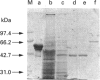
Abstract
1. NADPH binds to bovine catalase and to yeast catalases A and T, but not to Escherichia coli catalase HPII. The association was demonstrated using chromatography and fluorimetry. Bound NADPH fluoresces in a similar way to NADPH in solution. 2. Bound NADPH protects bovine and yeast catalases against forming inactive peroxide compound II either via endogenous reductant action or by ferrocyanide reduction during catalytic activity in the presence of slowly generated peroxide. 3. Bound NADPH reduces neither compound I nor compound II of catalase. It apparently reacts with an intermediate formed during the decay of compound I to compound II; this postulated intermediate is an immediate precursor of stable compound II either when the latter is formed by endogenous reductants or when ferrocyanide is used. It represents therefore a new type of hydrogen donor that is not included in the original classification of Keilin and Nicholls [Keilin, D. and Nicholls, P. (1958) Biochim. Biophys. Acta 29, 302-307] 4. A model for NADPH action is presented in which concerted reduction of the ferryl iron and of a neighbouring protein free radical is responsible for the observed NADPH effects. The roles of migrant radical species in mammalian and yeast catalases are compared with similar events in metmyoglobin and cytochrome c peroxidase reactions with peroxides.
Full text
PDF

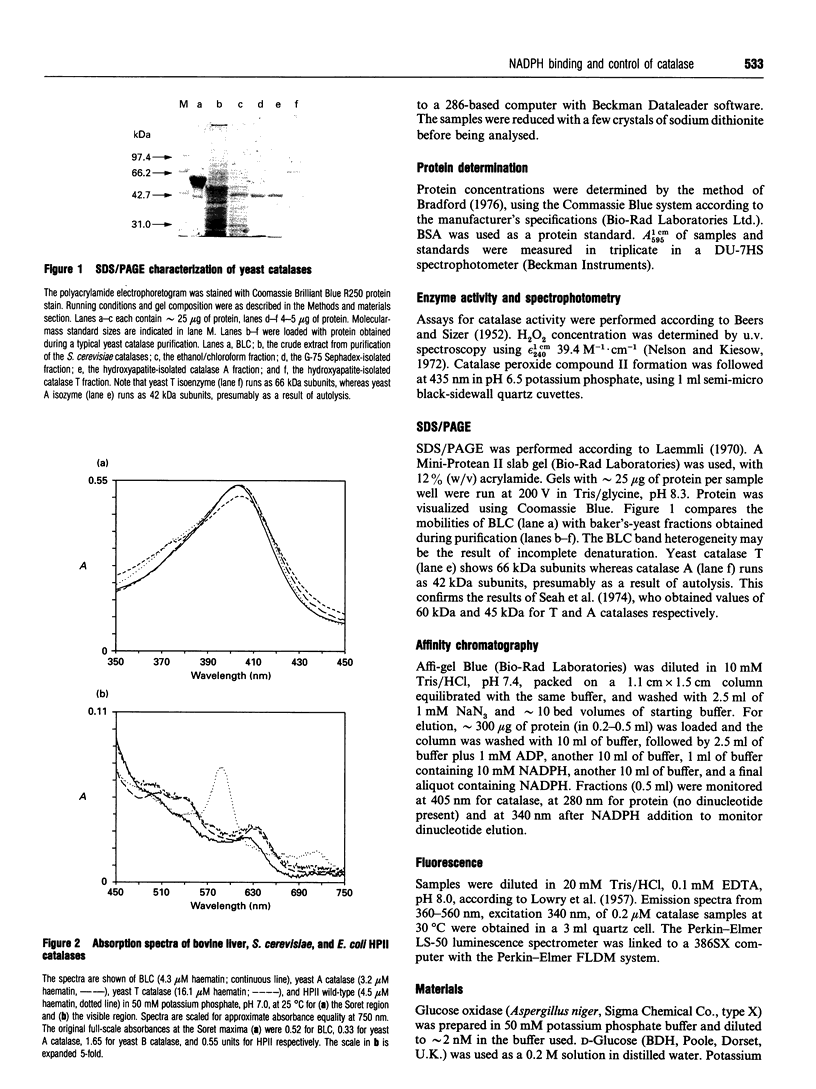
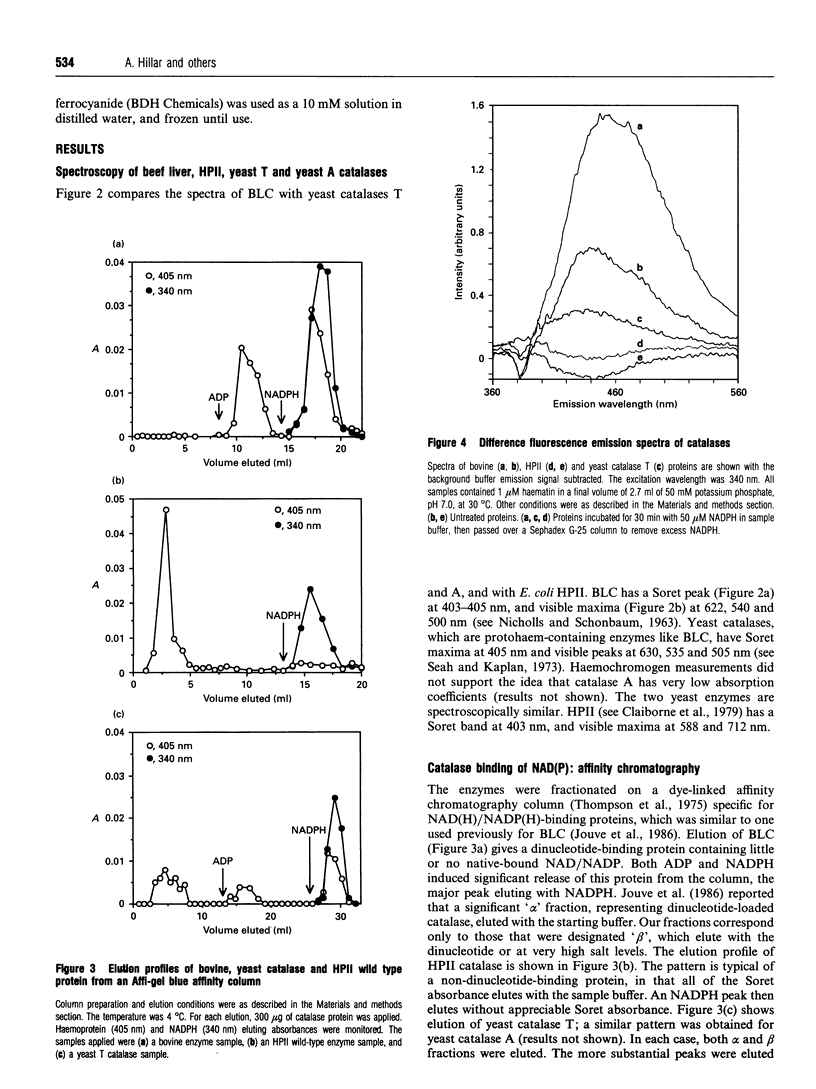
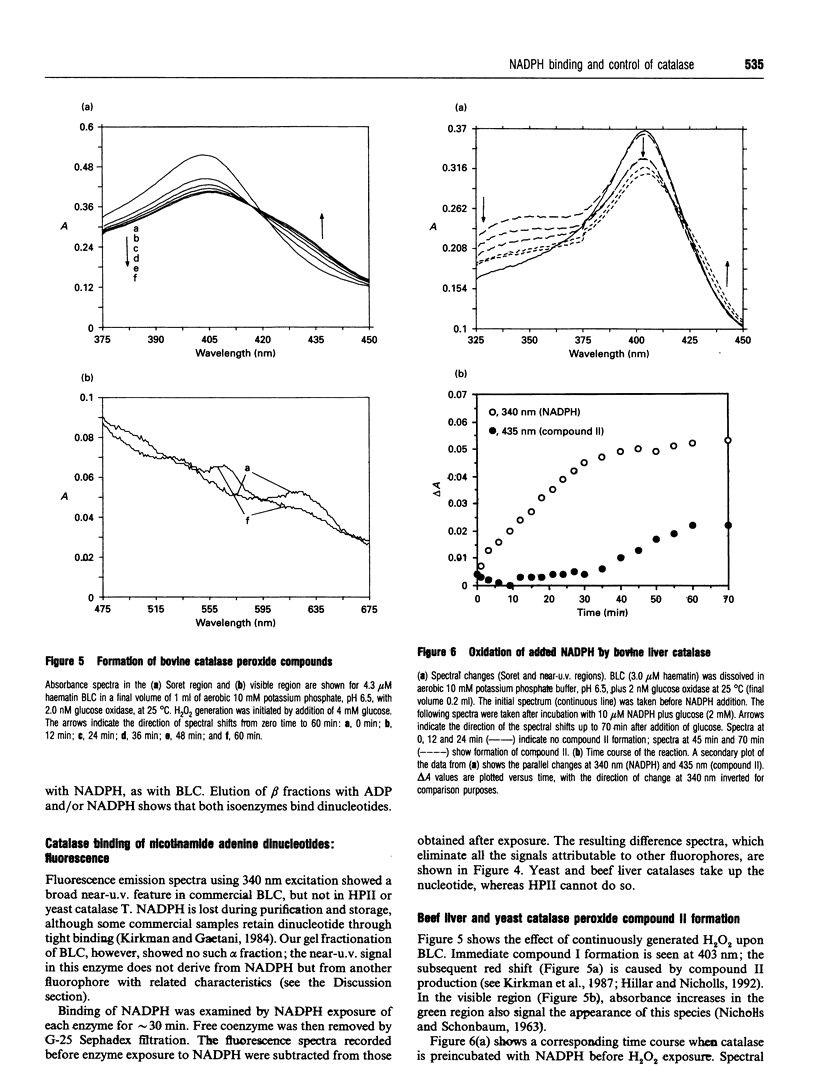
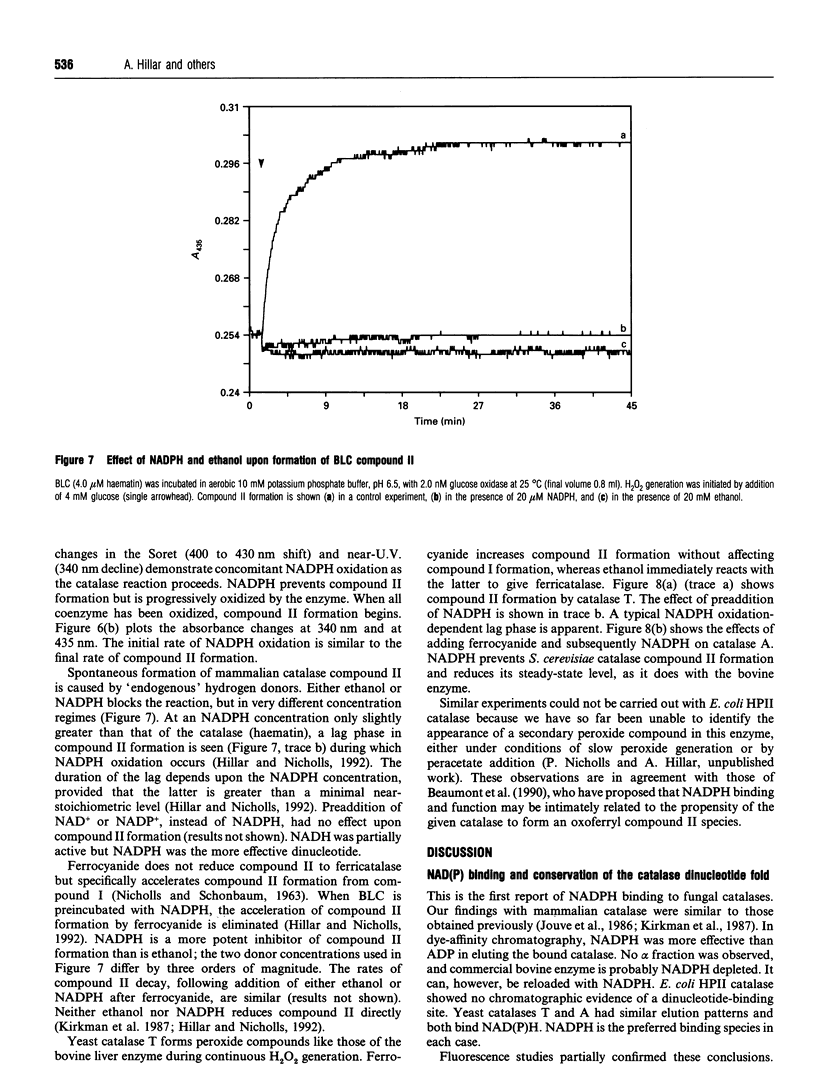
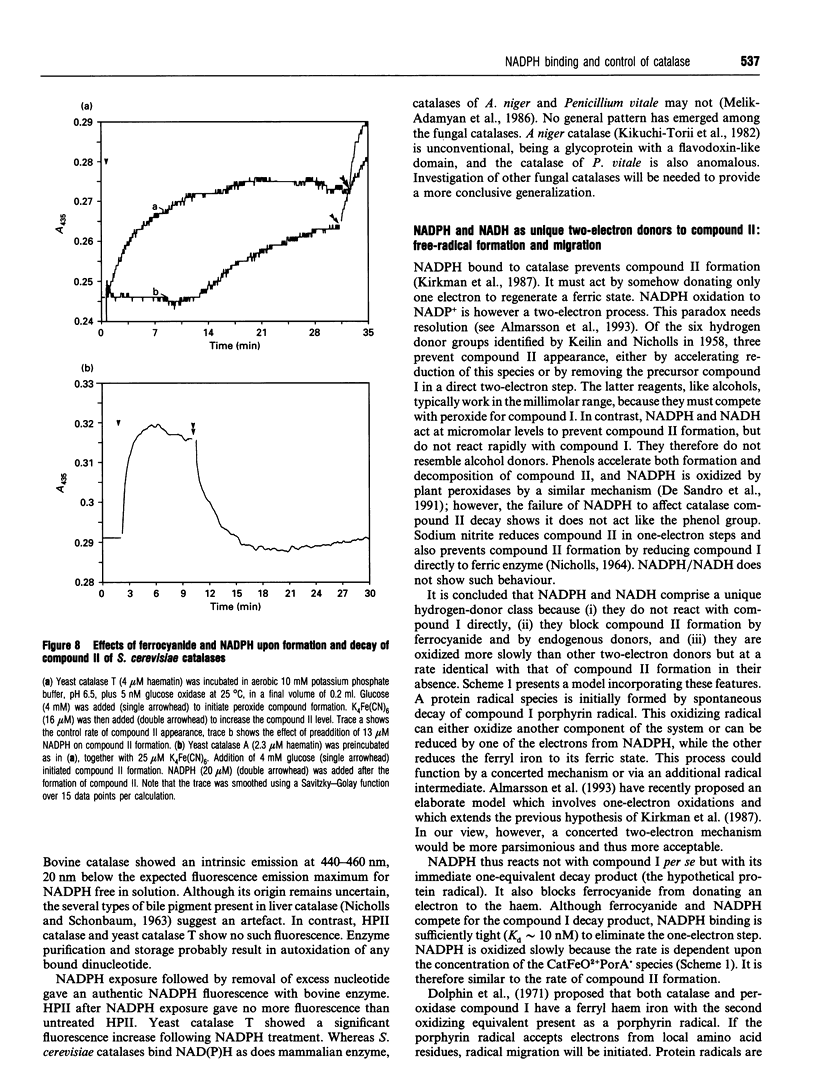


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi S., Nagano S., Ishimori K., Watanabe Y., Morishima I., Egawa T., Kitagawa T., Makino R. Roles of proximal ligand in heme proteins: replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry. 1993 Jan 12;32(1):241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- Adachi S., Nagano S., Watanabe Y., Ishimori K., Morishima I. Alteration of human myoglobin proximal histidine to cysteine or tyrosine by site-directed mutagenesis: characterization and their catalytic activities. Biochem Biophys Res Commun. 