Abstract
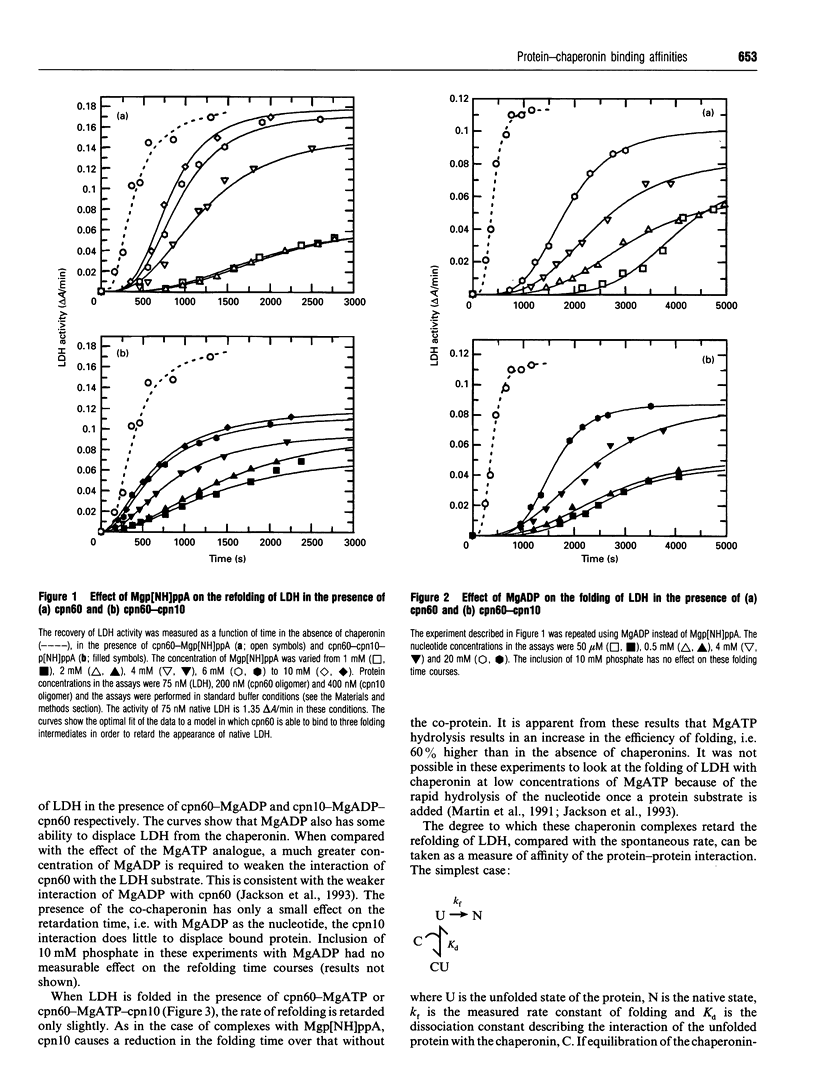
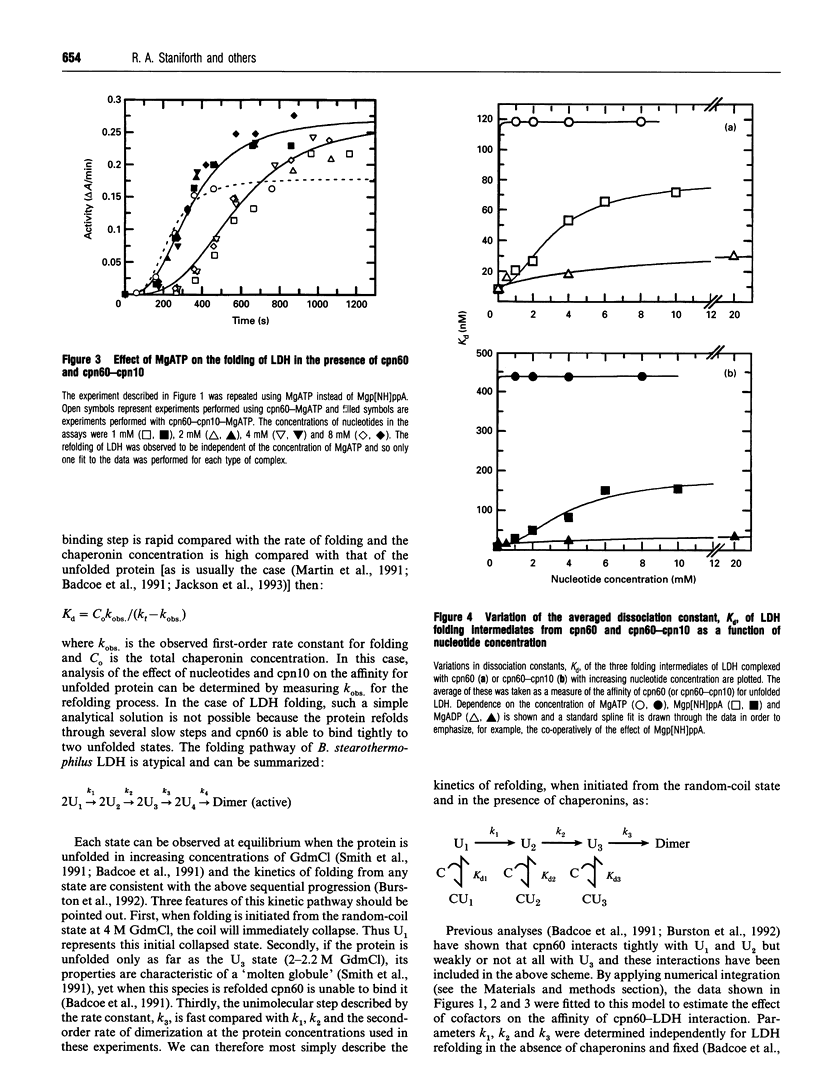
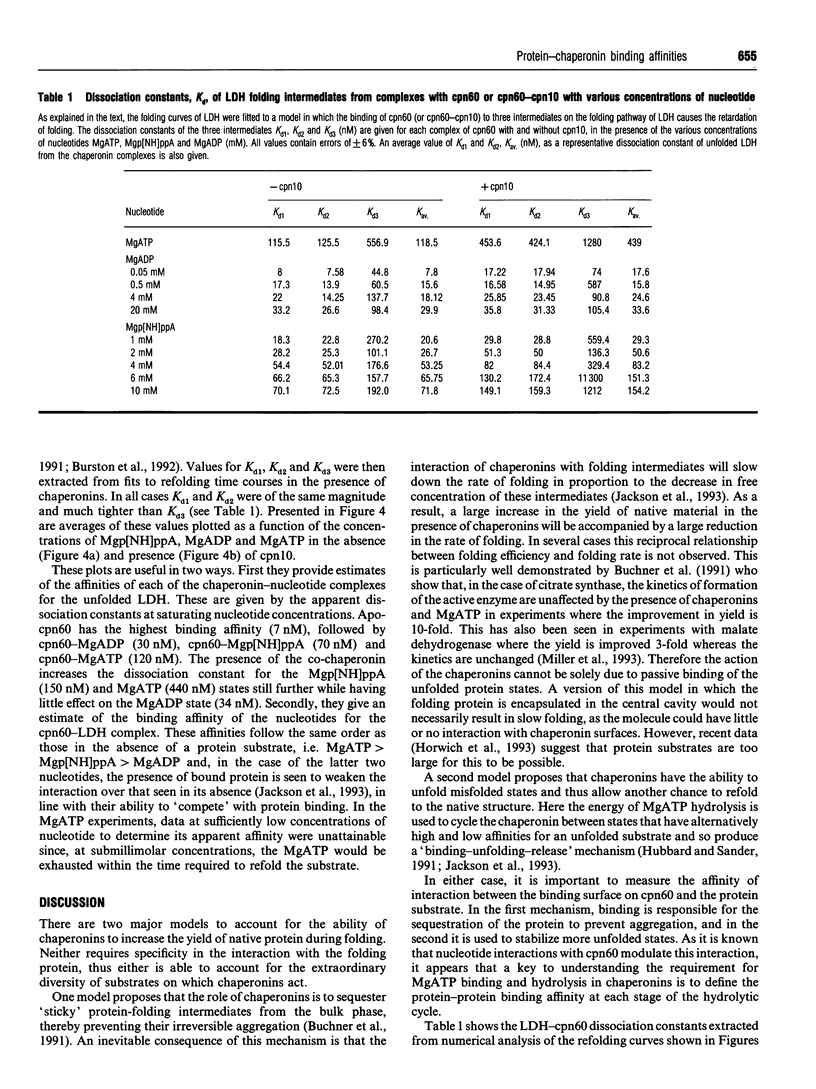
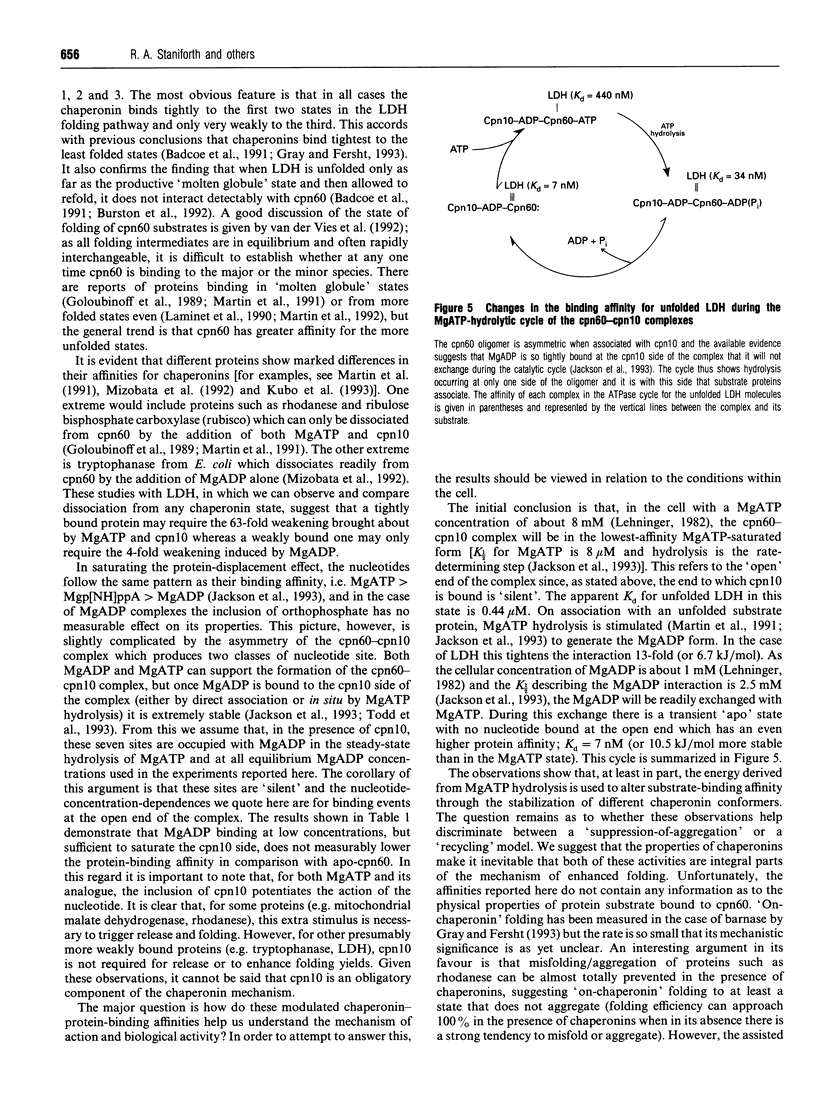
The refolding of lactate dehydrogenase fully unfolded in 4 M guanidinium chloride was initiated by dilution into assay buffer, and the emergence of active enzyme was recorded. This was performed in the presence of the following chaperonin complexes in the refolding medium: chaperonin-60 (cpn60), cpn60-MgATP, cpn60-Mgp[NH]ppA, cpn60-MgADP in both the presence and absence of chaperonin-10 (cpn10). For each nucleotide-chaperonin complex studied, the effect of nucleotide concentration was measured. Dissociation constants (Kd) for unfolded LDH bound to the various chaperonin complexes were derived directly from the ability of the complexes to retard the folding of the enzyme. Dissociation constants for the different complexes were found to be in the order: cpn60 < cpn60-MgADP-cpn10 (formed at low [MgADP]) < cpn60-MgADP < cpn60-MgADP-cpn10 < cpn60-Mgp[NH]ppA < cpn60-Mgp[NH]ppA-cpn10 < cpn60-MgATP < cpn60-MgATP-cpn10; i.e. the tightest complex is with cpn60 and the weakest with cpn60-MgATP-cpn10. Only when MgATP is the nucleotide do we see the yield of native enzyme increased on the time scale of 1 h. The results provide estimates of the change in binding energy between the chaperonin and a substrate protein through the cycle of MgATP binding, hydrolysis and dissociation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badcoe I. G., Smith C. J., Wood S., Halsall D. J., Holbrook J. J., Lund P., Clarke A. R. Binding of a chaperonin to the folding intermediates of lactate dehydrogenase. Biochemistry. 1991 Sep 24;30(38):9195–9200. doi: 10.1021/bi00102a010. [DOI] [PubMed] [Google Scholar]
