INTRODUCTION
The oral poliovirus vaccine (OPV) of Albert Sabin is nearly ideal for use in polio eradication [1]. This inexpensive vaccine is easily administered by mouth, facilitating its widespread use. OPV induces intestinal immunity, making recent OPV recipients resistant to infection by wild polioviruses, and effectively blocking wild poliovirus transmission when used in mass campaigns and routine immunizations. It provides long-term protection against poliomyelitis through durable humoral immunity. Because OPV is a live vaccine, vaccine virus can spread to and immunize unvaccinated contacts of vaccine recipients, increasing the impact of OPV beyond those actually immunized. Through effective use of this excellent vaccine, the World Health Organization’s Global Polio Eradication Initiative (GPEI) has brought wild polioviruses to the threshold of eradication [2, 3].
Despite its many advantages, the continued use of trivalent OPV was identified as a threat to maintaining eradication because use of OPV carries certain liabilities arising from the genetic instability of the live, attenuated vaccine strains during replication in the human intestine [4]. In 2016, the live type 2 component of the oral vaccine was removed from OPV and replaced with bivalent OPV (types 1 and 3) to reduce the risk of type 2 vaccine-derived polioviruses (VDPVs), though a stockpile of monovalent type 2 OPV (mOPV2) is maintained for outbreak response.
OPV and inactivated poliovirus vaccine (IPV) both induce neutralizing antibodies against poliovirus, especially after multiple vaccinations. The effectiveness of both vaccines led to the elimination of wild type polio with the exception of Pakistan, Afghanistan and Nigeria [4]. In order to successfully move forward with the eradication effort, we must adhere to the so-called pillars of the global polio eradication initiative: routine immunization, supplementary immunization, acute flaccid paralysis surveillance and targeted mop-up campaigns [5]. The ‘hallmark of efficacy’ for the poliovirus vaccines is a robust neutralizing antibody response against the viral capsid, with a 50% endpoint serum titer greater than 1:8 corresponding to protection [6].
While use of OPV has increased immunity to all three serotypes in populations, the attenuated vaccine virus can revert to a neurovirulent phenotype that is indistinguishable from that of wild polioviruses. VDPVs fall into three categories depending on the sampling source and epidemiologic data: iVDPV (derived from an individual with a known primary immunodeficiency) [5–8], circulating VDPV (cVDPV; at least two genetically related VDPVs derived from individuals who are not close contacts of one another, implying circulation in the community), and aVDPV (ambiguous—a single virus not genetically linked to any other VDPV, whether from a paralyzed case or an environmental source). In areas with low vaccine coverage, these VDPVs may emerge and circulate [7]. By definition, the VP1 nucleotide sequence of an OPV isolate must have diverged by more than 1% (0.6% for type 2 VDPV) from the reference OPV strain to be designated a VDPV [8].
Primary immune deficiencies affecting antibody production leave patients susceptible to infections, requiring regular treatment with intravenous immunoglobulin [9, 10]. In these patients, exposure to OPV through vaccination or contact with a vaccine recipient poses a higher risk of developing paralytic poliomyelitis [11, 12]. In patients with an antibody deficit, Sabin vaccine viruses can replicate for extended periods of time compared to immunocompetent vaccine recipients [13]. Over time, increasing numbers of nucleotide changes accumulate in the viral genome, with reversion to neurovirulence. Although extremely rare, some immunodeficient individuals can excrete VDPVs over prolonged periods (>6 months), posing a risk of infection in their community.
As these nucleotide substitutions are introduced, amino acid substitutions may accumulate in the proteins of VDPVs. These substitutions can occur in the capsid region within known antigenic sites, resulting in distinct antigenic profiles compared to the original OPV strains [14]. Where examined, sera from poliovirus recipients showed reduced neutralizing capacity against VDPVs, although titers were sufficiently high to confer protection [15–23]. However, little work has been done to determine the effect of increasing amino acid changes in iVDPVs on the neutralization capacity of sera from poliovirus vaccine recipients.
In the current study, we selected sera from a previous study investigating the effect of a combined OPV/IPV vaccine schedule in children from low income communities in the United States during the switch in immunization schedule from OPV to IPV [24]. The sera were tested for neutralizing antibodies against a panel of type 2 iVDPVs with increasing number of nucleotide changes in the viral capsid proteins. For each type 2 iVDPV, homology models of the capsid were generated for mapping antigenic sites and comparing the structures to the Sabin 2 reference.
MATERIALS AND METHODS
Patient Sera
Sera were obtained from children aged 19–35 months, who participated in a 4-year U.S. poliovirus immunity surveillance study from 1997–2001 conducted as part of the Childhood Immunization Demonstration Project of Community Health Networks [24]. The study protocol was reviewed and approved by the Centers for Disease Control and Prevention Institutional Review Board. Study participants consented to serological testing against poliovirus antigens, allowing for use of the specimens in the current study. This study was originally designed to evaluate the effects of the change in poliovirus vaccination schedule in the United States, from OPV to a combined OPV/ IPV, and IPV-only schedule. Immunity to poliovirus was measured in these children by determining the presence of neutralizing antibodies to poliovirus types 1, 2, and 3 using the poliovirus microneutralization assay [25]. Thirty serum specimens were randomly selected from each of the three sites in the 1997–2001 serosurvey: Detroit, Michigan; Manhattan, New York and Denver, Colorado. All sera were stored at −20°C at the Centers for Disease Control and Prevention, Atlanta, Georgia, until the cross neutralization study began in July 2012.
Poliovirus Isolates
The seven isolates used in this study are listed in Table 1. Isolates 18277 and 18278 were collected from the same patient. Viral stocks were grown in HEp2-C cells and incubated at 35°C with 5% CO2, in a humidified atmosphere, until full cytopathic effect (CPE) was observed. The virus was then harvested by performing three freeze-thaw cycles at −70°C, centrifugation to remove cell debris, and collection of the supernatant, which was stored in aliquots at −20°C until used in the assay.
Table 1.
iVPDV isolates analyzed in this study
Each virus was titrated on HEp2-C cells, as described previously [25], to ensure that all the antigens used had the same starting concentrations, expressed in 50% cell culture infectious dose (CCID50).