1991 Oct 15;180(1):138–144. doi: 10.1016/s0006-291x(05)81266-5. [DOI] [PubMed] [Google Scholar]
- Arduini A., Mancinelli G., Radatti G. L., Damonti W., Hochstein P., Cadenas E. Reduction of sperm whale ferrylmyoglobin by endogenous reducing agents: potential reducible loci of ferrylmyoglobin. Free Radic Biol Med. 1992 Oct;13(4):449–454. doi: 10.1016/0891-5849(92)90185-j. [DOI] [PubMed] [Google Scholar]
- BEERS R. F., Jr, SIZER I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952 Mar;195(1):133–140. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- CHANCE B. The reactions of catalase in the presence of the notatin system. Biochem J. 1950 Apr;46(4):387–402. doi: 10.1042/bj0460387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C. E., Choe Y. S., Ortiz de Montellano P. R. Reactions of the protein radical in peroxide-treated myoglobin. Formation of a heme-protein cross-link. J Biol Chem. 1989 Jun 25;264(18):10534–10541. [PubMed] [Google Scholar]
- Chuang W. J., Johnson S., Van Wart H. E. Resonance Raman spectra of bovine liver catalase: enhancement of proximal tyrosinate vibrations. J Inorg Biochem. 1988 Nov;34(3):201–219. doi: 10.1016/0162-0134(88)85030-x. [DOI] [PubMed] [Google Scholar]
- Chuang W. J., Van Wart H. E. Resonance Raman spectra of horseradish peroxidase and bovine liver catalase compound I species. Evidence for predominant 2A2u pi-cation radical ground state configurations. J Biol Chem. 1992 Jul 5;267(19):13293–13301. [PubMed] [Google Scholar]
- Claiborne A., Malinowski D. P., Fridovich I. Purification and characterization of hydroperoxidase II of Escherichia coli B. J Biol Chem. 1979 Nov 25;254(22):11664–11668. [PubMed] [Google Scholar]
