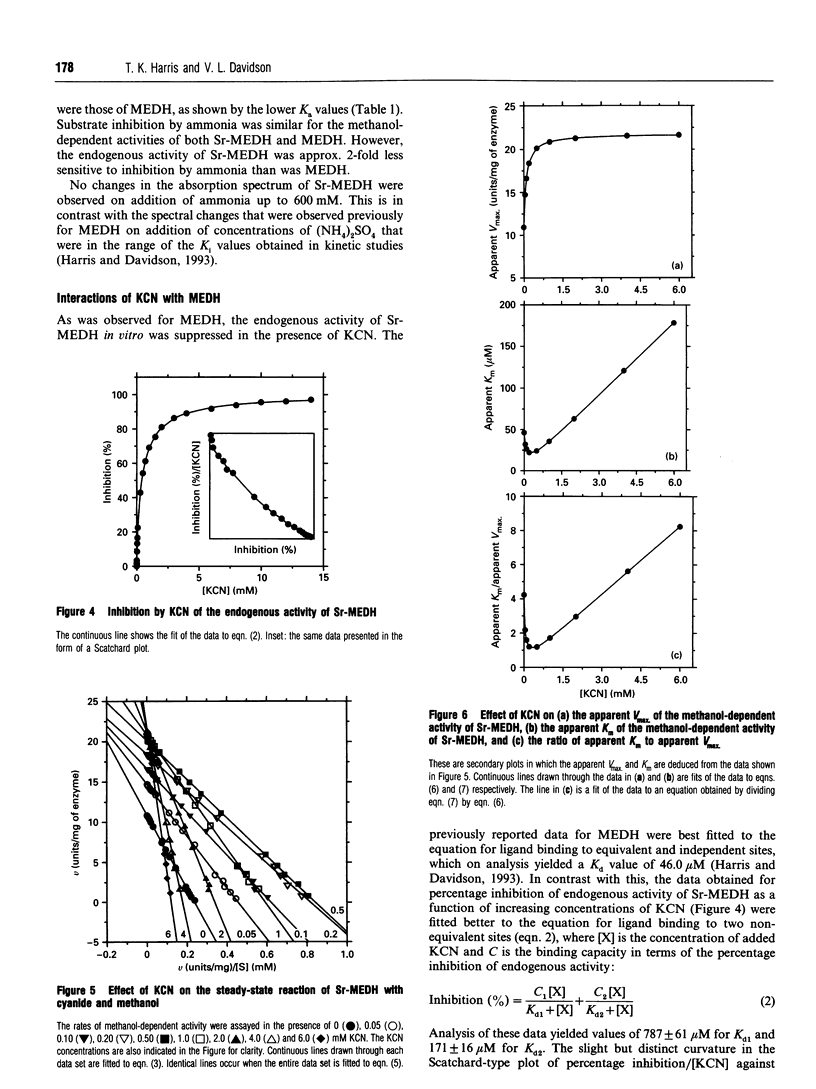
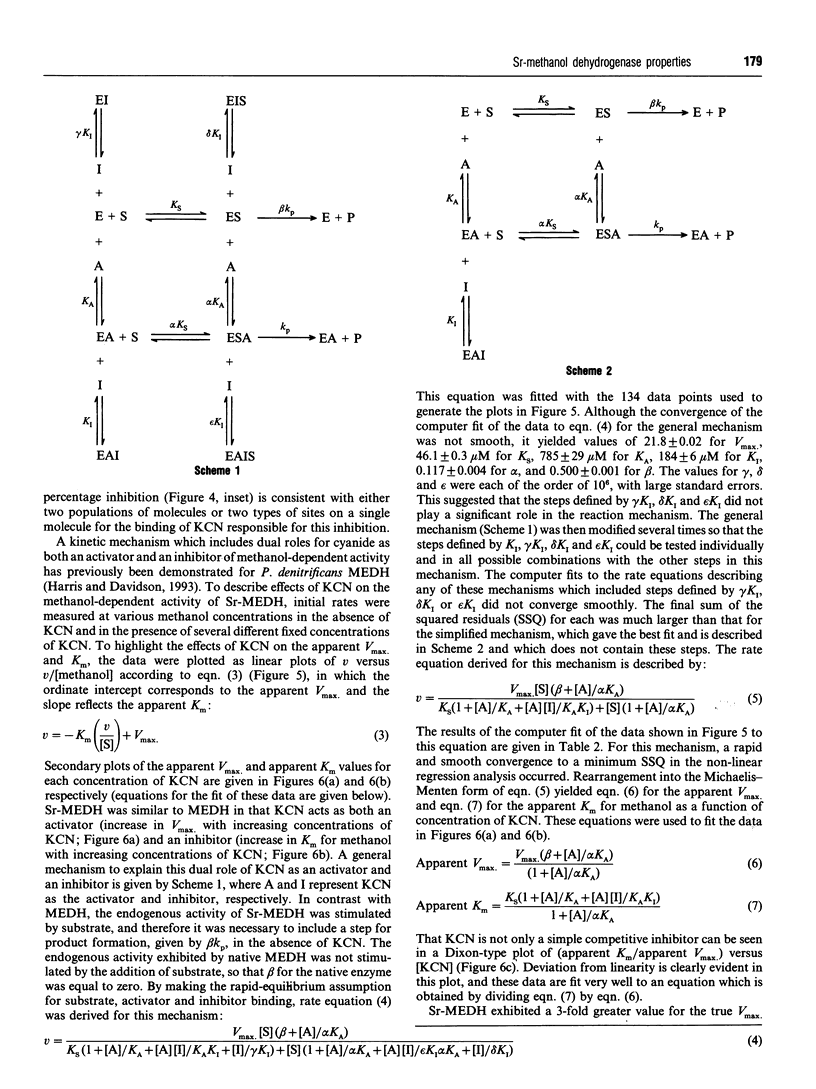
PDF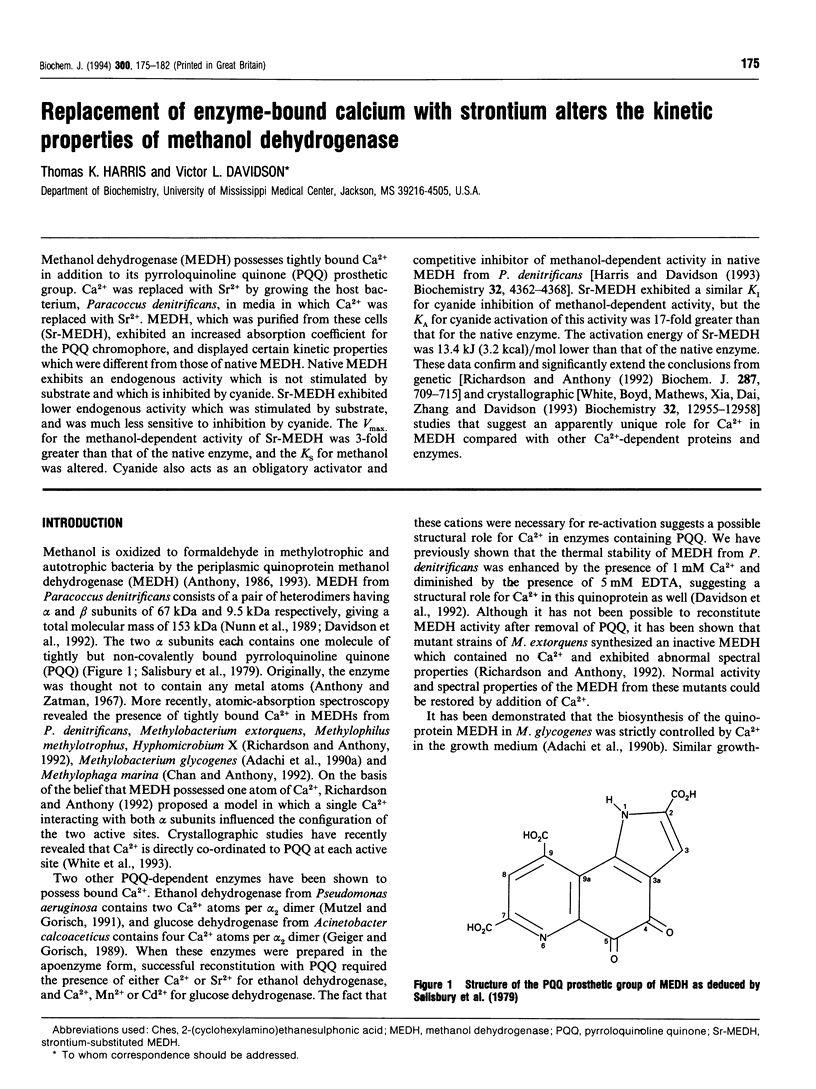



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J. 1964 Sep;92(3):614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. Purification and properties of the alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1967 Sep;104(3):953–959. doi: 10.1042/bj1040953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Bajorath J., Raghunathan S., Hinrichs W., Saenger W. Long-range structural changes in proteinase K triggered by calcium ion removal. Nature. 1989 Feb 2;337(6206):481–484. doi: 10.1038/337481a0. [DOI] [PubMed] [Google Scholar]
- Bode W., Schwager P. The refined crystal structure of bovine beta-trypsin at 1.8 A resolution. II. Crystallographic refinement, calcium binding site, benzamidine binding site and active site at pH 7.0. J Mol Biol. 1975 Nov 15;98(4):693–717. doi: 10.1016/s0022-2836(75)80005-2. [DOI] [PubMed] [Google Scholar]
- Boel E., Brady L., Brzozowski A. M., Derewenda Z., Dodson G. G., Jensen V. J., Petersen S. B., Swift H., Thim L., Woldike H. F. Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus. Biochemistry. 1990 Jul 3;29(26):6244–6249. doi: 10.1021/bi00478a019. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Legg M. J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3',5'-bisphosphate-calcium ion complex at 1.5-A resolution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson V. L. Methylamine dehydrogenases from methylotrophic bacteria. Methods Enzymol. 1990;188:241–246. doi: 10.1016/0076-6879(90)88040-h. [DOI] [PubMed] [Google Scholar]
