Abstract
Cardiovascular diseases are a prominent cause of mortality, emphasizing the need for early prevention and diagnosis. Utilizing artificial intelligence (AI) models, heart sound analysis emerges as a noninvasive and universally applicable approach for assessing cardiovascular health conditions. However, real-world medical data are dispersed across medical institutions, forming “data islands” due to data sharing limitations for security reasons. To this end, federated learning (FL) has been extensively employed in the medical field, which can effectively model across multiple institutions. Additionally, conventional supervised classification methods require fully labeled data classes, e.g., binary classification requires labeling of positive and negative samples. Nevertheless, the process of labeling healthcare data is time-consuming and labor-intensive, leading to the possibility of mislabeling negative samples. In this study, we validate an FL framework with a naive positive-unlabeled (PU) learning strategy. Semisupervised FL model can directly learn from a limited set of positive samples and an extensive pool of unlabeled samples. Our emphasis is on vertical-FL to enhance collaboration across institutions with different medical record feature spaces. Additionally, our contribution extends to feature importance analysis, where we explore 6 methods and provide practical recommendations for detecting abnormal heart sounds. The study demonstrated an impressive accuracy of 84%, comparable to outcomes in supervised learning, thereby advancing the application of FL in abnormal heart sound detection.
Introduction
Cardiovascular diseases (CVDs) are the leading cause of death worldwide, surpassing other causes in annual fatalities [1,2]. The importance of early diagnosis and preventive measures in cardiovascular healthcare cannot be overstressed. Due to its universal and noninvasive nature, heart sound analysis offers a promising avenue in medical care for assessing an individual’s cardiovascular status. Leveraging machine learning models for abnormal heart sound detection in digital healthcare provides a practical approach for early diagnosis and effective prevention of CVDs [3–6].
However, the issues of privacy protection and data silos seriously impede the exploration of medical data and the application of medical artificial intelligence (AI) models [7]. First, variations exist among medical institutions. Some institutions have limited resources and records that hinder effective medical machine learning modeling. Second, pertinent laws and regulations, including the Health Insurance Portability and Accountability Act (HIPAA) [8], restrict data exchange between medical institutions for security and privacy protection. Consequently, healthcare data become fragmented and scattered across medical institutions, causing the phenomenon of “data islands.”
Federated learning (FL) is a distributed machine learning paradigm that enables collaborative modeling among participants without sharing their private data [9–11]. It serves as a viable method to address the “data island” issue in the medical field through collaborative modeling across multiple centers. Consequently, it provides a certain degree of protection for data security and patient privacy. Our studies are based on SecureBoost [12], a federated ensemble learning framework embedded in FATE. [FATE (Federated AI Technology Enabler [13]) supports the FL architecture, as well as the secure computation and development of various machine learning algorithms; https://github.com/FederatedAI/FATE.] In this study, we practically applied the vertical-SecureBoost (Vertically Federated XGBoost) model on a multi-institutional heart sound database. [XGBoost (eXtreme Gradient Boosting [14]) provides an optimized distributed gradient boosting tree-based ensemble model designed to be highly efficient, flexible, and portable; https://xgboost.readthedocs.io.] We propose corresponding federated optimization strategies for the requirements of real-world healthcare scenarios with label scarcity.
In real-life medical scenarios, we consider 3 key issues: (a) Accurately labeling all heart sound records is resource-intensive, leading to only a fraction of the dataset being labeled [15–17]. Semisupervised FL is considered suitable, involving a few “positive” labeled samples and a large volume of “unlabeled” samples, which may contain both positive and negative samples. (b) The widely studied horizontal-FL, also known as sample-partitioned FL [18], requires data from institutions to have the same feature space and different sample spaces. Horizontal-FL is devised to facilitate collaboration among medical institutions with varied patient populations, given the inability to share data across institutions. Therefore, horizontal-FL data partitioning is recommended when developing models with limited sample size variability of FL participants. However, in real medical scenarios, the same patient may receive treatment at different hospitals, allowing for the use of records from multiple sources in diagnosis. Consequently, multiple healthcare institutions may serve the same patient population. Vertical-FL, akin to feature-partitioned FL [19], has recently garnered attention from researchers in cases where medical institutions participating in FL share the same user community but have different medical record feature spaces. This study centers on vertical-FL, aiming to model collaboration across multiple institutions with distinct medical record spaces to provide comprehensive insights into the same patient population. (c) Leveraging the high-dimensional features extracted from heart sound records, it is necessary to select an effective feature importance analysis scheme to retain the most influential feature set [20]. This enhances the efficiency of FL modeling and is anticipated to sustain comparable performance while achieving a reduction in feature dimensionality. Therefore, the contributions of our work can be summarized as follows:
• Our study uniquely shifts from traditional data-centric centralized learning to embrace the FL paradigm in the analysis of the PhysioNet/CinC heart sound database. (Classification of Normal/Abnormal Heart Sound Recordings [21,22]: the PhysioNet/Computing in Cardiology Challenge; https://physionet.org/content/challenge-2016/1.0.0.) We adopt a vertical data partitioning approach and leverage the vertical-SecureBoost FL framework for multi-medical center collaboration modeling to address data islands and privacy concerns in healthcare.
• To meet the demands of real medical scenarios, we promote an FL framework with a naive positive-unlabeled (PU) semisupervised learning strategy. In specific medical contexts, semisupervised FL emphasizes the integration of positive and unlabeled training strategies. The approach achieves a remarkable 84% accuracy, comparable to the outcomes of supervised learning, representing an important exploration of FL in the realm of abnormal heart sound detection.
• In our study practice, we explore 6 distinct methods for feature importance analysis. Utilizing the ensemble learning paradigm based on XGBoost, we compare 5 methods, namely, “gain, total_gain, cover, total_cover, weight,” with the SHAP method. [SHAP (SHapley Additive exPlanations [23]) is a game-theoretic method to explain the output of machine learning models. The method is used to determine the importance of an individual by calculating the contribution of that individual in the cooperation; https://shap.readthedocs.io.] Based on comparative experiments, we provide practical recommendations for feature selection in the context of abnormal heart sound detection.
The rest of the paper is organized as follows: The “Related Works” section introduces the related work. The “Materials and Methods” section describes data preprocessing methods, experimental design, and evaluation metrics. The “Experiment and Results” section presents our comparative experiments and results. The “Discussion” section provides a detailed discussion. Finally, we conclude the paper in the “Conclusion” section.
Related works
In the realm of healthcare, FL has emerged as a pivotal research area, addressing the challenges associated with collaborative modeling across diverse medical institutions. Recent studies emphasize its application in multicenter settings, enabling model training without raw data exchange, thus preserving privacy and adhering to data security regulations. Researchers have investigated federated approaches for tasks such as predictive modeling, disease diagnosis, and personalized treatment recommendations. Examples of noteworthy work include the following:
• Privacy-preserving patient data sharing: Pioneering studies have focused on preserving patient privacy while enabling collaborative model training [24,25]. Techniques such as federated averaging and secure aggregation have been employed to facilitate model updates without raw data sharing. This ensures that FL complies with data protection regulations such as HIPAA.
• Decentralized disease prediction models: Some researchers have applied FL to construct disease prediction models using data across multiple healthcare institutions [26–28]. This approach allows each institution to contribute to the model without sharing patient-specific information, enabling the development of robust and generalizable models.
• Real-world federated systems: Emerging research involves the implementation of FL systems in real-world healthcare settings [29–31]. These systems consider challenges like data heterogeneity, communication efficiency, and model convergence across multiple institutions.
A practical concern often overlooked in healthcare is the limited availability of labeled data. We study the real-world setting of FL medical applications, where assuming fully labeled data in each FL client is less practical. Two related areas include federated unsupervised representation learning and federated semisupervised learning. In scenarios with limited labeled data, semisupervised FL becomes crucial [15–17]. This paradigm involves training models using a combination of labeled and unlabeled data, making it particularly relevant for medical applications with limited annotated datasets. In terms of semisupervised FL, some studies explore cross-institutional transfer learning strategies to transfer knowledge between institutions with varying degrees of labeled data [32]. Therefore, models can leverage labeled data from one institution to enhance the performance on different institution datasets, contributing to better generalization. Additionally, some studies incorporate active learning techniques within FL frameworks to intelligently select and query instances for annotation [33]. This ensures efficient utilization of labeling resources and enhances model performance in scenarios with limited labeled samples. There are few FL studies directly addressing federated PU learning. Study [34] proposes a novel framework called Federated Learning with Positive and Unlabeled Data (FedPU). FedPU considers that each client can label only a limited amount of data for some classes. The work [35] introduces the FedMatch algorithm, a state-of-the-art federated semisupervised model based on consistency regularization training. FedMatch addresses scenarios where clients have both labeled and unlabeled data. We study the problem of learning from positive and unlabeled (PU) data in the federated setting. In contrast to the previous scenario, we focus on situations where some clients exclusively have positive and unlabeled samples, while others have only unlabeled samples.
To sum up, FL in healthcare is developing rapidly, with a focus on preserving privacy and addressing data distribution challenges. The incorporation of semisupervised learning techniques further extends the applicability of federated approaches, especially in scenarios with imbalanced or limited labeled data. These developments set the stage for tackling complex tasks like abnormal heart sound detection across multiple federated care institutions.
