SUMMARY
Endosymbiont gene transfer and import of host-encoded proteins are considered hallmarks of organelles necessary for stable integration of two cells. However, newer endosymbiotic models have challenged the origin and timing of such genetic integration during organellogenesis. Epithemia diatoms contain diazoplasts, closely related to recently-described nitrogen-fixing organelles, that are also stably integrated and co-speciating with their host algae. We report genomic analyses of two species, freshwater E.clementina and marine E.pelagica, which are highly divergent but share a common endosymbiotic origin. We found minimal evidence of genetic integration: nonfunctional diazoplast-to-nuclear DNA transfers in the E.clementina genome and 6 host-encoded proteins of unknown function in the E.clementina diazoplast proteome, far fewer than in other recently-acquired organelles. Epithemia diazoplasts are a valuable counterpoint to existing organellogenesis models, demonstrating that endosymbionts can be stably integrated and inherited absent significant genetic integration. The minimal genetic integration makes diazoplasts valuable blueprints for bioengineering endosymbiotic compartments de novo.
Keywords: Cyanobacteria, diatom, endosymbiosis, evolution, genome, genomics, horizontal gene transfer, nitrogen fixation, organelle, organellogenesis, symbiosis
INTRODUCTION
Endosymbiotic organelles are uniquely eukaryotic innovations that facilitated the acquisition of complex cellular functions, including aerobic respiration, photosynthesis, and nitrogen fixation. These events also resulted in expansive eukaryotic diversity1. An important question in cell evolution and engineering is: how do intermittent, facultative interactions evolve into persistent, stably integrated endosymbioses? As the oldest known endosymbiotic organelles, mitochondria and chloroplasts defined our initial understanding of organellogenesis. A series of genomic changes was proposed to be critical: First, the bacterial endosymbiont undergoes extensive genome reduction, streamlining gene content. Second, endosymbiont genes are not simply purged but transferred from the endosymbiont genome to the host eukaryotic nucleus in a process called endosymbiont gene transfer (EGT). Finally, these and other gene products, now under the control of host gene expression, are imported back into the endosymbiotic compartment to regulate endosymbiont growth and division. In this traditional model of endosymbiotic evolution, genetic integration resulting from EGT and/or import of host-encoded gene products is essential for maintaining the endosymbiont as a stable, integral cellular compartment, i.e. an organelle2–4.
Increased sampling of eukaryotic diversity has uncovered evidence that, amongst microbes, endosymbioses are ongoing and a common strategy for acquisition of new functions. New organelles have been recognized: the chromatophore in Paulinella chromatophora5–8 and the nitroplast (formerly called UCYN-A) in Braarudosphaera bigelowii9. Genome reduction, EGT, and host protein import have also been observed in obligate, vertically-inherited nutritional endosymbionts of the parasite Angomonas deanei and insects, which are not formally recognized as organelles10,11. With the benefit of these newer models, our understanding of genetic integration has become more nuanced. For example, the majority of host proteins imported into the Paulinella chromatophore do not originate from EGT but rather horizontal gene transfer (HGT) from other bacteria or eukaryotic genes12, showing that a host’s repertoire of pre-existing genes may play an important role in facilitating genetic integration13. There have been bigger surprises: Several organisms temporarily acquire plastids from partially digested prey algae14–16. The retained chloroplasts, called kleptoplasts, perform photosynthesis but cannot replicate and must be continuously acquired, far from the stability associated with organelles. Yet imported host proteins contribute to kleptoplast metabolic pathways in several species, indicating that genetic integration is not sufficient to achieve stable integration17–19. These findings highlight the importance of studying biodiverse organisms to inform new hypotheses for endosymbiotic evolution.
Amongst new model systems, Epithemia spp. diatoms offer a unique perspective on organellogenesis. First, these photosynthetic microalgae contain diazotroph endosymbionts (designated diazoplasts) that perform nitrogen fixation, a biological reaction that converts inert atmospheric nitrogen to bioavailable ammonia20–25. Nitrogen is often a limiting nutrient for primary producers, so the ability to fix both carbon and nitrogen fulfills a unique niche in ecosystems. Epithemia spp. are widespread in freshwater habitats and have recently been isolated from marine environments26–28. Second, the Epithemia endosymbiosis is very recent relative to mitochondria and chloroplasts, having originated ~35 Mya based on fossil records29. Nonetheless, diazoplasts are stably integrated with their hosts and retained in diverse Epithemia species described so far. Finally, Epithemia diazoplasts are closely related to the nitroplast, the diazotroph endosymbiont of B. bigelowii which was recently designated a nitrogen-fixing organelle based on evidence of host protein import into the endosymbiont9,25,30. Both diazoplasts and nitroplasts evolved from free-living Crocosphaera cyanobacteria that have engaged in endosymbioses with several host microalgae. The shared origin of these two independent diazotroph endosymbioses facilitates comparisons that can lead to more powerful insights.
If the significance of organelles is their functioning as stable, integral cellular compartments, then diazoplasts show robust metabolic and cellular integration at least on par with the nitroplast. Nitrogen fixation requires large amounts of ATP and reducing power, energy that can be supplied by photosynthesis. Yet nitrogenase, the enzyme that catalyzes nitrogen fixation, is exquisitely sensitive to oxygen produced during oxygenic photosynthesis. In free-living Crocosphaera, photosynthesis and nitrogen fixation must be temporally separated such that fixed carbon from daytime photosynthesis is stored as glycogen to fuel exclusively nighttime nitrogen fixation. Diazoplasts have lost all photosystem genes and depend entirely on host photosynthesis24,31. Recently, we showed that host and diazoplast metabolism are tightly coupled to support continuous nitrogenase activity throughout the day-night cycle: Diatom photosynthesis is required for daytime nitrogenase activity in the diazoplast, while nighttime nitrogenase activity also depends on diatom, rather than cyanobacteria, carbon stores25. In comparison, the nitroplast has lost only photosystem II and is dependent on both host photosynthesis and its own photosystem I, restricting it to daytime nitrogen fixation32,33 Both the diazoplast and the nitroplast are obligate endosymbionts and are vertically inherited during asexual cell division of their respective hosts. Diazoplasts have further been shown to be uniparentally inherited during sexual reproduction, similar to mitochondria and chloroplasts34. In laboratory cultures of Epithemia, we have observed that diazoplasts are retained by the host even when grown in nitrogen-supplemented media when its nitrogen fixation activity is downregulated35. Interestingly, the nitroplast is often lost from host cells during isolation and in culture, suggesting a less stable relationship at least in laboratory conditions36.
A pressing question that emerges from these observations is whether EGT and/or host protein import contributes to the stable integration of diazoplasts in Epithemia. Based on the traditional organellogenesis model and the similarity of diazoplasts to the nitroplast, the assumption would be yes. However, there is the intriguing possibility that, if genetic integration is not sufficient for stable integration (as shown by kleptoplasts), it is also not essential for stable integration. We previously established freshwater E. clementina as a laboratory model for functional studies and herein performed de novo assembly and annotation of its genome. The genome sequences for E. pelagica, a recently-discovered marine species, was publicly released by the Wellcome Sanger Institute27,37. To facilitate comparison between these species, we also performed de novo genome annotation of E. pelagica. Notably, no genomes of B. bigelowii (which hosts the nitroplast) nor the eukaryotic host in any other diazotroph endosymbiosis have been available. We report genome analyses of these two Epithemia species with the goals of 1) providing a necessary resource to accelerate investigation of this model and 2) elucidating the role of genetic integration in this very recent but stably integrated endosymbiosis.