- Barstow D. A., Clarke A. R., Chia W. N., Wigley D., Sharman A. F., Holbrook J. J., Atkinson T., Minton N. P. Cloning, expression and complete nucleotide sequence of the Bacillus stearothermophilus L-lactate dehydrogenase gene. Gene. 1986;46(1):47–55. doi: 10.1016/0378-1119(86)90165-4. [DOI] [PubMed] [Google Scholar]
- Braig K., Simon M., Furuya F., Hainfeld J. F., Horwich A. L. A polypeptide bound by the chaperonin groEL is localized within a central cavity. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3978–3982. doi: 10.1073/pnas.90.9.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F. X., Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991 Feb 12;30(6):1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- Burston S. G., Sleigh R., Halsall D. J., Smith C. J., Holbrook J. J., Clarke A. R. The influence of chaperonins on protein folding. A mechanism for increasing the yield of the native form. Ann N Y Acad Sci. 1992 Nov 30;672:1–9. doi: 10.1111/j.1749-6632.1992.tb32651.x. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar G. N., Tilly K., Woolford C., Hendrix R., Georgopoulos C. Purification and properties of the groES morphogenetic protein of Escherichia coli. J Biol Chem. 1986 Sep 15;261(26):12414–12419. [PubMed] [Google Scholar]
- Clarke A. R., Waldman A. D., Munro I., Holbrook J. J. Changes in the state of subunit association of lactate dehydrogenase from Bacillus stearothermophilus. Biochim Biophys Acta. 1985 Apr 29;828(3):375–379. doi: 10.1016/0167-4838(85)90319-x. [DOI] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Fayet O., Louarn J. M., Georgopoulos C. Suppression of the Escherichia coli dnaA46 mutation by amplification of the groES and groEL genes. Mol Gen Genet. 1986 Mar;202(3):435–445. doi: 10.1007/BF00333274. [DOI] [PubMed] [Google Scholar]