Materials and Methods
Dataset description and preprocessing
In this work, heart sound data are obtained from the PhysioNet/CinC [21,22] challenge, a high-quality, authentic public database. As shown in Table 1, it comprises 6 sub-databases, each independently gathered by diverse institutions in clinical and nonclinical environments. Samples labeled as “normal” originate from healthy subjects, whereas “abnormal” samples are derived from patients with various conditions like heart valve disease and coronary artery disease. We use openSMILE [36,37], a widely used open-source toolkit for audio-signal processing, to extract features. openSMILE provides features commonly used in traditional acoustic signal processing methods, including mel frequency cepstrum coefficients (MFCCs), physiological acoustic features, and energy spectrum features. Initially, it extracts low-level descriptor (LLD) features from the audio signal and then re-extracts statistical features from these frame-based LLD features. We use the ComParE [38] feature set in openSMILE, extracting a total of 6,373 dimensional features, which include 65 acoustic LLD features and their associated statistical features. The data preprocessing procedure is summarized in Fig. 1, and the specific steps are outlined below.
Table 1.
Summary of the sub-databases used in the PhysioNet/CinC Challenge
| Sub-set/source | Recordings | # Raw recordings | # Recording length (s) | |||||
|---|---|---|---|---|---|---|---|---|
| Abnormal/proportion (%) | Normal/proportion (%) | Min | Median | Max | ||||
| Dataseta (MIT) | 409 | 292 | 67.5 | 117 | 28.4 | 9.3 | 35.6 | 36.5 |
| Datasetb (AAD) | 490 | 104 | 14.9 | 386 | 60.2 | 5.3 | 8 | 8 |
| Datasetc (AUTH) | 31 | 24 | 64.5 | 7 | 22.6 | 9.6 | 44.4 | 122.0 |
| Datasetd (UHA) | 55 | 28 | 47.3 | 27 | 47.3 | 6.6 | 12.3 | 48.5 |
| Datasete (DLUT) | 2,141 | 183 | 7.1 | 1,958 | 86.7 | 8.1 | 21.1 | 101.7 |
| Datasetf (SUA) | 114 | 34 | 27.2 | 80 | 68.4 | 29.4 | 31.7 | 59.6 |
| Validation | 300 | 150 | 50.0 | 150 | 50.0 | 5.3 | 21.1 | 122.0 |
| Total/average | 3,240 | 665 | 18.1 | 2,575 | 73.0 | 5.3 | 20.8 | 122.0 |
MIT, Massachusetts Institute of Technology; AAD, Aalborg University; AUTH, Aristotle University of Thessaloniki; UHA, University of Haute Alsace; DLUT, Dalian University of Technology; SUA, Shiraz University
Fig. 1.
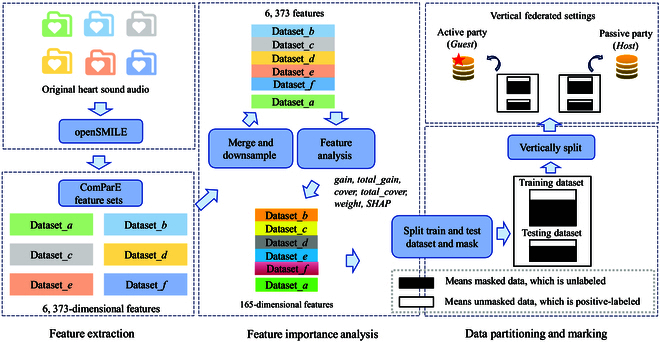
Illustration of the data preprocessing process.
Step 1: Due to the original databases collected by each institution, multiple sets of heart sound records may have been obtained from the same subjects. To ensure subject independence, the experiment combined the data from 5 medical institutions (Dataset{b − f}) as the training set, while the database Dataseta was designated separately as the public test set. Additionally, we implemented a downsampling strategy using the RandomUnderSampler function in Python to address the data imbalance problem. After balancing the samples, there are 665 positive samples and 665 negative samples. The training set to test set ratio is approximately 7:3. The validation set is derived from the officially provided “validation” dataset, comprising 150 positive and 150 negative samples, each.
Step 2: Further selecting the subset of features that have the most impact on the model benefits resource-constrained federated clients, as it is expected to improve model performance while reducing feature dimensionality. As the FL model in this paper is a novel privacy-preserving gradient tree boosting framework, it conducts FL by constructing boosting trees across multiple federated parties. Using the 6,373-dimensional ComParE feature set, we apply 5 tree-based feature importance analysis methods: gain, total_gain, cover, total_cover, and weight, along with a SHAP-based method to assess their individual contributions to the model. Subsequently, the selected 165 features will be used in the hyperparameter experiments of this study.
Step 3: Since accurate labels exist for all samples in the dataset, to assess the effectiveness of the semisupervised FL algorithm, we introduce the assumption that labels for some samples are absent. Following the PU scenario, we designate all negative samples as unlabeled, while also masking a portion of the positive samples as unlabeled. This approach, inspired by a previous study [39], involves randomly selecting 20% of the positive data as labeled positive examples, treating the rest of the data as unlabeled examples. The mask strategy is visually depicted in Fig. 4A, where the unmasked part represents positive samples, and the masked part is unlabeled. This masking strategy is applied to both the training and testing datasets.
Step 4: Following the completion of step 3, we vertically partition the preprocessed dataset, gearing up for the vertical-SecureBoost model with PU learning. In vertical-FL, datasets across institutions share the same sample space but exhibit different feature spaces. To adhere to this condition, vertical partitioning in this study involves vertically dividing the dataset. Let us consider a dataset D = (X, Y) consisting of a feature set X and a label set Y, partitioned into guest = (X1, Y) and host = (X2), where guest represents the federated participant with labels, host denotes the unlabeled participant, and X = X1 ∪ X2. The classifier’s objective is to label the unlabeled samples within the masked segment and accurately classify the unmasked positive samples.
Experimental design
Vertically federated XGBoost (vertical-SecureBoost)
FL is an emerging machine learning paradigm that leverages decentralized data and distributed learning. It offers a novel solution for collaborative modeling across multiple healthcare institutions. In the traditional horizontal-FL approach, participating institutions initially train their models using local data. Subsequently, they transfer the parameters of these local models, such as the gradients of neural networks, to a central server for aggregation. This process enables the construction of robust global models without sharing raw data. Horizontal-FL requires alignment of feature spaces among participants, which is an ideal scenario. This paper considers medical institutions as federated participants and studies the same patient population with different medical record feature spaces, which is consistent with the vertical-FL scenario. Vertical FL, also known as feature-partitioned FL, is suitable for scenarios where medical institutions share the same patient population. In other words, the data of these institutions have the same sample space but different feature spaces.
In this study, we employ a vertical-FL model named vertical-SecureBoost for semisupervised FL learning. In the vertical-SecureBoost setting, only one client has labels, while other clients only have features. The client with labels is referred to as the guest party, and the others are termed host parties. The role of the guest party is analogous to the central server in horizontal-FL. In real medical scenarios, some FL participants have unlabeled data and only serve as feature providers. In response, the semisupervised FL of this paper aims to address the problem of missing and unlabeled labels in federated medical institutions.
The guest party, holding the class labels, is responsible for computing gradient values for all samples and transmitting them to all host parties. Additionally, the guest party is tasked with aggregating feature bins from host parties, decrypting gradient histograms, traversing them, and determining the optimal split point along with the corresponding feature. For host parties, the main function is to compute their own feature bins and local gradient histograms based on the encrypted gradient values of all samples transmitted by the guest party. Upon receiving the broadcast from the guest party regarding the optimal splitting feature, the host party holding that feature must determine the corresponding threshold value. The node-splitting mechanism of the tree model in vertical-SecureBoost is illustrated in Fig. 2.
Fig. 2.
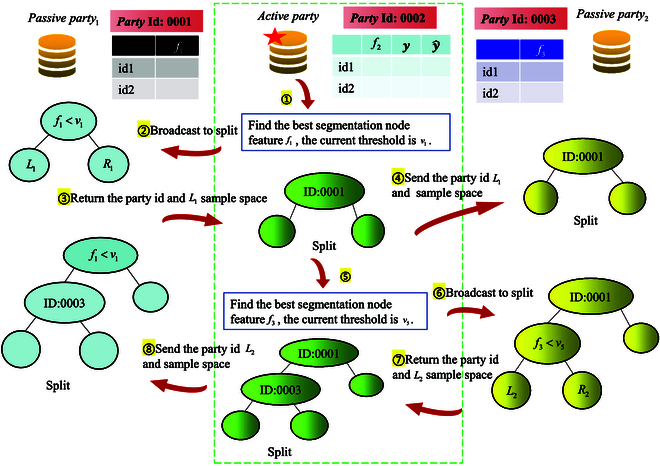
The splitting mechanism for privacy preservation. Vertical-SecureBoost guarantees the privacy and security in the process when multiple parties jointly build the tree model. When the guest party figures out the best split feature, it will notify the party that holds the feature, denoted as host. Then, the host will search for its threshold value, split the local model, and get the left children and right children. After splitting the local model, the host will transfer its party id and the sample space in the left children node to the guest party, since the sample space in the right children can be inferred from the left children. The guest party then records the party id in the current node and splits the local tree model. Then, the guest party will send party id and the sample space in the left children node to the remaining party. In this way, although all the parties share the same tree model, the recorded information of each node of each party’s tree model may be different. Each party can only have the authority to see its own data information.
