Abstract
Rationale:
Low density cholesterol receptor (LDLR) in the liver is critical for the clearance of low-density lipoprotein cholesterol (LDL-C) in the blood. In atherogenic conditions, proprotein convertase subtilisin/kexin 9 (PCSK9) secreted by the liver, in a nonenzymatic fashion, binds to LDLR on the surface of hepatocytes, preventing its recycling and enhancing its degradation in lysosomes, resulting in reduced LDL-C clearance. Our recent studies demonstrate that epsins, a family of ubiquitin-binding endocytic adaptors, are critical regulators of atherogenicity. Given the fundamental contribution of circulating LDL-C to atherosclerosis, we hypothesize that liver epsins promote atherosclerosis by controlling LDLR endocytosis and degradation.
Objective:
We will determine the role of liver epsins in promoting PCSK9-mediated LDLR degradation and hindering LDL-C clearance to propel atherosclerosis.
Methods and Results:
We generated double knockout mice in which both paralogs of epsins, namely, epsin-1 and epsin-2, are specifically deleted in the liver (Liver-DKO) on an ApoE −/− background. We discovered that western diet (WD)-induced atherogenesis was greatly inhibited, along with diminished blood cholesterol and triglyceride levels. Mechanistically, using scRNA-seq analysis on cells isolated from the livers of ApoE−/− and ApoE−/− /Liver-DKO mice on WD, we found lipogenic Alb hi hepatocytes to glycogenic HNF4α hi hepatocytes transition in ApoE−/− /Liver-DKO. Subsequently, gene ontology analysis of hepatocyte-derived data revealed elevated pathways involved in LDL particle clearance and very-low-density lipoprotein (VLDL) particle clearance under WD treatment in ApoE−/− /Liver-DKO, which was coupled with diminished plasma LDL-C levels. Further analysis using the MEBOCOST algorithm revealed enhanced communication score between LDLR and cholesterol, suggesting elevated LDL-C clearance in the ApoE−/− Liver-DKO mice. In addition, we showed that loss of epsins in the liver upregulates of LDLR protein level. We further showed that epsins bind LDLR via the ubiquitin-interacting motif (UIM), and PCSK9-triggered LDLR degradation was abolished by depletion of epsins, preventing atheroma progression. Finally, our therapeutic strategy, which involved targeting liver epsins with nanoparticle-encapsulated siRNAs, was highly efficacious at inhibiting dyslipidemia and impeding atherosclerosis.
Conclusions:
Liver epsins promote atherogenesis by mediating PCSK9-triggered degradation of LDLR, thus raising the circulating LDL-C levels. Targeting epsins in the liver may serve as a novel therapeutic strategy to treat atherosclerosis by suppression of PCSK9-mediated LDLR degradation.
Introduction
Atherosclerosis is the major contributor to many cardiovascular diseases (CVDs), including coronary artery disease, stroke and peripheral vascular disease1. CVDs are the leading causes of death globally, in the United States alone, approximately 610,000 individuals succumb to heart-related ailments annually, constituting nearly one-quarter of all deaths2. This chronic disease initiates with lipid accumulation in the subendothelial space of arterial walls, followed by an inflammatory response that accelerates atherosclerotic plaque formation3. Understanding the molecular mechanisms responsible for the initiation, growth, and destabilization of atheroma is essential for the development of more effective and targeted therapies to prevent ischemic injury, disability, or death in patients with CVDs4.
Modern lifestyles have rendered millions susceptible to hyperlipidemia, a key risk factor for atherosclerosis. In addition to diet-induced hyperlipidemia, familial hypercholesterolaemia (FH) is the most common inherited metabolic diseases that featured as markedly elevated plasma levels of low-density lipoprotein cholesterol (LDL-C)5. Mutations in low-density lipoprotein receptor (LDLR) are the main genetic cause of elevated LDL-C levels in familial hypercholesterolemia6. In the intima, LDL-C undergoes oxidative modifications by reactive oxygen species, and the oxidized LDL cholesterol (oxLDL-C) is then taken up by macrophages for the formation of foam cells7. Cholesterol-laden foam cells trigger the secretion of proinflammatory cytokines, as the inflammatory master cytokine, IL-1β activates the expression of many proinflammatory cytokines8. The infiltration of circulating macrophages, leukocytes, and monocytes into the atherosclerotic lesion pave the way for atherosclerosis progression9. Hence, strategies targeting LDL-C reduction are pivotal in combating inflammation-induced atherosclerosis.
The regulation of LDL-C involves its clearance by LDLRs, predominantly expressed in hepatocytes. LDLR is a cell surface protein predominantly expressed in hepatocytes and is the primary mechanism whereby excess LDL-C is removed from the circulation10. Proprotein convertase subtilisin/kexin type 9 (PCSK9) is primarily expressed in the liver, which promotes LDLR degradation and results in higher levels of circulating LDL-C11. PCSK9 inhibitors were widely reported to be good candidates for lipid-lowering since they can prevent the degradation of LDLR by inhibition of the interaction between PCSK9 and LDLR. LDLR deficient mice and LDLR-KO rabbits are good model for atherosclerosis study by inducing hyperlipidemia12,13. In addition, Keeter et al. reported that overexpression of PCSK9 that mediated by Adeno-associated virus-8 (AAV8) induces hyperlipidemia and promotes atherosclerosis14. A recent in vivo CRISPR based-editing strategy, namely, VERV101, achieved potent and durable inactivation of the expression of PCSK9 in the liver, resulting in significant reduction of LDL-C in nonhuman primates, making it promising strategy to treat hyperlipidemia15.
Despite these advancements, the mechanistic details of PCSK9-mediated LDLR degradation remain insufficiently understood, posing a challenge in identifying dual-action targets that both reduce cholesterol synthesis and prevent LDLR degradation. Epsins, a family of endocytic adaptor proteins, have recently gained attention for their role in atherosclerosis16. We previously showed that epsins 1 and 2 are upregulated in atherosclerotic plaques in apolipoprotein E-deficient (ApoE−/−) mice fed a western diet (WD), endothelial cell (EC)-specific epsin deficiency resulted in marked attenuation of atherogenesis in ApoE−/− mice fed a WD4. Mechanistically, epsin-deficiency reduced arterial inflammation by dampening expression of adhesion molecules and hindering macrophage recruitment. We also showed that myeloid-restricted epsin deficiency in ApoE−/− mice fed a WD retarded atherogenesis by eradicating foam cell formation and augmenting efferocytosis in the lesion17,18. Given that the liver is one of the main sites for cholesterol and lipid synthesis19,20, whether epsins in the liver regulate circulating LDL-C levels through modulating LDLR stability in atherosclerotic mice has not been investigated.
In this study, we investigated the potential role epsins in the liver play in regulating atherosclerosis using novel ApoE−/− mice harboring liver-specific deficiency of epsins (ApoE−/− /Liver-DKO). We discovered that WD-induced atherogenesis was greatly inhibited and accompanied with diminished blood cholesterol and triglyceride levels. Mechanistically, scRNA-seq analysis identified a transition from lipogenic Alb hi hepatocytes to glycogenic HNF4α hi hepatocytes in ApoE−/− /Liver-DKO mice. Gene Ontology (GO) enrichment analysis revealed upregulated pathways for LDL particle clearance in ApoE−/− /Liver-DKO mice. Additively, we further showed that epsins bind LDLR via the ubiquitin-interacting motif (UIM), PCSK9-triggered LDLR degradation was abolished by depletion of epsins that prevent the atheroma progression. Furthermore, our findings uncovered liver epsins mediated PCSK9-triggered LDLR ubiquitination for degradation. Intriguingly, we found elevated liver epsins expression in the hepatocytes from gain-of-function PCSK9 D374Y mutation mice that promote LDLR degradation. Finally, our therapeutic study by targeting liver epsins with nanoparticle-encapsulated siRNAs inhibit dyslipidemia and impede atherosclerosis. Thus, liver epsins could be potentially novel therapeutic targets for combating atherosclerosis.
MATERIALS and METHODS
Animal models
In this study, all animal procedures were performed in compliance with institutional guidelines and mouse protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital, MA, USA. Both male and female mice were used. C57BL/6 mice (stock #00664), ApoE −/− mice (stock #002052), Alb-Cre delete mice (stock #035593) were all purchased from Jackson Research Laboratory. As double knockout mice of Epsin 1 and 2 (Epsin 1−/−; Epsin 2−/−) lead to embryonic lethality, we generated conditional Epsin1 fl/fl; Epsin2 −/− mice previously described21–23. ApoE −/− mice, Alb-Cre+/− mice and Epsin1 fl/fl; Epsin2 −/− mice were backcrossed to C57BL/6 background. We bred Epsin1 fl/fl; Epsin2 −/− mice with Alb-Cre+/− mice to generate Epsin1 fl/fl; Epsin2 −/−; Alb-Cre+/− liver-specific Epsins deficient (Liver-DKO) mice (Fig. S1).
The detailed information of all the mice used in this study were described in Fig. S1. In addition, we bred Epsin1 fl/fl; Epsin2 −/−; Alb-Cre+/− mice with ApoE −/− (C57BL/6) background to generate Epsin1 fl/fl; Epsin2 −/−; Alb-Cre+/−; ApoE −/− mice (Liver-DKO/ApoE −/−) (Fig. S1).
The control mice for Epsin1 fl/fl; Epsin2 −/−; Alb-Cre+/− (Liver-DKO) mice were Epsin +/+; Epsin2 +/+; Alb-Cre +/− mice (WT) (Figure S1). The control mice for Epsin1 fl/fl; Epsin2 −/−; Alb-Cre+/−; ApoE −/− mice (Liver-DKO/ApoE −/−) were Epsin +/+; Epsin2 +/+; Alb-Cre +/−; ApoE−/− (WT/ApoE−/−) (Fig. S1).
To induce atherosclerosis, mice were fed Western diet (WD, Protein 17% kcal, Fat 40% kcal, Carbohydrate 43% kcal; D12079B, Research Diets, New Brunswick, USA) starting at the age of 6–8 weeks for 8–16 weeks. Mice were sacrificed at different time points based on experimental design and liver, blood, heart, aorta were harvested.
For control mice, in addition to ApoE−/−; Epsin 1+/+; Epsin 2 +/+ mice, we also used ApoE−/−; Epsin 1+/+; Epsin 2 +/+ mice with a single copy of Alb-Cre, and ApoE −/− mice; Epsin1 fl/fl; Epsin2 −/− littermates lacking the single copy of Alb-Cre. To simplify the terminology, we refer to these control mice as ApoE−/−, as results were not different in any of the analyses we performed.
For each experimental model and time point, 6–10 mice were analyzed and both male and female mice were used in separate groups. In the current study, we did not exclude any mice when analyzing.
Liver single-cell preparation and single-cell RNA (scRNA) sequencing
For liver cell isolation, the ApoE−/− and ApoE−/− /Liver-DKO mice (n=3) were anesthetized and restrained and the skin sprayed with 70% ethanol. The liver and other inner organs were revealed by cutting through the skin and peritoneum. A 24G needle was carefully inserted into the inferior vena cava and secured with a clamp, and chelating solution (0.05M HEPES, pH 7.2, 10 mM EGTA in HBSS without CaCl2 and MgCl2) was run at a low speed (1.5 – 2mL/minute). The portal vein was then cut and perfusion speed was increased to a flow rate of 7mL/minute. After that, the diaphragm was cut and the anterior vena cava clamped. The chelating perfusion was run for 7 minutes and then switched to collagenase solution (0.05 M HEPES, pH 7.2, 4.7mM CaCl2, 20 μg/mL Liberase, Sigma LIBTM-RO) at a flow rate of 7mL/minute for 7 minutes. The liver was then removed and passed through a 70 μm cell strainer with 10 ml ice-cold HBSS without CaCl2 and MgCl2. The resulting single-cell suspension was centrifuged at 300 g for 5 minutes at 4 °C and washed twice with ice-cold HBSS.
The isolated liver cells have been counted and diluted into 1000 cells/μL. The single cell RNA-seq library construction was performed according to the 10 x genomics protocol (GEMs, Single cell 3’ Reagent Kits v3.1, 10 x Genomics). The prepared libraries were sequenced on the HiSeq 2500 Sequencing System (Illumina, San Diego, CA) with 100-bp paired end reads.
ScRNA-seq data analysis
Firstly, we employed Cell Ranger (version 7.1.0) to map the raw reads of RNA sequencing data to the mouse genome (version mm10) and count the UMI for each gene. We then proceeded with the resulting UMI count matrix using the Seurat R package (version 4.3.0)24. We retained high-quality cells expressing between 200 and 2500 genes, excluding those with over 20% mitochondrial reads. Additionally, we filtered out rarely expressed genes detected in fewer than 3 cells. After filtering, the high-quality data was normalized, cell-level scaled with 10k reads, and natural-log transformed by adding 1. The normalized data underwent further processing steps: scaling (ScaleData function), principal component analysis (PCA) (RunPCA function, npcs =30), Uniform Manifold Approximation and Projection (UMAP) (RunUMAP function, reduction = “pca”, dims = 1:30), shared nearest neighbor graph (SNNG) construction (FindNeighbors function, reduction = “pca”, dims = 1:30), and cell clustering (FindCluster function, resolution=0.1). Further, We conducted Differentially Expressed Genes (DEGs) analysis in one cluster versus other clusters using the FindAllMarkers function. The Wilcoxon test method was used by default, with a minimum percentage of expressed cells set to 25% and a minimum log2 fold change of 0.25. Cell types were annotated based on known marker genes from PanglaoDB25, cell-Taxonomy26, disco27 databases, and relevant literature. Marker gene expressions were visualized by DotPlot and VlnPlot functions. For cell fate transition, trajectory analysis, and cell rank for directed single-cell transition mapping, we utilized scvelo (version 0.2.5)28,29, monocle3 (version 1.3.1)30,31, and cellrank (version 2.0.4)32 with default parameters. For trajectory analysis, we kept Albhi hepatocytes as the initial cell type and for cell rank for directed single-cell transition mapping we used the function of all states terminal states and initial state with n_states=[4,5]. The metabolite-mediated cell-cell communication was analyzed by MEBOCOST (version 1.0.0)33. The data were analyzed combined for both conditions following the tutorial on the MEBOCOST website. The prediction of sender-metabolite-sensor-receiver communication events was visualized by the bar and lollypop plots, using the ggplot2 library. Additionally, we performed cell-cell communication analysis using Cellchat (version 1.5.0)34. The communication probability of each condition was analyzed to highlight differences between conditions, and communication events were visualized using bar, flow, and circle plots. Finally, Gene Ontology (GO) functional enrichment analysis was performed using the clusterProfiler R package (version 3.18.1)35, and visualized by bar, lollipop, and cnet plots.