- Cohen G., Fessl F., Traczyk A., Rytka J., Ruis H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet. 1985;200(1):74–79. doi: 10.1007/BF00383315. [DOI] [PubMed] [Google Scholar]
- Cohen G., Rapatz W., Ruis H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988 Sep 1;176(1):159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- Davies M. J. Identification of a globin free radical in equine myoglobin treated with peroxides. Biochim Biophys Acta. 1991 Mar 8;1077(1):86–90. doi: 10.1016/0167-4838(91)90529-9. [DOI] [PubMed] [Google Scholar]
- Davies M. J., Puppo A. Direct detection of a globin-derived radical in leghaemoglobin treated with peroxides. Biochem J. 1992 Jan 1;281(Pt 1):197–201. doi: 10.1042/bj2810197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson J. H., Bracete A. M., Huff A. M., Kadkhodayan S., Zeitler C. M., Sono M., Chang C. K., Loewen P. C. The active site structure of E. coli HPII catalase. Evidence favoring coordination of a tyrosinate proximal ligand to the chlorin iron. FEBS Lett. 1991 Dec 16;295(1-3):123–126. doi: 10.1016/0014-5793(91)81401-s. [DOI] [PubMed] [Google Scholar]
- De Sandro V., Dupuy C., Kaniewski J., Ohayon R., Dème D., Virion A., Pommier J. Mechanism of NADPH oxidation catalyzed by horse-radish peroxidase and 2,4-diacetyl-[2H]heme-substituted horse-radish peroxidase. Eur J Biochem. 1991 Oct 15;201(2):507–513. doi: 10.1111/j.1432-1033.1991.tb16310.x. [DOI] [PubMed] [Google Scholar]
- Dolphin D., Forman A., Borg D. C., Fajer J., Felton R. H. Compounds I of catalase and horse radish peroxidase: pi-cation radicals. Proc Natl Acad Sci U S A. 1971 Mar;68(3):614–618. doi: 10.1073/pnas.68.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton J. W., Boraas M., Etkin N. L. Catalase activity and red cell metabolism. Adv Exp Med Biol. 1972;28:121–131. doi: 10.1007/978-1-4684-3222-0_8. [DOI] [PubMed] [Google Scholar]