PU classification scenario
PU classification is prevalent in real-world applications such as healthcare and bioinformatics. The data consist of an incomplete set of positive samples and a set of unlabeled samples that may be either positive or negative.
Stated formally, let y∈ {0, 1} be a binary label, x be the feature matrix, s = 1 if the sample is labeled, and s = 0 if the sample is not labeled. If y = 1, then s = 1. But if s = 0, y can be either 1 or 0. So, we have p(s = 1| x, y = 0) = 0, which means that the probability that a negative sample x appears in the labeled set is zero.
Theoretical basis of the naive PU training strategy
In this study, we adopt a naive PU training strategy, modeling only from positive and unlabeled data. This strategy initially treats all unlabeled samples as negative sample and then trains the model accordingly. High-scoring initial samples are identified as positive label, while the rest are labeled negative. Subsequently, the second classifier is trained. This process is repeated until the unlabeled samples yield the desired result.
The naive PU training strategy has been proved to be reasonable by the work [39]. It shows that a classifier trained on positive and unlabeled examples predicts probabilities that differ by only a constant factor from the true conditional probabilities of being positive. Let f(x) = p(y = 1| x), g(x) = p(s = 1| x). f is a traditional probabilistic classifier, while g is a nontraditional one. It can be proved that p(y = 1| x) = p(s = 1| x)/c, where c is a constant. The proof is that p(s = 1| x) = p(y = 1, s = 1| x) = p(y = 1| x)p(s = 1| y = 1, x) = p(y = 1| x)p(s = 1| y = 1); according to the definition of the PU scenario, p(s = 1| y = 1) is a constant. It can be noticed that f is an increasing function of g. This means that if the classifier f is only used to rank examples x according to the chance that they belong to class y = 1, then classifier g can be used directly instead of f, which verifies the rationality of the naive PU training strategy. The description of relevant variables is shown in Table 2.
Table 2.
List of notations used in the semisupervised vertical-SecureBoost model
| List of notations | Explanations |
|---|---|
| P | Dataset with positive labels |
| U | Unlabeled dataset |
| X1 ∈ Rn×a | Feature matrix of the labeled dataset for the guest party |
| X2 ∈ Rm×a | Feature matrix of the unlabeled dataset for the guest party |
| X3 ∈ R(n+m)×b | Feature matrix of the host party |
| s ∈ {0,1} | s = 1 means the sample is labeled, s = 0 means the sample is unlabeled. |
| y | True labels of the samples |
| ŷ | The predicted label of the sample in the previous round |
| i | Sample identification number |
| j | Client identification number |
| k | Feature identification number |
| q | Feature bin identification number |
| dj | Number of features for the jth client |
| u kq | The splitting value of the qth feature bin for the kth feature |
| gi | The first-order derivative of the loss function with respect to the predicted labels of the previous round, denoted as |
| hi | The second-order derivative of the loss function with respect to the predicted labels of the previous round, denoted as |
Workflow of PU vertical-SecureBoost
PU is applicable to classification tasks in the vertical-FL scenario. The constructed semisupervised FL model can be trained using positive samples and unlabeled samples, and the prediction of unlabeled samples is completed based on the trained model. As the labels change, the data distribution also undergoes alterations, requiring the model to rely on the updated data for continued training. The iterative process continues for multiple rounds until the labels in the dataset converge under predefined rules. Due to the absence of overlapping users among medical institutions, we merged data from five institutions for building the vertical-FL model. Specifically, the multi-dimensional table data extracted after merging are partitioned into 2 segments based on the feature columns, representing the feature spaces for the federated participants—guest and host, respectively. The FL participants, guest and host, meet the requirement that the sample space is the same but the feature space is different, thus enabling vertical-FL modeling. In this study, we designate the medical institution data warehouse as the federated client and establish 2 federated parties for vertical-FL modeling: the guest party and the host party.
Figure 3 illustrates the workflow of semisupervised vertical-SecureBoost with a naive PU training strategy, providing additional details on each component. As the guest participant in the FL, the guest holds 2 types of data: positive samples and unlabeled samples. In the data preprocessing stage, unlabeled samples are treated as negative samples, and the process incorporates the vertical-SecureBoost FL algorithm. The trained federated model is used to predict the unlabeled intersection data of the guest participant. Subsequently, these data are sorted based on their predicted probabilities, and those exceeding a predefined threshold are selected. Positive labels are then assigned to these selected high-probability unlabeled intersection data. Figure 4A illustrates the masking strategy used in our experiments with the selected dataset. Figure 4B provides a simple concrete example to illustrate the training process. Algorithm 1 describes the pseudocodes detailing the basic principles and workflow of semisupervised vertical-SecureBoost with a naive PU training strategy.
Fig. 3.
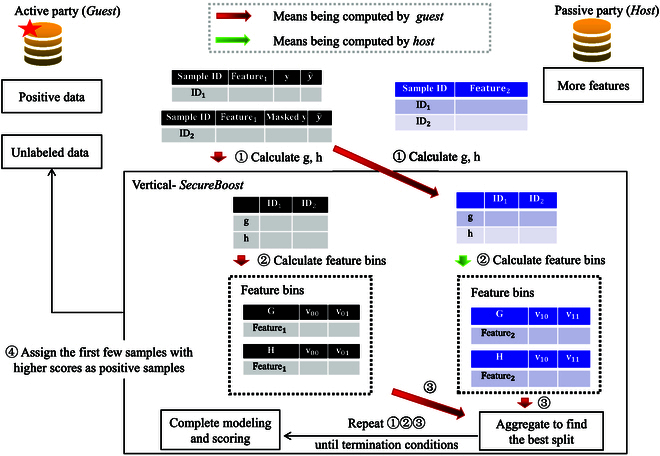
Rough outline of the workflow of semisupervised vertical-SecureBoost. The illustration shows 2 federation participants, a host party and a guest party. In the guest side, ID1 represents labeled samples, ID2 represents unlabeled samples. Masked y refers to our treatment of unlabeled samples based on the PU learning strategy. y represents the predictions of the samples from the previous round. The host side does not have labels and only provides features. In stage 1, the guest side calculates the first-order derivative (gi) and the second-order derivative (hi) of the loss function for each sample ID based on the real or masked labels and the predictions from the previous round, and sends this information to the host side. In stage 2, all parties calculate feature bins based on the information from gi and hi, and this relevant information is transmitted to the guest side. In stage 3, the guest side aggregates all the feature bin information from the participating parties and iteratively calculates the best split points for the tree. In stage 4, the algorithm ranks the samples based on the scoring values obtained using the PU learning strategy.
Fig. 4.
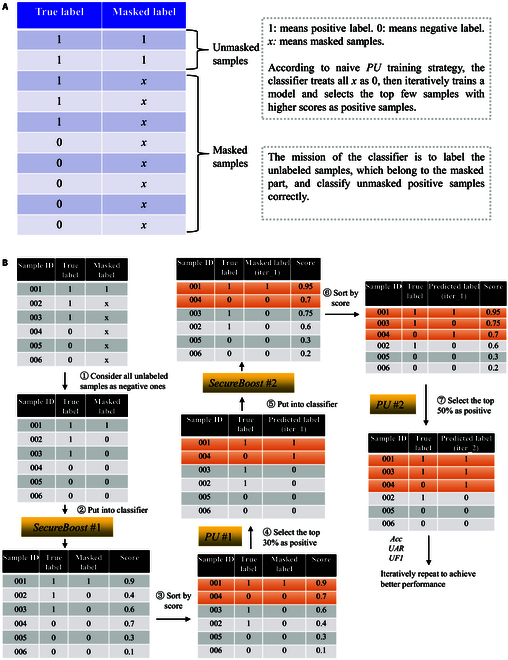
Rough outline of the workflow of semisupervised vertical-SecureBoost. (A) Mask strategy on the dataset. (B) Simple concrete example of the training process.
Evaluation metrics
The multi-institutional heart sound database reflects imbalances in sample size and class distribution across institutions. This study uses the following evaluation metrics, in addition to traditional methods such as accuracy (Acc), to measure model performance. Given are C classes, true positives (TP), false negatives (FN), false positives (FP), and true negatives (TN).
We utilize the unweighted average recall (UAR) and the unweighted F1-Score (UF1) to evaluate the performance of the diagnostic model. The importance of the UAR metric lies in its ability to give equal importance to the performance of each class. Therefore, UAR is especially valuable for evaluating models on datasets where some classes are under-represented. UAR is calculated as:
| (1) |
and UF1 can be formulated as:
| (2) |
Results
Our main objective is to investigate the hyperparameter configurations of the vertical-SecureBoost model with native PU learning. Subsequently, we will conduct comparative experiments to assess model performance using various feature importance analysis methods, aiming to provide valuable insights into abnormal heart sound detection. The experiment includes essential parameters within the interactive learning processes of both the SecureBoost and PU components, along with the selection of 165 features determined by feature importance analysis methods.