Polio neutralization assay
All sera had been previously randomized using a balanced block scheme. Since results from previous tests run on these sera were readily available, 30 specimens having positive titers for Sabin 1, 2, or 3 were chosen from the final year of each study[25]. The existing randomization scheme was followed, but specimens with volumes less than 200 µl were excluded. The sera were tested in multiple runs, with sera from all three sites being tested against the same two or three polioviruses each day. Therefore, each run contained 90 test sera plus an internal reference standard of pooled sera, used 10 times. All sera were randomly assigned to positions on 96-well cell culture plates (Corning Inc., Corning, NY). Each test plate contained four sera in triplicate in two-fold dilution increments ranging from 1:8 to 1:1024.
A virus titration control plate was prepared for each antigen to ensure that test plates were inoculated with the correct concentration of antigen. In addition, a cell control plate consisting only of cells and media was used to monitor cell viability. Serum dilutions were incubated with virus for 3 hours at 35°C before the addition of 25 µl of HEp2-C cell suspension at a concentration of 3×105 cells/ml to each well of the test plates, back titration plates, and cell control plate.
After incubation for five days, the plates were stained with 0.05% crystal violet solution and the optical density measured at 570 nm by plate reader. Endpoint titers were calculated using the Spearman-Kärber formula [26]. Seropositivity was defined as the detection of neutralizing antibody at a titer of 1:8 or higher (log(titer) ≥ 3).
Sequence analysis
Complete capsid sequence analysis was performed as described previously from cell culture isolates [27, 28]. Sequences have been deposited in GenBank (Table 1).
Structural modeling and antigenic site sequence comparisons
Polio protomer homology models were generated for each strain using the SWISS-model server (http://swissmodel.expasy.org), using the template PDB file 1EAH (MEF-1) as a reference, with amino acid similarity ranged from 94.3% for the 10 nt change isolate to 90.2% for the 115 nt change isolate. Amino acid changes and antigenic sites were highlighted using UCSF Chimera (Version 1.6.1). Each protomer was assembled into a pentamer configuration using the biological assembly data in the 1EAH crystal structure metadata file.
Protomer models for each iVDPV capsid (P1 region) were compared to the Sabin 2 capsid model using the MatchMaker tool in UCSF Chimera. The root mean square deviation (RMSD) was calculated across the entire capsid sequence for each iVDPV capsid.
To determine if there was a correlation between the degree of amino acid conservation across an antigenic site and the median neutralization titer for each iVDPV isolate, a substitution score was assigned to each amino acid identified as a key residue in each antigenic site by monoclonal antibody escape [21, 29–31]. For each antigenic site, the rate of substitutions per site was calculated using the JTT model of peptide substitution and then plotted versus the corresponding median neutralizing antibody titer for each virus isolate [32].
Data Analysis
All statistical analyses were conducted using SAS v. 9.2 (SAS Institute, Cary, NC). The Wilcoxon signed rank test was used to compare the antibody titers for each iVDPV2 with the Sabin 2 titer, as iVDPV2 and Sabin 2 titers were assumed to be dependent variables for a given serum.
RESULTS
Seropositivity rates in vaccinated children decrease as P1 nucleotide substitutions increase in iVDPVs.
To measure the neutralization capacity of seven type 2 iVDPVs with varying degrees of nucleotide substitution in P1. Isolates ranged in divergence from 10 nt changes (6 amino acid changes) to 115 nt changes in P1 (53 amino acid changes) (Table 1). Sera from 90 low-income children in New York, Detroit, and Denver, were used in the gold-standard poliovirus microneutralization assay. Sera with a log2 neutralization titer ≥ 3 are considered seropositive and protective against paralytic disease. The rates of seropositivity for the children’s sera were determined for all seven iVDPVs. We observed seropositivity rates for strains 18279, 18280, 18281 and 18282 of 98.9% each and for strains 18277 and 18278 of 94.4% each compared to 97.8% for Sabin 2 (p>0.05). Strain 18276, with 115 nucleotide changes from Sabin 2, was the exception to these very high seropositivity rates against the iVDPV2 panel, with only 28.9% of sera containing antibodies capable of neutralizing this antigen (p<0.001) (Table 2).
Table 2.
Seropositivity rates decrease significantly as the number of nucleotide substitutions increases in VP1 of type 2 iVDPVs. Neutralization titers ≥ 3.0 (-log2) were considered seropositive. For each study site, 30 sera were tested for neutralizing antibodies against all type 2 iVDPVs and Sabin 2.
| Isolate | # nt. changes from Sabin 2 | % Seropositivity at each site | |||
|---|---|---|---|---|---|
| NYC, NY | Denver, CO | Detroit, MI | Total | ||
| 18276 | 115 | 50.0 | 20.0 | 16.7 | 28.9 |
| 18277 | 107 | 96.7 | 93.3 | 93.3 | 94.4 |
| 18278 | 98 | 96.7 | 93.3 | 93.3 | 94.4 |
| 18279 | 20 | 100 | 100 | 96.7 | 98.9 |
| 18280 | 16 | 100 | 100 | 96.7 | 98.9 |
| 18281 | 14 | 100 | 100 | 96.7 | 98.9 |
| 18282 | 10 | 100 | 100 | 96.7 | 98.9 |
| Sabin 2 | 0 | 100 | 96.7 | 96.7 | 97.8 |
Because the sera used for analysis were collected from multiple sites, we determined the seropositivity rates for each site (n=30 per site) (Table 2). For strains 18279, 18280, 18281, and 18282, the seropositivity rates for each strain were 100%, 100%, and 96.7% for New York City, Denver and Detroit, respectively (p > 0.05, compared to Sabin 2). For strains 18277 and 18278, we observed a reduced seropositivity rate compared to Sabin 2 for New York City, Denver, and Detroit of 96.7%, 93.3%, and 93.3%, respectively (p > 0.05). The lowest seropositivity rates were seen against strain 18276 in Denver and Detroit (20% and 16.7%, respectively) (p < 0.001). Sera from the New York City site had the highest seropositivity against 18276 compared to Sabin 2, at 50% (p < 0.001).