RESULTS
Highly divergent E. clementina and E. pelagica genomes share many unique gene families
Epithemia spp are raphid, pennate diatoms composed of at least 50 freshwater species and 2 reported marine species26,27. Isolation and characterization of freshwater E. clementina was previously reported25 (Figure S1A). We isolated high molecular weight DNA from axenic E. clementina cultures and performed sequencing by long-read Nanopore and short-read Illumina, yielding a 418 Mbp haploid assembly of the diploid genome with a high level of heterozygosity of 1.48% (Figure S1B). The final reported haploid assembly is complete, contiguous, and of high sequence quality (Figure S1C, Table 1). A chromosome-level 60 Mbp genome assembly (GCA_946965045) of E. pelagica, a marine species, was reported by the Sanger Institute37. Whole-genome alignments of E. clementina and E. pelagica did not show significant syntenic blocks in their nuclear genomes (Figure S1E). In contrast, their diazoplast genomes showed 5 major and 2 minor syntenic blocks (Figure S1F), similar to the synteny reported between diazoplast genomes of other Epithemia species30,31. These observations are consistent with there being greater selection on diazoplast genomes than on the nuclear genomes in Epithemia species.
Table 1.
Epithemia genome assembly statistics
| E. Clementina | E. pelagica | |
|---|---|---|
| Genome size (bp) | 418,007,894 | 60,195,788 |
| GC | 44.3% | 48.19% |
| QV | 38.52 | - |
| Contig/chromosome # | 642 | 15 |
| N50 | 1,108,441 | - |
| L90 | 412 | - |
| Gene # | 26,453 | 20,203 |
| Repeat % | 80% | 27.36% |
| BUSCOgenome | 100% | 100% |
| BUSCOprotein | 99% | 94% |
| Diazoplast genome size (bp) | 3,072,807 | 2,483,960 |
| Diazoplast gene # | 1,910 | 1,679 |
Summary of assembly statistics for E. clementina and, where applicable, E. pelagica. Quality value (QV) represents a log-scaled estimate of the base accuracy across the genome, where a QV of 40 is 99.99% accurate. N50 and L90 are measures of genome contiguity. N50 represents the contig length (bp) such that 50% of the genome is contained in contigs ≥ N50. L90 represents the minimum number of contigs required to contain 90% of the genome. Finally, BUSCO (Benchmarking of Single Copy Orthologues) is an estimate of completeness of the genome (BUSCOgenome) and proteome (BUSCOprotein) of E. clementina and E. pelagica.
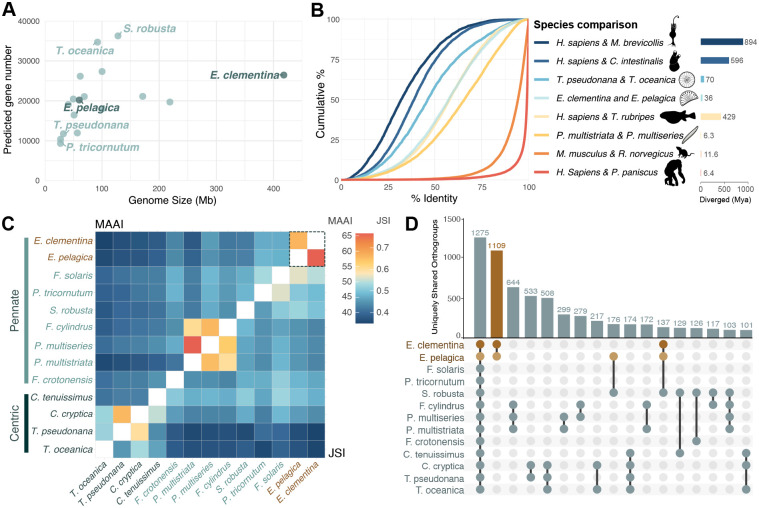
Both E. clementina and E. pelagica genomes were annotated using evidence from protein orthology and transcriptome profiling. The nuclear genomes were predicted to contain 20,203 genes in E. pelagica and 26,453 genes in E. clementina (Figure 1A). The completeness of their predicted proteomes was assessed based on the presence of known single-copy orthologs in stramenopiles, yielding BUSCOprotein scores of 99% for E. clementina and 94% for E. pelagica (Table 1). The larger predicted proteome and high BUSCOprotein score for E. clementina is likely because more growth conditions were used to obtain transcriptomes used for gene predictions in E. clementina than for E. pelagica (Figure S1D). We compared the amino acid identity between orthologs across proteomes of several pairs of representative diatom and metazoan species, as a measure of their divergence (Figures 1B and S2A). Despite their estimated 35 Mya of speciation, E. pelagica and E. clementina showed a similar distribution of identity across protein orthologs as humans and pufferfish (Homo sapiens and Takifugu rubripes), which are estimated to have shared a common ancestor 429 Mya38. This rapid divergence, relative to age, is also observed in other diatom species, T. pseudonada/ T. oceanica (70 Mya) and P. multistriata/ P. multiseries (6.3 Mya)38. The loss of synteny and lower protein ortholog identity suggest that E. pelagica and E. clementina have diverged substantially during speciation, reflecting the rapid evolution rates of diatoms39,40 (Figure 1C).
Figure 1. Highly divergent E. clementina and E. pelagica genomes share many unique gene families.
(A) Genome size and total gene number for published diatom genomes compared with Epithemia species (dark blue). (See also Figure S1, Table S1.)
(B) Cumulative distribution of amino acid identity between pairwise orthologs from reference species. Estimated divergence time of species pair is indicated (right bar graph).
(C) Asymmetrical heatmap of ortholog comparisons between diatom species pairs, showing mean amino acid identity (MAAI) of pairwise orthologs (top) and Jaccard similarity index (JSI) of orthogroups (bottom). (See also Figure S2.)
(D) UpSet plot depicting the number of uniquely shared orthogroups between all diatom species (first column) or subsets of 2–4 species. Orthogroups shared by E. pelagica and E. clementina are highlighted in brown. Columns are ranked by the number of uniquely shared orthogroups.
Because rapid divergence is common across diatoms, we evaluated the gene content of E. pelagica and E. clementina in comparison with other diatom species that have complete genomes available. Gene families, defined by orthogroups, were identified for each species. The overlap in gene families between all possible pairs of diatom species was quantified by Jaccard Similarity Index (JSI). We also quantified the number of uniquely shared gene families between subsets of diatom species, i.e. gene families only shared between that group of species and not found in any other diatoms. Of 10,740 and 10,612 gene families identified in E. clementina and E. pelagica respectively, they share 8,942 gene families, a greater overlap than is observed between any other pair of diatom species (Figures 1C and S2B). Of these, 1109 gene families are uniquely shared between E. clementina and E. pelagica, also more than any other species grouping including the more recently speciated Pseudo-nitzchia species (Figure 1D). In comparison, the core set of gene families shared by all diatoms is 1275. The enrichment in unique Epithemia gene families indicates genus-specific selection, possibly related to the endosymbiosis. Because HGT is known to be a source of genes for endosymbiont functions expressed by the host8 and 3–5% of diatom proteomes have been attributed to bacterial HGT41, a significantly greater proportion than detected in other eukaryotic proteomes42, we identified HGTs within gene families uniquely shared between E. clementina and E. pelagica, resulting in 51 Epithemia-specific HGT candidates shared between the two species (Table S3). Overall, the uniquely shared features of the divergent nuclear genomes of Epithemia genus are valuable for identifying potential signatures of endosymbiotic evolution.