Human samples
Human healthy control and diseased aortic arch samples from atherosclerosis patients were purchased from Maine Medical Center Biobank. In addition to aorta samples, liver samples from human healthy control and non-alcoholic fatty liver disease (NAFLD) patients were purchased from Maine Medical Center Biobank. The medical information of the atherosclerotic patients and healthy people samples, and NAFLD patients and healthy people samples is in Table S1. The paraffin sections were de-paraffined and performed antigen retrieval to unmask the antigenic epitope with 10 mM Sodium Citrate, pH 6.0, with 0.5% Tween 20 at 95 °C for 30 minutes. Allow the slides to cool for 30 minutes before proceeding with staining procedure. Immunofluorescence staining of the slides was performed with the standard protocol described below.
Synthesis of DSPE-PEG-GalNAc, preparation and characterization of targeted siRNA nanoparticles (NPs)
To further improve siRNA delivery to the liver, we propose to develop targeted hybrid NPs by surface modification with galactose-based ligands that can specifically bind to the ASGPR receptor exclusively expressed on hepatocytes36,37. Then, a robust self-assembly method was used to prepare the targeted polymer-lipid hybrid NPs for siRNA delivery38,39. In brief, G0-C14 and PLGA were dissolved separately in anhydrous dimethylformamide (DMF) to form a homogeneous solution at the concentration of 2.5 mg/mL and 5 mg/mL, respectively. DSPE-PEG-OCH3 (DSPE-mPEG) and DSPE-PEG-GalNAc were dissolved in HyPure water (GE Healthcare Life Sciences, catalog no. SH30538) at the concentration of 0.1 mg/mL. 0.75 nmol Epsin1 siRNA and 0.75 nmol Epsin2 siRNA were gently mixed with 100 μL of the G0-C14 solution. The mixture of siRNA and G0-C14 was incubated at room temperature for 15 minutes to ensure the full electrostatic complexation. Next, 500 μL of PLGA polymers were added and mixed gently. The resultant solution was subsequently added dropwise into 10 mL of HyPure water containing 1 mg lipid-PEGs (i.e., 50% DSPE-PEG-GalNAc and 50% DSPE-mPEG hybrids for the GalNAc-targeted siRNA NPs, or 100% DSPE-mPEG for the non-targeted siRNA NPs) under magnetic stirring (1,000 rpm) for 30 minutes. The siRNA NPs were purified by an ultrafiltration device (EMD Millipore, MWCO 100 kDa) to remove the organic solvent and free excess compounds via centrifugation at 4 °C. After washing 3 times with HyPure water, the siRNA NPs were collected and finally resuspended in pH 7.4 PBS buffer. The NPs were used freshly or stored at −80 °C for further use. The physicochemical properties (particle size and surface charge) of GalNAc-siEpsin1/2 were characterized by dynamic light scattering (DLS, Brookhaven Instruments Corporation). The GalNAc-siEpsin1/2 was ~ 89 nm in size as measured by DLS, and their surface charge was determined to be ~ −5.3 mV.
siRNA transfection
The siRNA transfection was performed according to the manufacturer’s instructions. Briefly, HepG2 cells were transfected by RNAiMAX (Invitrogen) with either scrambled siRNA duplex or epsin 1 (GAACUGGAGGCACGUCUACAAUU) or epsin 2 siRNA duplexes (GCAGUGCCGUGAGAACAUCUUUU) designed by Dharmacon™ (Horizon Discovery). At 48 hours post transfection, cells were processed for western blot assays.
RNA isolation and Real-time quantitative PCR
Total RNA was extracted from the liver tissue with Qiagen RNeasy Mini Kit based on manufacturer’s instruction including the optional step to eliminate genomic DNA. The extracted RNA was used for RT-qPCR according to the experimental designs.
For RT-qPCR, mRNA was reverse transcribed to cDNA with the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA, United States). 2 μL of 5 fold diluted cDNA product was subjected to RT-qPCR in StepOnePlus Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Master Mix reagent as the detector. PCR amplification was performed in triplicate on 96-well optical reaction plates and replicated in at least three independent experiments. The ΔΔCt method was used to analyze qPCR data. The Ct of β-actin cDNA was used to normalize all samples. Primers are listed in Major Resource Table.
Analysis of plasma triglyceride and cholesterol levels
Blood was removed from the right atrium of the mouse heart after sacrifice with isoflurane. Blood was allowed to clot for 30 minutes at room temperature followed by centrifugation at 3000 × g at 4 °C for 20 minutes. Serum was transferred to a new tube and stored at −20 °C. Serum cholesterol and lipid levels were determined using the Cholesterol Assay Kit for HDL and LDL/VLDL and Triglyceride Assay Kit from Abcam.
Atherosclerotic lesion characterization
The whole aortas were collected and fixed with 4% paraformaldehyde. Then, the aortas were stained with Oil Red O for en face analysis. Hearts and BCA were embedded in O.C.T compound and sectioned at 8 microns. Lesion area of the aortic root was quantified by hematoxylin and eosin (H&E) staining. Neutral lipids deposition was determined by Oil Red O staining. Aortic lesion size and lipid content of each animal were obtained by an average of three sections from the same mouse.
En face Oil Red O staining
Whole aortas were dissected symmetrically, pinned to parafilm to allow the en face exposed and fixed in formalin for 12 hours. Aortas were washed in PBS for 3 times, and rinsed in 100% propylene glycol followed by staining with 0.5% Oil Red O solution for 20 minutes at 65 °C. Aortas were then put in 85% propylene glycol for 2 minutes, followed by three washes in DD Water. Slides were next incubated with hematoxylin for 30 seconds, rinsed in running tap water. Imaging was performed using a Nikon SMZ1500 stereomicroscope, SPOT Insight 2Mp Firewire digital camera, and SPOT Software 5.1.
Oil Red O staining of cryostat sections
Cryostat sections of mouse aortic root and BCA were washed in PBS for 5 minutes, then fixed in 4% paraformaldehyde for 15 minutes. Slices were washed in PBS followed by staining with freshly prepared 0.5% Oil Red O solution in isopropanol for 10 minutes at 37 °C. Slices were then put in 60% isopropanol for 30 seconds, followed by 3 washes in water. Slices were next incubated with hematoxylin for 30 seconds, rinsed in running tap water, and mounted with 90% Glycerin.
H&E staining
Cryostat sections of mouse aortic root and BCA were washed in PBS for 5 minutes, then fixed in 4% paraformaldehyde for 15 minutes. Next, slides were stained with 0.1 hematoxylin for 3 minutes followed by running tap water washes for 10 minutes. Slices were then dipped in Eosin working solution for 30 seconds, quickly rinsed with tap water, dehydrated using graded ethanol (95% and 100% ethanol), followed by transparentizing by xylene: ethanol absolute (1:1) solution and 100% xylene for 1 hour. Slices were mounted in synthetic resin.
Van Gieson’s staining
Van Gieson’s staining were performed based on manufacturer’s instruction. In brief, Cryostat sections of mouse aortic root and BCA were washed in PBS for 5 minutes, then fixed in 4% paraformaldehyde for 15 minutes. Slices were placed in Elastic Stain Solution (5% hematoxylin + 10% ferric chloride + lugol’s Iodine Solution) for 20 minutes, then rinsed in running tap water. Then, slices were dipped in differentiating solution 20 times and in sodium thiosulfate solution for 1 min, following with rinsing in running tap water. Slices were dehydrated in 95% and 100% alcohol once, respectively. Slides were cleared and mounted in synthetic resin.
Immunofluorescence staining
The liver tissue or aorta from both ApoE−/− and ApoE−/− Liver-DKO were subjected for cryosections, and sections were further fixed in 4% paraformaldehyde for 15 minutes. The slides were blocked by blocking buffer (PBS/3% BSA/3% donkey serum/0.3% triton) for 30 minutes, and were further incubated by primary antibodies (epsin1, epsin2, CD68, αSMA) for overnight at 4°C. The slides were washed 3 times for 10 minutes each wash by PBS/0.3% triton buffer, and were further incubated with secondary antibodies at room temperature for 1 hour. Then the slides were washed 3 times for 10 minutes each wash by PBS/0.3% triton buffer. After the second wash, DAPI was used for nuclei stain. The slides were mounted Fluoroshield™ histology mounting medium. Imaging was performed using Zeiss LSM 880 Confocal Acquisition & Analysis.
Immunoprecipitation and Western Blotting
For immunoprecipitation, HepG2 cells were lysed with RIPA buffer (50 mM Tris, pH 7.4, with 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholic acid, 0.1% sodium dodecyl sulfate, 5 mM N-ethylmaleimide and protease inhibitor cocktail). For LDLR ubiquitination experiments, HepG2 cells were lysed using denaturing buffer (1% SDS in 50 mM Tris, pH 7.4) and boiled at 95 °C for 10 minutes to denature protein complexes. Lysates were renatured using nine volumes of ice-cold RIPA buffer then prepared for immunoprecipitation as follows. Cell lysates were pre-treated with Protein A/G Sepharose beads at 4 °C for 2 hours to remove nonspecific protein followed by centrifugation at 12000 rpm for 5 minutes at 4 °C. Supernatant was transferred to a new tube, incubated with Protein A/G Sepharose beads and antibodies against Epsin1 or LDLR or ubiquitin at 4 °C overnight. Mouse IgG was used as negative control. Protein A/G beads were washed with RIPA buffer for 2 times, followed by PBS for 1 time. Then, beads were suspended with 80 μL 2x loading buffer and heated at 95 °C for 10 minutes. After centrifugation, precipitated proteins were visualized by Western blot. Proteins were resolved by SDS-PAGE gel and electroblotted to nitrocellulose membranes. NC membranes were blocked with 5% milk (w/v) and blotted with antibodies. Western blots were quantified using NIH Image J software.
PCSK9 Adeno-associated Virus 8 (AAV8) Tail Vein Injection
Eight-week old male C57BI/6J mice, both WT and Liver-DKO mice (n=4), were intravenously injected via tail vein with a single dose of 2 × 1011 viral PCSK9-AAV8 and fed a WD for 8 weeks and 16 weeks. The serum samples collected from both WT and Liver-DKO mice were subjected for triglyceride and cholesterol measurement. The liver tissue from both WT and Liver-DKO mice were collected for further histology analysis and protein lysate preparation.
Nanoparticle-encapsulated epsin1/2 siRNAs Tail Vein Injection
Eight-week old male C57BI/6J mice, ApoE −/− mice were fed a WD for 8 weeks, and further divided into two groups (n=4). The control group mice were intravenously injected via tail vein with control siRNA NPs, and the experimental group mice were injected with 0.75 nmoles epsin1/2 siRNA NPs for continuous three weeks. Two doses injection per week. After injection, the serum samples collected from both control siRNA and epsin1/2 siRNA NPs injected groups were subjected for triglyceride and cholesterol measurement. The aortas were isolated En face ORO staining and histology analysis, and the liver tissue from both control siRNA and epsin1/2 siRNA NPs were collected for protein lysate preparation.
Cell culture and plasmids transfection
The HepG2 cell line (ATCC no. HB-8065) was used for plasmid transfection to map the binding sites of Epsin1 to LDLR. Flag-tagged Epsin1WT, Epsin1ΔUIM, Epsin1ΔENTH, Epsin1-DPW truncation constructs, and pcDNA vector were prepared previously in our laboratory. HepG2 cells were cultured in DMEM (10% FBS and 1% Pen-Strep) at 37°C in humidified air containing 5% CO2 atmosphere and transfected using Lipofectamine 2000 as instructed by the manufacturer.
Epsin1/2 siRNAs transfection and PCSK9 adeno-associated virus 8 (AAV8) infection
HepG2 cells were cultured in DMEM (10% FBS and 1% Pen-Strep) at 37°C in humidified air containing 5% CO2 atmosphere. One day before transfection, plate cells in 1 mL of growth medium without antibiotics such that they will be 30–50% confluent at the time of transfection. Prepare RNAi duplex-Lipofectamine ™ RNAiMAX complexes by mixture of Epsin1/2 siRNAs and Lipofectamine™ RNAiMAX, keep the mixture at room temperature for 20 minutes, and add the complexes to each well containing cells. Incubate the cells 48 hours at 37°C in a CO2 incubator.
The PCSK9-AAV8 virus stock (1013 GC/ml) was diluted by culture media into 1010 GC for the infection. Remove the original cell culture media, and add the PCSK9-AAV8 virus containing media to cell culture. Collect the cells 3 days after the PCSK9-AAV8 virus infection.
Hepatocyte Primary Culture, MG132 treatment, PCSK9-AAV8 virus infection
The anaesthetized animals (WT and Liver-DKO) were restrained and the skin sprayed with 70% ethanol. The liver and other inner organs were revealed by cutting through the skin and peritoneum. A 24G needle was carefully inserted into the inferior vena cava and secured with a clamp, and chelating solution (0.05 M HEPES pH 7.2, 10 mM EGTA in HBSS without CaCl2 and MgCl2) was run at a low speed (1.5–2 mL/min). The portal vein was then cut and perfusion speed was increased to a flow rate of 7 mL/min. After that, the diaphragm was cut and the anterior vena cava clamped. The chelating perfusion was run for 7 minutes and then switched to collagenase solution (0.05 M HEPES pH 7.2, 4.7 mM CaCl2 1 mg/mL Liberase, Sigma LIBTM-RO) at a flow rate of 2–4 mL/minute for 15 minutes. The liver was transferred to a 10 cm plate with plating media (DMEM low glucose, 5% FBS, 1% Penicillin-Streptomycin Solution), the liver cells were gently released with fine tip forceps. The liver cells suspension was filtered through a 70 μm cell strainer into a 50 mL tube. Spin at 50 × g for 2 minutes at 4 °C. While the samples are spinning, prepare fresh Percoll solution (90% Percoll in 1xHBSS). Aspirate the supernatant, add 10 mL plating media and resuspend by gentle swirling, and further add 10 mL Percoll solution and mix thoroughly by inverting the tube several times. Spin at 200 × g for 10 minutes at 4 °C. Aspirate the supernatant, and add 20 mL plating media, and then spin at 50 × g for 2 minutes at 4 °C. Aspirate supernatant, and add 20 mL plating media. Hepatocytes were counted and plated on collagen-coated cell culture 6-well plates. After 3 hours, change medium to warm maintenance media (Williams E media, 1% Glutamine, 1% Penicillin-Streptomycin Solution).
After 24 hours, proteasome inhibitor MG132 solution was added into the cultured primary hepatocyte cells. After 6 hours, the primary culture hepatocytes were subjected to PCSK9-AAV8 virus infection as mentioned above. The primary culture hepatocytes were collected and lysed for protein preparation.