Model hyperparameter experiment
We explore crucial hyperparameter settings in semisupervised FL models through 2 sets of experiments. The first set involves configuring the proportion parameter in the PU component and determining the number of trees in the SecureBoost component. The second set focuses on establishing the optimal numbers of SecureBoost and PU components in semisupervised FL. The “proportion in PU” refers to the percentage of top samples considered as positive when executing the current PU, determined based on the sorted scores of samples labeled by the preceding SecureBoost classifier.
Relationship between the first PU (PU1) proportion and model performance
The PU learning strategy enables the FL model to directly learn from a limited set of positive samples and a large pool of unlabeled samples. A control group experiment is conducted to analyze the impact of different proportions in PU1 on FL global model performance while keeping other settings fixed. Since the preprocessed data class is balanced with equal proportion of positive and negative samples, we set the final PU (PU2) proportion to 0.5. This helps the model’s predictions for the samples converge to an equal distribution of positive and negative outcomes. As depicted in Table 3, the model’s performance improves with increasing proportions of PU1. However, when the proportion exceeds 30%, the model metrics start to decline. The model achieves optimal values (Acc: 84.36%, UAR: 84.33%, UF1: 84.35%) when the proportion in PU1 is 30%.
Table 3.
Mean testing performance (in [%]) of 50 repetitions of the semisupervised FL model. Exploring the relationship between the proportion in the first PU (PU1) and model performance. Fixed parameters: The proportion in the second PU (PU2) is 0.5. The number of trees in SecureBoost{1, 2, 3} is 10, 20, and 30, respectively, and the depth of the trees is 3.
|
PU1
proportion |
PU2
proportion |
Acc | UAR | UF1 |
|---|---|---|---|---|
| 0.1 | 0.5 | 80.601 | 80.687 | 80.600 |
| 0.2 | 0.5 | 81.353 | 81.411 | 81.350 |
| 0.3 | 0.5 | 84.360 | 84.338 | 84.351 |
| 0.35 | 0.5 | 82.105 | 82.108 | 82.096 |
| 0.4 | 0.5 | 82.105 | 82.136 | 82.099 |
| 0.5 | 0.5 | 82.105 | 82.136 | 82.099 |
Another control group experiment involves varying the number of tree models in the SecureBoost component. This pertains to the impact of the SecureBoost model complexity on the performance of the semisupervised FL model. The experiment fixed 3 SecureBoost components, each with relevant parameters, and examined the performance variation of the FL model with 10, 20, 30, and 40 trees within each component. As indicated in Table 4, the semisupervised FL model achieved its best performance (Acc: 84.36%, UAR: 84.33%, UF1: 84.35%) when SecureBoost1 has 10 trees, SecureBoost2 has 20 trees, and SecureBoost3 has 30 trees.
Table 4.
Mean testing performance (in [%]) of 50 repetitions of the semisupervised FL model. Exploring the impact of the number of tree models in the SecureBoost component on the performance of semisupervised FL. Fixed parameters: proportion 0.3 in the first PU (PU1), proportion 0.5 in the second PU (PU2). The depth of the tree in SecureBoost is 3.
| FL components | Number of trees | Acc | UAR | UF1 |
|---|---|---|---|---|
| SecureBoost {1, 2, 3} | 10, 10, 10 | 80.977 | 80.936 | 80.959 |
| SecureBoost {1, 2, 3} | 20, 20, 20 | 81.353 | 81.326 | 81.338 |
| SecureBoost {1, 2, 3} | 30, 30, 30 | 82.105 | 82.164 | 82.102 |
| SecureBoost {1, 2, 3} | 40, 40, 40 | 81.062 | 81.034 | 81.058 |
| SecureBoost {1, 2, 3} | 10, 20, 30 | 84.360 | 84.338 | 84.351 |
Relationship between the number of PU and model performance
Figure 4 illustrates the interactive learning process between the SecureBoost and PU components in the FL model based on PU. The number of SecureBoost and PU components determines the iterations or rounds of the learning process. In the control experiment, we varied the number of PUs from 1 to 3, and the corresponding number of SecureBoost components from 2 to 4. As indicated in Table 5, the semisupervised FL model achieves its optimal performance with 2 PU components and 3 SecureBoost components. Experimental results, in conjunction with tree models, demonstrate that we can achieve higher classification performance of the semisupervised FL model with relatively lower model complexity.
Table 5.
Mean testing performance (in [%]) of 50 repetitions of the semisupervised FL model. Exploring the impact of the number of SecureBoost and PU components on model performance. Fixed parameters: The proportion for PU1 is 0.3, and for PU2, it is 0.5. The number of trees in SecureBoost{1, 2, 3} is 10, 20, and 30 respectively, and the depth of the trees is 3.
| Number of PU | Number of SecureBoost | Acc | UAR | UF1 |
|---|---|---|---|---|
| 1 (PU{1}) | 2 (SecureBoost{1, 2}) | 80.225 | 80.240 | 80.216 |
| 2 (PU{1, 2}) | 3 (SecureBoost{1, 2, 3}) | 80.601 | 80.630 | 80.594 |
| 3 (PU{1, 2, 3}) | 4 (SecureBoost{1, 2, 3, 4}) | 80.601 | 80.622 | 80.590 |

Comparative experiment on feature selection methods
This study has 2 main objectives for the semisupervised FL classification model. First, it should perform well, accurately predicting the output of given input features. Second, the model should be interpretable, providing an understanding of the relationship between input features and output. This is crucial when using auxiliary diagnostic models in the sensitive field of healthcare. For instance, in a cardiac auscultation model, it is vital to predict the patient’s diagnosis and understand which features contribute to the result.
Feature importance analysis is a widely used method for interpreting classification models. It quantifies the individual contributions of specific features to a given classifier. Thus, the importance of input data features is model-dependent. In this study, we compared the effects of various feature importance analysis methods on the classification performance of our model, utilizing a high-dimensional feature set extracted from the original heart sound recordings.
In the vertical-FL framework, each federated participant has a distinct feature space. Furthermore, we aim to identify which features contribute most to the performance of the semisupervised FL model in this study. Since vertical-SecureBoost is implemented based on the XGBoost model, we employed 5 tree-based feature selection methods: gain, total_gain, cover, total_cover, and weight. Additionally, we conducted comparative experiments using the SHAP method. Although they are technically related and partially overlap, there is a distinction between feature importance and feature selection. The experiments show that these methods consistently filter the same set of 165 contributing heart sound features (including LLD features and statistical features), with only differences in the importance ranking of these features. The table in the Appendix presents the computed results (feature coefficients and importance values) for the 165 features, sorted by feature importance from the SHAP method. In the comparative experiments, selecting the top 165 features based on the SHAP method yielded optimal model performance (Acc: 84.36%, UAR: 84.33%, UF1: 84.35%). The model results for other feature selection methods under the same conditions are compared in Table 6. Moreover, our optimal model performance closely matches that of the supervised SecureBoost model when using 30 trees and a tree depth of 3. Comparative experimental demonstrate that the semisupervised SecureBoost model efficiently identifies the heart sound features that contribute most, particularly when employing the SHAP feature importance analysis method. The advantage lies in selecting fewer features to achieve superior classification performance, providing clear benefits over other methods. To further demonstrate the superiority of the proposed method, we conduct a comparison with the semisupervised FL algorithms. FedPU (https://github.com/littleSunlxy/FedPU-torch) [34] and FedMatch (https://github.com/wyjeong/FedMatch) [35], compared with the optimization models in this paper, represent the most comparable and state-of-the-art semisupervised FL models. Table 6 presents the performance comparison among FedPU, FedMatch, and the proposed method. Given the limited data resources in this study, the proposed method achieves state-of-the-art performance on the multi-institutional heart sound database. This also demonstrates that our method outperforms other semisupervised FL methods under low-resource conditions.
Table 6.
Mean testing performance (in [%]) of 50 repetitions of semisupervised and supervised FL models. Performance comparison of semisupervised FL models when utilizing different feature importance analysis methods. Fixed parameters: In semisupervised learning, the proportion for PU1 is 0.3, and for PU2, it is 0.5. The number of trees in SecureBoost{1, 2, 3} is 10, 20, and 30, respectively. In supervised learning, the number of trees is 30 and the depth of the trees is 3.
| Methods | Acc | UAR | UF1 |
|---|---|---|---|
| SecureBoost (Supervised) | 84.628 | 84.971 | 85.107 |
| FedPU [34](SHAP feature set) | 75.103 | 68.330 | 68.001 |
| FedMatch [35] (SHAP feature set) | 65.007 | 65.791 | 62.205 |
| SecureBoost with naive PU | - | - | - |
| gain | 81.353 | 81.383 | 81.347 |
| total_gain | 80.977 | 80.964 | 80.964 |
| cover | 79.473 | 79.600 | 79.473 |
| total_cover | 81.353 | 81.383 | 81.347 |
| weight | 80.601 | 80.743 | 80.601 |
| SHAP | 84.360 | 84.338 | 84.351 |
Discussion
We will now discuss 3 aspects: the application of the semisupervised FL model in heart sound classification, the identification of the most important features, and whether the crucial features vary depending on the technique used.