Highly diverged type 2 iVDPVs are poorly neutralized
iVDPV2 isolates 18282, 18281, 18280 and 18279 were fully neutralized, with median titers of 9.34, 9.17, 8.50 and 8.50, respectively, compared to a median titer of 8.17 to the Sabin 2 control (p = 0.0561 for all compared to Sabin 2). The data for the iVDPV isolates 18278, 18277 and 18276 showed median titers of 7.17, 7.00 and 2.50, respectively, indicating a significant drop in neutralizing capacity at the 98 nucleotide substitution threshold (Figure 1) (p < 0.001 for all compared to Sabin 2). Interestingly, Sabin 2 did not always appear to correspond to the highest titers. iVDPVs with 10, 14, 16 and 20 nucleotide substitutions showed higher median neutralizing capability than Sabin 2 in this study.
Figure 1.
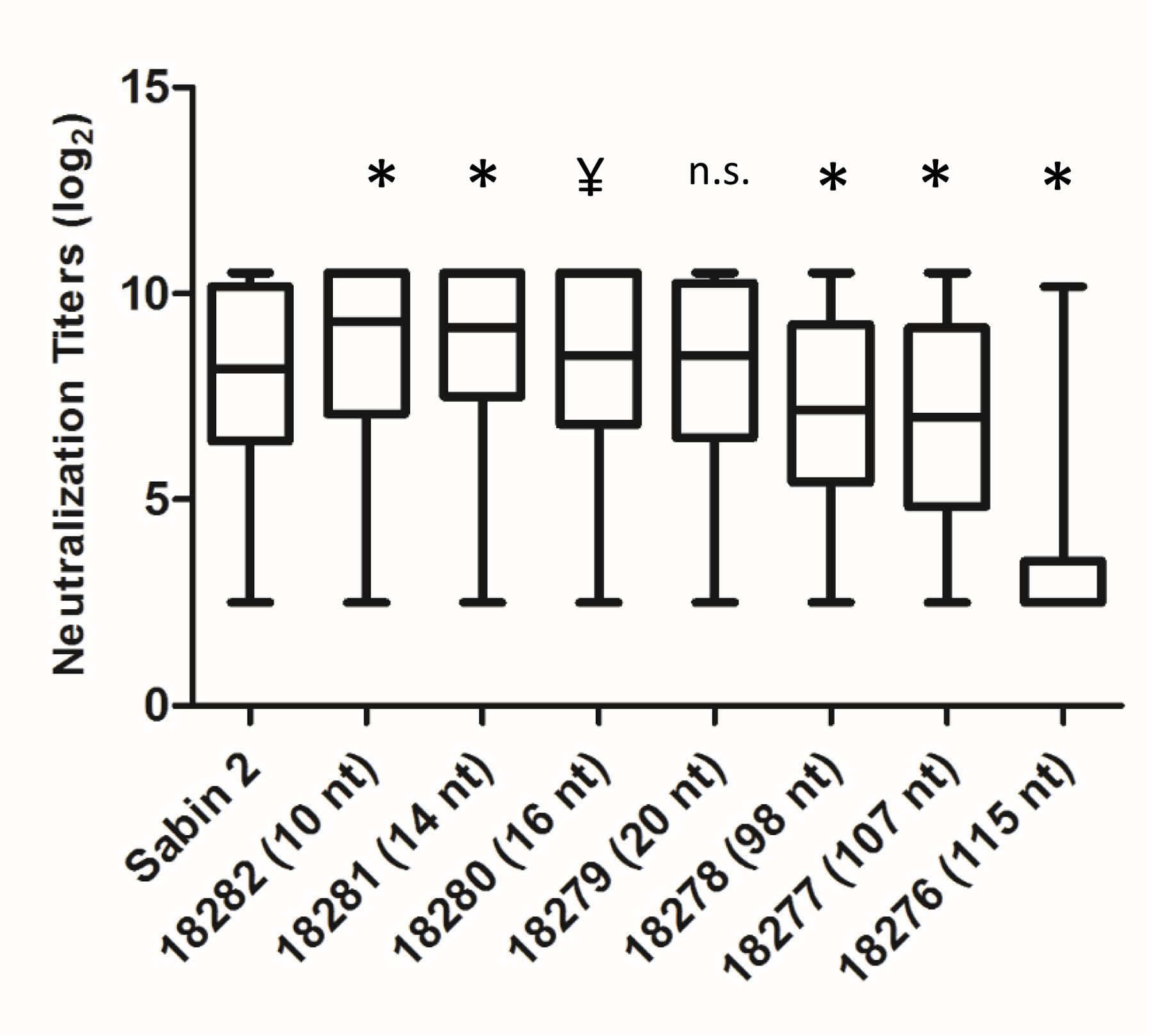
Highly diverged type 2 iVDPVs are poorly neutralized by sera from OPV/IPV vaccinated children. Box plots of neutralization titers against each type 2 iVDPV and Sabin 2, indicating median, upper and lower quartiles, and minimum/maximum titers. All titers are represented in a -log2 format.
For a given serum, the neutralizing titer for Sabin 2 was compared to that of each iVDPV. To quantify that statistical relationship, we conducted a Spearman correlation test (Figure 2). The highest coefficients (0.95, 0.93, 0.95, and 0.93) were seen in the iVDPV2 variants with 10, 14, 20 and 26 changes (18282, 18281, 18280 and 18279, respectively). There was a noticeable reduction in the value of the statistic as the number of mutations increased. Isolate 18276, with 115 nucleotide changes in VP1 showed the weakest correlation coefficient value of 0.59. Therefore, as the number of nucleotide and amino acid changes in VP1 increases, the less Sabin-like the VDPV becomes in terms of neutralizing properties.
Figure 2.