- Fisher M. T. Promotion of the in vitro renaturation of dodecameric glutamine synthetase from Escherichia coli in the presence of GroEL (chaperonin-60) and ATP. Biochemistry. 1992 Apr 28;31(16):3955–3963. doi: 10.1021/bi00131a010. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989 Dec 21;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Gray T. E., Fersht A. R. Refolding of barnase in the presence of GroE. J Mol Biol. 1993 Aug 20;232(4):1197–1207. doi: 10.1006/jmbi.1993.1471. [DOI] [PubMed] [Google Scholar]
- Hendrix R. W. Purification and properties of groE, a host protein involved in bacteriophage assembly. J Mol Biol. 1979 Apr 15;129(3):375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- Hohn T., Hohn B., Engel A., Wurtz M., Smith P. R. Isolation and characterization of the host protein groE involved in bacteriophage lambda assembly. J Mol Biol. 1979 Apr 15;129(3):359–373. doi: 10.1016/0022-2836(79)90501-1. [DOI] [PubMed] [Google Scholar]
- Horwich A. L., Low K. B., Fenton W. A., Hirshfield I. N., Furtak K. Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell. 1993 Sep 10;74(5):909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- Hubbard T. J., Sander C. The role of heat-shock and chaperone proteins in protein folding: possible molecular mechanisms. Protein Eng. 1991 Oct;4(7):711–717. doi: 10.1093/protein/4.7.711. [DOI] [PubMed] [Google Scholar]
- Ishii N., Taguchi H., Sumi M., Yoshida M. Structure of holo-chaperonin studied with electron microscopy. Oligomeric cpn10 on top of two layers of cpn60 rings with two stripes each. FEBS Lett. 1992 Mar 9;299(2):169–174. doi: 10.1016/0014-5793(92)80240-h. [DOI] [PubMed] [Google Scholar]
- Jackson G. S., Staniforth R. A., Halsall D. J., Atkinson T., Holbrook J. J., Clarke A. R., Burston S. G. Binding and hydrolysis of nucleotides in the chaperonin catalytic cycle: implications for the mechanism of assisted protein folding. Biochemistry. 1993 Mar 16;32(10):2554–2563. doi: 10.1021/bi00061a013. [DOI] [PubMed] [Google Scholar]