Application of the semisupervised FL model. We study the problem of learning from positive and unlabeled (PU) data in the federated setting. Specifically, we concentrate on scenarios where some clients have only positive and unlabeled samples, while others have only unlabeled samples. The semisupervised FL model can effectively learn from different institutions with a limited pool of positive samples and unlabeled samples. We validated the effectiveness of this framework on real-world heart sound recordings through a series of experiments. Additionally, this framework demonstrates the ability to achieve better classification performance with relatively low model complexity. When utilizing the SHAP feature importance analysis method, all metrics consistently reach above 84%. The semisupervised FL model can conduct multi-institutional federated modeling without sharing local medical institution data. This helps address the issue of medical data silos and partially safeguards patient privacy. However, it is worth noting that the limited data and the relatively simple PU strategy mean that the performance of the FL model in medical diagnosis needs to be improved. To this end, we are collaborating with multiple medical institutions to build a larger, high-quality multi-institutional heart sound database, such as https://www.vob-bit.org, as part of our current work. In practical applications, assessing the performance of the proposed model necessitates considering the diverse environments of each medical institution. Future work should explore various factors in practical applications, such as the number of federated participants, communication costs, data distribution, and FL modeling based on multimodal data [40,41].
What features are the most important? To refine effective representations of heart sounds from the 6,373 features in the ComParE feature set for the model, we employed various feature importance methods. Consistently, these methods identified the same 165 features contributing to the model, albeit with differences in importance ranking. The key statistical findings are as follows: The most influential features encompass 73 related “udSpec” features, with 57 related to “udSpec_Rfilt” and 6 related to “udspecRasta.” Additionally, there are 45 features associated with “fcc_sma” and 36 features linked to “cm_fftMag,” including 30 features tied to “cm_fftMag_spectral” and 5 features associated with “cm_RMSenergy.” This implies that distinct methods can identify the same effective features for the same classification model. Furthermore, the features extracted from the heart sound data exhibit high correlation, making the classification task straightforward. Thus, different feature importance analysis methods can enable the FL model to achieve better classification accuracy.
Do the most important features differ depending on the technique? The most important features indeed depend on the method used. Our experiments indicate that the SHAP method provides better results, as the model’s performance is optimal and stable when the first 165 SHAP features are selected. By selecting fewer features and achieving optimal performance in the analyzed cases, SHAP has a clear advantage over other methods. Ultimately, this study provides insights into screening one-dimensional acoustic signal features for abnormal heart sound examination. It is noteworthy that this framework, rooted in traditional machine learning, is designed for processing one-dimensional tabular data rather than phonocardiogram (PCG) images. Although model interpretability was not the primary focus, the feature importance analysis in this paper lays the foundation for future FL research on feature-based interpretability.
Conclusion
This study was motivated by 2 primary objectives. First, we assessed the classification performance of the semisupervised FL model using real-world heart sound recordings. Second, we investigated the influence of various feature importance methods on the model’s classification performance. Utilizing the classical ComParE feature set, we identified 165 features contributing to the model. Notably, we observed superior performance in heart sound classification with the SHAP-based method, which selected fewer features in the analyzed cases while meeting the model’s performance criteria.
The framework employed a naive PU learning strategy, one of the most basic semisupervised learning methods. In future work, we will explore more complex PU training strategies to enhance the performance of the FL model. Moreover, we intend to replicate the proposed analytical scheme on a larger scale, particularly aiming to implement the techniques utilized in neural network-based FL frameworks. The synergy of advanced nonlinear FL models and sophisticated PU learning strategies is expected to demonstrate significant potential for extensive PCG signals.
Appendix
Table A1.
Based on the ComParE feature set, we present the selected 165 heart sound features and their corresponding computational results
| Feature name | SHAP value | Weight | Gain | Cover | Total gain | Total cover |
|---|---|---|---|---|---|---|
| udspecRasta_lengthL1norm_sma_de_stddevRisingSlope numeric | 0.1749088 | 4 | 59.07317352 | 1330 | 236.2926941 | 5320 |
| fcc_sma[5]_peakMeanRel numeric | 0.066608705 | 2 | 33.78628922 | 908 | 67.57257843 | 1816 |
| fcc_sma[4]_percentile99.0 numeric | 0.027021766 | 1 | 9.735995293 | 1330 | 9.735995293 | 1330 |
| cm_fftMag_spectralSkewness_sma_meanFallingSlope numeric | 0.01917158 | 1 | 5.615310669 | 1330 | 5.615310669 | 1330 |
| cm_fftMag_spectralSlope_sma_risetime numeric | 0.016092975 | 1 | 3.445723057 | 1330 | 3.445723057 | 1330 |
| udSpec_Rfilt_sma_de[22]_quartile3 numeric | 0.015471268 | 2 | 6.0129776 | 374 | 12.0259552 | 748 |
| cm_fftMag_spectralFlux_sma_de_quartile2 numeric | 0.014841603 | 1 | 6.25514555 | 747 | 6.25514555 | 747 |
| udspec_lengthL1norm_sma_de_lpc0 numeric | 0.014638417 | 1 | 13.98928738 | 328 | 13.98928738 | 328 |
| udSpec_Rfilt_sma[11]_risetime numeric | 0.014299023 | 3 | 1.715965867 | 611 | 5.14789772 | 1833 |
| udSpec_Rfilt_sma_de[23]_quartile3 numeric | 0.014214776 | 1 | 9.673725128 | 679 | 9.673725128 | 679 |
| fcc_sma[4]_iqr1-3 numeric | 0.013853458 | 2 | 7.019974709 | 430 | 14.03994942 | 860 |
| oicingFinalUnclipped_sma_flatness numeric | 0.013512484 | 1 | 8.57629776 | 498 | 8.57629776 | 498 |
| udSpec_Rfilt_sma[0]_quartile2 numeric | 0.013094406 | 1 | 2.308807135 | 1330 | 2.308807135 | 1330 |
| oicingFinalUnclipped_sma_lpc0 numeric | 0.01280389 | 1 | 7.025602341 | 596 | 7.025602341 | 596 |
| cm_fftMag_spectralHarmonicity_sma_percentile1.0 numeric | 0.012099205 | 1 | 11.92963409 | 258 | 11.92963409 | 258 |
| cm_fftMag_spectralCentroid_sma_skewness numeric | 0.011240978 | 2 | 0.81099081 | 942 | 1.621981621 | 1884 |
| fcc_sma[3]_peakMeanAbs numeric | 0.011181817 | 1 | 14.97934723 | 451 | 14.97934723 | 451 |
| udSpec_Rfilt_sma_de[13]_stddevRisingSlope numeric | 0.010769798 | 1 | 1.756378412 | 1330 | 1.756378412 | 1330 |
| fcc_sma[3]_iqr2-3 numeric | 0.010016562 | 1 | 5.267727852 | 738 | 5.267727852 | 738 |