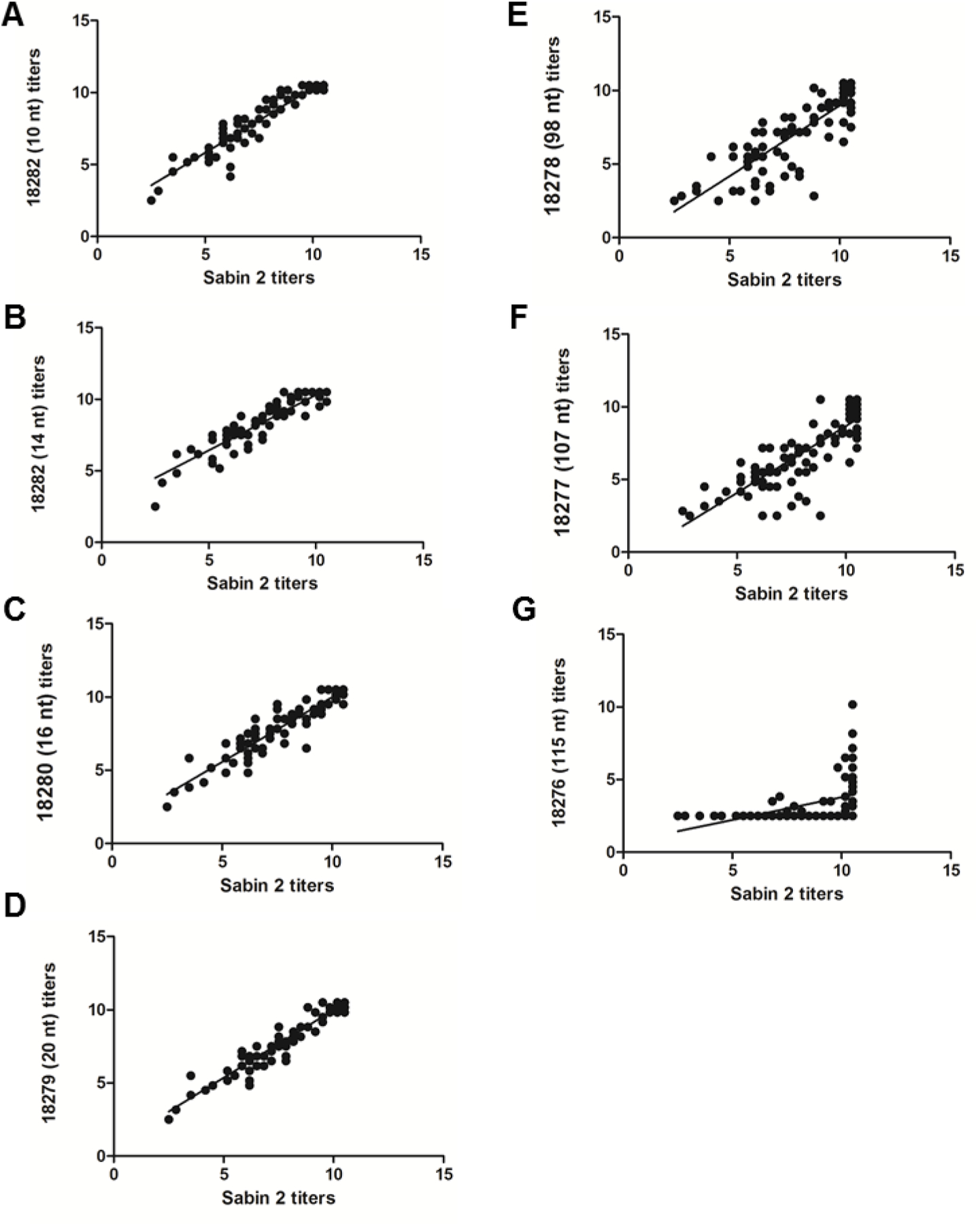
Neutralizing antibody titers to highly divergent type 2 iVDPVs do not correlate with Sabin 2 titers. The neutralization titers of 90 sera from children vaccinated using a combined OPV/IPV schedule against iVDPV2 isolates with increasing numbers of nt substitutions were correlated with the neutralization titers against Sabin 2. All titers are represented in a -log2 format. (A) 10 nt changes, 18282; (B) 14 nt changes, 18281; (C) 16 nt changes, 18280; (D) 20 nt changes, 18279; (E) 98 nt changes, 18278; (F) 107 nt changes, 18277; (G) 115 nt changes, 18276.
Highly diverged iVDPVs accumulate a higher number of amino acid changes in neutralizing antigenic sites
Because we observed a trend of decreasing neutralization titers against iVDPVs with increasing VP1 nucleotide substitutions, we performed multiple sequence alignments of the neutralizing antigenic sites in the translated capsid proteins (Figure 3). The alignment of the neutralizing antigenic sites indicated that as nucleotide changes accumulate in P1, there is a corresponding increase in amino acid changes in the capsid neutralizing antigenic sites. For iVDPVs with fewer than 98 nucleotide changes in VP1, we observed that the majority of amino acid substitutions occurred in the NAg3a and NAg3b. In the isolate with 10 nt changes (18282), we observed non-conservative substitutions at VP3-T73→A (symbolized as T3073A),. For the isolate with 14 nt change (18281), there were two non-conservative substitutions at S3058G and K3061M. The sequence data also indicated a mixed base at amino acid residue 1221 of VP1 for the isolate with 14 nt changes (18281), resulting in either a Sabin-like Ala or a Thr substitution. In the isolate with 16 nt changes (18280), we identified two non-conservative mutations at Q3059K and T3073A. In the isolate with 20 nt change (18279), we observed a non-conservative substitution at T3073M.
Figure 3.
Amino acid substitutions in neutralizing antigenic (NAg) sites occur more frequently in highly diverged type 2 iVDPVs. Multiple sequence alignments for each neutralizing antigenic site (NAg1, NAg2, NAg3a, and NAg3b) were generated for the type 2 iVDPVs using the Sabin 2 sequence as a reference. Numbering system begins with each chain of the poliovirus capsid (VP4, VP2, VP3, or VP1), e.g. 2069 is VP2 residue 69. Residues numbers highlighted in yellow are known to be involved in recognition of the epitope by antigenic-site-specific monoclonal antibodies.
As the number of accumulated nucleotide substitutions in VP1 reached 98 or more, we observed a higher frequency of non-conservative substitutions in all neutralizing antigenic sites. In the isolate with 98 nt change (18278), we observed substitutions in NAg2 at A1221T, T1223A, A2170E, and N2172K. We also observed non-conservative substitutions in NAg3a/b at Q3059G, E3069M, R2070S, H3075P and T3078E. In the 107 nt change isolate (18277), we observed non conservative substitutions in NAg2 at N2172E. We also observed a non-conservative substitution in NAg3a/b at H3075P.
