Abstract
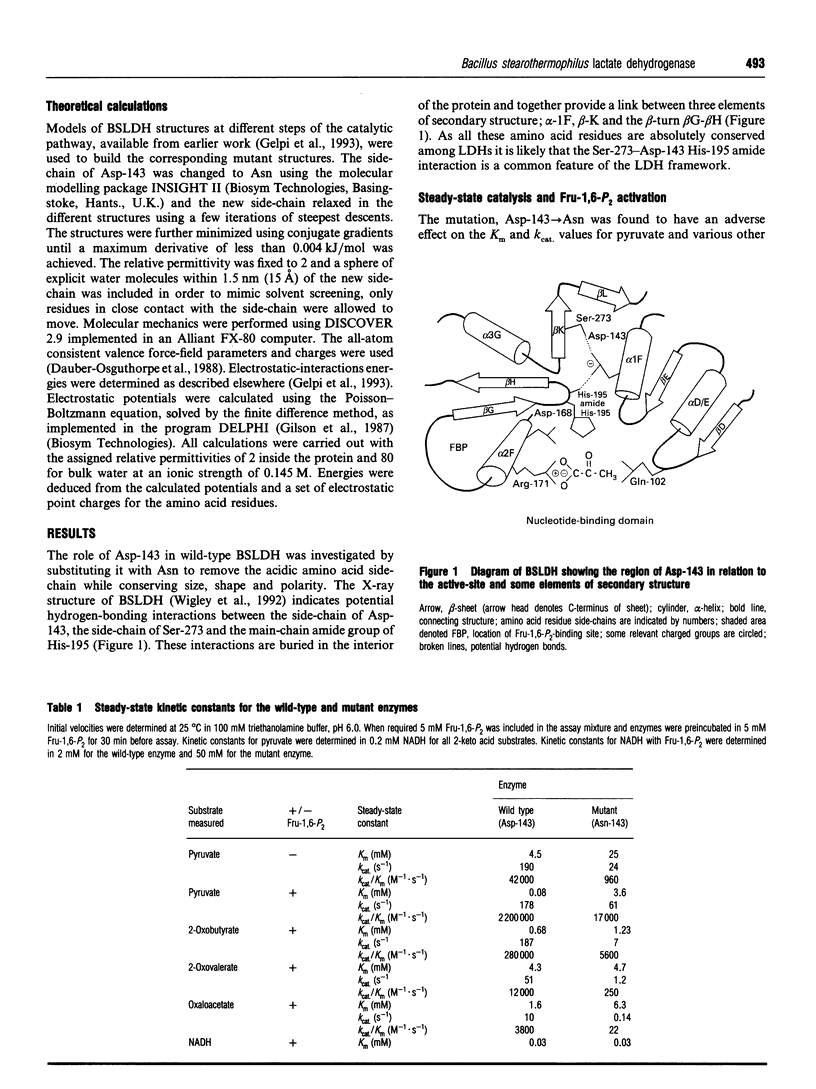
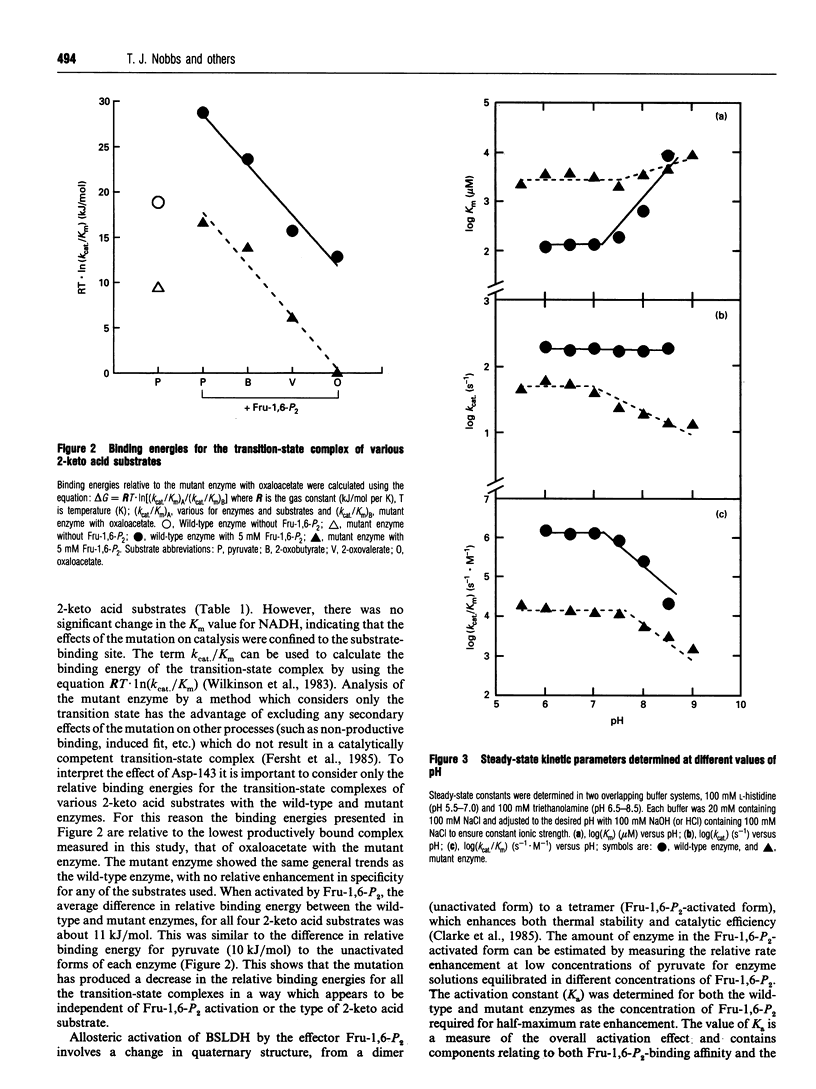
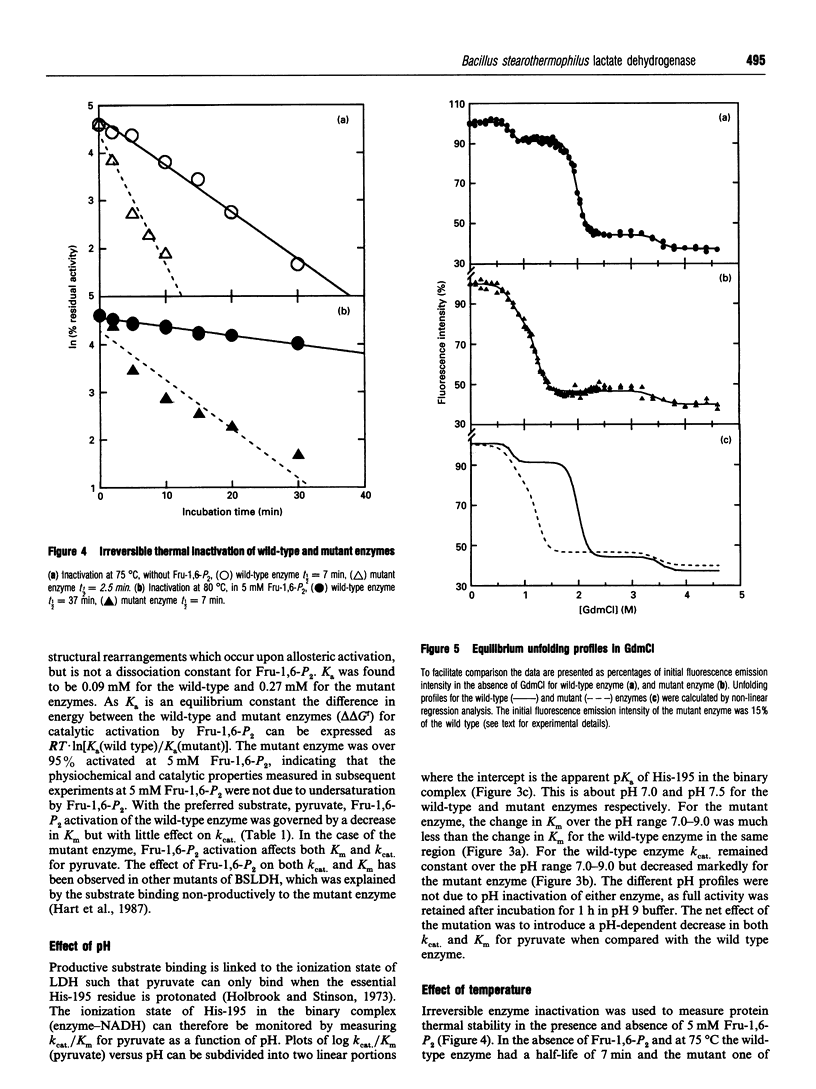
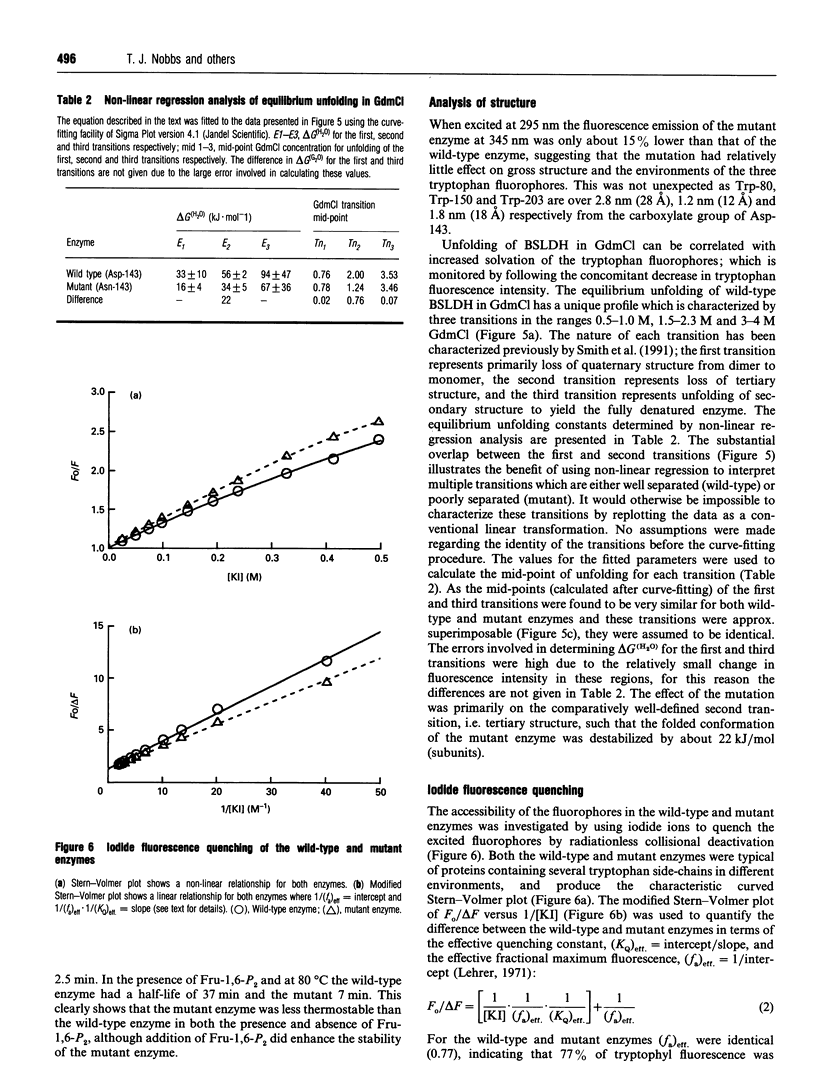
The X-ray structure of lactate dehydrogenase (LDH) shows the side-chain carboxylate group of Asp-143 to be buried in the hydrophobic interior of the enzyme, where it makes hydrogen-bonding interactions with both the side-chain hydroxyl group of Ser-273 and the main-chain amide group of His-195. This is an unusual environment for a carboxylate side-chain as hydrogen bonding normally occurs with water molecules at the surface of the protein. A charged hydrogen-bonding interaction in the interior of a protein would be expected to be much stronger than a similar interaction on the solvent-exposed exterior. In this respect the side-chain carboxylate group of Asp-143 appears to be important for maintaining tertiary structure by providing a common linkage point between three discontinuous elements of the secondary structure, alpha 1F, beta K and the beta-turn joining beta G and beta H. The contribution of the Asp-143 side-chain to the structure and function of Bacillus stearothermophilus LDH was assessed by creating a mutant enzyme containing Asn-143. The decreased thermal stability of both unactivated and fructose-1,6-diphosphate (Fru-1,6-P2)-activated forms of the mutant enzyme support a structural role for Asp-143. Furthermore, the difference in stability of the wild-type and mutant enzymes in guanidinium chloride suggested that the carboxylate group of Asp-143 contributes at least 