- Fishel L. A., Farnum M. F., Mauro J. M., Miller M. A., Kraut J., Liu Y. J., Tan X. L., Scholes C. P. Compound I radical in site-directed mutants of cytochrome c peroxidase as probed by electron paramagnetic resonance and electron-nuclear double resonance. Biochemistry. 1991 Feb 19;30(7):1986–1996. doi: 10.1021/bi00221a036. [DOI] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The NADPH binding site on beef liver catalase. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1604–1608. doi: 10.1073/pnas.82.6.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The active center of catalase. J Mol Biol. 1985 Sep 5;185(1):21–37. doi: 10.1016/0022-2836(85)90180-9. [DOI] [PubMed] [Google Scholar]
- Hartig A., Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986 Nov 3;160(3):487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- Heimberger A., Eisenstark A. Compartmentalization of catalases in Escherichia coli. Biochem Biophys Res Commun. 1988 Jul 15;154(1):392–397. doi: 10.1016/0006-291x(88)90698-5. [DOI] [PubMed] [Google Scholar]
- Hillar A., Nicholls P. A mechanism for NADPH inhibition of catalase compound II formation. FEBS Lett. 1992 Dec 14;314(2):179–182. doi: 10.1016/0014-5793(92)80969-n. [DOI] [PubMed] [Google Scholar]
- Jouve H. M., Gouet P., Boudjada N., Buisson G., Kahn R., Duee E. Crystallization and crystal packing of Proteus mirabilis PR catalase. J Mol Biol. 1991 Oct 20;221(4):1075–1077. doi: 10.1016/0022-2836(91)90918-v. [DOI] [PubMed] [Google Scholar]
- Jouve H. M., Pelmont J., Gaillard J. Interaction between pyridine adenine dinucleotides and bovine liver catalase: a chromatographic and spectral study. Arch Biochem Biophys. 1986 Jul;248(1):71–79. doi: 10.1016/0003-9861(86)90402-9. [DOI] [PubMed] [Google Scholar]
- KEILIN D., HARTREE E. F. Catalase, peroxidase and metmyoglobin as catalysts of coupled peroxidatic reactions. Biochem J. 1955 Jun;60(2):310–325. doi: 10.1042/bj0600310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEILIN D., NICHOLLS P. Reactions of catalase with hydrogen peroxide and hydrogen donors. Biochim Biophys Acta. 1958 Aug;29(2):302–307. doi: 10.1016/0006-3002(58)90189-6. [DOI] [PubMed] [Google Scholar]