- Davidson V. L., Wu J., Miller B., Jones L. H. Factors affecting the stability of methanol dehydrogenase from Paracoccus denitrificans. FEMS Microbiol Lett. 1992 Jul 1;73(1-2):53–58. doi: 10.1016/0378-1097(92)90582-9. [DOI] [PubMed] [Google Scholar]
- Dijkstra B. W., Kalk K. H., Hol W. G., Drenth J. Structure of bovine pancreatic phospholipase A2 at 1.7A resolution. J Mol Biol. 1981 Mar 25;147(1):97–123. doi: 10.1016/0022-2836(81)90081-4. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jr Studies on methanol dehydrogenase from Hyphomicrobium X. Isolation of an oxidized form of the enzyme. Biochem J. 1980 Apr 1;187(1):213–219. doi: 10.1042/bj1870213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Jr, Dijkstra M., Duine J. A., Balny C. Kinetic and spectral studies on the redox forms of methanol dehydrogenase from Hyphomicrobium X. Eur J Biochem. 1988 Jun 1;174(2):331–338. doi: 10.1111/j.1432-1033.1988.tb14102.x. [DOI] [PubMed] [Google Scholar]
- Geiger O., Görisch H. Reversible thermal inactivation of the quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Ca2+ ions are necessary for re-activation. Biochem J. 1989 Jul 15;261(2):415–421. doi: 10.1042/bj2610415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Quayle J. R. Purification and properties of the methanol dehydrogenase from Methylophilus methylotrophus. Biochem J. 1981 Oct 1;199(1):245–250. doi: 10.1042/bj1990245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Gros P., Betzel C., Dauter Z., Wilson K. S., Hol W. G. Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content. J Mol Biol. 1989 Nov 20;210(2):347–367. doi: 10.1016/0022-2836(89)90336-7. [DOI] [PubMed] [Google Scholar]