| cm_fftMag_spectralSkewness_sma_de_lpgain numeric | 0.009126852 | 1 | 1.210110903 | 1330 | 1.210110903 | 1330 |
| oicingFinalUnclipped_sma_lpgain numeric | 0.007829903 | 1 | 2.564095974 | 611 | 2.564095974 | 611 |
| udSpec_Rfilt_sma_de[2]_risetime numeric | 0.007798587 | 1 | 1.246006727 | 1330 | 1.246006727 | 1330 |
| cm_RMSenergy_sma_peakRangeAbs numeric | 0.007496908 | 1 | 7.285607815 | 734 | 7.285607815 | 734 |
| cm_fftMag_spectralCentroid_sma_minRangeRel numeric | 0.007475868 | 1 | 0.660296619 | 1330 | 0.660296619 | 1330 |
| cm_fftMag_spectralVariance_sma_flatness numeric | 0.007420569 | 1 | 2.622623205 | 775 | 2.622623205 | 775 |
| fcc_sma[3]_amean numeric | 0.006865831 | 1 | 3.782421112 | 393 | 3.782421112 | 393 |
| udSpec_Rfilt_sma[2]_linregerrQ numeric | 0.006835031 | 1 | 0.912325859 | 1326 | 0.912325859 | 1326 |
| cm_RMSenergy_sma_flatness numeric | 0.006573637 | 1 | 6.422821045 | 749 | 6.422821045 | 749 |
| udSpec_Rfilt_sma[5]_quartile3 numeric | 0.006294543 | 1 | 0.478050798 | 1321 | 0.478050798 | 1321 |
| udSpec_Rfilt_sma[5]_iqr1-2 numeric | 0.005954946 | 1 | 1.598445177 | 1111 | 1.598445177 | 1111 |
| fcc_sma[12]_iqr2-3 numeric | 0.005753407 | 1 | 6.725850105 | 583 | 6.725850105 | 583 |
| fcc_sma[13]_lpgain numeric | 0.005692956 | 1 | 0.502746999 | 1298 | 0.502746999 | 1298 |
| cm_fftMag_fband250-650_sma_de_peakDistStddev numeric | 0.005576141 | 1 | 0.389642864 | 1330 | 0.389642864 | 1330 |
| udSpec_Rfilt_sma_de[24]_quartile2 numeric | 0.005573068 | 2 | 1.020026922 | 1330 | 2.040053844 | 2660 |
| udSpec_Rfilt_sma[6]_quartile3 numeric | 0.005310398 | 1 | 5.443786621 | 434 | 5.443786621 | 434 |
| udspecRasta_lengthL1norm_sma_de_iqr1-2 numeric | 0.005278943 | 1 | 1.099442482 | 1018 | 1.099442482 | 1018 |
| fcc_sma[5]_lpgain numeric | 0.005124319 | 1 | 1.139160156 | 424 | 1.139160156 | 424 |
| udSpec_Rfilt_sma_de[13]_meanRisingSlope numeric | 0.005056385 | 1 | 0.394382507 | 1280 | 0.394382507 | 1280 |
| fcc_sma_de[3]_kurtosis numeric | 0.004974036 | 1 | 2.690096855 | 819 | 2.690096855 | 819 |
| fcc_sma_de[2]_percentile1.0 numeric | 0.00495234 | 1 | 0.621264398 | 790 | 0.621264398 | 790 |
| cm_fftMag_fband250-650_sma_linregc1 numeric | 0.004844865 | 1 | 0.281745851 | 1312 | 0.281745851 | 1312 |
| fcc_sma_de[2]_skewness numeric | 0.004828318 | 1 | 0.945549786 | 770 | 0.945549786 | 770 |
| udspec_lengthL1norm_sma_meanSegLen numeric | 0.004793007 | 1 | 1.342338324 | 979 | 1.342338324 | 979 |
| udSpec_Rfilt_sma[6]_meanSegLen numeric | 0.004744658 | 1 | 0.997637093 | 1169 | 0.997637093 | 1169 |
| udSpec_Rfilt_sma[0]_risetime numeric | 0.004732204 | 1 | 3.225561857 | 74 | 3.225561857 | 74 |
| fcc_sma[2]_maxSegLen numeric | 0.004728207 | 1 | 0.519239247 | 1327 | 0.519239247 | 1327 |
| udSpec_Rfilt_sma_de[14]_stddevRisingSlope numeric | 0.00470226 | 1 | 1.475333691 | 391 | 1.475333691 | 391 |
| fcc_sma_de[2]_quartile2 numeric | 0.004379132 | 1 | 1.170669794 | 1303 | 1.170669794 | 1303 |
| fcc_sma_de[11]_peakDistStddev numeric | 0.004351366 | 1 | 0.770152211 | 1330 | 0.770152211 | 1330 |
| fcc_sma_de[9]_peakDistStddev numeric | 0.004338105 | 1 | 0.388319731 | 1325 | 0.388319731 | 1325 |
| cm_fftMag_spectralVariance_sma_linregc2 numeric | 0.004324389 | 1 | 2.174813986 | 879 | 2.174813986 | 879 |
| fcc_sma[1]_quartile1 numeric | 0.004088204 | 2 | 2.74508667 | 351.5 | 5.49017334 | 703 |
| udSpec_Rfilt_sma[6]_iqr2-3 numeric | 0.004022839 | 1 | 1.191879869 | 1294 | 1.191879869 | 1294 |
| cm_fftMag_psySharpness_sma_minRangeRel numeric | 0.003896863 | 1 | 0.401606768 | 1062 | 0.401606768 | 1062 |
| udspecRasta_lengthL1norm_sma_meanSegLen numeric | 0.003853667 | 1 | 0.445445478 | 1295 | 0.445445478 | 1295 |
| udSpec_Rfilt_sma_de[3]_leftctime numeric | 0.003800792 | 1 | 2.882632017 | 592 | 2.882632017 | 592 |
| fcc_sma[10]_peakRangeRel numeric | 0.003772016 | 2 | 1.125457048 | 1041.5 | 2.250914097 | 2083 |
| fcc_sma_de[5]_lpc1 numeric | 0.003659269 | 1 | 2.361129761 | 923 | 2.361129761 | 923 |
| cm_fftMag_spectralSkewness_sma_lpc0 numeric | 0.003499466 | 1 | 3.718276978 | 523 | 3.718276978 | 523 |
| udSpec_Rfilt_sma_de[12]_stddevFallingSlope numeric | 0.003484988 | 1 | 1.881630421 | 407 | 1.881630421 | 407 |
| fcc_sma[2]_upleveltime50 numeric | 0.003321341 | 1 | 0.834810019 | 983 | 0.834810019 | 983 |
| cm_fftMag_spectralSlope_sma_de_quartile3 numeric | 0.003269716 | 1 | 2.736748695 | 569 | 2.736748695 | 569 |
| fcc_sma_de[4]_pctlrange0-1 numeric | 0.003264664 | 1 | 1.344154358 | 555 | 1.344154358 | 555 |
| udSpec_Rfilt_sma[10]_meanSegLen numeric | 0.003213854 | 1 | 0.999613822 | 1074 | 0.999613822 | 1074 |
| cm_fftMag_spectralSkewness_sma_de_flatness numeric | 0.003093224 | 1 | 3.515030384 | 127 | 3.515030384 | 127 |
| cm_fftMag_spectralSkewness_sma_de_percentile99.0 numeric | 0.00309266 | 1 | 1.360512137 | 950 | 1.360512137 | 950 |
| udSpec_Rfilt_sma[10]_lpc1 numeric | 0.003090039 | 1 | 0.669033051 | 888 | 0.669033051 | 888 |
| fcc_sma[7]_linregerrQ numeric | 0.003059623 | 1 | 0.362798691 | 1302 | 0.362798691 | 1302 |
| fcc_sma[13]_iqr2-3 numeric | 0.00305221 | 1 | 2.921410799 | 89 | 2.921410799 | 89 |
| fcc_sma_de[9]_quartile2 numeric | 0.002958984 | 1 | 0.74704951 | 901 | 0.74704951 | 901 |
| fcc_sma[8]_range numeric | 0.002835295 | 1 | 0.767194033 | 1327 | 0.767194033 | 1327 |
| fcc_sma_de[13]_risetime numeric | 0.002822805 | 1 | 1.911473036 | 60 | 1.911473036 | 60 |
| cm_fftMag_spectralSlope_sma_linregc1 numeric | 0.002801212 | 1 | 1.542387009 | 347 | 1.542387009 | 347 |
| udSpec_Rfilt_sma[11]_segLenStddev numeric | 0.002742412 | 1 | 0.933002472 | 385 | 0.933002472 | 385 |
| fcc_sma_de[7]_pctlrange0-1 numeric | 0.002739604 | 1 | 1.344755292 | 506 | 1.344755292 | 506 |
| udSpec_Rfilt_sma_de[6]_percentile1.0 numeric | 0.002661523 | 1 | 1.024646759 | 1330 | 1.024646759 | 1330 |
| udSpec_Rfilt_sma[15]_peakRangeRel numeric | 0.001648004 | 1 | 0.53542912 | 263 | 0.53542912 | 263 |
| fcc_sma[2]_linregc1 numeric | 0.001599217 | 1 | 0.394384265 | 600 | 0.394384265 | 600 |
| cm_fftMag_psySharpness_sma_linregc1 numeric | 0.001595959 | 1 | 0.334819168 | 1324 | 0.334819168 | 1324 |
| udSpec_Rfilt_sma[7]_leftctime numeric | 0.001591305 | 1 | 1.373440266 | 44 | 1.373440266 | 44 |
| fcc_sma[3]_upleveltime90 numeric | 0.001587785 | 1 | 2.038755417 | 760 | 2.038755417 | 760 |
| fcc_sma[5]_rqmean numeric | 0.001532889 | 2 | 0.726613462 | 1266.5 | 1.453226924 | 2533 |
| fcc_sma[10]_skewness numeric | 0.001477398 | 1 | 0.527558625 | 324 | 0.527558625 | 324 |
| cm_fftMag_spectralKurtosis_sma_de_flatness numeric | 0.001469919 | 1 | 0.467760682 | 1241 | 0.467760682 | 1241 |
| udSpec_Rfilt_sma[25]_upleveltime50 numeric | 0.001460258 | 1 | 0.302553505 | 1326 | 0.302553505 | 1326 |
| fcc_sma_de[5]_lpc4 numeric | 0.001403587 | 1 | 0.539424777 | 1291 | 0.539424777 | 1291 |
| udSpec_Rfilt_sma_de[10]_upleveltime90 numeric | 0.001380694 | 1 | 1.193989992 | 23 | 1.193989992 | 23 |
| cm_fftMag_spectralRollOff75.0_sma_de_stddevFallingSlope numeric | 0.001334538 | 1 | 0.547998667 | 798 | 0.547998667 | 798 |
| oicingFinalUnclipped_sma_range numeric | 0.00132699 | 1 | 1.335298538 | 31 | 1.335298538 | 31 |
