Abstract
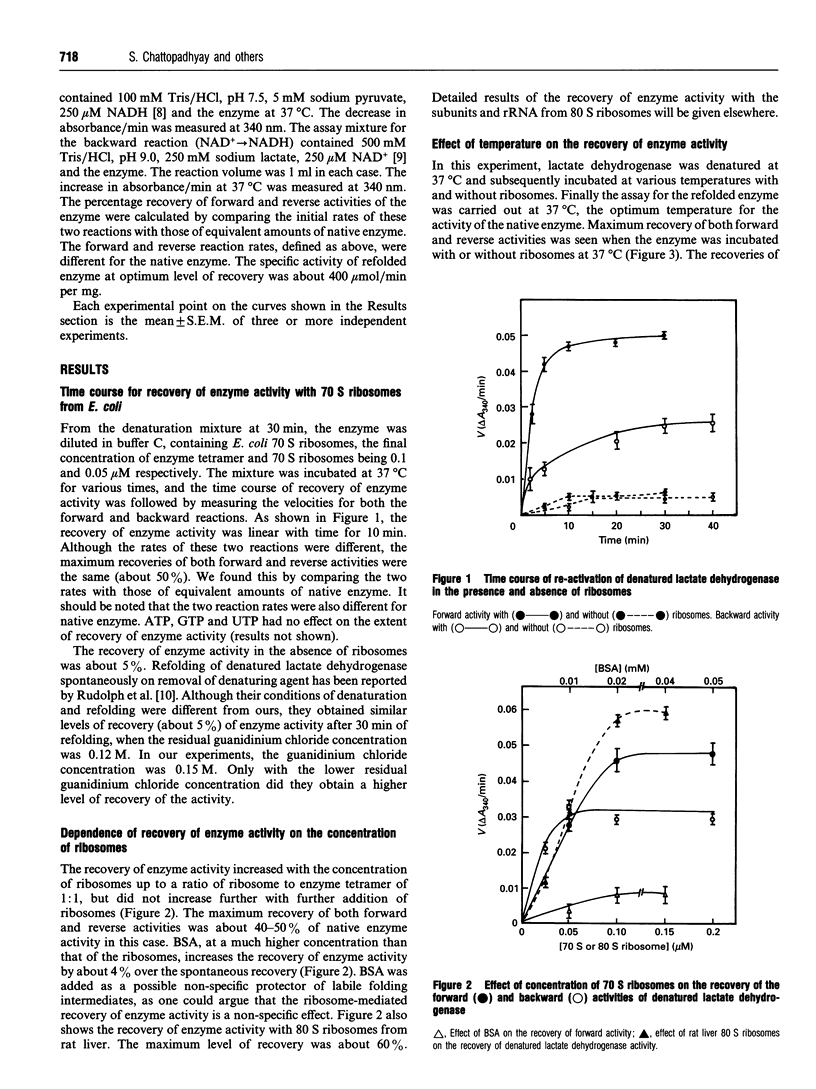
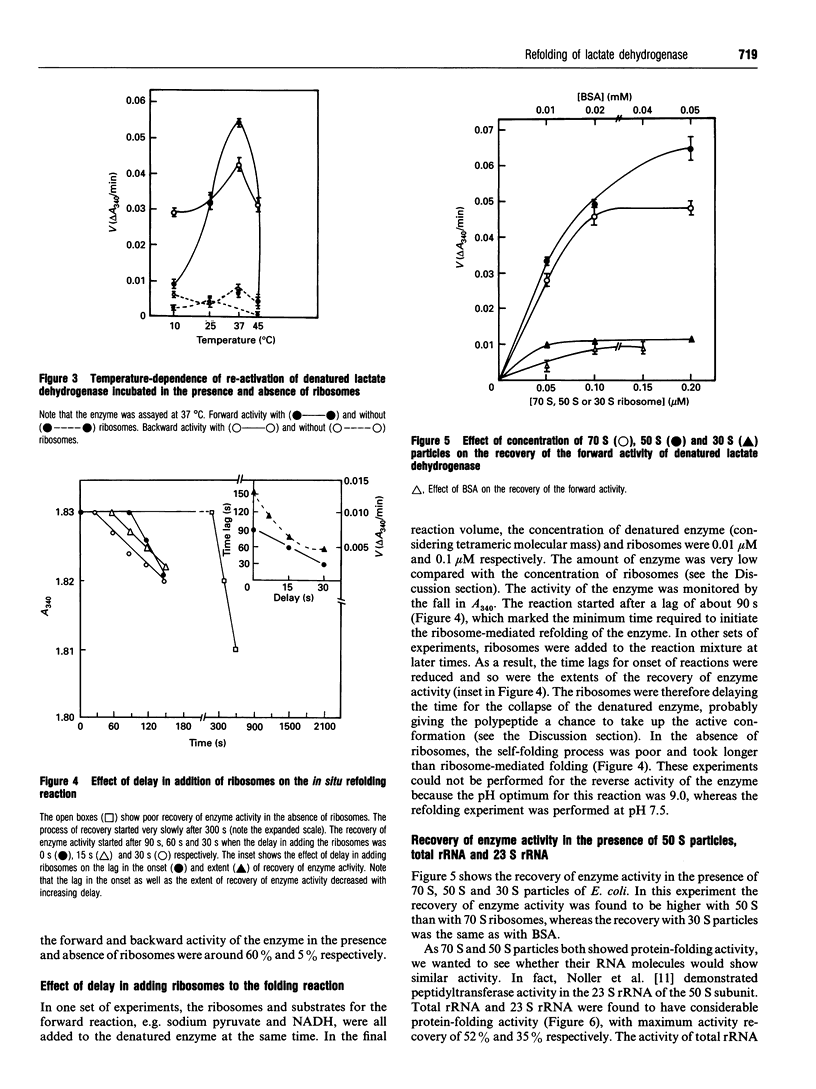
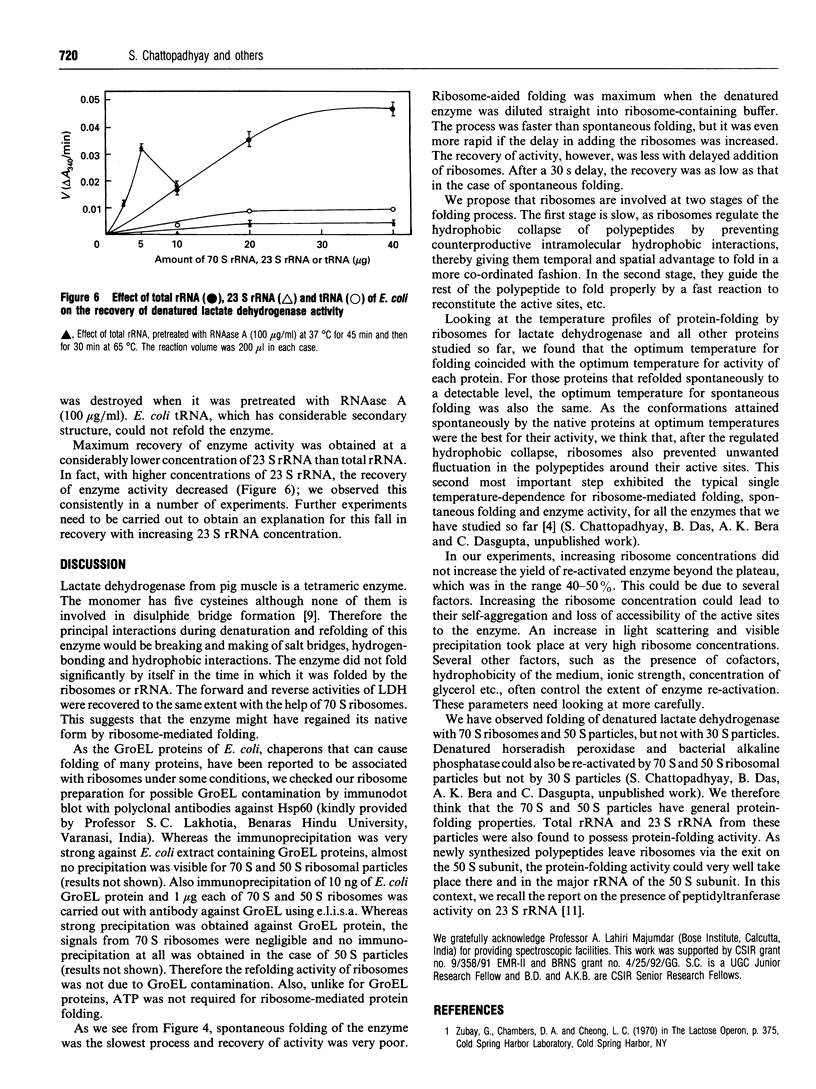
Escherichia coli ribosomes were used to refold denatured lactate dehydrogenase from porcine muscle. This activity of ribosomes, unlike most of the chaperons, did not require the presence of ATP. The molar concentration of ribosomes required for this refolding was comparable with that of the enzyme. Restoration of the enzyme activity was demonstrated using assays for both the forward and backward reactions. Binding of the denatured enzyme to ribosomes and its refolding were fairly rapid processes as revealed by the time course of the reaction and inhibition of folding when the denatured enzyme was allowed to refold spontaneously for short times before the addition of ribosomes. This protein-folding activity was detected in 70 S ribosomes as well as its RNA, in 50 S particles and in 23 S rRNA. However, 30 S particles failed to refold the enzyme.