- Harms N., de Vries G. E., Maurer K., Hoogendijk J., Stouthamer A. H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987 Sep;169(9):3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. K., Davidson V. L. A new kinetic model for the steady-state reactions of the quinoprotein methanol dehydrogenase from Paracoccus denitrificans. Biochemistry. 1993 Apr 27;32(16):4362–4368. doi: 10.1021/bi00067a028. [DOI] [PubMed] [Google Scholar]
- Herzberg O., James M. N. Structure of the calcium regulatory muscle protein troponin-C at 2.8 A resolution. Nature. 1985 Feb 21;313(6004):653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- Hulette C. M., Earl N. L., Anthony D. C., Crain B. J. Adult onset Niemann-Pick disease type C presenting with dementia and absent organomegaly. Clin Neuropathol. 1992 Nov-Dec;11(6):293–297. [PubMed] [Google Scholar]
- KORNBERG H. L., MORRIS J. G. THE UTILIZATION OF GLYCOLLATE BY MICROCOCCUS DENITRIFICANS: THE BETA-HYDROXYASPARTATE PATHWAY. Biochem J. 1965 Jun;95:577–586. doi: 10.1042/bj0950577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V. D., Lee L., Edwards B. F. Refined crystal structure of calcium-liganded carp parvalbumin 4.25 at 1.5-A resolution. Biochemistry. 1990 Feb 13;29(6):1404–1412. doi: 10.1021/bi00458a010. [DOI] [PubMed] [Google Scholar]
- McPhalen C. A., Schnebli H. P., James M. N. Crystal and molecular structure of the inhibitor eglin from leeches in complex with subtilisin Carlsberg. FEBS Lett. 1985 Aug 19;188(1):55–58. doi: 10.1016/0014-5793(85)80873-5. [DOI] [PubMed] [Google Scholar]