Statistical analysis
Gene expression was assessed by quantifying mRNA levels of target genes via qPCR, with normalization to the internal control, β-actin. Quantitative data were analyzed using either one-way ANOVA or Student’s t-test, as appropriate, with Prism software (GraphPad Software, San Diego, CA) running on Apple OS X. All data are presented as are the mean ± SEM.
Data availability
The scRNA-seq data (GSE273386) of the ApoE−/− and ApoE−/− /Liver-DKO are available at the Gene Expression Omnibus. The scRNA-seq data (GSE254971) for D374Y mCherry-APOB mice is available in published paper entitled “Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult” (https://doi.org/10.1038/s44161-024-00448-6). Source data are provided with this paper.
Results
Elevated Epsin1 and Epsin2 Expression in Atherosclerotic Patients.
In this study, we measured the protein expression of epsin1 and epsin2 between healthy control and atherosclerotic patients by immunofluorescence staining. Intriguingly, we found significantly elevated expression of epsin1 and epsin2 protein in diseased aortic arch samples (Fig.S2A–C). As expected, we found dramatically increased CD68 protein expression in atherosclerotic patients (Fig.S2A, 2B), suggests more macrophages accumulation and more atherosclerotic lesion in patients. Especially, we discovered significantly higher colocalization between epsin1 and CD68, and higher overlay percentage between epsin2 and CD68 in the macrophages in atherosclerotic patients than healthy control (Fig.S2A–C).
Diminished LDLR and HNF4α but Elevated Epsin1 and Epsin2 Expression in the Liver from WD-fed Mice and NAFLD Patients
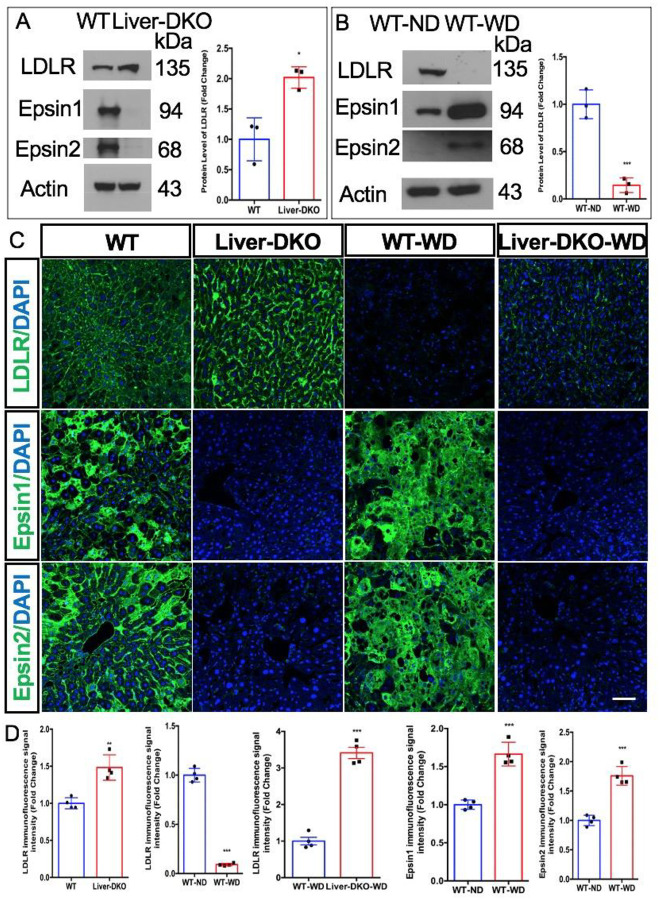
In addition to aorta, we found significantly elevated expression of both epsin1 and epsin2 proteins in the liver from WD-fed mice (Fig.4B, 4C), but with significantly diminished expression of LDLR protein in the liver from WD-fed mice (Fig.4B, 4C). It is reported that atherosclerosis and NAFLD are two sides of the same coin3, therefore, we have evaluated the protein expression of epsin1, epsin2, LDLR and HNF4α in the liver between healthy control and NAFLD patients. Intriguingly, we also found significantly elevated epsin1 and epsin2 expression in the liver from NAFLD patients (Fig.S3A, 3B), but with dramatically diminished LDLR expression in the liver from NAFLD patients (Fig.S3A, 3B). In addition, we detected markedly diminished HNF4α in NAFLD patients both at protein and mRNA levels (Fig.S4 A–C).
Fig. 4: Elevated LDLR expression in the liver of liver specific epsin deficiency mice and diminished LDLR expression in the liver of WD-fed mice.
(A) Western blot (WB) analysis of liver tissue harvested from WT and Liver-DKO mice revealed elevated LDLR expression in Liver-DKO (left). Data quantification of LDLR expression (right) (n=3, *p< 0.05). (B) Western blot (WB) analysis of liver tissue harvested from WT (normal diet, ND) and WT (western diet, WD) mice showed diminished LDLR expression and elevated epsin1 and epsin2 expression in WT-WD (left). Data quantification of LDLR expression (right) (n=3, ***p<0.001). (C) Immunofluorescence (IF) analysis of liver cryosections from WT, Liver-DKO mice, and WT-WD revealed elevated LDLR expression in Liver-DKO (left), but diminished LDLR expression in the liver from WD-fed WT mice (left), however, the diminished LDLR expression in the liver from WD-fed is inhibited in WD-fed Liver-DKO (left). Data quantification of LDLR, Epsin1, Epsin2 expression (right) (n=4, **p<0.01).
Epsins Depletion in the Liver Inhibits Atherogenesis and Reduces Lipid Levels
In this study, we explored the role of hepatic epsins in atherosclerosis by employing liver-specific epsins-deficient mice (ApoE−/−/Liver-DKO) and compared their phenotypic outcomes to ApoE−/− controls. Our results demonstrated that epsins depletion significantly inhibited western diet (WD)-induced atherogenesis (Fig. 5A). Specifically, the ApoE−/−/Liver-DKO mice exhibited markedly reduced atherosclerotic lesion formation, as evidenced by decreased plaque size and lipid accumulation in the arterial walls compared to the ApoE−/− controls (Fig. 5B). These findings indicate that liver epsins play a crucial role in the pathogenesis of atherosclerosis.
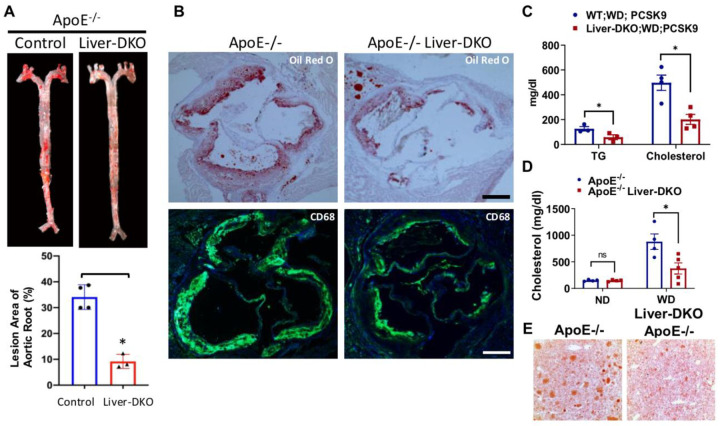
Fig.5: Liver-deficiency of epsins inhibits atherosclerotic lesion formation and macrophage accumulation.
(A) En face ORO staining of aortas (top) from ApoE−/− or ApoE−/− / Liver-DKO mice fed a WD, and unpaired t-test (bottom) for the lesion areas (n=4, ***p< 0.001) (B) Aortic roots from WD-fed ApoE−/− or ApoE−/− Liver-DKO mice stained with ORO or the CD68 macrophage marker. Scale bars=500 um. (C) Plasma triglyceride (TG) and cholesterol levels in WD-fed WT and Liver-DKO mice treated with AAV8-PCSK9 (n=4, *<0.05) (D) Cholesterol in ApoE−/− and ApoE−/− Liver-DKO mice after 8 weeks on a WD (n=4, p*<0.05) (E) ORO staining of liver tissue (n=4).
Plasma cholesterol and triglyceride (TG) levels, measured by Wako enzymatic and TG Infinity kits, showed decreases resulting from loss of hepatic epsins in mice injected with PCSK9-AAV8 (2×1011 genomes) and fed a WD for 8 weeks. We found significantly diminished TG and plasma cholesterol levels in WD-fed Liver-DKO mice after PCSK9-AAV8 injection (Fig.5C). Additively, quantitative analysis revealed that the ApoE−/−/Liver-DKO mice had significantly lower plasma cholesterol and triglyceride levels than those in ApoE−/− mice (Fig. 5D). These reductions in lipid levels are indicative of improved lipid metabolism and clearance in the absence of hepatic epsins, suggesting that liver epsins contribute to hyperlipidemia in atherosclerosis. In addition, we found significantly reduced hepatic lipid accumulation in ApoE−/−/Liver-DKO mice by Oil Red O staining (Fig.5E). The reduced lipid levels were also associated with decreased systemic inflammation, as shown by lower levels of pro-inflammatory cytokines in the serum of ApoE−/−/Liver-DKO mice (Data not shown).
Single-Cell RNA Sequencing Identified Lipogenic Albhi Hepatocyte and Glycogenic HNF4α hi Hepatocytes
To understand the cellular mechanisms underlying the observed phenotypic changes, we performed single-cell RNA sequencing (scRNA-seq) on liver tissues from both ApoE−/− and ApoE−/−/Liver-DKO mice (Fig. 1A). The Uniform Manifold Approximation and Projection (UMAP) visualization revealed different cell types in the liver based on cell type-specific gene markers and their corresponding expression between ApoE−/− and ApoE−/−/Liver-DKO mice (Fig.S6A, S6B). Further, the scRNA-seq analysis revealed distinct hepatocyte populations in the livers between ApoE−/− and ApoE−/−/Liver-DKO mice, highlighting the heterogeneity of hepatocyte cell populations. UMAP visualization of hepatocytes illustrated a clear separation of different subcluster hepatocytes between of ApoE−/− and ApoE−/−/Liver-DKO mice (Fig. 1B), including HC HNF4α hi, HC1 Albhi, HC2 Albhi, HC3 Albhi, indicating significantly transcriptional reprogramming in the absence of epsins. The hepatocyte markers for hepatocytes clustering are highlighted in the dotplot (Fig.1C).
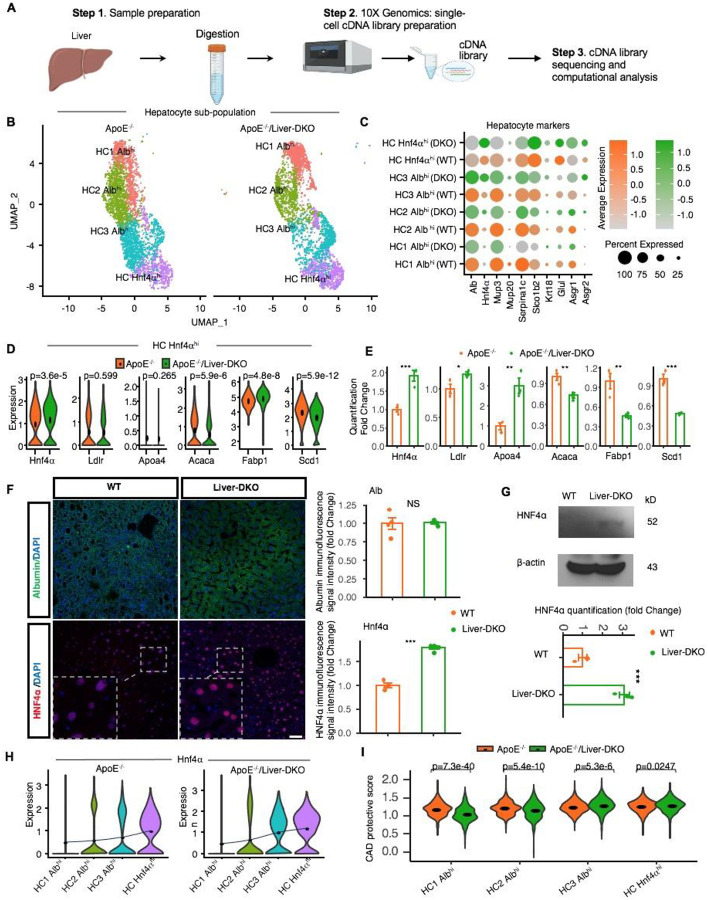
Fig. 1: Single-cell RNA-sequencing reveals gene expression dynamics in Liver-DKO mice on an ApoE−/− background.
A: Schematic representation of the single-cell RNA-sequencing (snRNA-seq) process performed on liver tissues from ApoE−/− and ApoE−/−/Liver-DKO mice. B: UMAP visualization illustrating the heterogeneity of hepatocyte cell populations indicating distinct clustering patterns of hepatocytes in ApoE−/−/Liver-DKO mice compared to ApoE−/− controls. C: Dot plot showing the expression of gene markers in respective sub-cell types. D-E: Elevated gene expression related to LDL particle clearance and decreased gene expression related to fatty acid synthesis in ApoE−/−/Liver-DKO as compared to ApoE−/−, shown through single-cell analysis (D) and real-time quantitative PCR (qPCR) (E). F: Elevated HNF4α expression level in Liver-DKO mice. Immunofluorescence stain of HNF4α, Albumin in the liver from both WT and Liver-DKO mice (left), HNF4α is in red color, Albumin is in green color (marker of hepatocytes), and DAPI is used for nuclei stain. Quantification of HNF4α immunofluorescence signal intensity between WT and Liver-DKO (right). G: Western blot of HNF4α for the liver lysates from WT and Liver-DKO, beta-Actin is used as internal reference (left), the quantification of HNF4α. H: Hnf4α expression increasing from HC1 Albhi to HC3 Albhi in both conditions ApoE−/− and ApoE−/−/Liver-DKO. I: Coronary artery disease (CAD) protective score comparing ApoE−/−/Liver-DKO to ApoE−/−. Note: We used the two-tailed t- test to compare the samples in panel (E). expression in both WT and Liver-DKO (right). n=3, **p<0.01, ***p<0.001.