- Kikuchi-Torii K., Hayashi S., Nakamoto H., Nakamura S. Properties of Aspergillus niger catalase. J Biochem. 1982 Nov;92(5):1449–1456. doi: 10.1093/oxfordjournals.jbchem.a134069. [DOI] [PubMed] [Google Scholar]
- Kirkman H. N., Gaetani G. F. Catalase: a tetrameric enzyme with four tightly bound molecules of NADPH. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4343–4347. doi: 10.1073/pnas.81.14.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman H. N., Galiano S., Gaetani G. F. The function of catalase-bound NADPH. J Biol Chem. 1987 Jan 15;262(2):660–666. [PubMed] [Google Scholar]
- LOWRY O. H., ROBERTS N. R., KAPPHAHN J. I. The fluorometric measurement of pyridine nucleotides. J Biol Chem. 1957 Feb;224(2):1047–1064. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loewen P. C., Switala J., Triggs-Raine B. L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985 Nov 15;243(1):144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- Melik-Adamyan W. R., Barynin V. V., Vagin A. A., Borisov V. V., Vainshtein B. K., Fita I., Murthy M. R., Rossmann M. G. Comparison of beef liver and Penicillium vitale catalases. J Mol Biol. 1986 Mar 5;188(1):63–72. doi: 10.1016/0022-2836(86)90480-8. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Bandyopadhyay D., Mauro J. M., Traylor T. G., Kraut J. Reaction of ferrous cytochrome c peroxidase with dioxygen: site-directed mutagenesis provides evidence for rapid reduction of dioxygen by intramolecular electron transfer from the compound I radical site. Biochemistry. 1992 Mar 17;31(10):2789–2797. doi: 10.1021/bi00125a020. [DOI] [PubMed] [Google Scholar]
- NICHOLLS P. THE FORMATION AND CATALYTIC ROLE OF CATALASE PEROXIDE COMPOUND II. Biochim Biophys Acta. 1964 Mar 9;81:479–495. doi: 10.1016/0926-6569(64)90133-6. [DOI] [PubMed] [Google Scholar]