| udSpec_Rfilt_sma[6]_pctlrange0-1 numeric | 0.001314231 | 1 | 0.401833385 | 1185 | 0.401833385 | 1185 |
| cm_fftMag_fband250-650_sma_de_range numeric | 0.001303879 | 1 | 0.647293746 | 507 | 0.647293746 | 507 |
| oicingFinalUnclipped_sma_de_quartile3 numeric | 0.001280073 | 1 | 0.825291157 | 49 | 0.825291157 | 49 |
| udSpec_Rfilt_sma[17]_lpc3 numeric | 0.001216713 | 1 | 0.705320001 | 55 | 0.705320001 | 55 |
| fcc_sma[6]_qregc1 numeric | 0.001205926 | 1 | 0.807898402 | 100 | 0.807898402 | 100 |
| cm_fftMag_fband1000-4000_sma_de_minPos numeric | 0.00111533 | 1 | 0.484175861 | 522 | 0.484175861 | 522 |
| udSpec_Rfilt_sma_de[25]_minSegLen numeric | 0.001072463 | 1 | 1.136362314 | 33 | 1.136362314 | 33 |
| udSpec_Rfilt_sma_de[6]_lpc4 numeric | 0.001038906 | 1 | 0.51261425 | 532 | 0.51261425 | 532 |
| cm_fftMag_spectralSlope_sma_minPos numeric | 0.001018498 | 1 | 1.17227602 | 775 | 1.17227602 | 775 |
| udSpec_Rfilt_sma_de[22]_skewness numeric | 0.000986683 | 1 | 0.63786608 | 796 | 0.63786608 | 796 |
| udspec_lengthL1norm_sma_de_pctlrange0-1 numeric | 0.00096522 | 1 | 0.421422035 | 44 | 0.421422035 | 44 |
| udSpec_Rfilt_sma_de[3]_quartile2 numeric | 0.000914101 | 1 | 0.789074838 | 27 | 0.789074838 | 27 |
| cm_fftMag_fband1000-4000_sma_qregc3 numeric | 0.000913065 | 1 | 0.399933308 | 712 | 0.399933308 | 712 |
| udSpec_Rfilt_sma_de[7]_upleveltime75 numeric | 0.000890098 | 1 | 0.495113492 | 1330 | 0.495113492 | 1330 |
| fcc_sma[12]_stddevFallingSlope numeric | 0.000889177 | 1 | 0.514449418 | 41 | 0.514449418 | 41 |
| cm_fftMag_spectralFlux_sma_lpc0 numeric | 0.000887822 | 1 | 0.672494352 | 83 | 0.672494352 | 83 |
| udSpec_Rfilt_sma_de[17]_peakRangeRel numeric | 0.000886993 | 1 | 0.509827614 | 34 | 0.509827614 | 34 |
| udSpec_Rfilt_sma_de[0]_maxPos numeric | 0.000876428 | 1 | 0.861174822 | 16 | 0.861174822 | 16 |
| cm_RMSenergy_sma_de_stddevFallingSlope numeric | 0.000875804 | 1 | 0.281979769 | 1330 | 0.281979769 | 1330 |
| udSpec_Rfilt_sma[9]_percentile1.0 numeric | 0.000866589 | 1 | 0.848887801 | 93 | 0.848887801 | 93 |
| cm_fftMag_spectralFlux_sma_lpc4 numeric | 0.000853217 | 1 | 0.259635895 | 117 | 0.259635895 | 117 |
| cm_fftMag_spectralFlux_sma_peakMeanRel numeric | 0.000839713 | 1 | 0.686322689 | 433 | 0.686322689 | 433 |
| udSpec_Rfilt_sma_de[4]_quartile2 numeric | 0.000838053 | 1 | 0.496914715 | 32 | 0.496914715 | 32 |
| cm_fftMag_spectralEntropy_sma_peakDistStddev numeric | 0.000832948 | 1 | 0.411569834 | 208 | 0.411569834 | 208 |
| udSpec_Rfilt_sma_de[2]_quartile2 numeric | 0.000829766 | 1 | 0.520026982 | 24 | 0.520026982 | 24 |
| udSpec_Rfilt_sma[12]_upleveltime90 numeric | 0.000786718 | 1 | 0.829185367 | 27 | 0.829185367 | 27 |
| cm_zcr_sma_de_peakRangeRel numeric | 0.00076202 | 1 | 0.883337021 | 1330 | 0.883337021 | 1330 |
| udspecRasta_lengthL1norm_sma_quartile1 numeric | 0.000733587 | 1 | 0.555594325 | 16 | 0.555594325 | 16 |
| cm_fftMag_spectralSkewness_sma_qregc3 numeric | 0.000690544 | 1 | 0.295283973 | 27 | 0.295283973 | 27 |
| udSpec_Rfilt_sma_de[12]_peakRangeRel numeric | 0.00067913 | 1 | 0.487163782 | 35 | 0.487163782 | 35 |
| udSpec_Rfilt_sma[13]_lpgain numeric | 0.00065719 | 1 | 0.308226794 | 1295 | 0.308226794 | 1295 |
| cm_fftMag_spectralVariance_sma_range numeric | 0.000638415 | 1 | 0.73640269 | 1119 | 0.73640269 | 1119 |
| cm_fftMag_spectralKurtosis_sma_peakMeanMeanDist numeric | 0.000616125 | 1 | 0.413334399 | 1330 | 0.413334399 | 1330 |
| udSpec_Rfilt_sma[19]_minPos numeric | 0.0005893 | 1 | 0.760017276 | 1330 | 0.760017276 | 1330 |
| udspec_lengthL1norm_sma_de_meanRisingSlope numeric | 0.000573997 | 1 | 0.358050197 | 1330 | 0.358050197 | 1330 |
| cm_fftMag_spectralKurtosis_sma_linregc1 numeric | 0.00056523 | 1 | 0.284590483 | 1330 | 0.284590483 | 1330 |
| udspecRasta_lengthL1norm_sma_de_lpc0 numeric | 0.000546952 | 1 | 0.879844904 | 12 | 0.879844904 | 12 |
| fcc_sma[2]_lpc2 numeric | 0.000519563 | 1 | 0.439446568 | 23 | 0.439446568 | 23 |
| udSpec_Rfilt_sma[0]_maxPos numeric | 0.000489432 | 1 | 0.514279604 | 1330 | 0.514279604 | 1330 |
| udspec_lengthL1norm_sma_leftctime numeric | 0.000487373 | 1 | 0.960110188 | 23 | 0.960110188 | 23 |
| udSpec_Rfilt_sma[8]_minRangeRel numeric | 0.000476519 | 1 | 0.255015016 | 26 | 0.255015016 | 26 |
| cm_RMSenergy_sma_iqr1-2 numeric | 0.000443091 | 1 | 0.301709265 | 1330 | 0.301709265 | 1330 |
| cm_RMSenergy_sma_upleveltime90 numeric | 0.000413161 | 1 | 0.188143015 | 19 | 0.188143015 | 19 |
| cm_fftMag_spectralRollOff75.0_sma_upleveltime75 numeric | 0.000366831 | 1 | 0.074845433 | 31 | 0.074845433 | 31 |
| udspecRasta_lengthL1norm_sma_maxPos numeric | 0.000324066 | 1 | 0.175449252 | 10 | 0.175449252 | 10 |
| fcc_sma[6]_minPos numeric | 0.000319397 | 1 | 0.101224005 | 18 | 0.101224005 | 18 |
| udspec_lengthL1norm_sma_lpc0 numeric | 0.000275772 | 2 | 0.0727164 | 6.5 | 0.1454328 | 13 |
| udspec_lengthL1norm_sma_percentile99.0 numeric | 0.000275314 | 1 | 0.221500084 | 8 | 0.221500084 | 8 |
| udSpec_Rfilt_sma[7]_lpc4 numeric | 0.000273154 | 1 | 0.122567415 | 14 | 0.122567415 | 14 |
| udSpec_Rfilt_sma[4]_lpc0 numeric | 0.000215668 | 1 | 0.021040797 | 14 | 0.021040797 | 14 |
| udspec_lengthL1norm_sma_de_risetime numeric | 0.000187452 | 1 | 0.08378467 | 9 | 0.08378467 | 9 |
| udSpec_Rfilt_sma[16]_percentile1.0 numeric | 0.000168175 | 1 | 0.068053588 | 9 | 0.068053588 | 9 |
| udspec_lengthL1norm_sma_maxPos numeric | 9.77E−05 | 1 | 0.018804565 | 8 | 0.018804565 | 8 |
| udspec_lengthL1norm_sma_risetime numeric | 8.27E−05 | 1 | 0.017634902 | 8 | 0.017634902 | 8 |
Acknowledgments
Funding: This work was partially supported by the National Natural Science Foundation of China (grant number 62272044); the Ministry of Science and Technology of the People’s Republic of China with the STI2030-Major Projects (grant number 2021ZD0201900); the Teli Young Fellow Program from the Beijing Institute of Technology, China; the Grants-in-Aid for Scientific Research (grant number 20H00569) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; the JSPS KAKENHI (grant number 20H00569), Japan; the JST Mirai Program (grant number 21473074), Japan; the JST MOONSHOT Program (grant number JPMJMS229B), Japan; and the BIT Research and Innovation Promoting Project (grant number 2023YCXZ014).
Author contributions: W.Q. and C.Q. led the investigation, conceptualization, experimentation, and drafting of the initial manuscript. Y. Yu and E.K. conducted data analysis and co-developed the program. K.Q. and B.H. revised the intellectual content and contributed to writing, review, editing, and funding acquisition. B.W.S. and Y. Yamamoto supervised the writing process and provided support for manuscript editing and revision. All authors reviewed and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data Availability
This study utilizes heart sound recordings from the PhysioNet/Computing in Cardiology Challenge, which provides a publicly available heart sound database. The database can be accessed at: https://physionet.org/content/challenge-2016/1.0.0 . Additional preprocessed data relevant to this paper may be requested from the author via email.