Intriguingly, we identified significantly elevated HNF4α expression in the HNF4α hi hepatocytes in ApoE−/−/Liver-DKO mice (Fig.1D, 1E). In addition, we found higher mean HNF4α expression in HC HNF4α hi, HC1 Albhi, HC2 Albhi, HC3 Albhi in ApoE−/−/Liver-DKO than ApoE−/− (Fig.1F). Especially, we discovered that Albhi hepatocytes have lower expression of apolipoprotein genes, such as Apob, Apoa4, but with their higher expression in HNF4α hi hepatocytes (Fig.1F–H, Fig.S11), indicating more effective LDL cholesterol clearance in HNF4α hi hepatocytes that transported by ApoB and ApoA4 proteins. The lipogenic genes, such as Acaca, Scd1, have their higher expression in HC3 Albhi hepatocytes but lower expression in HNF4α hi hepatocytes (Fig.S11). Especially, HNF4α hi hepatocytes have higher expression of glycogenic genes, such as Pgm1, Gys2, Ugp2, but with their lower expression in Albhi hepatocytes (Fig.S11), suggests increased glycogenesis in HNF4α hi hepatocytes. Therefore, Albhi hepatocytes prone to lipogenesis in the liver, while HNF4α hi hepatocytes have preference for glycogenesis. In addition, in the whole liver, the elevated glycogenic genes expression and diminished lipogenic genes expression in ApoE−/−/Liver-DKO that have been validated by RT-qPCR (Fig.S10A, S10B).
Notably, the ApoE−/−/Liver-DKO mice exhibited elevated gene expression related to LDL particle clearance and decreased expression related to fatty acid synthesis (Fig. 1D, 2E). Real-time quantitative PCR (RT-qPCR) further validated the single-cell RNA sequencing findings (Fig. 1E, 2F). These transcriptional changes reflect a shift towards improved lipid metabolism and clearance in the absence of hepatic epsins. Especially, we found elevated cardiovascular disease (CAD) protective score that from lipogenic Albhi hepatocytes to glycogenic HNF4α hi hepatocytes (Fig.1G).
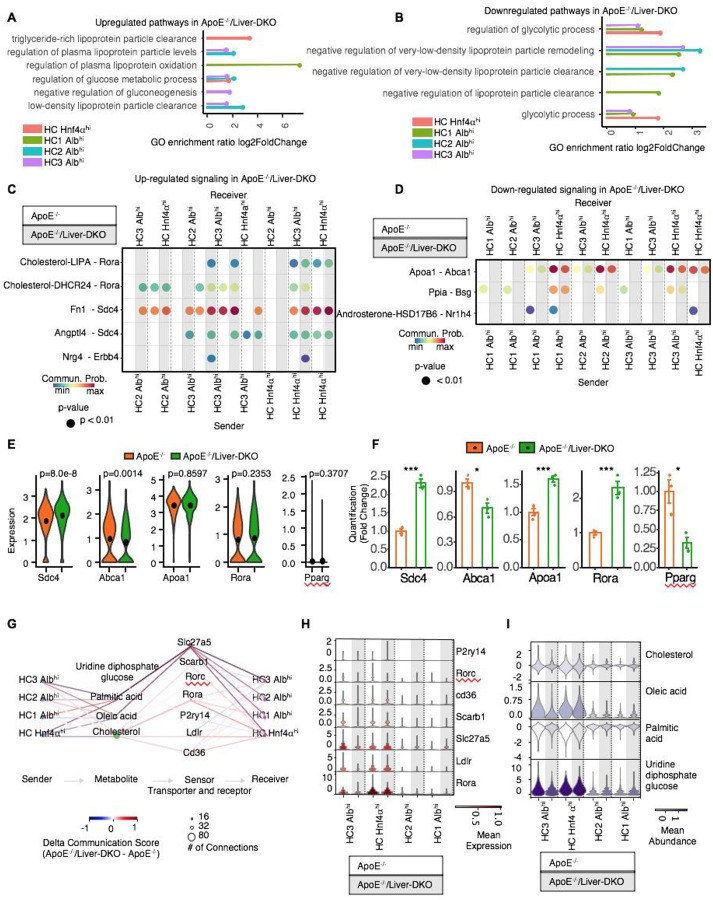
Fig. 2: Enhanced LDL particle clearance and LDLR-cholesterol communication elucidate improved LDL-C clearance and reduced atherogenesis.
A-B: Gene Ontology (GO) analysis showing significantly enriched pathways for LDL particle clearance that are upregulated (A) and downregulated (B) in ApoE−/−/Liver-DKO relative to ApoE−/−. C-D: Illustration of LDLR-cholesterol communication pathways with increased signaling interactions (C) and decreased signaling (D) in ApoE−/−/Liver-DKO relative to ApoE−/−, indicating enhanced LDL-C particle clearance in ApoE−/−/Liver-DKO. E-F: Elevated gene expression related to LDLR-cholesterol interactions and decreased gene expression related to fatty acid synthesis in ApoE−/−/Liver-DKO mice, shown through single-cell analysis (E) and real-time quantitative PCR (qPCR) (F). G: Quantitative analysis demonstrating reduced blood cholesterol and triglyceride levels in ApoE−/−/Liver-DKO mice, suggesting improved lipid metabolism and clearance. H: Relative receptor expression in ApoE−/−/Liver-DKO compared to ApoE−/−. i: Relative metabolite abundance in ApoE−/−/Liver-DKO compared to ApoE−/−. Note: We used the CellChat default method for the permutation test to calculate significant communication in panels (C) and (D). We also used the two-tailed t- test to compare the samples in panel (E).
Lipogenic Alb hi Hepatocytes to Glycogenic HNF4α hi Hepatocytes Transition in ApoE−/− Liver-DKO, and HNF4α hi Hepatocytes are Protective that with Higher CAD Protective Score under Western Diet Treatment.
We performed single cell transcriptome analysis for the liver cells from both ApoE−/− and ApoE−/− Liver-DKO under western diet treatment, and the workflow for libraries preparation of scRNA-seq has been illustrated (Fig.1A). After hepatocyte-derived data analysis, we isolated four different hepatocyte clusters, including HC1 Albhi, HC2 Albhi, HC3 Albhi, and HC HNF4α hi (Fig.1B), and their cell proportions in the liver are highlighted (Fig.S1A). Intriguingly, we found lower proportion of lipogenic Albhi hepatocytes but higher proportion of glycogenic HNF4α hi hepatocytes in ApoE−/−/Liver-DKO mice (Fig.S1A). Correspondingly, Gene Ontology (GO) enrichment analysis revealed upregulated pathways for LDL particle clearance and downregulated pathways for glycolytic process in different type of hepatocytes in the ApoE−/−/Liver-DKO mice (Fig. 2A, 2B; Fig.S8A–G). These pathways were significantly enriched compared to the ApoE−/− controls, suggesting improved LDL-C clearance in the absence of hepatic epsins. Correspondingly, LDLR-cholesterol communication pathways were also enhanced, as evidenced by increased signaling interactions in the ApoE−/−/Liver-DKO mice (Fig. 2C, 2D). This enhancement in LDL-C clearance mechanisms likely contributes to the reduced atherogenesis observed in these mice. Consequently, we found elevated expression of genes involved in LDL-C clearance in the liver in ApoE−/−/Liver-DKO mice (Fig.2E, 2F). The hepatocyte markers for hepatocyte subcluster are shown in dotplot (Fig.1C). Intriguingly, in HNF4α hi hepatocytes, we also discovered significantly elevated HNF4α expression in ApoE−/− Liver-DKO that have been validated at both mRNA and protein levels (Fig.1D–G), also diminished expression of lipogenesis genes, such as Acaca and Scd1, and gene for lipid uptake, Fabp1, but elevated gene expression of lipoprotein clearance, including Apoa4 and Ldlr (Fig.1D, 1E). Especially, we found HNF4α hi hepatocytes are protective in ApoE−/− Liver-DKO, the CAD protective score is positively associated with HNF4α expression in ApoE−/− Liver-DKO (Fig.1G).
Further analysis identified a transition from lipogenic Albhi hepatocytes to glycogenic HNF4α hi hepatocytes in the ApoE−/−/Liver-DKO mice (Fig. S1B–F). Correlate to this transition, lipogenic genes, such as Acaca, Scd1, Acly, Hmgcr, Fasn, show diminished expression in the hepatocytes in ApoE−/−/Liver-DKO (Fig.S7A), while show elevated expression of apolipoprotein genes, such as Apoa4 and Apob, which is positively associated with HNF4α expression (Fig.S7B). RNA velocity and CellRank analyses supported these dynamic shifts, demonstrating an increased propensity for hepatocyte differentiation towards a glycogenic state in the absence of hepatic epsins (Fig. S1B–F). This transition is likely a compensatory mechanism to enhance glucose metabolism and reduce lipid synthesis, contributing to the reduced lipid levels observed in the ApoE−/−/Liver-DKO mice. In addition, by single cell RNA-seq, under normal chow, we also discovered downregulated genes involved in lipogenesis and lipid uptake, such as Acaca, Scd1, and Fabp1, but genes respond for lipoprotein clearance, including Apoa4, Apob, Apoc1, are significantly upregulated in Liver-DKO (Data not shown). Similarly, by comparison of cardiovascular diseases (CAD) susceptible genes expression that were reported by GWAS analysis expression between WT and Liver-DKO, with particularly emphasize on genes that participate in low-density lipoprotein particles removal40. We found significantly higher CAD protective score in the hepatocytes in Liver-DKO compared with WT (Data not shown). Furthermore, both the RNA velocity and CellRank analyses showed that higher probability from lipogenic Alb hi hepatocytes to glycogenic HNF4α hi hepatocytes transition in Liver-DKO than WT (Data not shown).
HNF4α hi Hepatocytes in ApoE−/− Liver-DKO have Upregulated Low-density Lipoprotein Particle Clearance and Glycogen Biosynthesis Compared with ApoE−/− under Western Diet Treatment.
Mechanistically, by Gene Ontology (GO) analysis, we found upregulated plasma lipoprotein oxidation and elevated low-density lipoprotein particle clearance, but with downregulated glycolytic process in ApoE−/− Liver-DKO (Fig2A, 2B). Similarly, we also found upregulated of pathways involved in LDL-C particle clearance in Liver-DKO under normal chow (Data not shown). Furthermore, we performed cell-cell communication analysis, we found significantly upregulated Rorα and Sdc4 associated pathways in ApoE−/− Liver-DKO, which inhibit lipogenesis in the liver41 (Fig.2C; Fig.S9A). Similarly, under normal chow, by cell-cell communication analysis, and found Sdc4 and Nr1h3 pathways, which are reported to reduce steatosis42,43, are upregulated in Liver-DKO under normal chow (Data not shown). We also found downregulation of Ppia and Nr1h4 pathways, which suppress lipogenesis in the liver44,45 (Fig.2D; Fig.S9B). The representative genes for inhibition of lipogenesis, such as Sdc4, Rorα, are significantly elevated in ApoE−/− Liver-DKO (Fig.2E, 2F), and the lipogenic Pparγ is significantly diminished in ApoE−/− Liver-DKO (Fig2E, 2F). Likewise, under normal chow, we also discovered downregulated of lipogenic pathways in Liver-DKO, such as Pparγ and Ppia pathways44,46 (Data not shown). The metabolites analysis by MEBOCOST algorithm showed enhanced communication interactions for cholesterol/LDLR and cholesterol/Rorα pathways in HNF4α hi hepatocytes in ApoE−/− Liver-DKO (Fig.3G), and resulted in lower intensity of cholesterol in ApoE−/−/Liver-DKO mice (Fig.2I). However, we found no significantly difference for the mean expression of Rorα gene between ApoE−/− and ApoE−/− Liver-DKO (wFig.2H). Similarly, We also found significant elevated communication interactions for cholesterol/LDLR and cholesterol/Rorα pathways in Albhi HNF4α hi hepatocytes in Liver-DKO under normal chow, and with higher mean expression of Rorα gene in Liver-DKO (Data not shown), which promote the cholesterol clearance. Intriguingly, we found significantly lower mean abundance of cholesterol but higher abundance of uridine diphosphate glucose (UDPG) as intermediate metabolite for glycogenesis in HNF4α hi hepatocytes in ApoE−/− Liver-DKO (Fig.2I), suggests elevated glycogenesis in the ApoE−/−/Liver-DKO mice. Similarly, we also found lower mean abundance of cholesterol but higher abundance of uridine diphosphate glucose (UDPG) as intermediate metabolite for glycogenesis in Albhi HNF4α hi hepatocytes in Liver-DKO under normal chow (Data not shown).
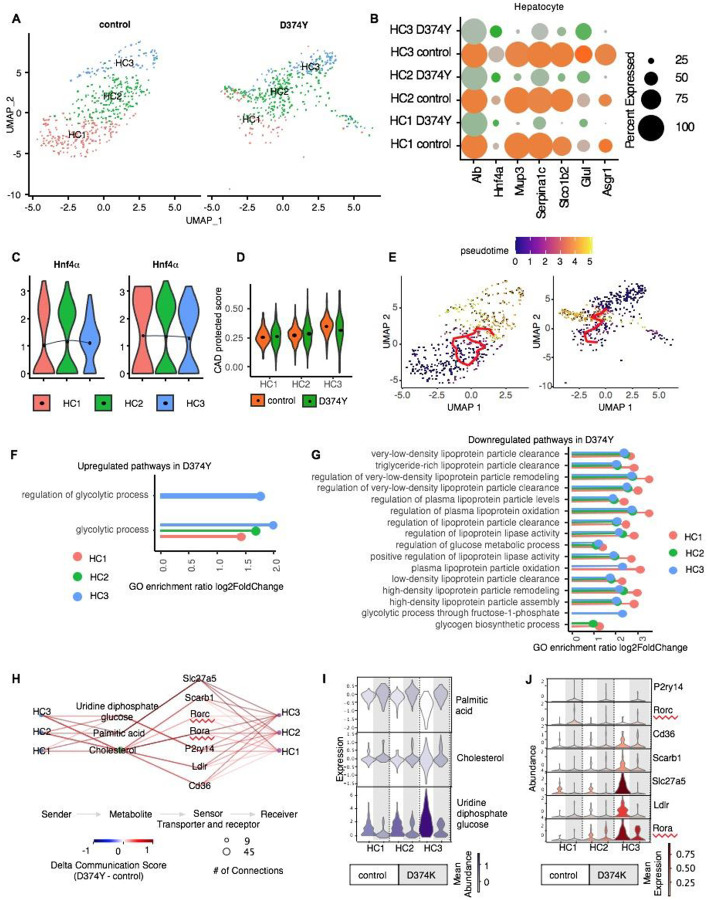
Fig. 3: Single-cell RNA-sequencing reveals gene expression dynamics in PCSK9 D374Y mutated mice.