- Nelson D. P., Kiesow L. A. Enthalpy of decomposition of hydrogen peroxide by catalase at 25 degrees C (with molar extinction coefficients of H 2 O 2 solutions in the UV). Anal Biochem. 1972 Oct;49(2):474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- Orii Y., Sakai Y., Ozawa K. Ubiquitous formation of catalase compound II in hemoglobin-free perfused rat liver and detection of novel spectral species. Biochem Biophys Res Commun. 1989 Aug 15;162(3):1272–1278. doi: 10.1016/0006-291x(89)90811-5. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano P. R. Catalytic sites of hemoprotein peroxidases. Annu Rev Pharmacol Toxicol. 1992;32:89–107. doi: 10.1146/annurev.pa.32.040192.000513. [DOI] [PubMed] [Google Scholar]
- Oshino N., Chance B. Properties of glutathione release observed during reduction of organic hydroperoxide, demethylation of aminopyrine and oxidation of some substances in perfused rat liver, and their implications for the physiological function of catalase. Biochem J. 1977 Mar 15;162(3):509–525. doi: 10.1042/bj1620509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy M. E. Catalase: an old enzyme with a new role? Can J Biochem Cell Biol. 1984 Oct;62(10):1006–1014. doi: 10.1139/o84-129. [DOI] [PubMed] [Google Scholar]
- Reid T. J., 3rd, Murthy M. R., Sicignano A., Tanaka N., Musick W. D., Rossmann M. G. Structure and heme environment of beef liver catalase at 2.5 A resolution. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah T. C., Bhatti A. R., Kaplan J. G. Novel catalatic proteins of bakers' yeast. I. An atypical catalase. Can J Biochem. 1973 Nov;51(11):1551–1555. doi: 10.1139/o73-208. [DOI] [PubMed] [Google Scholar]
- Seah T. C., Kaplan J. G. Purification and properties of the catalase of bakers' yeast. J Biol Chem. 1973 Apr 25;248(8):2889–2893. [PubMed] [Google Scholar]