22 kJ/mol to the conformational stability of the wild-type enzyme. However, there was no alteration in the amount of accessible tryptophan fluorescence in the mutant enzyme, indicating that the mutation caused a structural weakness rather than a gross conformational change. Comparison of the wild-type and mutant enzyme steady-state parameters for various 2-keto acid substrates showed the mutation to have a general effect on catalysis, with an average difference in binding energy of 11 kJ/mol for the transition-state complexes. The different effects of pH and Fru-1,6-P2 on the wild-type and mutant enzymes also confirmed a perturbation of the catalytic centre in the mutant enzyme. As the side-chain of Asp-143 is not sufficiently close to the active site to be directly involved in catalysis or substrate binding it is proposed that the effects on catalysis shown by the mutant enzyme are induced either by a structural change or by charge imbalance at the active site.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barstow D. A., Clarke A. R., Chia W. N., Wigley D., Sharman A. F., Holbrook J. J., Atkinson T., Minton N. P. Cloning, expression and complete nucleotide sequence of the Bacillus stearothermophilus L-lactate dehydrogenase gene. Gene. 1986;46(1):47–55. doi: 10.1016/0378-1119(86)90165-4. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Fernley R. T., Bradshaw R. A., Banaszak L. J. Amino acid sequence homology among the 2-hydroxy acid dehydrogenases: mitochondrial and cytoplasmic malate dehydrogenases form a homologous system with lactate dehydrogenase. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6166–6170. doi: 10.1073/pnas.79.20.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P., Roberts V. A., Osguthorpe D. J., Wolff J., Genest M., Hagler A. T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1988;4(1):31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Hart K. W., Clarke A. R., Wigley D. B., Waldman A. D., Chia W. N., Barstow D. A., Atkinson T., Jones J. B., Holbrook J. J. A strong carboxylate-arginine interaction is important in substrate orientation and recognition in lactate dehydrogenase. Biochim Biophys Acta. 1987 Aug 21;914(3):294–298. doi: 10.1016/0167-4838(87)90289-5. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Stinson R. A. The use of ternary complexes to study ionizations and isomerizations during catalysis by lactate dehydrogenase. Biochem J. 1973 Apr;131(4):739–748. doi: 10.1042/bj1310739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S. S. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971 Aug 17;10(17):3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- Nicholls D. J., Wood I. S., Nobbs T. J., Clarke A. R., Holbrook J. J., Atkinson T., Scawen M. D. Dissecting the contributions of a specific side-chain interaction to folding and catalysis of Bacillus stearothermophilus lactate dehydrogenase. Eur J Biochem. 1993 Mar 1;212(2):447–455. doi: 10.1111/j.1432-1033.1993.tb17681.x. [DOI] [PubMed] [Google Scholar]
- Piontek K., Chakrabarti P., Schär H. P., Rossmann M. G., Zuber H. Structure determination and refinement of Bacillus stearothermophilus lactate dehydrogenase. Proteins. 1990;7(1):74–92. doi: 10.1002/prot.340070108. [DOI] [PubMed] [Google Scholar]