- Jenkins A. J., March J. B., Oliver I. R., Masters M. A DNA fragment containing the groE genes can suppress mutations in the Escherichia coli dnaA gene. Mol Gen Genet. 1986 Mar;202(3):446–454. doi: 10.1007/BF00333275. [DOI] [PubMed] [Google Scholar]
- Kubo T., Mizobata T., Kawata Y. Refolding of yeast enolase in the presence of the chaperonin GroE. The nucleotide specificity of GroE and the role of GroES. J Biol Chem. 1993 Sep 15;268(26):19346–19351. [PubMed] [Google Scholar]
- Laminet A. A., Ziegelhoffer T., Georgopoulos C., Plückthun A. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the beta-lactamase precursor. EMBO J. 1990 Jul;9(7):2315–2319. doi: 10.1002/j.1460-2075.1990.tb07403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Horwich A. L., Hartl F. U. Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science. 1992 Nov 6;258(5084):995–998. doi: 10.1126/science.1359644. [DOI] [PubMed] [Google Scholar]
- Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate. Nature. 1991 Jul 4;352(6330):36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Mendoza J. A., Rogers E., Lorimer G. H., Horowitz P. M. Chaperonins facilitate the in vitro folding of monomeric mitochondrial rhodanese. J Biol Chem. 1991 Jul 15;266(20):13044–13049. [PubMed] [Google Scholar]
- Miller A. D., Maghlaoui K., Albanese G., Kleinjan D. A., Smith C. Escherichia coli chaperonins cpn60 (groEL) and cpn10 (groES) do not catalyse the refolding of mitochondrial malate dehydrogenase. Biochem J. 1993 Apr 1;291(Pt 1):139–144. doi: 10.1042/bj2910139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. C., Kelly S. M., Thomson G. J., Coggins J. R., Wood S., auf der Mauer A. The unfolding and attempted refolding of the bacterial chaperone protein groEL (cpn60). Biochim Biophys Acta. 1993 Jan 15;1161(1):52–58. doi: 10.1016/0167-4838(93)90195-w. [DOI] [PubMed] [Google Scholar]