- Sharma K. D., Andersson L. A., Loehr T. M., Terner J., Goff H. M. Comparative spectral analysis of mammalian, fungal, and bacterial catalases. Resonance Raman evidence for iron-tyrosinate coordination. J Biol Chem. 1989 Aug 5;264(22):12772–12779. [PubMed] [Google Scholar]
- Spevak W., Fessl F., Rytka J., Traczyk A., Skoneczny M., Ruis H. Isolation of the catalase T structural gene of Saccharomyces cerevisiae by functional complementation. Mol Cell Biol. 1983 Sep;3(9):1545–1551. doi: 10.1128/mcb.3.9.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T. Structure of myoglobin refined at 2-0 A resolution. I. Crystallographic refinement of metmyoglobin from sperm whale. J Mol Biol. 1977 Mar 5;110(3):537–568. doi: 10.1016/s0022-2836(77)80111-3. [DOI] [PubMed] [Google Scholar]
- Thompson S. T., Cass K. H., Stellwagen E. Blue dextran-sepharose: an affinity column for the dinucleotide fold in proteins. Proc Natl Acad Sci U S A. 1975 Feb;72(2):669–672. doi: 10.1073/pnas.72.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein B. K., Melik-Adamyan W. R., Barynin V. V., Vagin A. A., Grebenko A. I., Borisov V. V., Bartels K. S., Fita I., Rossmann M. G. Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution. J Mol Biol. 1986 Mar 5;188(1):49–61. doi: 10.1016/0022-2836(86)90479-1. [DOI] [PubMed] [Google Scholar]
- Wilks A., Ortiz de Montellano P. R. Intramolecular translocation of the protein radical formed in the reaction of recombinant sperm whale myoglobin with H2O2. J Biol Chem. 1992 May 5;267(13):8827–8833. [PubMed] [Google Scholar]
- Zimniak P., Hartter E., Woloszczuk W., Ruis H. Catalase biosynthesis in yeast: formation of catalase A and catalase T during oxygen adaptation of Saccharomyces cerevisiae. Eur J Biochem. 1976 Dec 11;71(2):393–398. doi: 10.1111/j.1432-1033.1976.tb11126.x. [DOI] [PubMed] [Google Scholar]
- von Ossowski I., Mulvey M. R., Leco P. A., Borys A., Loewen P. C. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991 Jan;173(2):514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]