- Smith C. J., Clarke A. R., Chia W. N., Irons L. I., Atkinson T., Holbrook J. J. Detection and characterization of intermediates in the folding of large proteins by the use of genetically inserted tryptophan probes. Biochemistry. 1991 Jan 29;30(4):1028–1036. doi: 10.1021/bi00218a021. [DOI] [PubMed] [Google Scholar]
- Waldman A. D., Hart K. W., Clarke A. R., Wigley D. B., Barstow D. A., Atkinson T., Chia W. N., Holbrook J. J. The use of genetically engineered tryptophan to identify the movement of a domain of B. stearothermophilus lactate dehydrogenase with the process which limits the steady-state turnover of the enzyme. Biochem Biophys Res Commun. 1988 Jan 29;150(2):752–759. doi: 10.1016/0006-291x(88)90455-x. [DOI] [PubMed] [Google Scholar]
- Wigley D. B., Gamblin S. J., Turkenburg J. P., Dodson E. J., Piontek K., Muirhead H., Holbrook J. J. Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution. J Mol Biol. 1992 Jan 5;223(1):317–335. doi: 10.1016/0022-2836(92)90733-z. [DOI] [PubMed] [Google Scholar]
- Wilkinson A. J., Fersht A. R., Blow D. M., Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983 Jul 19;22(15):3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- Winter G., Fersht A. R., Wilkinson A. J., Zoller M., Smith M. Redesigning enzyme structure by site-directed mutagenesis: tyrosyl tRNA synthetase and ATP binding. Nature. 1982 Oct 21;299(5885):756–758. doi: 10.1038/299756a0. [DOI] [PubMed] [Google Scholar]