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amils R., Matthews E. A., Cantor C. R. An efficient in vitro total reconstitution of the Escherichia coli 50S ribosomal subunit. Nucleic Acids Res. 1978 Jul;5(7):2455–2470. doi: 10.1093/nar/5.7.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badcoe I. G., Smith C. J., Wood S., Halsall D. J., Holbrook J. J., Lund P., Clarke A. R. Binding of a chaperonin to the folding intermediates of lactate dehydrogenase. Biochemistry. 1991 Sep 24;30(38):9195–9200. doi: 10.1021/bi00102a010. [DOI] [PubMed] [Google Scholar]
- Das B., Chattopadhyay S., Das Gupta C. Reactivation of denatured fungal glucose 6-phosphate dehydrogenase and E. coli alkaline phosphatase with E. coli ribosome. Biochem Biophys Res Commun. 1992 Mar 16;183(2):774–780. doi: 10.1016/0006-291x(92)90550-5. [DOI] [PubMed] [Google Scholar]
- De Crombrugghe B., Chen B., Anderson W., Nissley P., Gottesman M., Pastan I., Perlman R. Lac DNA, RNA polymerase and cyclic AMP receptor protein, cyclic AMP, lac repressor and inducer are the essential elements for controlled lac transcription. Nat New Biol. 1971 Jun 2;231(22):139–142. doi: 10.1038/newbio231139a0. [DOI] [PubMed] [Google Scholar]
- Fedorov A. N., Friguet B., Djavadi-Ohaniance L., Alakhov Y. B., Goldberg M. E. Folding on the ribosome of Escherichia coli tryptophan synthase beta subunit nascent chains probed with a conformation-dependent monoclonal antibody. J Mol Biol. 1992 Nov 20;228(2):351–358. doi: 10.1016/0022-2836(92)90825-5. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992 Jun 5;256(5062):1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Rudolph R., Zettlmeissl G., Jaenicke R. Reconstitution of lactic dehydrogenase. Noncovalent aggregation vs. reactivation. 2. Reactivation of irreversibly denatured aggregates. Biochemistry. 1979 Dec 11;18(25):5572–5575. doi: 10.1021/bi00592a008. [DOI] [PubMed] [Google Scholar]