The isolate with 115 nt change (18276) had 13 non-conservative amino acid substitutions, distributed among all antigenic sites. For NAg3b and NAg2, we observed substitutions at R2070S, T2164K, A2166T, T2167D, and N2172K. For NAg3a/3b, substitutions occurred at T3073A, H3075N, S3076L, and T3078E. For NAg1, NAg2 and NAg3a, substitutions occurred at K1099E, S1102D, T1223A, and G1228E. Most notably, the alignment of the NAg1 site indicates that the A1101 residue was deleted in this virus.
Linear regression analysis was used to determine whether a correlation exists between the rate of substitution per site and the neutralization titer; a negative slope and a high R2 would suggest that the substitutions in a given antigenic site correlate with the neutralization titers. The linear regression analysis suggests that the neutralization titers have the strongest correlation with the rate of substitution per site for NAg1 (slope = −0.030, R2 = 0.866) (Figure 4A). In addition, NAg2 (Figure 4B) had a good correlation between the rate of substitution per site and neutralizing antibody titers (slope = −0.069, R2 = 0.600), while only a weak correlation exists for NAg3b (slope = −0.129, R2 = 0.513) (Figure 4D). No correlation was observed between neutralization titers and rate of substitutions per site for NAg3a (Figure 4C). These data suggests that NAg1, 2, and 3b might play a role in the reduced neutralization capacity for the highly divergent iVDPV2 isolates.
Figure 4.
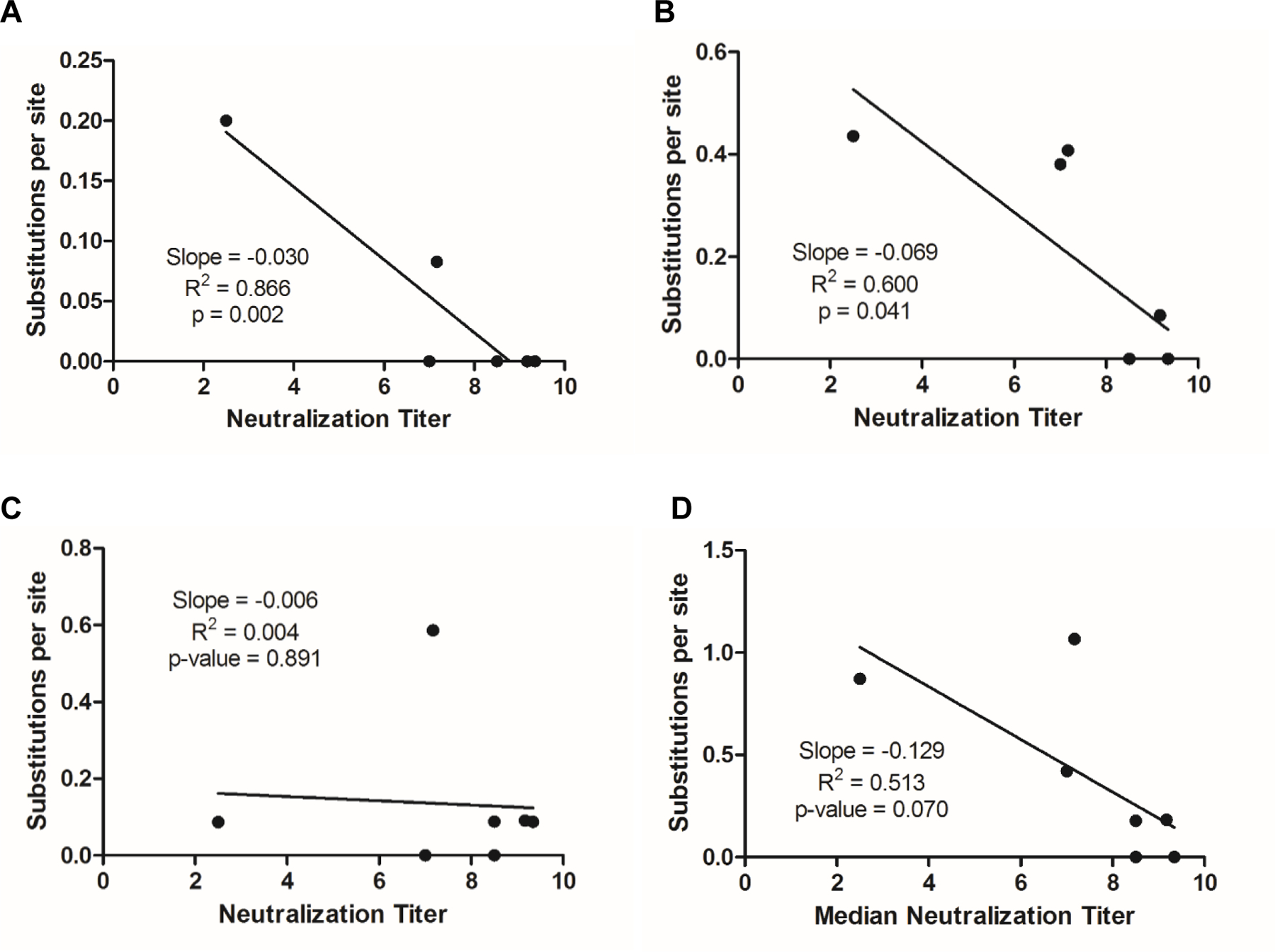
Correlation between increased amino acid substitutions in NAg1 and decreasing median neutralization titers for highly divergent type 2 iVDPVs. A substitution matrix based on the JTT model was used to compare amino acid residues mapped to (A) NAg1, (B) NAg2, (C) NAg3a, and (D) NAg3b.
Predicted structural displacement within NAg1 of highly diverged iVDPVs containing an amino acid deletion in the BC loop of VP1
We sought to investigate further the alterations in the NAg sites that may have contributed to the reduced capacity of vaccine-induced antibodies to neutralize isolate 18276. Therefore, we generated models of the capsid protomer, assembled five copies into the pentameric structure for the 18276 isolate, and compared it with the structure of Sabin 2. The structure similarity was determined using the root mean square difference (RMSD) method and visualized on the pentameric unit (Figure 5). Not surprisingly, the highest degree of structure variation occurred near the deletion site in the BC loop of VP1 (which corresponds to NAg1) and also at NAg2 in VP2. The structure comparison of the homology models suggests a role for both NAg sites in the reduced capacities of sera from vaccinated individuals to neutralize the highly divergent iVDPV.