Extensive repeat expansion in E. clementina occurred during speciation after the endosymbiotic event
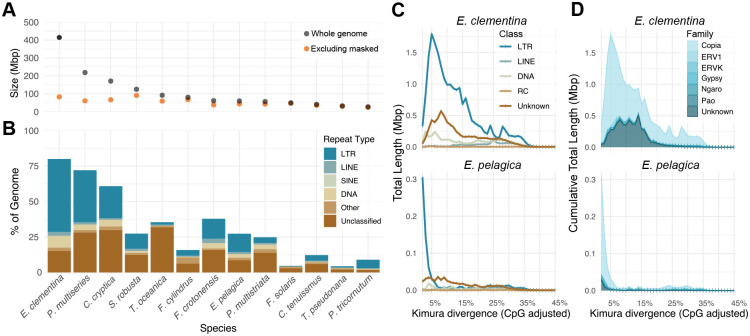
While the gene numbers between E. clementina and E. pelagica are similar and typical for diatoms, the total genome size of E. clementina is 7 times larger (Figures 1A and 2A, Table 1). The increased genome size is due to a substantial repeat expansion unique to freshwater E. clementina: 80% of the E. clementina genome is composed of repeat elements, compared to only 26% of the E. pelagica genome (Figure 2B). This high repeat percent is unusual amongst sequenced diatoms genomes. Notably, the differences in genome size observed amongst diatoms is largely due to repeat content (Figure 2A). In both Epithemia genomes, the dominant repeat type is LTR retrotransposons, in particular the Ty-1-copia family (Figures 2C and 2D). However, multiple LTR families and DNA transposons show expansions that contribute to the high repeat percent in E. clementina (Figure 2C, Table S2). This cross-family expansion indicates broadly relaxed selection on the repeatome of E. clementina.
Figure 2. Extensive repeat expansion in E. clementina occurred during speciation after the endosymbiotic event.
(A) Comparison of repeat content in diatom genomes showing size of the whole genome (grey dots) or the genome excluding masked repeat elements (orange dots). X-axis is the same as 2B.
(B) Breakdown of repeat types in diatom genomes showing amount in Mbp of the genome occupied by repeat elements of specific class, indicated by color.
(C) Repeat landscape of E. clementina (top) and E. pelagica (bottom) showing the amount in Mbp of the genome occupied by classes of repeat elements as a function of their divergence from the inferred ancestral repeat sequence, a proxy for age since insertion. (See also Figure S3A.)
(D) Same as B, showing only LTRs and plotted cumulatively, colored by family. (See also Figure S3B.)
We evaluated the repeat landscapes of E. clementina and E. pelagica to detect ancestral repeat expansions that may be associated with the endosymbiotic event. A consensus sequence was derived from all identified repeat elements in each genome to approximate the ancestral repeat elements. The divergence of individual repeat elements from the consensus served as a proxy for age of the insertion: the longer the time since the insertion, the more mutations accumulate43. Comparing the repeat landscapes of E. clementina and E. pelagica, the majority of LTR retrotransposons in E. pelagica were 0–5% diverged indicating relatively recent repeat expansion in the genome, while E. clementina repeats showed higher divergence with a wider distribution more consistent with accumulation of repeat elements over a longer time period (Figure 2C). This difference was consistent across retrotransposon families (Figure 2D). We ran the repeat masker pipeline de novo on all available unmasked or soft-masked genomes for the diatoms and generated repeat landscapes for each (Table S1). We were unable to detect any ancestral repeat expansions in diatoms, even in more recently speciated pairs such as P. multiseries and P. multistriata (Figures S3A and S3B). Repeat landscapes can, in principle, reconstruct ancient repeat expansions predating the estimated divergence time of E. pelagica and E. clementina species (35 Mya) and of raphid diatoms (80 Mya)44–46. However, the rapid diversification of diatoms combined with poor recovery of highly divergent sequences using consensus identification, such as repeat identification, may limit the timescale for detecting repeat expansion in diatoms. We were unable to detect repeat expansion associated with endosymbiotic acquisition. Rather, significant repeat expansion observed in E. clementina is likely related to its speciation, such as the transition to freshwater habitats.
Diazoplast to nuclear transfer of DNA is actively occurring in E. clementina
Having broadly characterized the Epithemia genomes for shared and distinct features, we turned to specifically interrogate genetic integration between Epithemia and the diazoplast. EGT entails the transfer of functional genes from the endosymbiont to the host nucleus, a rare event believed to occur in the background of frequent, ongoing endosymbiont-to-nuclear transfers of DNA. Indeed, it has been shown that nuclear integrations of organellar DNA originating from mitochondria (designated NUMT) and plastids (NUPT) still occur47,48. Given the significantly younger age of the diazoplast, it is not clear whether nuclear integrations of diazoplast DNA (which we will refer to as NUDT) and/or functional transfers of genes (EGT) have occurred.
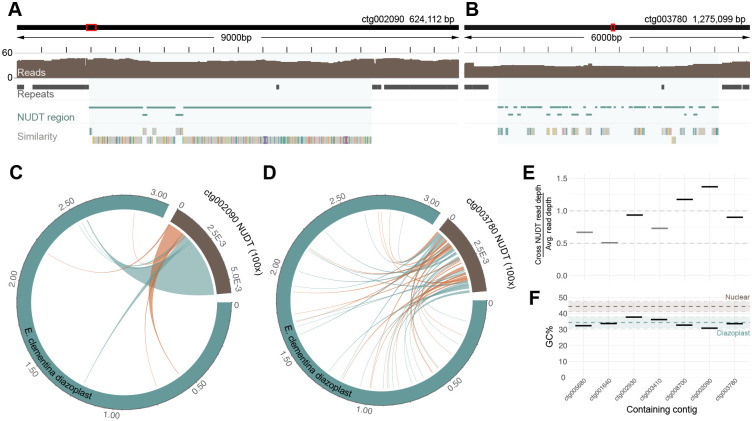
To identify transfers of endosymbiont DNA to the host nucleus, we performed homology searches against the nuclear assemblies of E. clementina and E. pelagica. As queries, we used the diazoplast genomes of 4 Epithemia species (including E. clementina and E. pelagica) and 5 related free-living cyanobacteria species for which whole genomes were available (Table S1). To prevent spurious identifications, alignments were excluded if they were <500 contiguous base pairs in length. In E. clementina, we identified seven segments, ranging from 1700–6400bp, with homology to the E. clementina diazoplast (Figures 3A and 3B, Data S1A–S1G). No homology to free-living cyanobacteria genomes was detected. The E. coli genome and a reversed sequence of the E. pelagica diazoplast were used as negative control queries and yielded no alignments. Finally, no regions of homology to any of the queries were detected in the nuclear genome of E. pelagica.
Figure 3. Detection of nuclear integrations of diazoplast DNA (NUDTs).
(A) A representative, NUDT containing E. clementina nuclear genome locus on contig ctg002090. Tracks shown from top to bottom: nuclear sub-region being viewed (red box) within the contig (black rectangle); length of the sub-region, with ticks every 500bp; nanopore sequencing read pileup, showing long read support across the NUDT; location of repeat masked regions (dark grey bars); locations of homology to E. clementina diazoplast identified by BLAST, demarcating the NUDT (blue shade); regions of homology to the E. clementina diazoplast identified by minimap2 alignment, colors represent SNVs between the diazoplast and nuclear sequence. (See also, Data S1F.)
(B) Same as A, for the NUDT on contig ctg003780. (See also, Data S1G.)
(C) Circlize plot depicting the fragmentation and rearrangement of NUDTs. The diazoplast genome (blue) and the NUDT on contig ctg002090 (brown) with chords connecting source diazoplast regions to their corresponding nuclear region, inversions in red. The length of the NUDT is depicted at 100x true relative length for ease of visualization. (See also, Figure S4A–S4E.)