A: UMAP visualization illustrating the heterogeneity of hepatocyte cell populations indicating distinct clustering patterns of hepatocytes in control mice compared to D374Y mutated. B: Dot plot showing the expression of gene markers in respective sub-cell types. C: Hnf4α expression increased from HC1 Albhi to HC3 Albhi in both control and D374Y mutated conditions. D: Coronary artery disease (CAD) protective score comparing D374 mutated to control. E: Pseudotime trajectory and RNA velocity analysis mapping the transition pathway from lipogenic Albhi hepatocytes (HC1 Albhi) to glucogenic Hnf4αhi hepatocytes (HC3 Albhi) in control, in contrast to D374Y mutated. F-G: Gene Ontology (GO) analysis showing significantly enriched pathways for upregulated glycolytic process (F) and downregulated LDL particle clearance (G) and in D374Y mutated relative to control. H: Quantitative analysis demonstrating metabolite communication score in D374Y mutated mice compared to control. I: Relative metabolite expression abundance in D374Y mutation compared to control. J: Relative metabolite receptor expression abundance in D374Y mutated compared to control.
Elevated Lipogenic Gene Expression but Diminished Glycogenic Gene Expression in Albhi HNF4α hi Hepatocytes and with Lower CAD Protective Score in hPCSK9 D374Y Mutant.
In addition to ApoE−/− atherosclerotic mouse model, we reanalyzed liver single cell RNA-seq data from hPCSK9 D374Y mutation mice. PCSK9-D374Y gain-of-function mutant has a markedly increased affinity for LDLR and promote its degradation47. As expected, we identified diminished Ldlr expression in hPCSK9 D374Y mutants, especially in HC3_Albhi HNF4α hi hepatocytes (Fig.S13). Intriguingly, we found reduced proportion of HC3_Albhi HNF4α hi hepatocytes in hPCSK9 D374Y mutants (Fig.3A), and the HC1, HC2, and HC3 hepatocytes have been clustered and evaluated by the expression of HNF4α and other hepatocyte markers (Fig.3B). Subsequently, we found significantly lower CAD protective score in Albhi HNF4α hi hepatocytes in hPCSK9 D374Y mutant than control. Intriguingly, the CAD protective score in both control and hPCSK9 D374Y mutant is positively correlated with HNF4α expression level, with highest CAD protective score in HC3_Albhi HNF4α hi hepatocytes among three different hepatocytes (Fig.3D). Especially, we discovered significantly diminished HNF4α expression and with dramatically elevated epsin1 expression in HC3_Albhi HNF4α hi hepatocytes in hPCSK9 D374Y mutant (Fig.3C, Fig.S13). Consequently, lipogenic genes, such as Acly and Fasn, are significantly induced in hPCSK9 D374Y mutant (Fig.S14A), suggests activated lipogenesis in hPCSK9 D374Y mutant. On the opposite, glycogenic genes, such as Gys2, Ugp2, are potently inhibited in hPCSK9 D374Y mutant (Fig.S14B), indicates diminished glycogenesis in hPCSK9 D374Y mutant. Almost all the apolipoprotein genes, such as Apoa1, Apoa2, Apoa4, Apob, Apoc1, Apoc2, and Apoc3, are dramatically inhibited in hPCSK9 D374Y mutant (Fig.S12), reveals diminished LDL cholesterol transportation for its clearance.
Diminished Low-density Lipoprotein Particle Clearance and Glycogen Biosynthesis and Hindered Alb hi Hepatocytes to Alb hi HNF4α hi Hepatocytes Transition in the Liver of hPCSK9 D374Y Mice
Mechanistically, by Gene Ontology (GO) analysis, the pathways for the regulation of low-density lipoprotein particle clearance and the regulation of glycogen biosynthetic process are significantly downregulated for enrichment in hPCSK9 D374Y mutant (Fig.3G), while the pathways involved in regulation of glycolytic process are significantly enriched that upregulated in hPCSK9 D374Y mutant (Fig.3F), suggests diminished low-density lipoprotein clearance and inhibited glycogenesis in hPCSK9 D374Y mutant. The further metabolites analysis by MEBOCOST algorithm showed weakened communication interactions for cholesterol/LDLR and cholesterol/Rorα pathway in Alb hi HNF4α hi Hepatocytes in hPCSK9 D374Y mutant (Fig.3H, 3J). Consequently, we found significantly higher mean abundance of cholesterol but lower abundance of uridine diphosphate glucose (UDPG) as intermediate metabolite for glycogenesis in Alb hi HNF4α hi in hPCSK9 D374Y (Fig.3I).
Subsequently, the pseudotime trajectory analysis showed that significantly inhibited cell fate transition from HC1_lipogenic Alb hi hepatocytes to glycogenic HC3_Alb hi HNF4α hi in hPCSK9 D374Y mutant than control (Fig.3E). Correlate to this transition, lipogenic genes, such as Acly, Fasn, show elevated fatty acid synthesis in the hepatocytes in hPCSK9 D374Y mutant (Fig.S14A), while show diminished expression of apolipoprotein genes, such as Apoa1, Apoa2, Apoa4, Apob, Apoc1, Apoc2, and Apoc3, suggest impaired low-density lipoprotein cholesterol clearance in hPCSK9 D374Y mutant (Fig.S12). The glycogenic genes exhibit diminished expression, such as Gys2, Ugp2, indicate inhibited glycogenesis in the hepatocytes in hPCSK9 D374Y mutant (Fig.S14B). RNA velocity and CellRank analyses supported these dynamic shifts, demonstrating an inhibited propensity for hepatocyte differentiation towards a glycogenic state in the presence of elevated hepatic epsins in hPCSK9 D374Y (Data not shown). This transition is likely a pathological mechanism in hPCSK9 D374Y mutant by inhibition of glycogenesis and enhanced lipid synthesis, contributing to the hyperlipidemia in hPCSK9 D374Y mutant.
Elevated LDLR Expression in the Liver of Liver Specific Epsins Deficiency Mice and Diminished LDLR Expression in the Liver from WD-fed Mice
Western blot analysis of liver tissue lysates from both WT and Liver-DKO mice revealed elevated LDLR expression in Liver-DKO, and the absence of Epsin1 and Epsin2 expression in Liver-DKO (Fig.4A). The protein levels of LDLR in WT and Liver-DKO have been quantified (Fig.4A). Intriguingly, we found significantly diminished LDLR expression in the livers of western diet (WD)-fed WT mice compared to normal diet (ND)-fed WT mice (Fig.4B). However, we discovered that both Epsin1 and Epsin2 proteins are dramatically elevated in the livers of WD-fed WT mice compared with ND-fed WT mice (Fig.4B). The protein levels of LDLR in both ND-fed WT and WD-fed WT have been quantified (Fig.4B). In addition to WB analysis, we performed immunofluorescence (IF) analysis of liver cryosections from both WT and Liver-DKO mice. We found elevated LDLR immunofluorescence signal in the liver of Liver-DKO (Fig.4C,4D). Notably, the LDLR immunofluorescence signal in the liver from WD-fed WT is markedly diminished compared with ND-fed WT (Fig.4C,4D). However, the LDLR immunofluorescence signal intensity in WD-fed Liver-DKO is partially maintained compared with ND-fed Liver-DKO (Fig.4C,4D), suggests epsins deficiency in the liver prevention of the degradation of LDLR under western diet treatment. The LDLR immunofluorescence signal intensity have been quantified (Fig.4D). The absence of both Epsin1 and Epsin2 immunofluorescence signals in the livers of Liver-DKO mice compared to WT mice (Fig.4C).
Liver-deficiency of Epsins Inhibits Atherosclerotic Lesion Formation and Macrophage Accumulation
En face Oil Red O staining of aortas from ApoE −/− and ApoE −/− / Liver-DKO mice fed a WD revealed significantly diminished atherosclerotic lesion in ApoE −/− / Liver-DKO mice compared with ApoE −/− mice (Fig.5A). The lesion areas of aortic root in both ApoE −/− and ApoE −/− / Liver-DKO mice have been quantified (Fig.5A). In addition, aortic roots cryosections from both ApoE −/− and ApoE −/− / Liver-DKO mice were stained with Oil Red O (ORO), and we found significantly reduced lesion size in ApoE −/− / Liver-DKO mice than ApoE −/− mice (Fig.5B). We also discovered significantly diminished CD68 immunofluorescence signals in ApoE −/− / Liver-DKO mice than ApoE −/− mice, indicating fewer macrophage accumulation in the atherosclerotic lesions (Fig.5B). We further measured plasma triglyceride (TG) and cholesterol levels in WD-fed WT and Liver-DKO mice injected with PCSK9-AAV8, and we detected significantly reduced TG and cholesterol levels in WD-fed Liver-DKO; PCSK9-AAV8 mice compared to WD-fed WT; PCSK9-AAV8 mice (Fig.5C). In addition, we also measured plasma cholesterol level from both ApoE −/− and ApoE −/− / Liver-DKO mice under either ND or WD treatment, and we found significantly diminished plasma cholesterol in ApoE −/− / Liver-DKO mice compared to ApoE −/− mice (Fig.5D). Oil Red O staining of liver cryosections were performed in both ApoE −/− and ApoE −/− / Liver-DKO mice, we found significantly reduced hepatic lipids in the livers of ApoE −/− / Liver-DKO mice compared to ApoE −/− mice (Fig.5E).
LDLR is Resistant to PCSK9-induced Proteasomal Degradation in Liver-DKO Mice and LDLR Directly Binds to Epsin1 UIM Domain.
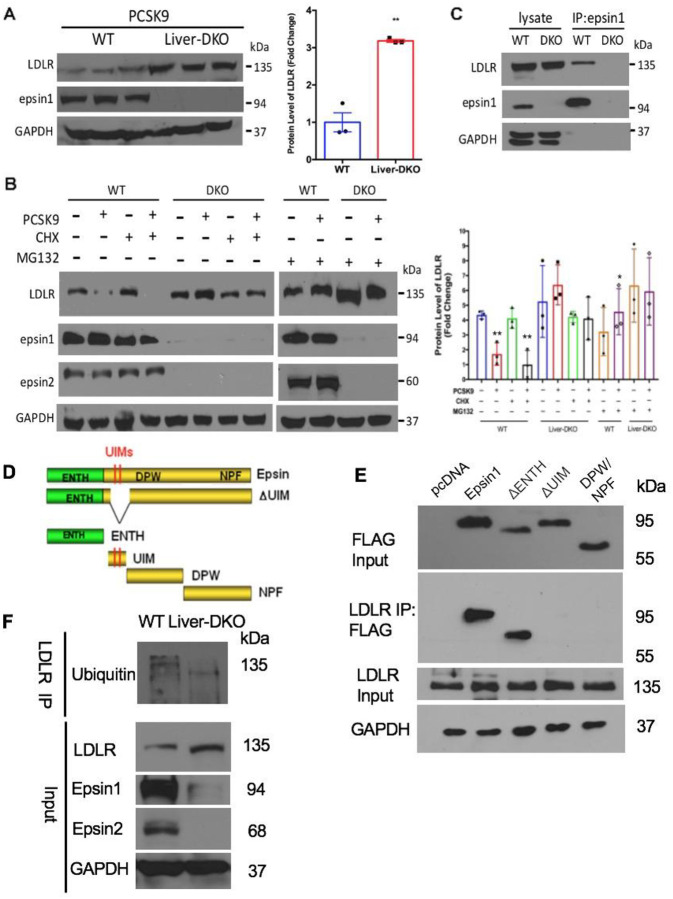
Western blot (WB) analysis of liver tissue harvested from WT and Liver-DKO mice injected with PCSK9-AAV8 virus (2×1011 genomes) revealed LDLR was markedly degraded by PCSK9 administration in WT, but not in Liver-DKO livers (Fig.6A). The protein levels of LDLR in WT and Liver-DKO mice were quantified, with significantly enhanced LDLR expression in the liver from Liver-DKO mice (Fig.6A). In addition to liver tissue lysate analysis, we also performed WB analysis of lysate from primary hepatocytes isolated from WT and Liver-DKO mice, treated with PCSK9, cycloheximide (CHX) and MG132. We found PCSK9-induced LDLR degradation occurred independent of new protein synthesis (in the presence of CHX) but was blocked by either loss of epsins or proteasomal inhibitor MG132 (Fig.6B). The protein levels of LDLR were quantified, with significantly higher LDLR expression in the hepatocytes from Liver-DKO under either with or without MG132 treatment (Fig.6B). To study the interaction between LDLR and Epsin1, we performed anti-epsin1 co-immunoprecipitation (co-IPs) analysis. We found LDLR directly binds epsin1 in WT, but not in Liver-DKO primary hepatocytes (Fig.6C). To further study which epsin1 motif can bind to LDLR, we transfected different FLAG-tagged epsin1 deletion mutants plasmids into HepG2 cells (Fig.6D), including pcDNA (empty plasmid), full length epsin1 plasmid, epsin1-ΔENTH plasmid, epsin1-ΔUIM plasmid, epsin1-DPW/NPF plasmid. Intriguingly, we found LDLR can specific bind to both the full length epsin1 and epsin1-ΔENTH, but not bind to epsin1-ΔUIM and epsin1-DPW/NPF, indicating the epsin1-UIM domain is the binding motif for the interaction between epsin1 and LDLR (Fig.6E). In addition, we discovered that significantly diminished ubiquitinated LDLR in the liver lysate from Liver-DKO by testing ubiquitin expression after LDLR antibody for immunoprecipitation (IP) (Fig.6F). In summary, by preventing PCSK9-triggered LDLR degradation, hepatic epsin depletion enhances LDL-C clearance and ameliorates dyslipidemia in atherosclerosis (Fig.8).
Fig.6: LDLR is resistant to PCSK9-induced proteasomal degradation in Liver-DKO mice and Liver-DKO primary hepatocytes and directly bind to epsin1 UIM.
(A) Western blot (WB) analysis of liver tissue harvested from WT and Liver-DKO mice injected with PCSK9-AAV8 virus revealed PCSK9-triggered LDLR degradation is inhibited in Liver-DKO mice (left). Data quantification (right) (n=3, **<0.01). (B) WB analysis of lysate from primary hepatocytes isolated from WT and Liver-DKO mice, treated with PCSK9, cycloheximide (CHX) and MG132 showed PCSK9-induced LDLR degradation occurred independent of new protein synthesis (in the presence of CHX) but was blocked by either loss of epsins or proteasomal inhibitor MG132 (left). Data quantification (right) (n=3, **p<0.01 vs lane 1, *p<0.05 vs lane 2). (C) Anti-epsin1 co-IPs showed LDLR directly binds epsin 1 in WT, but not Liver-DKO mouse primary hepatocytes (n=4). (D) Epsin deletion mutants and individual protein domains (E) LDLR antibody immunoprecipitation with lysates from HepG2 cell that were transfected by Flag-fused plasmids, including pcDNA full length epsin1, ΔENTH, ΔUIM, DPW/NPF. Anti-FLAG antibody was used for detect the binding between LDLR and epsin1, ΔUIM is essential for LDLR bind to epsin1. F: LDLR antibody immunoprecipitation with lysates from liver tissues in both WT and Liver-DKO, Anti-ubiquitin antibody was used for detect the ubiquitinated LDLR between WT and Liver-DKO. Liver lysates from both WT and Liver-DKO were also used as Input control for testing anti-LDLR, Epsin1, Epsin2, and GAPDH antibodies.