- Meyer E., Cole G., Radhakrishnan R., Epp O. Structure of native porcine pancreatic elastase at 1.65 A resolutions. Acta Crystallogr B. 1988 Feb 1;44(Pt 1):26–38. doi: 10.1107/s0108768187007559. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Day D., Anthony C. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem J. 1989 Jun 15;260(3):857–862. doi: 10.1042/bj2600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner C., Suck D. Crystallographic refinement and structure of DNase I at 2 A resolution. J Mol Biol. 1986 Dec 5;192(3):605–632. doi: 10.1016/0022-2836(86)90280-9. [DOI] [PubMed] [Google Scholar]
- Richardson I. W., Anthony C. Characterization of mutant forms of the quinoprotein methanol dehydrogenase lacking an essential calcium ion. Biochem J. 1992 Nov 1;287(Pt 3):709–715. doi: 10.1042/bj2870709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury S. A., Forrest H. S., Cruse W. B., Kennard O. A novel coenzyme from bacterial primary alcohol dehydrogenases. Nature. 1979 Aug 30;280(5725):843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- Van Spanning R. J., Wansell C. W., De Boer T., Hazelaar M. J., Anazawa H., Harms N., Oltmann L. F., Stouthamer A. H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991 Nov;173(21):6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S., Boyd G., Mathews F. S., Xia Z. X., Dai W. W., Zhang Y. F., Davidson V. L. The active site structure of the calcium-containing quinoprotein methanol dehydrogenase. Biochemistry. 1993 Dec 7;32(48):12955–12958. doi: 10.1021/bi00211a002. [DOI] [PubMed] [Google Scholar]