(D) Same as C, for the NUDT on contig ctg003780. (See also, Figure S4A–S4E.)
(E) Ratio of long read depth of NUDT compared to average read depth for the containing contig. Heterozygous insertions (light grey bars) show approximately 0.5x depth; homozygous insertions (black bars) show approximately 1.0x depth.
(F) GC content of NUDTs, compared to mean GC content for 5kb sliding windows of the diazoplast genome (blue dashed line) and the nuclear genome (brown dashed line). Shaded regions represent mean ± 1 SD.
NUDTs showed features suggesting they were distinct from diazoplast genomic sequences and unlikely to be assembly errors. First, 4 of the 7 NUDTs were supported by long reads equivalent to 1x coverage of the genome indicating the insertions were homozygous. 3 NUDTs contained on ctg003410, ctg001640, ctg005680 showed the equivalent of 0.5x genome coverage, consistent with a heterozygous insertion in the diploid eukaryotic genome (Figure 3E, Data S1A, S1B, and S1D). Second, NUDTs had low GC content similar to that of the diazoplast but contain many single nucleotide variants (SNVs) with mean identity of 98.4% to their source sequences, indicative of either neutral or relaxed selection (Figures 3F and 4B). Finally, each NUDT was composed of multiple fragments corresponding to distal regions in the endosymbiont genome, ranging from as few as 8 distal fragments composing the NUDT on ctg002090 to as many as 42 on ctg003780 (Figures 3C, 3D, and S4A–S4E). This composition of NUDTs indicates either that fragmentation and rearrangement of the diazoplast genome occurred prior to insertion into the eukaryotic genome or that NUDTs were initially large insertions that then underwent deletion and recombination. Overall, the detection of NUDTs suggests that, in this very recent endosymbiosis, diazoplast-to-nuclear DNA transfer is occurring.
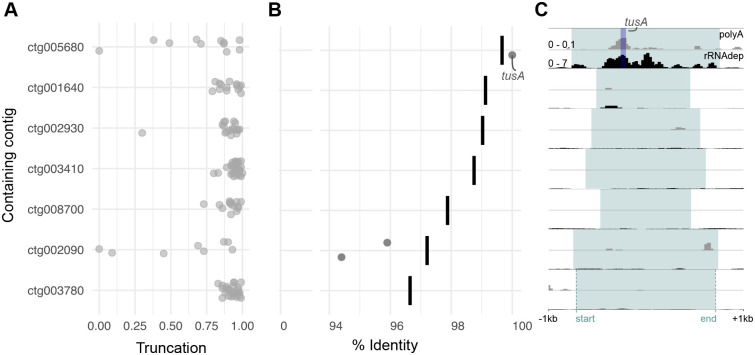
Figure 4. Most NUDTs are decaying and non-functional.
(A) Truncation of diazoplast genes contained within each NUDT relative to the full-length diazoplast gene.
(B) Nucleotide identity of diazoplast genes that are <30% truncated (points) contained within each NUDT compared to identity of the full containing NUDT sequence (bars). (See also, Figures S4F and S4G.)
(C) Normalized expression across each NUDT (blue highlight) +/− 1kb of the genomic region surrounding the NUDT. For each NUDT, a pair of tracks shows RNA-seq reads after polyA enrichment of whole RNA plotted within background signal range, from 0 – 0.1 BPM (top, grey) and RNA-seq reads after rRNA depletion of whole RNA, plotted from 0 – 7 BPM (bottom, black).). The region corresponding to the tusA gene in ctg005680 is highlighted in dark blue (See also Data S1A–S1G.)
Most NUDTs are decaying and non-functional
To determine whether any of the identified NUDTs have resulted in EGT, we identified diazoplast genes present in NUDTs and evaluated their potential for function. A total of 124 diazoplast genes and gene fragments were carried over into the NUDTs (Figure 4A). (A few of these diazoplast genes have conserved eukaryotic homologs and were also predicted as eukaryotic genes in the E. clementina genome annotation (Data S1A–S1G).) 121 diazoplast genes detected in NUDTs are truncated >30% compared to the full-length diazoplast gene (Figure 4A). Of the three remaining, two genes contained on ctg002090 showed accumulation of SNVs that resulted in a premature stop codon and a nonstop mutation (Figures 4B, S4F and S4G). We performed transcriptomics to assess the expression from NUDTs. Neither of the two genes on ctg002090 showed appreciable expression. All except one NUDT showed <2 bins per million mapped reads (BPM), equivalent to background transcription levels within the region (Figures 4C and Data S1A–S1G). The truncation, mutation accumulation, and lack of appreciable expression of diazoplast genes encoded in NUDTs suggest that most are not functional.
Only a single EGT candidate was detected contained on ctg005680: an intact sulfotransferase (tusA) gene that is 100% identical to the diazoplast-encoded gene (Figures 4A and 4B, Data S1A). The NUDT that contains this candidate appears to be very recent as it is heterozygous and shows 99.7% identity to the source diazoplast sequence (Figure 4B). Interestingly, tusA is implicated in Fe-S cluster regulation that could be relevant for nitrogenase function. Due to the high sequence identity, it is not possible to distinguish transferred tusA from that of tusA encoded in the diazoplast genome by sequence alone. However, transcript abundance above background levels was only detected in rRNA depleted samples that contain diazoplast transcripts and not in polyA-selected samples that remove diazoplast transcripts, indicating that the observed expression is largely attributed to diazoplast-encoded tusA (Figure 4C). Moreover, host proteins imported to endosymbiotic compartments often use N-terminal (occasionally C-terminal) targeting sequences. We were unable to identify any added sequences in the transferred tusA indicative of a targeting sequence; the sequence immediately surrounding consisted only of native diazoplast sequence carried over with the larger fragment (Data S1A). Though there is no evidence for gene function, the transfer of this intact gene indicates that the conditions for EGT are present in E. clementina.
Few host-encoded proteins are detected in the diazoplast proteome
The critical step in achieving genetic integration is evolution of pathways for importing host proteins into the endosymbiont. While EGT and HGT from other bacteria can expand the host’s genetic repertoire, neither transferred genes nor native eukaryotic genes can substitute for or regulate endosymbiont functions unless the gene products are targeted to the endosymbiotic compartment. Abundant host-encoded proteins were detected in the proteomes of recently-acquired endosymbionts that have been designated organelles: 450 in the chromatophore of Paulinella8 and 368 in the nitroplast of B. bigelowii9. In both the chromatophore and nitroplast, several host-encoded proteins detected in the endosymbiont fulfill missing functions that complete endosymbiont metabolic pathways, providing further support for the import of host-encoded proteins.