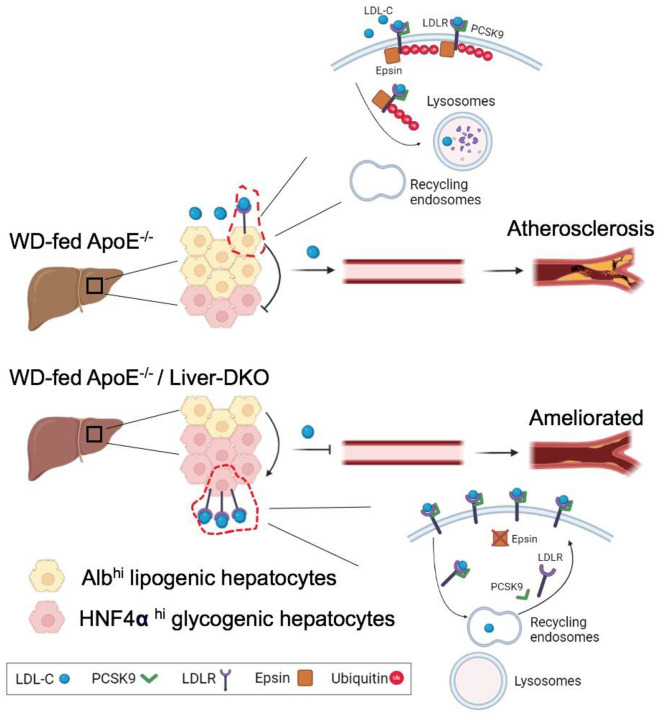
Fig.8. Western diet (WD)-fed ApoE−/− /Liver-DKO mice have elevated low-density lipoprotein cholesterol (LDL-C) clearance compared with WD-fed ApoE−/− mice that is attributable to promoted the cell fate transition from Albhi lipogenic hepatocytes to HNFα hi glycogenic hepatocytes.
In the liver, the glycogenesis inhibits lipogenesis. Consequently, the progression of atherosclerotic plaques are significantly ameliorated in WD-fed ApoE−/− /Liver-DKO mice. Mechanistically, loss of epsins protein in the liver prevent ubiquitination-driven LDLR degradation. The expressed LDLR in hepatocyte cell membrane uptakes LDL-C from the circulation. In the presence of epsins protein in the liver (top), in WT mice, PCSK9 bind to LDLR, epsins protein mediate LDLR ubiquitination, and the ubiquitinated LDLR is directed to lysosomes for protein degradation. As a result, elevated circulating LDL-C due to PCSK9-mediated LDLR degradation. In the absence of epsins protein in the liver (bottom), in epsin1/2 Liver-DKO mice, PCSK9 bind to LDLR, but the LDLR ubiquitination is abolished thanks to the deficiency of epsins protein. The LDLR is directed to recycling endosomes, and LDLR protein can be recycled and back to the membrane of hepatocytes. As a result, diminished circulating LDL-C thanks to the recycling LDLR.
Nanoparticle-mediated Delivery of Epsins siRNAs Potently Inhibits Lesion Development, Reduces Foam Cell Formation, and Decreases Cholesterol and TG Levels in ApoE−/− Mice.
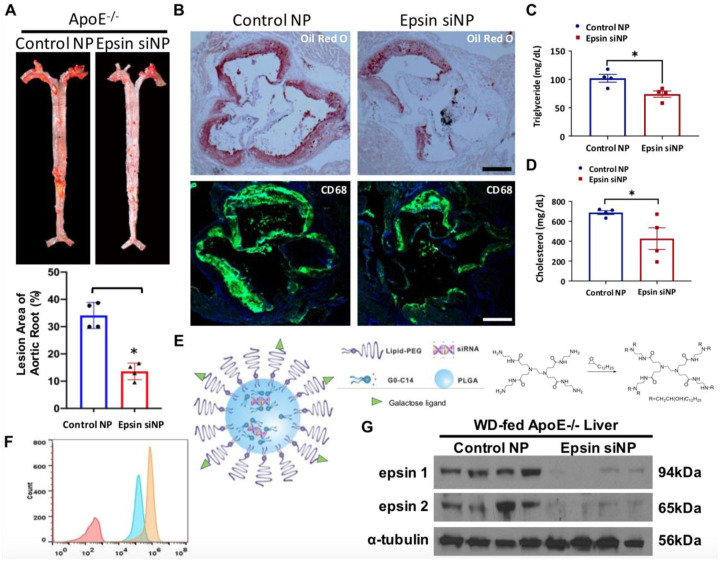
To explore the therapeutic potential of targeting hepatic epsins, we employed nanoparticle-encapsulated siRNAs specifically targeting epsins in the liver. Therapeutically, we applied galactose targeted - nanoparticles (NPs) with epsin1/2 siRNA (Fig.7E) to inject ApoE −/− mice under WD-treatment, control siRNA NPs were injected to WD-fed ApoE −/− mice as the control. Before injection of siRNA NPs into ApoE −/− mice, we have performed cytometry to test the efficiency of Gal-targeted Cy5.5-siRNA NPs and non-targeted Cy5.5-siRNA NPs in THLE-3 cells. We found significantly higher uptake efficiency in Gal-targeted Cy5.5-siRNA NPs compared to non-targeted Cy5.5-siRNA NPs in THLE-3 (Fig.7F). Western blots of liver lysates isolated from WD-fed ApoE −/− mice (8 weeks) that treated with control or epsins siRNA NPs revealed dramatically diminished epsin1 and epsin2 protein expression, indicating the highly efficiency of epsins siRNA NPs for knockdown of epsin1 and epsin2 proteins (Fig.7G). En face ORO staining of aortas from control siRNA NP-treated or epsin1/2 siRNA NP treated ApoE −/− mice fed a WD revealed significantly diminished atherosclerotic lesion in epsin1/2 siRNA NP treated ApoE −/− mice compared with control siRNA NPs ApoE −/− mice (Fig.7A). The lesion areas of aortic root in both control siRNA NPs treated ApoE −/− mice and epsin1/2 siRNA NPs treated ApoE −/− mice have been quantified (Fig.7A). In addition, aortic roots from control siRNA NPs treated ApoE −/− or epsin1/2 siRNA NP treated ApoE −/− mice were stained with Oil Red O or the CD68 macrophage marker CD68. We found significantly reduced lesion size in epsin1/2 siRNA NP treated ApoE −/− mice than control siRNA NPs treated ApoE −/− mice (Fig.7B). We also discovered significantly diminished CD68 immunofluorescence signals in epsin1/2 siRNA NP treated ApoE −/− mice than control siRNA NPs treated ApoE −/− mice, indicating fewer macrophage accumulation in the atherosclerotic lesions in epsin1/2 siRNA NP treated ApoE −/− mice (Fig.7B). We further measured plasma triglyceride (TG) and cholesterol levels in both control siRNA NPs treated ApoE −/− mice and epsin1/2 siRNA NPs treated ApoE −/− mice, and we detected significantly reduced TG (Fig.7C) and cholesterol levels (Fig.7D) in epsin1/2 siRNA NP treated ApoE −/− mice compared to control siRNA NPs treated ApoE −/− mice. These findings suggest that targeting liver epsins presents a novel and promising therapeutic strategy for the treatment of atherosclerosis.
Fig.7. Nanoparticles (NP) with epsin1/2 siRNA inhibits lesion formation and macrophage accumulation.
(A) En face ORO staining of aortas (top) from control siRNA NP-treated ApoE−/− or epsin1/2 siRNA NP treated ApoE−/− mice fed a WD, and unpaired t-test (bottom) for lesion areas (n=4, ***p<0.001). (B) Aortic roots from control siRNA NP-treated ApoE−/− or epsin1/2 siRNA NP treated ApoE−/− mice were stained with ORO or the CD68 macrophage marker CD68. Scale = 500 um. (C) Plasma triglyceride (TG) levels in WD-fed ApoE−/− (WT) and epsin siRNA-nanoparticle (NP) treated mice (n=4, *p<0.05). (D) Cholesterol levels in WD-fed ApoE−/− (WT) and epsin siRNA-nanoparticle (NP) treated mice (n=4, *p<0.05). (E) Schematic of the targeted hybrid siRNA NP platform composed of a lipid-PEG shell with a targeting ligand and a PLGA core containing G0-C14/siRNA complexes (left). Synthesis of G0-C14 by reacting alkyl epoxides with polyamidoamine generation 0 (PAMAM, G0) with a ratio of 7:1 through ring-opening chemistry (right). We will synthesize G0-C14 analogs by changing the G0 to C14 ratio and the alkyl chain length. (F) Cytometry of uptake of Gal-targeted Cy5.5-siRNA NPs vs non-targeted Cy5.5-siRNA NPs in THLE-3 cells. (G) Western blots of liver lysates isolated from WD-fed ApoE−/− mice (8 weeks) and treated with control or epsin siRNA NPs (0.75 nmoles) for 3 weeks (2 doses/week) (n=4).
In summary, our results demonstrate that liver-specific epsins depletion significantly inhibits atherogenesis and reduces lipid levels in a mouse model of atherosclerosis. The observed phenotypic changes are associated with transcriptional reprogramming of hepatocytes, lipogenic Alb hi Hepatocytes to glycogenic HNF4α hi Hepatocytes, enhanced LDL-C clearance pathways, and improved lipid metabolism. Mechanistically, the deficiency of liver epsins protect LDLR from PCSK9-triggered degradation. By targeting liver epsins with nanoparticle-encapsulated siRNAs, it has highly efficacious at inhibiting dyslipidemia and impeding atherosclerosis. These findings highlight the potential of targeting hepatic epsins as a therapeutic strategy for the treatment of atherosclerosis and related cardiovascular diseases.
Discussion
Our previous studies have elucidated an atheroprone function of epsins in both endothelial cells4, macrophages17,18, and vascular smooth muscle cells48 due to significantly elevated inflammation in the atherosclerotic plaque. However, atherosclerosis is initiated from the abnormal accumulation of lipid in the subendothelial layer of the arterial wall for hyperlipidemia49. The liver is the central organ for the control of lipid homeostasis50. The processes of de-novo lipogenesis (DNL) taking place in the liver and hepatic lipid metabolism are critical for regulation of the levels and distribution of lipids throughout the body20. LDLR is expressed in the liver that is essential to clearance of circulating LDL-C that play protective roles in prevention of atherosclerosis13,51. PCSK9 is dominantly expressed in the liver, and PCSK9 binds to LDLR that promote LDLR degradation52–54. Currently, PCSK9 antibody-based therapeutic to reduce circulating levels of LDL have been developed by several drug companies55,56; however, the mechanistic details of PCSK9-mediated LDLR degradation remain insufficiently understood.
In this study, we identified significantly lower plasma cholesterol and triglyceride in ApoE−/− / Liver-DKO mice or WD-fed Liver-DKO mice injected with PCSK9-AAV8 than those in ApoE−/− controls or WD-fed WT mice (Fig.5C, 5D). Intriguingly, the plasma cholesterol and triglyceride levels in epsins deficient in other different cell types, such as endothelial cell, macrophage, and vascular smooth muscle cells4,17,18,48, do not have significantly differences when comparing with ApoE−/− controls, indicating epsins in the hepatocytes participate in lipogenesis, which have been validated with significantly diminished gene expression that involved in lipogenesis in ApoE−/− / Liver-DKO mice (Fig.1D, 1E). The liver is the main organ that regulate circulating LDL-C homeostasis by LDLR for LDL-C clearance19. Our single cell RNA sequencing data showed that significantly elevated genes that are enriched in low-density lipoprotein particles and triglyceride-rich lipoprotein particles clearance gene ontology (GO) in the hepatocytes in ApoE−/− / Liver-DKO mice (Fig.2A), which is consistent to its reduced plasma cholesterol and triglyceride. Subsequently, we also showed evidences that with enhanced communication score between LDLR and cholesterol in the hepatocytes from ApoE−/− / Liver-DKO mice by MEBOCOST analysis (Fig.2G–I), indicating enhanced capacity of low-density lipoprotein cholesterol (LDL-C) clearance by LDLR in ApoE−/− / Liver-DKO mice. Consequently, we found significantly reduced atherosclerotic lesion area in ApoE−/− / Liver-DKO mice than those in in ApoE−/− controls (Fig.5A, 5B). In addition to diminished lipogenic genes expression, the enhanced UDPG communication score in HNF4α hi hepatocytes in ApoE−/− / Liver-DKO mice that resulted in elevated UDPG metabolite level promote hepatic glycogenesis (Fig.2I). Chen et al. recently reported that hepatic glycogenesis inhibits lipogenesis57. Consequently, the enhanced UDPG metabolite levels in the liver inhibits fatty acid synthesis that could also contribute to reduced cholesterol and triglyceride in plasma in ApoE−/− / Liver-DKO mice.
Currently, almost all studies involved in human liver single cell RNA-seq analysis are in the context of chronic liver diseases, such as nonalcoholic fatty liver disease (NAFLD)58,59, or acute liver failure (ALF)60. Unfortunately, no single cell RNA-seq study for dissecting of human liver transcriptome for patients in the context of coronary artery disease or other cardiovascular diseases, and most single cell RNA-seq studies for patients in the context of coronary artery disease are for exploring the transcriptome differences for atherosclerotic plaques61,62. Therefore, it is impossible to compare our liver single cell RNA-seq data with liver single cell RNA-seq data from human patients with coronary artery disease. However, the recently published article explored liver transcriptome by single cell RNA-seq for hPCSK9-D374Y mice with emphasized analysis on Kupffer cells63, this dataset empower us to analyze heterogeneity of hepatocytes in the liver between hPCSK9-D374Y mice and controls. The hPCSK9-D374Y mutation potently elevates the affinity between PCSK9 and LDLR interaction, and further promote the degradation of LDLR64. In this study, we reanalyzed the liver single cell RNA-seq dataset by emphasizing the analysis in the context of hepatocytes from hPCSK9-D374Y mice63. Intriguingly, in the hepatocytes of hPCSK9-D374Y mice, we discovered diminished glycogenic genes expression but elevated lipogenic genes expression in hPCSK9-D374Y mice, results in diminished glycogenesis and activated lipogenesis that is consistent with lower CAD protective score in hPCSK9-D374Y mice, especially for HC3_Albhi HNF4α hi hepatocytes (Fig.3D, Fig.S14). Therefore, there are highly phenotypic similarities between hPCSK9-D374Y mice and ApoE−/− mice. Mechanistically, we identified significantly elevated epsin1 expression in hPCSK9-D374Y mice that would contribute to downregulated LDL particles and VLDL particles clearance by reduction of LDLR expression (Fig.3G, Fig.S13). Subsequently, we found diminished communication score between LDLR and cholesterol in the hepatocytes from hPCSK9-D374Y mice by MEBOCOST analysis (Fig.3H–J), which is consistent to serum cholesterol accumulation due to reduced hepatic LDLR levels in hPCSK9-D374Y mice65. Similar to ApoE−/− mice, the decreased UDPG communication score in glycogenic HC3_Albhi HNF4α hi hepatocytes (Fig.3H, 3I), and together with diminished genes of glycogenesis that further contribute to elevated lipogenesis57. Like to ApoE−/− mice, the diminished tendency of cell fate transition from HC1_lipogenic Albhi hepatocytes into HC3_glycogenic Albhi HNF4α hi hepatocytes in hPCSK9-D374Y mice, which is consistent with its elevated lipogenic genes and diminished glycogenic genes (Fig.S14). In summary, by comparison of ApoE−/− and hPCSK9-D374Y mice, these two mouse model for atherosclerosis study shared similar hepatocyte heterogeneity and the common pathological signaling pathways for inducing atherosclerosis or dyslipidemia, which might be highly correlated with the common elevated epsins expression in the liver that mediate LDLR degradation.