Figure 5.
Predicted alterations in the antigenic surface of highly divergent type 2 iVDPV compared to Sabin 2. Protomer capsid structures for iVDPV isolate 18276 and Sabin 2 were threaded onto the protomer X-ray crystal structure for MEF-1 (type 2) (PDB: 1EAH). Each protomer was assembled into a pentamer and aligned to the Sabin 2 pentamer structure. Root-mean square deviation (RMSD) was calculated for each amino acid residue and the relative RMSD visualized on the isolate 18276 (115 nt difference) capsid space-fill model. White areas of the pentamer indicate regions structurally similar with Sabin 2 while the intensity of green shading indicates the degree of structural displacement.
Discussion
The primary drawback to OPV is the genetic instability of the vaccine strains, leading to reversion from an attenuated phenotype to the neurovirulent, wild phenotype [33, 34]. These vaccine-derived polioviruses have the capacity to transmit from person-to-person in regions with poor vaccine coverage and have led to outbreaks in Africa, Asia, and the Caribbean [35]. As a result, the Global Polio Eradication Initiative Strategic Plan 2013–2018 called for a switch from trivalent OPV to bivalent OPV and IPV during the pre-eradication phase in an effort to reduce the emergence of VDPVs [36] and the global switch occurred during April 2016.
The ability of OPV and IPV to induce neutralizing antibodies to VDPVs has not been investigated extensively. Previous studies have investigated the antigenic properties of circulating vaccine derived polioviruses (cVDPV) and iVDPV isolates using antigenic site-specific monoclonal antibodies or polyclonal sera from OPV or IPV recipients [17, 19, 21, 22, 28]. These approaches were limited by the number and divergence of VDPV isolates as well as the source and number of sera tested. In the present study, we have demonstrated that nucleotide substitutions in the capsid correlate with reduced neutralizing capacity of sera from immunized children. Furthermore, the substitutions were mapped to sites that are predicted to result in significant structural changes to important neutralization epitopes based on comparisons of pentamer structure models. It is expected that these structural changes contribute to reduced neutralization capacity.
The data from isolate 18276 show some regional differences in seropositivity rates for immunized children, with low rates observed in all three sites included in this study. Interestingly, the study reported that vaccine coverage for at least three doses was 93% in New York, 79% in Detroit and 88% in Denver [17]. However, the proportion of sera with a protective titer against isolate 18276 was 50% in the sera from New York and only 16.7% and 20% in the sera from Detroit and Denver, respectively. This is consistent with a relationship between high vaccine coverage rates and high seropositivity rates against these particular highly diverged iVDPVs. Data from a Sabin-based IPV clinical trial in China support the antigenic divergence of this isolate with an average seropositivity rate of ~24% [37].
The potential for iVDPVs to spread in a population with either high or low OPV/IPV coverage is unclear but wide circulation is considered unlikely. Two documented cases of iVDPVs suggest that it is possible for a VDPV from an immunocompromised individual to be transmitted to unvaccinated subjects. In a 2002 iVDPV case in Russia, a hospitalized infant exhibiting symptoms of acute flaccid paralysis transmitted a VDPV to contact children sharing a hospital room [15]. In a 2005 iVDPV event in the United States, the evidence did not support a clear source of the virus; however, the circulating VDPV was hypothesized to originate from another immunodeficient individual previously exposed to OPV outside the United States [38]. In the case report for isolate 18276 used in this study, there was no indication that household or hospital contacts were tested for poliovirus or for antibodies against the iVDPV, although it was noted that none exhibited neurologic symptoms of poliomyelitis [39]. VDPVs resembling strains isolated from immunodeficient excretors have been detected in environmental samples from countries that have not reported cases of poliomyelitis since 1985 [40–42].
The sequence and structural analysis of isolate 18276 showed that an amino acid deletion occurred at position 101 in the VP1-NAg1 region, resulting in prediction of a significant structural change in the BC loop of VP1. The antigenicity of this site and the flexibility of the BC loop has been proposed for the generation of chimeric vaccine virus, suggesting an important role for NAg1 in the immunogenicity of polioviruses [43]. Mulders et al. have reported a type 1 VDPV isolate with a two-amino acid deletion in the BC loop of VP1 which exhibited a unique neutralization pattern with antigenic-site-specific monoclonal antibodies [19]. However, sera from IPV and OPV vaccine recipients were able to neutralize the VDPV strain with titers against Sabin 1 equal to those of the Mahoney wild reference strain [19]. The BC loop of VP1, which makes up part of NAg1, clearly plays an important role in antibody binding and neutralization although other sites also influence the neutralization capacity of highly diverged VDPV isolates. VDPV type 2 isolates shed from a patient in the United Kingdom for over 30 years have been shown to have reduced binding with antigenic site specific monoclonal antibodies and sera from UK adults, although the most diverged isolate tested did not have a deletion in the BC loop as with isolate 18276 [13]. Due to the significant gap between the number of nucleotide changes in isolate 18279 (20 changes) and isolate 18278 (98 changes), it is not possible to know whether there is a sudden or gradual decrease in neutralizing capacity as the number of mutations increases.
Conclusions
Maintenance of population immunity to poliovirus through high vaccine coverage will prevent cVDPV emergence and spread. Apart from possible limited transmission within an unimmunized community of a VDPV that may have originated with an infection of an immunodeficient person [44], evidence is lacking for spread of iVDPVs in the general population. This observation suggests that other factors may limit transmission of even the most antigenically divergent iVDPVs. Possible contributing factors include: 1) residual cross-neutralizing immunity from immunization may protect immunocompetent individuals before induction of antigen-specific neutralizing antibodies; 2) improved hygiene of older individuals may reduce exposure of contacts to excreted virus; and 3) the highly divergent iVDPVs have distinctive biological properties (such as unusual antigenic structures) that may impair replicative fitness. However, to minimize the potential risks of iVDPV spread, it is important to maintain high levels of population immunity and sensitive poliovirus surveillance now and into the future. In addition, surveillance of immunodeficient patients exposed to OPV should be conducted to identify chronic or long-term excretors of iVDPVs [45]. Although no current treatment options exist, new antiviral drugs and monoclonal antibody treatments show some promise as treatments for these patients with the added benefit of reducing the risk of re-introduction of poliovirus post-eradication [46, 47].