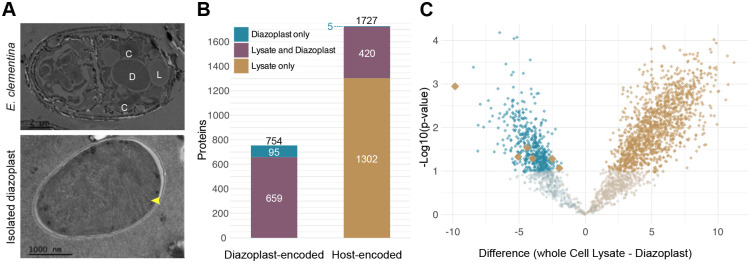
To determine whether host protein import is occurring in the diazoplast, we identified the proteome of the E. clementina diazoplast. We were unable to maintain long-term E. pelagica cultures to perform proteomics for comparison. Diazoplasts were isolated from E. clementina cells by density gradient centrifugation. The purity of isolated diazoplasts was evaluated by light microscopy. The protein content of isolated diazoplasts and whole E. clementina cells containing diazoplasts were determined by LC-MS/MS. A total of 2481 proteins were identified with ≥2 unique peptides: 754 proteins were encoded by the diazoplast genome (detected/total protein coding = 43% coverage) and 1727 proteins encoded by the nuclear genome (6.5% coverage) (Figure 5B, Table S4). Of note, TusA, the only EGT candidate identified, was not detected in either proteome. To identify proteins enriched in either the diazoplast or host compartments, we compared protein abundance in isolated diazoplasts and whole cell samples across 3 biological replicates (Figure 5C). 492 diazoplast-encoded proteins were significantly enriched in the diazoplast and none were enriched in whole cell samples, supporting the purity of the isolated diazoplast sample. Similarly, most host-encoded proteins (1281) were significantly enriched in whole cell samples, indicating localization in host compartments. Six unique host-encoded proteins were significantly enriched in diazoplast samples, suggesting possible localization to the diazoplast. Five were encoded by Ec_g00815, Ec_g12982, Ec_g13000, Ec_g13118, and Ec_g25610. The sixth protein was encoded by two identical genes, Ec_g24166 and Ec_g03819, resulting from an apparent short duplication of it and two neighboring genes. Because the duplication makes Ec_g24166 and Ec_g03819 indistinguishable by amino acid sequence, we considered them one import candidate. Of the 6 host protein import candidates, Ec_g12982 and Ec_g13000 were detected only in the diazoplast sample, while the rest were identified in both diazoplast and whole cell lysate samples. Neither genetics nor immunofluorescence are available in E. clementina to further validate their protein localization and rule out the possibility of nonspecific enrichment.
Figure 5. Few host-encoded proteins are detected in the diazoplast proteome.
(A) Electron micrographs of (top) E. clementina cells with diazoplast (D), chloroplast lobes (C), and lipid bodies (L) indicated and (bottom) diazoplasts following purification with thylakoids (yellow arrow) indicated.
(B) Number of diazoplast-encoded (left) and host-encoded (right) proteins identified by LC-MS/MS. Total number of proteins identified from each respective proteome is shown above each stacked bar. Colored bars and numbers indicate proteins identified in purified diazoplasts only, whole cell lysate only, or both.
(C) Volcano plot showing the enrichment of diazoplast-encoded (blue) and host-encoded (brown) proteins in whole cell lysate or purified diazoplasts, represented by the difference between log2-transformed iBAQ values. Proteins enriched in the diazoplast are on the left side of the graph while those enriched in the host are on the right; the darker shade of each color represents significantly enriched hits. Host-encoded proteins significantly enriched in the diazoplast are shown with larger brown markers.
We sought additional evidence to support the import of these host proteins by evaluating their potential functions49. No domains, GO terms, or BLAST hits (other than to hypothetical proteins found in other diatoms) were identified for any of the candidates except for Ec_g13118 which is annotated as an E3 ubiquitin ligase. Consistent with their being diatom-specific proteins, 3 candidates (Ec_g24166/Ec_g03819, Ec_g12982, and Ec_g13000) belonged to orthogroup OG0000250 which is uniquely shared with E. pelagica but no other diatoms (Figure 1D) and the other 3 belonged to separate orthogroups (OG0001966, OG0004498, and OG0009247) which are shared broadly among diatoms including E. pelagica. In contrast to the unclear functions of these candidates, several host proteins detected in the chromatophore and nitroplast proteome were assigned to conserved cyanobacterial growth, division, or metabolic pathways in these organelles. Moreover, none of the candidates for import into the diazoplast have significant homology to proteins encoded in diazoplast or free-living Crocosphaera genomes to suggest they might fulfill unidentified cyanobacterial functions. Our functional analysis suggests these import candidates are unlikely to have critical functions in conserved cyanobacterial pathways, even if they are targeted to the diazoplast.
Finally, the detection of ~100-fold fewer import candidates in the diazoplast indicate that host protein import, if occurring, is far less efficient than in the chromatophore or nitroplast. Since the sensitivity of proteomics is highly dependent on biomass, we estimated the coverage of the diazoplast proteome based on the ratio of diazoplast-encoded proteins detected (754) compared to the total diazoplast protein-coding genes (1727). The coverage of the diazoplast proteome (43%) was comparable to the coverage of the published chromatophore proteome (422/867 = 49%) and that of the nitroplast (609/1186 = 51%) and therefore does not account for the low number of import candidates. Overall, the number of host proteins detected in the diazoplast was significantly less and their functional significance unclear compared to host proteins detected in the chromatophore and nitroplast.
DISCUSSION
The triad of genome reduction, EGT, and host protein import has been held as a necessary progression to achieve the stable integration of organelles4,50,51. But this view of organellogenesis has been challenged by findings in recent endosymbionts from diverse organisms. We report analysis of two genomes of Epithemia diatoms and evaluate the extent of their genetic integration with their nitrogen-fixing endosymbionts (diazoplasts), thereby adding this very recent endosymbiosis to existing model systems that can elucidate the stable integration of two cells into one.
A window into the early dynamics of nuclear gene transfers.
Our first significant finding was the detection of active diazoplast-to-nuclear DNA transfers but, as yet, no functional gene transfer in Epithemia. Our observations support findings in the chromatophore and nitroplast that EGT is not necessary for genetic integration9,12,52. Given that functional EGT does not necessarily precede evolution of host protein import pathways, it may be a suboptimal solution for the inevitable genome decay in small asexual endosymbiont populations as a consequence of Muller’s ratchet53,54. Instead, the decayed nature of the NUDTs we detected in E. clementina is consistent with stochastic, transient, ongoing DNA transfer. Nonfunctional DNA transfers were previously only described from mitochondria or plastids with far more reduced genomes. The status of nuclear transfers from more recently acquired organelles is unknown, as only protein-coding regions were used as queries to identify chromatophore transfers in Paulinella and only a transcriptome is available for the nitroplast host, B. bigelowii. NUDTs in Epithemia genomes therefore provide a rare window into the early dynamics of DNA transfer. For example, using the same homology criteria, we identified 5 NUMTs but no NUPTs in E. clementina. The NUMTs were significantly shorter than NUDTs and did not show rearrangement, which may suggest different mechanisms of transfer for NUDTs, NUMTs, and NUPTs in the same host nucleus. In addition, between-species differences may identify factors that affect transfer rates. The lack of observed NUDTs in E. pelagica suggest constraints on diazoplast-to-nuclear transfers such as lower tolerance to DNA insertions in its comparatively smaller, non-repetitive genome. Finally, the lack of NUDT gene expression, even with transfer of a full-length unmutated tusA gene, points to barriers to achieving eukaryotic expression from bacterial gene sequence. Epithemia is an apt model system to interrogate how horizontal gene transfer impacts eukaryotic genome evolution with at least 20 species easily obtained from freshwater globally and consistently adaptable to laboratory cultures25,26,30,31.
Epithemia diazoplasts as a counterpoint to existing models of organellogenesis.