References
- 1.Timmis A, Vardas P, Townsend N, Torbica A, Katus H, De Smedt D, Gale CP, Maggioni AP, Petersen SE, Huculeci R, et al. European Society of Cardiology: Cardiovascular disease statistics 2021. Eur Heart J. 2022;43(8):716–799. [DOI] [PubMed] [Google Scholar]
- 2.Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the gbd 2019 study. J Am Coll Cardiol. 2020;76(25):2982–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winther S, Nissen L, Schmidt SE, Westra J, Andersen IT, Nyegaard M, Madsen LH, Knudsen LL, Urbonaviciene G, Larsen BS, et al. Advanced heart sound analysis as a new prognostic marker in stable coronary artery disease. Eur Heart J Digital Health. 2021;2(2):279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy JK, Roy TS, Mukhopadhyay SC. Heart sound: Detection and analytical approach towards diseases. In: Modern sensing technologies. Cham: Springer; 2019. p. 103–145. [Google Scholar]
- 5.Li S, Li F, Tang S, Xiong W. A review of computer-aided heart sound detection techniques. Biomed Res Int. 2020;2020: Article 5846191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial intelligence: Practical primer for clinical research in cardiovascular disease. J Am Heart Assoc. 2019;8(17): Article e012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao D, Liu J, Wang M, Zhang X, Zhou M. Epidemiology of cardiovascular disease in China: Current features and implications. Nat Rev Cardiol. 2019;16(4):203–212. [DOI] [PubMed] [Google Scholar]
- 8.Accountability Act. Health Insurance Portability and Accountability Act. Public Law. 2023.
- 9.McMahan B, Moore E, Ramage D, Hampson S, Arcas BAY. Communication-efficient learning of deep networks from decentralized data. In: Artificial intelligence and statistics. Fort Lauderdale (FL): PMLR; 2017. p. 1273–1282. [Google Scholar]
- 10.Xu J, Glicksberg BS, Su C, Walker P, Bian J, Wang F. Federated learning for healthcare informatics. J Healthc Inform Res. 2021;5(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen DC, Pham Q-V, Pathirana PN, Ding M, Seneviratne A, Lin Z, Dobre O, Hwang W-J. Federated learning for smart healthcare: A survey. ACM Comput Surv. 2022;55(3):1–37. [Google Scholar]
- 12.Cheng K, Fan T, Jin Y, Liu Y, Chen T, Papadopoulos D, Yang Q. Secureboost: A lossless federated learning framework. IEEE Intell Syst. 2021;36(6):87–98. [Google Scholar]
- 13.Liu Y, Fan T, Chen T, Xu Q, Yang Q. Fate: An industrial grade platform for collaborative learning with data protection. J Mach Learn Res. 2021;22(226):1–6. [Google Scholar]
- 14.Chen T, Guestrin C. Xgboost: A scalable tree boosting system, Proc ACM SIGKDD Int Conf Knowl Discov Data Min. 2016:785–794.
- 15.Das S, Pal S, Mitra M. Acoustic feature based unsupervised approach of heart sound event detection. Comput Biol Med. 2020;126: Article 103990. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan R, Rajpurkar P, Topol EJ. Self-supervised learning in medicine and healthcare. Nat Biomed Eng. 2022;6(12):1346–1352. [DOI] [PubMed] [Google Scholar]
- 17.Itahara S, Nishio T, Koda Y, Morikura M, Yamamoto K. Distillation-based semi-supervised federated learning for communication-efficient collaborative training with non-iid private data. IEEE Trans Mob Comput. 2021;22(1):191–205. [Google Scholar]
- 18.Huang W, Li T, Wang D, Du S, Zhang J, Huang T. Fairness and accuracy in horizontal federated learning. Inf Sci. 2022;589:170–185. [Google Scholar]
- 19.Qiu W, Quan C, Zhu L, Yu Y, Wang Z, Ma Y, Sun M, Chang Y, Qian K, Hu B, et al. Heart sound abnormality detection from multi-institutional collaboration: Introducing a federated learning framework. IEEE Trans Biomed Eng. 2024;1–12. [DOI] [PubMed] [Google Scholar]
- 20.Linardatos P, Papastefanopoulos V, Kotsiantis S. Explainable ai: A review of machine learning interpretability methods. Entropy. 2020;23(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clifford GD, Liu C, Moody B, Springer D, Silva I, Li Q, Mark RG. Classification of normal/abnormal heart sound recordings: The PhysioNet/Computing in Cardiology Challenge 2016. In: Proc Comput Cardiol. p. 2016:609–2016:612.
- 22.Liu C, Springer D, Li Q, Moody B, Juan RA, Chorro FJ, Castells F, Roig JM, Silva I, Johnson AE, et al. An open access database for the evaluation of heart sound algorithms. Physiol Meas. 2016;37(12):2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Proces Syst. 2017;30:4765–4774. [Google Scholar]
- 24.Zerka F, Barakat S, Walsh S, Bogowicz M, Leijenaar RT, Jochems A, Miraglio B, Townend D, Lambin P. Systematic review of privacy-preserving distributed machine learning from federated databases in health care. JCO Clin Cancer Inform. 2020;4:184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaissis GA, Makowski MR, Rückert D, Braren RF. Secure, privacy-preserving and federated machine learning in medical imaging. Nat Mach Intell. 2020;2(6):305–311. [Google Scholar]
- 26.Qiu W, Qian K, Wang Z, Chang Y, Bao Z, Hu B, Schuller BW, Yamamoto Y. A federated learning paradigm for heart sound classification. Paper presented at: 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); 2022; Glasgow, Scotland, UK. [DOI] [PubMed]
- 27.Liu JC, Goetz J, Sen S, Tewari A. Learning from others without sacrificing privacy: Simulation comparing centralized and federated machine learning on mobile health data. JMIR Mhealth Uhealth. 2021;9(3): Article e23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qayyum A, Ahmad K, Ahsan MA, Al-Fuqaha A, Qadir J. Collaborative federated learning for healthcare: Multi-modal covid-19 diagnosis at the edge. IEEE Open J Comput Soc. 2022;3:172–184. [Google Scholar]
- 29.Kumar Y, Singla R. Federated learning systems for healthcare: Perspective and recent progress. In: Federated learning systems: Towards next-generation AI. Cham: Springer; 2021. p. 141–156.
- 30.Li J, Meng Y, Ma L, Du S, Zhu H, Pei Q, Shen X. A federated learning based privacy-preserving smart healthcare system. IEEE Trans Industr Inform. 2021;18(3):1–12. [Google Scholar]
- 31.Tam P, Song I, Kang S, Kim S. Privacy-aware intelligent healthcare services with federated learning architecture and reinforcement learning agent. In: International Conference on Computer Science and its Applications and the International Conference on Ubiquitous Information Technologies and Applications. Singapore: Springer; 2022. p. 583–590. [Google Scholar]
- 32.Chen Y, Qin X, Wang J, Yu C, Gao W. Fedhealth: A federated transfer learning framework for wearable healthcare. IEEE Intell Syst. 2020;35(4):83–93. [Google Scholar]
- 33.Goetz J, Malik K, Bui D, Moon S, Liu H, Kumar A. Active federated learning. arXiv. 2019. 10.48550/arXiv.1909.12641 [DOI]
- 34.Lin X, Chen H, Xu Y, Xu C, Gui X, Deng Y, Wang Y. Federated learning with positive and unlabeled data. In: International Conference on Machine Learning. Baltimore (MD): PMLR; 2022. p. 13344–13355. [Google Scholar]
- 35.Jeong W, Yoon J, Yang E, Hwang SJ. Federated semi-supervised learning with inter-client consistency & disjoint learning. arXiv. 2020. 10.48550/arXiv.2006.12097 [DOI]
- 36.Eyben F, Wöllmer M, Schuller B. Opensmile: The munich versatile and fast open-source audio feature extractor. Paper presented at: Proceedings of the 18th ACM international conference on Multimedia; 2010; Firenze, Italy.
- 37.Eyben F, Weninger F, Gross F, Schuller B. Recent developments in opensmile, the Munich open-source multimedia feature extractor. Paper presented at: Proceedings of the 21st ACM International Conference on Multimedia; 2013; Barcelona, Spain.
- 38.Eyben F. Real-time speech and music classification by large audio feature space extraction. Cham: Springer; 2015. [Google Scholar]
- 39.Elkan C, Noto K. Learning classifiers from only positive and unlabeled data. Paper presented at: Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2008; Las Vegas, NV, USA.
- 40.Tao Y, Yang M, Li H, Wu Y, Hu B. Depmstat: Multimodal spatio-temporal attentional 531 transformer for depression detection. IEEE Trans Knowl Data Eng. 2024;36(7):2956–2966. [Google Scholar]
- 41.Yang M, Wu Y, Tao Y, Hu X, Hu B.. Trial selection tensor canonical correlation analysis (tstcca) for depression recognition with facial expression and pupil diameter. IEEE J Biomed Health Inform. 2023;1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study utilizes heart sound recordings from the PhysioNet/Computing in Cardiology Challenge, which provides a publicly available heart sound database. The database can be accessed at: https://physionet.org/content/challenge-2016/1.0.0 . Additional preprocessed data relevant to this paper may be requested from the author via email.