In this study, we firstly elaborated how liver epsins mediate the LDLR degradation that triggered by PCSK9 in the liver (Fig.8). In liver epsins-deficient mice (Liver-DKO), significantly elevated LDLR expression in the membrane of hepatocytes that empower upregulated LDL-C clearance (Fig.4A, 4C), which is resistant to WD-induced espins mediated LDLR degradation (Fig.4C). PCSK9 protein mediates LDLR protein degradation66. In this study, by injection of PCSK9-AAV8 into both WT controls and Liver-DKO mice for PCSK9 overexpression, LDLR is dramatically degraded in WT controls; however, the degradation of LDLR is significantly inhibited in Liver-DKO mice (Fig.6A, 6B), which strongly supports that liver epsins are essential for PCSK9-mediated LDLR degradation. Subsequently, we discovered that epsin1 can directly bind to LDLR protein that further trigger its degradation (Fig.6C). Our previous studies revealed that epsins are critical adaptor proteins that involved in endocytosis16,23,67–69. To explore which motif in epsin1 can specific bind to LDLR, such as ENTH, UIM, DPW and NPF (Fig.6D), FLAG-tagged epsin1 deletion mutants plasmids, including pcDNA control, full length epsin1 plasmid, epsin1-ΔENTH plasmid, epsin1-ΔUIM plasmid and epsin1-DPW/NPF plasmid have been transfected into HepG2 cells. Specifically, LDLR binds to UIM motif only but not binds to other motifs (Fig.6E), the ubiquitin-interacting motif (UIM) in epsins facilitated ubiquitination mediated protein degradation70. Consequently, diminished LDLR expression in the liver due to the interaction between LDLR and epsin1 UIM motif for the activation of ubiquitination of LDLR facilitate to its degradation in WT controls, but not in Liver-DKO mice. In addition, we revealed diminished ubiquitinated LDLR in the liver lysate in Liver-DKO by LDLR antibody immunoprecipitation (IP) assay, consequently, higher LDLR expression in the input lysate in Liver-DKO mice than WT controls (Fig.6F), supporting the loss of epsins enhances the stability of LDLR. MG132 proteasome inhibitors blocked the degradation of LDLR by inhibition of ubiquitination71. We further isolated primary hepatocytes from both WT and Liver-DKO for PCSK9, cycloheximide (CHX) and proteasomal inhibitor MG132 treatment, identified that PCSK9-induced LDLR degradation occurred independent of new protein synthesis, similar to MG132 treatment, and LDLR degradation was blocked by loss of epsins in the liver. In summary, epsins play gatekeeping roles in the PCSK9-mediated ubiquitination-driven LDLR degradation that by interaction between epsin1 UIM motif and LDLR. Therefore, the liver epsins might as potential targets to treatment of dyslipidemia or further atherosclerosis by protecting hepatic LDLR from degradation.
The liver is the primary organ of lipid nanoparticles accumulation following intravenous administration72,73, and lipid nanoparticle-mediated RNAs or siRNAs delivery holds great potential to treat liver diseases74,75. In this study, we engineer the hybrid siRNA NPs for better targeting of hepatocytes in vivo by surface modification with carbohydrate ligands (e.g. galactose) that can recognize the asialoglycoprotein receptor (ASGPR) predominately expressed on hepatocytes and minimally expressed by non-hepatic cells36,37. Finally, targeting liver epsins with lipid nanoparticle encapsulated siRNAs that with highly efficacious at inhibiting dyslipidemia and impeding atherosclerosis. We discovered significantly reduced atherosclerotic lesion in epsin1/2 siRNA NPs injected mice comparing to control NPs group (Fig.7A, 7B). As expected, we detected significantly lower cholesterol and triglyceride levels in epsin1/2 siRNA NPs injected group than control NPs group (Fig.7C, 7D). Therefore, we have tested the therapeutic efficiency for treatment of atherosclerosis by targeting liver epsins in mice. However, in this study, the lack of therapeutic data from large animals will be insufficient for evaluation of the efficiency by targeting liver epsins for inhibiting atherosclerotic progression. To fill in the gaps, in future, we will test the efficiency of epsin1/2 siRNA NPs targeting liver epsins in large animals, such as rabbit76 and pig77 for atherosclerotic regression studies. It has been widely applied for rabbit model to investigate familial hypercholesteromia (FH), and hypercholesteromia rabbits have very high LDL-C level due to dysfunction of LDLR, also with severe atherosclerosis12,78. By targeting epsins in the liver in hypercholesteromia rabbit with epsin1/2 siRNA NPs, we will measure the plasma LDL-C level and atherosclerotic lesion between control NPs group and epsin1/2 siRNA NPs group.
In this study, mechanistically, we identified significantly higher hepatocyte nuclear factor 4α (HNF4α) expression in ApoE−/− /Liver-DKO mice than ApoE−/− (Fig.1D–F), and HNF4α is reported as critical transcription factor that modulation of lipid homeostasis79. The hepatic HNF4α deficiency cause severe hepatic lipid accumulation, and overexpression of hepatic HNF4α lowers plasma cholesterol levels79. In addition, the accumulation of hepatic glycogen is disrupted by loss of hepatic HNF4α80. Intriguingly, we presented higher lipogenic genes expression in Albhi hepatocytes than HNF4αhi hepatocytes, while higher glycogenic genes expression in HNF4αhi hepatocytes than Albhi hepatocytes (Fig.S11). The higher percentage of glycogenic HNF4αhi hepatocytes but with lower lipogenic Albhi hepatocytes proportion in ApoE−/− /Liver-DKO mice than ApoE−/− (Fig.2A), which contribute to reduced cholesterol and triglyceride in plasma in ApoE−/− / Liver-DKO mice. Coincidently, the cardiovascular disease (CAD) protective score is positive associated with hepatic HNF4α expression in ApoE−/− / Liver-DKO mice, indicating glycogenic HNF4αhi hepatocytes are protective that attributable mainly to the inhibition of lipogenesis. Parviz et al. and Wu et al. demonstrated that HNF4α is essential for hepatic glycogen synthesis, and hepatic HNF4α deletion induces significantly impairment of hepatic glycogen accumulation in the liver80,81. In addition to glycogenesis, Bonzo et al. demonstrated that genetic deletion of HNF4α cause steatosis82, and both Xu et al. and Yang et al. clarified that overexpression of HNF4α significantly ameliorate hepatic steatosis by reducing of hepatic triglycerides and free fatty acids (FFA)83,84. In this study, we discovered that significantly diminished hepatic and serum lipids in ApoE−/− /Liver-DKO, but with elevated genes expression that involved in glycogenesis, indicating upregulated hepatic glycogenesis suppress lipogenesis in the liver that is consistent to the recently report that hepatic glycogenesis antagonizes lipogenesis57. Chen et al. demonstrated that elevated UDPG mediated glycogenesis that repressed the cleavage of premature SREBP1 and SREBP2 into mature SREBP1 and SREBP2, resulted in diminished hepatic lipogenesis57. In this study, MEBOCOST analysis revealed elevated UDPG metabolite, as intermediate of glycogenesis, in HNF4αhi hepatocytes in ApoE−/− /Liver-DKO, which promote hepatic glycogenesis that inhibition of hepatic lipogenesis. Intriguingly, there is a significantly higher tendency of cell fate transition from lipogenic Albhi hepatocytes into glycogenic HNF4αhi hepatocytes in ApoE−/− /Liver-DKO than ApoE−/− (Fig.S1 B–F), suggests the activated glycogenesis and inhibited lipogenesis in the liver in ApoE−/− /Liver-DKO. As HNF4αhi hepatocytes are positively regulation of hepatic glycogenesis80,81, in addition to elevated LDLR expression, the elevated hepatic glycogenesis and inhibited hepatic lipogenesis is a novel casual factor that contribute to the diminished atherosclerotic plaque in ApoE−/− /Liver-DKO mice.
In summary, in our study, we firstly demonstrated how hepatic epsins in mediating PCSK9-driven LDLR degradation (Fig.8). In the absence of hepatic epsins, PCSK9-mediated LDLR degradation is significantly repressed, and higher LDLR expression in the liver that promotes the LDL-C clearance in ApoE−/− /Liver-DKO mice and further inhibition of atherosclerotic plaque progression. In the presence of hepatic epsins, LDLR specially binds to the epsin1 UIM motif that further be processed for PCSK9-driven ubiquitination for proteosome degradation in ApoE−/− mice, which accelerated the LDL-C accumulation and promote the atherosclerotic plaque progression. Mechanistically, we elaborated that elevated pathways involved in LDL particle clearance and diminished genes of de novo lipogenesis in ApoE−/− /Liver-DKO. Especially, we firstly proposed that the cell fate transition from lipogenic Albhi hepatocytes into glycogenic HNF4αhi hepatocytes might be a novel protective mechanism for combating atherosclerosis in ApoE−/− /Liver-DKO. Finally, for therapeutic study, we synthesized lipid nanoparticles (LNP) encapsulated epsins siRNAs for treatment of WD-induced atherosclerotic ApoE−/− mice, which achieved significantly inhibited dyslipidemia and impeded atherosclerotic plaque progression. However, as the potentially therapeutic novel target for atherosclerosis, therapeutic studies on mice are not sufficient for convincing its medical application. In future, we will further evaluate the efficiency of LNP encapsulated epsins siRNAs in larger animals, such as rabbit or pig, for inhibition of dyslipidemia and atherosclerotic plaque progression.
Conclusion and Perspective
In this study, we discovered significantly elevated epsin1 and epsin2 expression in both WD-fed mice and atherosclerotic patients, but with dramatically diminished expression of LDLR protein in the liver from WD-fed mice and fatty liver disease patients. To study the roles of liver epsins in atherosclerosis, we specific deleted liver epsin1 and epsin2 using albumin Cre (Liver-DKO) on an ApoE −/− background. We discovered that WD-induced atherosclerosis was significantly inhibited, along with diminished blood cholesterol and triglyceride levels in ApoE −/− Liver-DKO mice.
Mechanistically, scRNA-seq analysis on hepatocyte-derived data revealed elevated pathways involved in LDL particle clearance under WD treatment in ApoE−/− /Liver-DKO mice, which was coupled with diminished plasma LDL-C levels. Further analysis using the MEBOCOST algorithm revealed enhanced communication score between LDLR and cholesterol, suggesting elevated LDL-C clearance in the ApoE−/− Liver-DKO mice. In addition, we showed that loss of epsins in the liver upregulates of LDLR protein level. We further showed that epsins bind LDLR via the ubiquitin-interacting motif (UIM), and PCSK9-triggered LDLR degradation was abolished by depletion of epsins, preventing atheroma progression.
Intriguingly, scRNA-seq analysis revealed the activated cell fate transition from lipogenic Alb hi hepatocytes to glycogenic HNF4α hi hepatocytes in the liver of ApoE−/− Liver-DKO mice, which is consistent with its diminished lipogenic genes expression but elevated glycogenic genes expression. The CAD protective score in HNF4α hi hepatocytes is higher than Alb hi hepatocytes, which is positively correlated with HNF4α expression level in ApoE−/− Liver-DKO mice. In addition to ApoE−/− mice, analysis of hepatocyte-derived data from hPCSK9-D374Y mice revealed similar pathological pathways involved in atherosclerosis or dyslipidemia as WD-fed ApoE−/− mice, with significantly elevated epsins expression in both hPCSK9-D374Y and WD-fed ApoE−/− mice that mediated LDLR degradation.
Finally, our therapeutic strategy, which involved targeting liver epsins with nanoparticle-encapsulated epsins siRNAs, was highly efficacious at inhibiting dyslipidemia and impeding atherosclerosis. Targeting epsins in the liver may serve as a novel therapeutic strategy to treat atherosclerosis by suppression of PCSK9-mediated LDLR degradation. In future, targeting epsins in the liver using nanoparticle-encapsulated epsins siRNAs, we will test its efficiency at inhibiting of dyslipidemia and impeding atherosclerosis in larger animals, such as rabbit or pig.
Supplementary Material
Acknowledgments
We thank the imaging core at Boston Children’s Hospital, and the Biopolymers Facility at Harvard Medical School for quality control analysis of DNA libraries prepared for scRNA-sequencing. We thank the animal core facility for the daily maintenance at Boston Children’s Hospital.
Sources of Funding
This work was supported in part by NIH grants Nos. R01HL137229, R01HL1563626, R01HL158097, and R01HL158097 to HC.
Funding Statement
This work was supported in part by NIH grants Nos. R01HL137229, R01HL1563626, R01HL158097, and R01HL158097 to HC.
Footnotes
Disclosure
None.