A second unexpected finding was the identification of only 6 host proteins in the diazoplast proteome, much fewer and with less clear functional significance than in comparable endosymbionts that have been designated organelles. Methods for validating the localization of these import candidates are unavailable in Epithemia. Even if confirmed to target to the diazoplast, the candidates lack conserved domains or homology with cyanobacterial proteins to indicate they replace or supplement diazoplast metabolic function, growth, or division. Our findings are not explained by current models of organellogenesis that propose import of host proteins as a necessary step to establish an integral endosymbiotic compartment. In the traditional organellogenesis model (as described in the introduction), host protein targeting is a “late” bottleneck step required for the regulation of the endosymbiont growth and division. More recently, “targeting-early” has been proposed to account for establishment of protein import pathways prior to stable integration as observed in kleptoplasts16,17. In this model, protein import is selected over successive transient endosymbioses, possibly driven by the host’s need to export metabolites from the endosymbiont via transporters or related mechanisms55. The establishment of protein import pathways then facilitates endosymbiont gene loss with metabolic functions fulfilled by host proteins leading to endosymbiont fixation. Contradicting both models, we observed minimal evidence for genetic integration despite millions of years of co-evolution resulting in diverse Epithemia species retaining diazoplasts, indicating that genetic integration is not necessary for its stable maintenance. Diazoplasts appear to be surrounded by a host-derived membrane; host proteins localized to this outer membrane (which was lost during diazoplast purification) may mediate interactions with diazoplasts without requiring protein import pathways (Figure 5A). At a minimum, the unclear functions of the few host proteins identified in the diazoplast proteome, if imported, suggest that the genesis of host protein import in Epithemia is very different than would be predicted by current models.
Diazotroph endosymbioses are fundamentally different from photosynthetic endosymbioses that are the basis for current organellogenesis models. First, the diazoplast is derived from a cyanobacteria that became heterotrophic by way of losing its photosynthetic apparatus. Regulation of endosymbiont growth and division by the availability of host sugars (without requiring an additional layer of regulation via import of host metabolic enzymes) may be more facile with heterotrophic endosymbionts maintained for a nonphotosynthetic function compared to autotrophic endosymbionts. It will be interesting to see how integration of the diazoplast differs from the endosymbiont of Climacodium freunfeldianum, another diazotrophic endosymbiont descended from Crocosphaera that likely retains photosynthesis56, or a non-cyanobacterial diazotroph endosymbiont recently discovered as a major contributor to marine nitrogen fixation57. Second, ammonia, the host-beneficial metabolite in diazotroph endosymbioses, can diffuse through membranes in its neutral form and does not require host transporters for efficient trafficking58. Previously, we observed efficient distribution of fixed nitrogen from diazoplasts into host compartments following 15N2 labeling25. Ammonia diffusion may have reduced early selection pressure for host protein import as posited by the targeting-early model. Finally, the eukaryotic hosts in most diazotroph endosymbioses are already photosynthetic, in contrast to largely heterotrophic hosts that acquired photosynthesis by endosymbiosis. For instance, cellular processes that enabled intracellular bacteria to take up residence in the ancestor of Epithemia spp. were likely different than those of the bacterivore amoeba ancestor of Paulinella chromatophora. Autotrophy and lack of digestive pathways would reduce the frequency by which bacteria might gain access to the host cell, such that the selection of host protein import pathways over successive transient interactions would be less effective. Overall, a universal model of organellogenesis is premature given the limited types of interaction that have been investigated in depth, highlighting instead the importance of increasing the diversity of systems studied.
Are diazoplasts “organelles”?
Diazoplasts share many features with recently-described nitrogen-fixing organelles, nitroplasts9. As detailed in the introduction, metabolic and cellular integration of diazoplasts with their host alga even exceeds that of nitroplasts in some respects36. However, hundreds of host proteins were detected in the nitroplast proteome including many likely to fill gaps in nitroplast metabolic pathways, compared with the few host proteins of unknown function in the diazoplast proteome. Based on the conventional definition which specifies genetic integration as the dividing line between endosymbionts and organelles, diazoplasts would not qualify4,50,51. However, over a decade ago, Keeling and Archibald59 suggested that “if we use genetic integration as the defining feature of an organelle, we will never be able to compare different routes to organellogenesis because we have artificially predefined a single route.” They further hypothesized that if an endosymbiont became fixed in its host absent genetic integration, “it might prove to be even more interesting… by focusing on how it did integrate, perhaps we will find a truly parallel pathway for the integration of two cells.” The diazoplast appears to be such a parallel case in which non-genetic interactions were sufficient to integrate two cells. It serves as another example that the current organelle definition does not account for observations in many biological systems and may be overdue for revision to reflect biological significance in the spectrum of endosymbiotic interactions.
Identifying parallel pathways to integrate cells is more than an academic exercise. The ability to engineer bacteria as membrane compartments to introduce new metabolic functions would be transformative60,61. For example, nitrogen-fixing crop plants that could replace fertilizers is a major goal for sustainable agriculture. But efforts to transfer the genes for nitrogen fixation to plant cells have been slow, hampered by the many genes required as well as the complex assembly, high energy requirements, and oxygen sensitivity of the process. We previously proposed an alternative strategy inspired by diazotroph endosymbioses: introducing nitrogen-fixing bacteria into plant cells as an integral organelle-like compartment. This approach has the advantage that diazotrophs express all required genes with intact regulation, coupled to respiration, and in a protected membrane. Diazoplasts, which achieve stable integration without significant genetic integration, is an important alternative to the nitroplast and other organelles, which are defined by their genetic integration, to inform this strategy. Identifying the nongenetic interactions that facilitated diazoplast integration with Epithemia will be critical for guiding bioengineering.
Ongoing genome reduction may drive genetic integration in diazotroph endosymbioses.
The fewer number of host protein candidates and their lack of clear function in diazoplasts versus the nitroplast is not associated with differences in their function as stable cellular compartments. Rather, an alternative explanation points towards differences in the extent of genome reduction in diazoplasts, which encode 1720–1900 genes, compared to nitroplasts, which encode 1159 −1200 genes62. Among the genes missing from the nitroplast genome are cyanobacterial IspD, ThrC, PGLS, and PyrE; for each, an imported host protein was identified that could substitute for the missing function. In contrast, these genes are retained in diazoplast genomes, including those of E. clementina and E. pelagica, obviating the need for host proteins to fulfill their functions (Figure S5). Consistent with the diazoplast and the nitroplast being at different stages of genome reduction, diazoplast genomes contain >150 pseudogenes compared to 57 detected in the nitroplast genome, suggesting diazoplasts are in a more active stage of genome reduction. Interestingly, even genes retained in the nitroplast, namely PyrC and HemE, have imported host-encoded counterparts9. The endosymbiont copies may have acquired mutations resulting in reduced function, necessitating import of host proteins to compensate. Alternatively, once efficient host protein import pathways were established, import of redundant host proteins may render endosymbiont genes obsolete, further accelerating genome reduction. Genetic integration may in fact be destabilizing for an otherwise stably integrated endosymbiont, at least initially, as it substitutes essential endosymbiont genes with host-encoded proteins that may not be functionally equivalent and require energy-dependent import pathways. Comparing these related but independent diazotroph endosymbioses yields valuable insight, which otherwise would not be apparent. Diazoplasts at 35 Mya may represent an earlier stage of the same evolutionary path as the ~140 Mya nitroplast, in which continued genome reduction will eventually select for protein import pathways. Alternatively, diazoplasts may have evolved unique solutions to combat destabilizing genome decay, for example through the early loss of mobile elements.24,30,63 Whether they represent an early intermediate destined for genetic integration or an alternative path, diazoplasts provide a valuable new perspective on cellular evolution.
METHODS
Resource availability
Requests for further information and resources should be directed to and will be fulfilled by the 149 lead contact, Ellen Yeh (ellenyeh@stanford.edu)
Lead contact
Requests for further information or resources should be directed to ellenyeh@stanford.edu
Materials availability
Cultures and reagents used in this study are available upon request from lead contact.