Acknowledgements
This work was supported by Federal appropriations to the Centers for Disease Control and Prevention. We greatly appreciate the contributions of Jackie Quay, Jane Iber and Renee Park in sequencing analysis, and Shohreh Shahmahmoodi and Dioselina Pelaez for laboratory testing of isolates from Iran and Colombia, respectively.
References
- [1].Sutter RW, Kew OM, Cochi SL, Aylward RB. Poliovirus vaccine—live. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 6 ed. London: W.B. Saunders; 2013. p. 598–645. [Google Scholar]
- [2].Moturi EK, Porter KA, Wassilak SGF, Tangermann RH, Diop OM, Burns CC, et al. Progress toward polio eradication — Worldwide, 2013–2014. Morbid Mortal Wkly Rep 2014;63:468–72. [PMC free article] [PubMed] [Google Scholar]
- [3].Jorba J, Diop OM, Iber J, Sutter RW, Wassilak SG, Burns CC. Update on Vaccine-Derived Polioviruses - Worldwide, January 2015-May 2016. MMWR Morbidity and mortality weekly report. 2016;65:763–9. [DOI] [PubMed] [Google Scholar]
- [4].Khan F, Datta SD, Quddus A, Vertefeuille JF, Burns CC, Jorba J, et al. Progress Toward Polio Eradication - Worldwide, January 2016-March 2018. MMWR Morbidity and mortality weekly report. 2018;67:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Centers for Disease C, Prevention. Tracking progress toward global polio eradication, 2010–2011. MMWR Morbidity and mortality weekly report. 2012;61:265–9. [PubMed] [Google Scholar]
- [6].Brown GC, Rabson AS, Schieble JH. The effect of gamma globulin on sub clinical infection in familial associates of poliomyelitis cases. II. Serological studies and virus isolations from pharyngeal secretions. Journal of immunology. 1955;74:71–80. [PubMed] [Google Scholar]
- [7].Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, et al. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science. 2002;296:356–9. [DOI] [PubMed] [Google Scholar]
- [8].Expanding contributions of the global laboratory network for poliomyelitis eradication, 2000–2001. Releve epidemiologique hebdomadaire / Section d’hygiene du Secretariat de la Societe des Nations = Weekly epidemiological record / Health Section of the Secretariat of the League of Nations. 2002;77:133–7. [PubMed] [Google Scholar]
- [9].Maarschalk-Ellerbroek LJ, Hoepelman IM, Ellerbroek PM. Immunoglobulin treatment in primary antibody deficiency. International journal of antimicrobial agents. 2011;37:396–404. [DOI] [PubMed] [Google Scholar]
- [10].Halliday E, Winkelstein J, Webster AD. Enteroviral infections in primary immunodeficiency (PID): a survey of morbidity and mortality. The Journal of infection. 2003;46:1–8. [DOI] [PubMed] [Google Scholar]
- [11].Strebel PM, Aubert-Combiescu A, Ion-Nedelcu N, Biberi-Moroeanu S, Combiescu M, Sutter RW, et al. Paralytic poliomyelitis in Romania, 1984–1992. Evidence for a high risk of vaccine-associated disease and reintroduction of wild-virus infection. American journal of epidemiology. 1994;140:1111–24. [DOI] [PubMed] [Google Scholar]
- [12].Driss N, Ben-Mustapha I, Mellouli F, Ben Yahia A, Touzi H, Bejaoui M, et al. High susceptibility for enterovirus infection and virus excretion features in Tunisian patients with primary immunodeficiencies. Clinical and vaccine immunology : CVI. 2012;19:1684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dunn G, Klapsa D, Wilton T, Stone L, Minor PD, Martin J. Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative. PLoS Pathog 2015;11:e1005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rezapkin G, Martin J, Chumakov K. Analysis of antigenic profiles of inactivated poliovirus vaccine and vaccine-derived polioviruses by block-ELISA method. Biologicals : journal of the International Association of Biological Standardization. 2005;33:29–39. [DOI] [PubMed] [Google Scholar]
- [15].Cherkasova EA, Yakovenko ML, Rezapkin GV, Korotkova EA, Ivanova OE, Eremeeva TP, et al. Spread of vaccine-derived poliovirus from a paralytic case in an immunodeficient child: an insight into the natural evolution of oral polio vaccine. Journal of virology. 2005;79:1062–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bellmunt A, May G, Zell R, Pring-Akerblom P, Verhagen W, Heim A. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology. 1999;265:178–84. [DOI] [PubMed] [Google Scholar]
- [17].Martin J, Odoom K, Tuite G, Dunn G, Hopewell N, Cooper G, et al. Long-term excretion of vaccine-derived poliovirus by a healthy child. Journal of virology. 2004;78:13839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shimizu H, Thorley B, Paladin FJ, Brussen KA, Stambos V, Yuen L, et al. Circulation of type 1 vaccine-derived poliovirus in the Philippines in 2001. Journal of virology. 2004;78:13512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mulders MN, Reimerink JH, Stenvik M, Alaeddinoglu I, van der Avoort HG, Hovi T, et al. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. The Journal of general virology. 1999;80 ( Pt 4):907–16. [DOI] [PubMed] [Google Scholar]
- [20].Blomqvist S, Bruu AL, Stenvik M, Hovi T. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. The Journal of general virology. 2003;84:573–80. [DOI] [PubMed] [Google Scholar]
- [21].Yakovenko ML, Cherkasova EA, Rezapkin GV, Ivanova OE, Ivanov AP, Eremeeva TP, et al. Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties. Journal of virology. 2006;80:2641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pliaka V, Achilleos C, Kyriakopoulou Z, Tsakogiannis D, Ruether IG, Gartzonica C, et al. Determination of antigenic properties of vaccine derived poliovirus strains. Vaccine. 2010;29:26–33. [DOI] [PubMed] [Google Scholar]