Supplemental Material
Extended Methods
Major Resources Table
Nonstandard Abbreviations and Acronyms
Novelty and Significance
References
- 1.Libby P. et al. Atherosclerosis. Nat Rev Dis Primers 5, 56, doi: 10.1038/s41572-019-0106-z (2019). [DOI] [PubMed] [Google Scholar]
- 2.Pahwa R. & Jialal I. in StatPearls (2024). [PubMed]
- 3.Zhu B. et al. Two sides of the same coin: Non-alcoholic fatty liver disease and atherosclerosis. Vascul Pharmacol 154, 107249, doi: 10.1016/j.vph.2023.107249 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Dong Y. et al. Epsin-mediated degradation of IP3R1 fuels atherosclerosis. Nat Commun 11, 3984, doi: 10.1038/s41467-020-17848-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito-Vicente A. et al. Familial Hypercholesterolemia: The Most Frequent Cholesterol Metabolism Disorder Caused Disease. Int J Mol Sci 19, doi: 10.3390/ijms19113426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaefer J. R., Kurt B., Sattler A., Klaus G. & Soufi M. Pharmacogenetic aspects in familial hypercholesterolemia with the special focus on FHMarburg (FH p.W556R). Clin Res Cardiol Suppl 7, 2–6, doi: 10.1007/s11789-012-0041-y (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batty M., Bennett M. R. & Yu E. The Role of Oxidative Stress in Atherosclerosis. Cells 11, doi: 10.3390/cells11233843 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markin A. M. et al. The Role of Cytokines in Cholesterol Accumulation in Cells and Atherosclerosis Progression. Int J Mol Sci 24, doi: 10.3390/ijms24076426 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soehnlein O. & Libby P. Targeting inflammation in atherosclerosis - from experimental insights to the clinic. Nat Rev Drug Discov 20, 589–610, doi: 10.1038/s41573-021-00198-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein J. L. & Brown M. S. The LDL receptor. Arterioscler Thromb Vasc Biol 29, 431–438, doi: 10.1161/ATVBAHA.108.179564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao X. et al. Targeting proprotein convertase subtilisin/kexin type 9 (PCSK9): from bench to bedside. Signal Transduct Target Ther 9, 13, doi: 10.1038/s41392-023-01690-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R. et al. Spontaneous severe hypercholesterolemia and atherosclerosis lesions in rabbits with deficiency of low-density lipoprotein receptor (LDLR) on exon 7. EBioMedicine 36, 29–38, doi: 10.1016/j.ebiom.2018.09.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H. et al. In Vivo AAV-CRISPR/Cas9-Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 141, 67–79, doi: 10.1161/CIRCULATIONAHA.119.042476 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Keeter W. C., Carter N. M., Nadler J. L. & Galkina E. V. The AAV-PCSK9 murine model of atherosclerosis and metabolic dysfunction. Eur Heart J Open 2, oeac028, doi: 10.1093/ehjopen/oeac028 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee R. G. et al. Efficacy and Safety of an Investigational Single-Course CRISPR Base-Editing Therapy Targeting PCSK9 in Nonhuman Primate and Mouse Models. Circulation 147, 242–253, doi: 10.1161/CIRCULATIONAHA.122.062132 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Chen H. et al. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394, 793–797, doi: 10.1038/29555 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Brophy M. L. et al. Myeloid-Specific Deletion of Epsins 1 and 2 Reduces Atherosclerosis by Preventing LRP-1 Downregulation. Circ Res 124, e6–e19, doi: 10.1161/CIRCRESAHA.118.313028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui K. et al. Epsin Nanotherapy Regulates Cholesterol Transport to Fortify Atheroma Regression. Circ Res 132, e22–e42, doi: 10.1161/CIRCRESAHA.122.321723 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan Y. et al. Regulation of cholesterol homeostasis in health and diseases: from mechanisms to targeted therapeutics. Signal Transduct Target Ther 7, 265, doi: 10.1038/s41392-022-01125-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders F. W. & Griffin J. L. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol Rev Camb Philos Soc 91, 452–468, doi: 10.1111/brv.12178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasula S. et al. Endothelial epsin deficiency decreases tumor growth by enhancing VEGF signaling. J Clin Invest 122, 4424–4438, doi: 10.1172/JCI64537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessneer K. L. et al. Genetic reduction of vascular endothelial growth factor receptor 2 rescues aberrant angiogenesis caused by epsin deficiency. Arterioscler Thromb Vasc Biol 34, 331–337, doi: 10.1161/ATVBAHA.113.302586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H. et al. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc Natl Acad Sci U S A 106, 13838–13843, doi: 10.1073/pnas.0907008106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzen O., Gan L. M. & Bjorkegren J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019, doi: 10.1093/database/baz046 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang S. et al. Cell Taxonomy: a curated repository of cell types with multifaceted characterization. Nucleic Acids Res 51, D853–D860, doi: 10.1093/nar/gkac816 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M. et al. DISCO: a database of Deeply Integrated human Single-Cell Omics data. Nucleic Acids Res 50, D596–D602, doi: 10.1093/nar/gkab1020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Manno G. et al. RNA velocity of single cells. Nature 560, 494–498, doi: 10.1038/s41586-018-0414-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergen V., Lange M., Peidli S., Wolf F. A. & Theis F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 38, 1408–1414, doi: 10.1038/s41587-020-0591-3 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Trapnell C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386, doi: 10.1038/nbt.2859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502, doi: 10.1038/s41586-019-0969-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange M. et al. CellRank for directed single-cell fate mapping. Nat Methods 19, 159–170, doi: 10.1038/s41592-021-01346-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rongbin Zheng Y. Z., Tsuji Tadataka, Gao Xinlei, Wagner Allon, Yosef Nir, Chen Hong, Zhang Lili, Tseng Yu-Hua, Chen Kaifu. MEBOCOST: Metabolite-mediated Cell Communication Modeling by Single Cell Transcriptome. doi:doi: 10.1101/2022.05.30.494067 (2022). [DOI] [Google Scholar]
- 34.Jin S. et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088, doi: 10.1038/s41467-021-21246-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141, doi: 10.1016/j.xinn.2021.100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanhueza C. A. et al. Efficient Liver Targeting by Polyvalent Display of a Compact Ligand for the Asialoglycoprotein Receptor. J Am Chem Soc 139, 3528–3536, doi: 10.1021/jacs.6b12964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang K. W. et al. Galactose Derivative-Modified Nanoparticles for Efficient siRNA Delivery to Hepatocellular Carcinoma. Biomacromolecules 19, 2330–2339, doi: 10.1021/acs.biomac.8b00358 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Tao W. et al. siRNA nanoparticles targeting CaMKIIgamma in lesional macrophages improve atherosclerotic plaque stability in mice. Sci Transl Med 12, doi: 10.1126/scitranslmed.aay1063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X. et al. Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc Natl Acad Sci U S A 112, 7779–7784, doi: 10.1073/pnas.1505629112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aragam K. G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat Genet 54, 1803–1815, doi: 10.1038/s41588-022-01233-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K. et al. RORalpha controls hepatic lipid homeostasis via negative regulation of PPARgamma transcriptional network. Nat Commun 8, 162, doi: 10.1038/s41467-017-00215-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Nardo W. et al. Proteomic analysis reveals exercise training induced remodelling of hepatokine secretion and uncovers syndecan-4 as a regulator of hepatic lipid metabolism. Mol Metab 60, 101491, doi: 10.1016/j.molmet.2022.101491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peet D. J. et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93, 693–704, doi: 10.1016/s0092-8674(00)81432-4 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Li Z., Zhang B., He H. & Bai Y. PPIA is a novel adipogenic factor implicated in obesity. Obesity (Silver Spring) 23, 2093–2100, doi: 10.1002/oby.21208 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Clifford B. L. et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab 33, 1671–1684 e1674, doi: 10.1016/j.cmet.2021.06.012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schadinger S. E., Bucher N. L., Schreiber B. M. & Farmer S. R. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab 288, E1195–1205, doi: 10.1152/ajpendo.00513.2004 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lagace T. A. et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest 116, 2995–3005, doi: 10.1172/JCI29383 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beibei Wang K. C., Zhu Bo, Dong Yunzhou, Wang Donghai, Singh Bandana, Lu Yao Wei, Wu Hao, Bhattacharjee Sudarshan, Shahram Eisa-Beygi, Wong Scott, Cowan Douglas B., Du Mulong, Chen Hong. Epsins promote atherosclerosis through VSMC phenotypic modulation. doi:doi: 10.1101/2024.01.08.574714 (2024). [DOI] [Google Scholar]
- 49.Linton M. F. et al. in Endotext (eds Feingold K. R. et al. ) (2000). [Google Scholar]
- 50.Alves-Bezerra M. & Cohen D. E. Triglyceride Metabolism in the Liver. Compr Physiol 8, 1–8, doi: 10.1002/cphy.c170012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Go G. W. & Mani A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med 85, 19–28 (2012). [PMC free article] [PubMed] [Google Scholar]
- 52.Lagace T. A. PCSK9 and LDLR degradation: regulatory mechanisms in circulation and in cells. Curr Opin Lipidol 25, 387–393, doi: 10.1097/MOL.0000000000000114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natarajan P. & Kathiresan S. PCSK9 Inhibitors. Cell 165, 1037, doi: 10.1016/j.cell.2016.05.016 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Narasimhan S. D. Beyond Statins: New Therapeutic Frontiers for Cardiovascular Disease. Cell 169, 971–973, doi: 10.1016/j.cell.2017.05.032 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Robinson J. G. et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med 372, 1489–1499, doi: 10.1056/NEJMoa1501031 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Sabatine M. S. et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N Engl J Med 376, 1713–1722, doi: 10.1056/NEJMoa1615664 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Chen J. et al. Hepatic glycogenesis antagonizes lipogenesis by blocking S1P via UDPG. Science 383, eadi3332, doi: 10.1126/science.adi3332 (2024). [DOI] [PubMed] [Google Scholar]
- 58.Gribben C. et al. Acquisition of epithelial plasticity in human chronic liver disease. Nature 630, 166–173, doi: 10.1038/s41586-024-07465-2 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fred R. G. et al. Single-cell transcriptome and cell type-specific molecular pathways of human non-alcoholic steatohepatitis. Sci Rep 12, 13484, doi: 10.1038/s41598-022-16754-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matchett K. P. et al. Multimodal decoding of human liver regeneration. Nature 630, 158–165, doi: 10.1038/s41586-024-07376-2 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y. et al. Single-Cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue. Circulation 142, 1374–1388, doi: 10.1161/CIRCULATIONAHA.120.046528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou E. L. et al. Aortic Cellular Diversity and Quantitative Genome-Wide Association Study Trait Prioritization Through Single-Nuclear RNA Sequencing of the Aneurysmal Human Aorta. Arterioscler Thromb Vasc Biol 42, 1355–1374, doi: 10.1161/ATVBAHA.122.317953 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Nunzio G., Hellberg S., Zhang Y. et al. Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult. Nat Cardiovasc Res 3, 356–371, doi: 10.1038/s44161-024-00448-6 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timms K. M. et al. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet 114, 349–353, doi: 10.1007/s00439-003-1071-9 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Herbert B. et al. Increased secretion of lipoproteins in transgenic mice expressing human D374Y PCSK9 under physiological genetic control. Arterioscler Thromb Vasc Biol 30, 1333–1339, doi: 10.1161/ATVBAHA.110.204040 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Maxwell K. N., Fisher E. A. & Breslow J. L. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc Natl Acad Sci U S A 102, 2069–2074, doi: 10.1073/pnas.0409736102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H., Polo S., Di Fiore P. P. & De Camilli P. V. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci U S A 100, 14908–14913, doi: 10.1073/pnas.2136625100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen H., Slepnev V. I., Di Fiore P. P. & De Camilli P. The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem 274, 3257–3260, doi: 10.1074/jbc.274.6.3257 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Chen H. & De Camilli P. The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc Natl Acad Sci U S A 102, 2766–2771, doi: 10.1073/pnas.0409719102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oldham C. E., Mohney R. P., Miller S. L., Hanes R. N. & O’Bryan J. P. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr Biol 12, 1112–1116, doi: 10.1016/s0960-9822(02)00900-4 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Yan H. et al. MG132, a proteasome inhibitor, enhances LDL uptake in HepG2 cells in vitro by regulating LDLR and PCSK9 expression. Acta Pharmacol Sin 35, 994–1004, doi: 10.1038/aps.2014.52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bottger R. et al. Lipid-based nanoparticle technologies for liver targeting. Adv Drug Deliv Rev 154–155, 79–101, doi: 10.1016/j.addr.2020.06.017 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Sato Y., Kinami Y., Hashiba K. & Harashima H. Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J Control Release 322, 217–226, doi: 10.1016/j.jconrel.2020.03.006 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Han X. et al. Ligand-tethered lipid nanoparticles for targeted RNA delivery to treat liver fibrosis. Nat Commun 14, 75, doi: 10.1038/s41467-022-35637-z (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J. et al. Liver-Targeted siRNA Lipid Nanoparticles Treat Hepatic Cirrhosis by Dual Antifibrotic and Anti-inflammatory Activities. ACS Nano 14, 6305–6322, doi: 10.1021/acsnano.0c02633 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Fan J. et al. Rabbit models for the study of human atherosclerosis: from pathophysiological mechanisms to translational medicine. Pharmacol Ther 146, 104–119, doi: 10.1016/j.pharmthera.2014.09.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamamdzic D. & Wilensky R. L. Porcine models of accelerated coronary atherosclerosis: role of diabetes mellitus and hypercholesterolemia. J Diabetes Res 2013, 761415, doi: 10.1155/2013/761415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shiomi M. The History of the WHHL Rabbit, an Animal Model of Familial Hypercholesterolemia (II) - Contribution to the Development and Validation of the Therapeutics for Hypercholesterolemia and Atherosclerosis. J Atheroscler Thromb 27, 119–131, doi: 10.5551/jat.RV17038-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin L., Ma H., Ge X., Edwards P. A. & Zhang Y. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol 31, 328–336, doi: 10.1161/ATVBAHA.110.217828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parviz F. et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34, 292–296, doi: 10.1038/ng1175 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Wu H. et al. A negative reciprocal regulatory axis between cyclin D1 and HNF4alpha modulates cell cycle progression and metabolism in the liver. Proc Natl Acad Sci U S A 117, 17177–17186, doi: 10.1073/pnas.2002898117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonzo J. A., Ferry C. H., Matsubara T., Kim J. H. & Gonzalez F. J. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J Biol Chem 287, 7345–7356, doi: 10.1074/jbc.M111.334599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang T. et al. Therapeutic HNF4A mRNA attenuates liver fibrosis in a preclinical model. J Hepatol 75, 1420–1433, doi: 10.1016/j.jhep.2021.08.011 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Xu Y. et al. Hepatocyte Nuclear Factor 4alpha Prevents the Steatosis-to-NASH Progression by Regulating p53 and Bile Acid Signaling (in mice). Hepatology 73, 2251–2265, doi: 10.1002/hep.31604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data (GSE273386) of the ApoE−/− and ApoE−/− /Liver-DKO are available at the Gene Expression Omnibus. The scRNA-seq data (GSE254971) for D374Y mCherry-APOB mice is available in published paper entitled “Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult” (https://doi.org/10.1038/s44161-024-00448-6). Source data are provided with this paper.