Method details
Cultivation and generation of axenic strain
Wild isolates of E. clementina were cultivated in CSi-N media in vented flasks under 10 μmole photon m −2 s-1 of white light at 20°C. Full cultivation procedures are detailed in25. Initially, cultures were started from a single cell of E. clementina and were thus clonal but xenic. To generate axenic cultures for sequencing, cells were incubated overnight with lysozyme to disrupt the cell walls of gram-positive bacteria, then treated to a 30-minute pulse of antibiotic cocktail (100 μg/mL Carbenicillin, 25 μg/mL Chloramphenicol, 5 mg/mL Levofloxacin, 50 mg/mL Rifampicin, 50 μg/mL Streptomycin). Immediately following, cultures were spray plated64. A small volume of dilute culture suspension was aspirated into a glass Pasteur pipette and held perpendicular to a stream of sterile air. The air atomizes the culture; the small droplets are then captured on a CSi-N agar plate. This process isolates single cells of E. clementina and disrupts their associated bacterial community. Cells on the agar plates were allowed to form colonies which were screened for any visual bacterial growth. Only those colonies that lacked bacterial growth were chosen for further cultivation. The resulting strain was expanded and confirmed axenic in subsequent sequencing experiments.
Scanning electron microscopy
Xenic cultures of E. clementina were resuspended, pelleted at 23°C at 1000 × g, rinsed with CSi-N media, and then resuspended in 250uL of PBS. Cells were transferred in a droplet to poly-L-lysine coated 12mm diameter glass cover slips and left to sit on a flat surface for 5min. The PBS was gently aspirated, and a droplet of 4% paraformaldehyde in PBS was added to coat the entire cover slip surface. Cells were fixed for 10min in the dark and then the cover slip was rinsed twice with PBS. An ethanol dehydration series was performed wherein cover slips were sequentially immersed in 60%, 70%, 80%, 90%, and 100% v/v ethanol in PBS. The cover slips were gently dried on a 42°C heat block. The cover slips were secured to a low-profile pin mount and sputter coated in a Leica ACE600 High Vacuum Sputter Coater with gold to a thickness of 6nm. Samples were imaged on a Zeiss Sigma FE-SEM.
High molecular weight DNA extraction
E. clementina were grown to a density of approximately 400,000 cells/mL and 20–30 million cells were used as input to HMW DNA extraction. Xenic cultures were first subjected to a round of centrifugation through a discontinuous Percoll gradient to deplete excess bacteria. E. clementina cells pellet out of the solution entirely, whereas a portion of their bacterial community stays suspended in various Percoll fractions. Centrifugation steps were performed at 23°C at 1000 × g. For both xenic and axenic cultures, HMW DNA was isolated using a nuclei extraction method65. Cells were suspended in a minimal volume of nuclear isolation buffer (NIB) and the transferred to a mortar where they were flash frozen and then ground with the pestle until a paste formed. This grinding process was repeated a total of three times. Cell homogenate was transferred to a 15mL falcon tube containing NIB, rinsing the mortar with NIB if necessary, and incubated at 4°C for 15min. No miracloth filtering step was performed. The cell homogenate was spun down at 4°C and 2900 × g. The resulting nuclei pellet was rinsed with 15mL NIB until the solution was clear of any photosynthetic pigments. The resulting nuclei/cellular compartment mix was used as input for the Nanobind plant nuclei big DNA kit from PacBio. Steps were followed as listed in the kit protocol except for large cell inputs, in which reagent volumes were doubled and the Proteinase K digestion step was extended to 2hrs. The isolated DNA from this protocol was processed with the Short Read Eliminator kit from PacBio to deplete DNA fragments < 25kb in length. The final, HMW DNA was used as input for nanopore library preparation.
Nanopore library preparation and sequencing
For all sequencing runs from axenic cultures of E. clementina, 1–2μg of HMW DNA was used as input to the Oxford Nanopore Technology sequencing by ligation kit (SQK-LSK112). The nanopore protocol (Version: GDE_9141_v112_revC_01Dec2021) was followed with the following minor modifications: end repair incubation was lengthened to 30min at 20°C and the adapter ligation incubation was lengthened to 60min at room temperature. Resulting libraries were loaded onto primed, high-accuracy MinION R10.4 flow cells (FLO-MIN112) at a target amount of 9 fmoles of 10kb DNA. In actuality, DNA sizes ranged within samples and between sequencing runs, but 9 fmole maintains a recommended loading amount for the flow cell at a range of fragment sizes. All sequencing of xenic cultures was performed similarly, but with previous iterations of the sequencing kit (SQK-LSK110) and the flow cell MinION R9.4.1 (FLO-MIN111). If pore occupancy dipped below roughly 1/3 of starting occupancy during the sequencing run, the run was paused, and the flow cell was washed with the Flow Cell Wash Kit (EXP-WSH004) from Nanopore according to the associated protocol. The same prepped library was then reloaded onto the flow cell and the sequencing run was restarted. Each run was left to sequence for 3–5 days, or until pore occupancy was near zero.
Isolation of genomic DNA for Illumina sequencing
DNA was extracted from axenic E. clementina cultures following the QIAGEN DNeasy Plant Pro Kit (69206) protocol. For the lysis step, cell suspension was transferred to the kit’s tissue-disrupting tubes included along with 100mg 0.5mm autoclaved glass beads added and placed in a bead-beater and shaken for one minute. 300ng of the isolated axenic E. clementina DNA was used as input to the NEBNext Ultra II FS DNA Library Kit for Illumina (E7805S). A fragmentation time of 16min was used for a target insert size of 200–450bp. Samples were indexed with NEBNext Multiplex Oligos for Illumina Dual Index Primers Set 1 (E7600S). DNA concentration of resulting libraries was determined with a Qubit dsDNA Quantification Assay High Sensitivity kit (Q32851). Final libraries were checked for quality and size-range using an Agilent Bioanalyzer High-Sensitivity DNA chip. The final mean insert size was 440bp, with a well-formed size distribution around the mean and minimal adapter dimers. The library was sequenced on an Illumina NextSeq 2000 P3 for 2 × 150bp reads. Raw reads were trimmed and paired with fastp (--qualified_quality_phred 20, --unqualified_percent_limit 20) for a final total of in 402 million read pairs from axenic E. clementina.
RNA isolation and sequencing
To capture a wide range of transcripts, axenic cultures of E. clementina were exposed to different nitrogen conditions and collected at different times in the day-night cycle. Axenic cultures of E. clementina were seeded in 175cm2 sterile vented flasks at a density of 1.2 million cells per flask. For conditions of nitrogen repletion, media in the flask contained 100μM of ammonium. Cells were kept in -N or +NH4+ conditions for 72 hours and harvested two hours into the day period. Cells in nitrogen depleted conditions were additionally collected two hours into the night period. All cells were scraped from the flask, centrifuged to concentrate, resuspended in trizol, and flash frozen. Each condition was collected in triplicate for each experiment, and the whole experiment was performed twice. To lyse, the trizol suspended cells were held on ice and a sonicator probe was submerged at the center of the tube. Sonication was performed with a microtip at 50/50 on/off pulses for one minute at an intensity setting of six on a Branson 250 Sonifier (B250S). RNA was isolated using the QIAGEN RNeasy Plus Universal Mini Kit (74134) following the included protocol. 500ng of RNA per condition per replicate collected from the first experiment was used as input to the NEBNext poly(A) mRNA Magnetic Isolation module (E7490L) to enrich for mRNA and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760L) and indexes from NEBNext Multiplex Oligos for Illumina Dual Index Primers Set 1 (E7600S) were used for library preparation. 350ng of RNA per condition per replicate collected from the second experiment was used as input to the Zymo-Seq RiboFree Universal cDNA Kit (R3001) and indexed with Zymo-Seq UDI Primer Set (Indexes 1–12) (D3008). For each experiment, libraries were pooled and sequenced on an Illumina NextSeq 2000 P3 for 2 × 150bp reads.