- Saibil H., Dong Z., Wood S., auf der Mauer A. Binding of chaperonins. Nature. 1991 Sep 5;353(6339):25–26. doi: 10.1038/353025b0. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Buchner J. Interaction of GroE with an all-beta-protein. J Biol Chem. 1992 Aug 25;267(24):16829–16833. [PubMed] [Google Scholar]
- Smith C. J., Clarke A. R., Chia W. N., Irons L. I., Atkinson T., Holbrook J. J. Detection and characterization of intermediates in the folding of large proteins by the use of genetically inserted tryptophan probes. Biochemistry. 1991 Jan 29;30(4):1028–1036. doi: 10.1021/bi00218a021. [DOI] [PubMed] [Google Scholar]
- Tilly K., Murialdo H., Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd M. J., Viitanen P. V., Lorimer G. H. Hydrolysis of adenosine 5'-triphosphate by Escherichia coli GroEL: effects of GroES and potassium ion. Biochemistry. 1993 Aug 24;32(33):8560–8567. doi: 10.1021/bi00084a024. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Lubben T. H., Reed J., Goloubinoff P., O'Keefe D. P., Lorimer G. H. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990 Jun 19;29(24):5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- Zeilstra-Ryalls J., Fayet O., Georgopoulos C. The universally conserved GroE (Hsp60) chaperonins. Annu Rev Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- van der Vies S. M., Viitanen P. V., Gatenby A. A., Lorimer G. H., Jaenicke R. Conformational states of ribulosebisphosphate carboxylase and their interaction with chaperonin 60. Biochemistry. 1992 Apr 14;31(14):3635–3644. doi: 10.1021/bi00129a012. [DOI] [PubMed] [Google Scholar]