- [23].Horie H, Yoshida H, Matsuura K, Miyazawa M, Wakabayashi K, Nomoto A, et al. Isolation of vaccine-derived type 1 polioviruses displaying similar properties to virulent wild strain Mahoney from sewage in Japan. Journal of medical virology. 2002;68:445–51. [DOI] [PubMed] [Google Scholar]
- [24].Prevots DR, Pascual FB, Angellili ML, Brayden R, Irigoyen M, Larussa P, et al. Population immunity to polioviruses among preschool children from four urban underserved low income communities, United States, 1997–2001. The Pediatric infectious disease journal. 2004;23:1130–6. [PubMed] [Google Scholar]
- [25].Weldon WC, Oberste MS, Pallansch MA. Standardized Methods for Detection of Poliovirus Antibodies. Methods Mol Biol 2016;1387:145–76. [DOI] [PubMed] [Google Scholar]
- [26].Archiv GK fuer Experimentelle Pathologie und Pharmakologie. Archiv fuer Experimentelle Pathologie und Pharmakologie. 1931;162:480–3. [Google Scholar]
- [27].Burns CC, Shaw J, Jorba J, Bukbuk D, Adu F, Gumede N, et al. Multiple independent emergences of type 2 vaccine-derived polioviruses during a large outbreak in northern Nigeria. Journal of virology. 2013;87:4907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shaw J, Jorba J, Zhao K, Iber J, Chen Q, Adu F, et al. Dynamics of Evolution of Poliovirus Neutralizing Antigenic Sites and Other Capsid Functional Domains during a Large and Prolonged Outbreak. Journal of virology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hogle JM, Filman DJ. The antigenic structure of poliovirus. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 1989;323:467–78. [DOI] [PubMed] [Google Scholar]
- [30].Patel V, Ferguson M, Minor PD. Antigenic sites on type 2 poliovirus. Virology. 1993;192:361–4. [DOI] [PubMed] [Google Scholar]
- [31].La Monica N, Kupsky WJ, Racaniello VR. Reduced mouse neurovirulence of poliovirus type 2 Lansing antigenic variants selected with monoclonal antibodies. Virology. 1987;161:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences : CABIOS. 1992;8:275–82. [DOI] [PubMed] [Google Scholar]
- [33].Agol VI. Vaccine-derived polioviruses. Biologicals : journal of the International Association of Biological Standardization. 2006;34:103–8. [DOI] [PubMed] [Google Scholar]
- [34].Henderson DA, Witte JJ, Morris L, Langmuir AD. Paralytic Disease Associated with Oral Polio Vaccines. JAMA : the journal of the American Medical Association. 1964;190:41–8. [DOI] [PubMed] [Google Scholar]
- [35].Burns CC, Diop OM, Sutter RW, Kew OM. Vaccine-derived polioviruses. J Infect Dis 2014;210:S283–S93. [DOI] [PubMed] [Google Scholar]
- [36].Initiative GPE. Polio Eradication and Endgame Strategic Plan 2013–2018. 2013. [Google Scholar]
- [37].Sun M, Li C, Xu W, Liao G, Li R, Zhou J, et al. Immune Serum From Sabin Inactivated Poliovirus Vaccine Immunization Neutralizes Multiple Individual Wild and Vaccine-Derived Polioviruses. Clin Infect Dis 2017;64:1317–25. [DOI] [PubMed] [Google Scholar]
- [38].Alexander JP, Ehresmann K, Seward J, Wax G, Harriman K, Fuller S, et al. Transmission of imported vaccine-derived poliovirus in an undervaccinated community in Minnesota. The Journal of infectious diseases. 2009;199:391–7. [DOI] [PubMed] [Google Scholar]
- [39].DeVries AS, Harper J, Murray A, Lexau C, Bahta L, Christensen J, et al. Vaccine-derived poliomyelitis 12 years after infection in Minnesota. N Engl J Med 2011;364:2316–23. [DOI] [PubMed] [Google Scholar]
- [40].Blomqvist S, Savolainen C, Laine P, Hirttio P, Lamminsalo E, Penttila E, et al. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolated from sewage in Estonia. Journal of virology. 2004;78:4876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roivainen M, Blomqvist S, Al-Hello H, Paananen A, Delpeyroux F, Kuusi M, et al. Highly divergent neurovirulent vaccine-derived polioviruses of all three serotypes are recurrently detected in Finnish sewage. Euro surveillance : bulletin europeen sur les maladies transmissibles = European communicable disease bulletin. 2010;15:pii/19566. [PubMed] [Google Scholar]
- [42].Shulman LM, Manor Y, Handsher R, Delpeyroux F, McDonough MJ, Halmut T, et al. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. Journal of clinical microbiology. 2000;38:3729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stirk HJ, Thornton JM. The BC loop in poliovirus coat protein VP1: an ideal acceptor site for major insertions. Protein engineering. 1994;7:47–56. [DOI] [PubMed] [Google Scholar]
- [44].Alexander JP, Ehresmann K, Seward J, Wax G, Harriman K, Fuller S, et al. Transmission of imported vaccine-derived poliovirus in an under-vaccinated community—Minnesota, USA. J Infect Dis 2009;199:391–7. [DOI] [PubMed] [Google Scholar]
- [45].de Silva R, Gunasena S, Ratnayake D, Wickremesinghe GD, Kumarasiri CD, Pushpakumara BA, et al. Prevalence of prolonged and chronic poliovirus excretion among persons with primary immune deficiency disorders in Sri Lanka. Vaccine. 2012;30:7561–5. [DOI] [PubMed] [Google Scholar]
- [46].Chen Z, Chumakov K, Dragunsky E, Kouiavskaia D, Makiya M, Neverov A, et al. Chimpanzee-human monoclonal antibodies for treatment of chronic poliovirus excretors and emergency postexposure prophylaxis. Journal of virology. 2011;85:4354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Collett MS, Hincks JR, Benschop K, Duizer E, van der Avoort H, Rhoden E, et al. Antiviral Activity of Pocapavir in a Randomized, Blinded, Placebo-Controlled Human Oral Poliovirus Vaccine Challenge Model. The Journal of infectious diseases. 2017;215:335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]