E. pelagica cultures provided by courtesy of Chris Schvarcz and Kelsey McBeain proved to be unculturable long-term in lab conditions after shipment. Therefore, RNA was extracted upon receipt of overnight shipment from University of Hawaii at Manoa, HI. Otherwise, the same method of poly(A)-enrichment and Illumina sequencing was used as for E. clementina.
Data filtering and genome assembly of E. clementina
Initial genome size, ploidy, and repeat content estimates were made by counting k-mers in the axenic Illumina reads with jellyfish v2.2.10 (-C -m -k 35 -s 5G) and plotting with GenomeScope66,67. The raw fast5 sequencing files were basecalled with guppy v1.1.alpha13–0-g1ec7786. Reads were filtered based on minimum length 3kb and quality 20 with Nanofilt v2.8.068. Read statistics were calculated with NanoPlot v1.30.1. Basic quality checks were performed with fastqc v0.11.969. Post filtering, 19.5Gb of sequence from axenic cultures of E. clementina and 30.2Gb of sequencing data from xenic cultures were used for a two-step assembly process. First, axenic reads were assembled with NextDenovo v2.5.070. Then, xenic nanopore sequencing data was aligned to the axenic assembly using minimap2 (-ax map-ont) v2.24-r1122 to identify probable diatom reads in the xenic data71. Finally, axenic and diatom-mapped xenic nanopore reads were combined and assembled with NextDenovo (using default or machine-specific options, except read_cutoff=5k, genome_size=350M). Axenic Illumina data was mapped to the assembly with BWA v0.7.17-r118872. Contigs in the assembly were removed if less than 70% of the contig was covered by the axenic Illumina reads or if those reads mapped at significantly lower depth than to the rest of the contigs (< 4% of mean depth). The axenic Illumina reads were then used as input for 3 rounds of polishing with Racon v1.5.0 and one round of Polca (part of MaSuRCA v4.0.5)73–75. Further contamination analysis of the assembly and reads was performed with blobtools v1.1.176. Organellar genomes for the diazoplast, chloroplast, and mitochondria were assembled and annotated as previously reported25. All contigs in the assembly were aligned to the organellar genomes and to the diazoplast genome to check for remaining organellar contaminants in the assembly. Any remaining organellar contigs contaminating the nuclear assembly were identified and removed if they aligned end-to-end to the already assembled organellar and endosymbiont genomes. Basic assembly statistics were extracted with QUAST v5.2.077. Final assembly completeness and consensus quality (QV) was assessed with the k-mer spectra tool Merqury v1.378. The QV of our final assembly was 38.52. BUSCO v5.3.2 in genome mode was also used to estimate completeness at the gene level79.
Repeat masking of E. clementina and E. pelagica
The final nuclear assembly of E. clementina and the publicly released37 but raw nuclear sequence of E. pelagica (uoEpiScrs1.2 GCA_946965045.2) were used as input to the RepeatModeler2 and RepeatMasker pipelines. To identify and classify the repeat elements for both Epithemia genomes, the workflow for RepeatModeler v2.0.2 with built-in LTR detection and classification was run80 (BuildDatabase -engine rmblast, RepeatModeler -LTRStruct, RepeatClassifier -engine rmblast). Since the repeat models for the organisms are de novo and repeat data for diatoms in the source databases may be limited, the repeat families classified as ‘Unknown’ were further interrogated to ensure no protein-coding genes were annotated as repeats. To do this, the Unknown repeat families were used as input to ncbi-blast+ against the NCBI non-redundant (NR) protein database81,82 (November 3rd 2022). Approximately 8% of Unknown repeats had significant similarity to eukaryotic and Bacillariophyta proteins. Out of caution, these regions annotated as Unknown repeats with protein hits were removed from the repeat database to be kept unmasked. Finally, RepeatMasker v4.1.2-p1 (-engine rmblast, -s no_is -norna -gff -xsmall) was run on the genomes to soft-mask all repeat regions. The ParseRM tool by Aurelie Kapusta was used to extract repeat type and divergence from consensus from the raw .classified and .align output files from RepeatMasker43.
Gene annotation of E. clementina and E. pelagica
For both E. pelagica and E. clementina, the masked nuclear genome of each organism was annotated in two independent runs of BRAKER2 v2.1.6, which applied installs of GeneMark-ETP v1.0, AUGUSTUS v3.4.0, and ProtHint v2.6.0. First, the BRAKER2 pipeline was given extrinsic protein evidence as input. Protein sequences were sourced from the orthoDB v10 protozoa database which was manually edited to include diatom proteins from recent annotations. Second, the BRAKER2 pipeline was given transcriptomic evidence from the source organism83–91. To produce the aligned RNA-seq evidence, the RNAseq reads were quality filtered, trimmed and paired with fastp92 v0.22.0 (--qualified_quality_phred 20, --unqualified_percent_limit 20), and then aligned to the source genome with hisat2 v2.1.093 (--rnastrandness RF). Alignment files were sorted and converted to binary alignment files with samtools v1.16.194. For E. pelagica, a single 280 million read Illumina run from polyA-enriched RNA was used as input. For E. clementina, actively maintained lab cultures enabled more extensive sequencing of the transcriptome. RNA from 30 samples and five different conditions using both polyA-enrichment and rRNA-depletion methods of isolation were used as input. The outputs of these two independent protein-based and transcriptome-based annotations were merged using TSEBRA v1.0.3 into a single annotation86. Both the input and output general transfer format (GTF) file were fixed with the fix_gtf_ids.py script included with TSEBRA. The output GTF files were converted to multi-isoform fasta files, removing any pseudo genes or genes interrupted by stop codons using gffread v0.12.795 (-J --no-pseudo -y). Completeness of the final annotation was assessed with BUSCO v5.3.2 in proteins mode. To inspect isoforms, the AGAT v1.0.0 agat_sp_keep_longest_isoform.pl tool was used96.
Orthologue analysis
Curated species proteomes and genomeswere downloaded from NCBI or associated online repositories37,40,97–105 (Table S1). The agat package was used to remove short isoforms (agat_sp_keep_longest_isoforms.pl). Where necessary, gene feature files were reformatted96 (agat_sp_manage_attributes.pl -p gene -att transcript_id). Finally, longest isoform proteomes were produced from the gene feature files and the corresponding species genome with gffread, removing genes without a complete, valid coding sequence and removing pseudo-genes95 (gffread -J –no-pseudo -y). The resulting proteomes were used as input for Orthologue analysis.
Orthogroups were identified with orthofinder v2.5.4 (-M msa -T iqtree) and orthogroup overlaps between species were extracted from Orthogroups_SpeciesOverlaps106. In order to quantify shared orthologues between species without biasing for total proteome size differences, the Jaccard similarity coefficient for each species pair was calculated according to the standard Jaccard index formula where A and B are the total number of self-orthologues identified for each organism and A ∩ B is the number of orthologues identified between the organisms as contained in the OrthologuesStats_one-to-one file. To identify uniquely overlapping orthogroups (e.g. orthogroups shared between two species and not by any other species), orthogroup sets from Orthogroups.GeneCount.tsv were parsed and plotted with UpSetR107. To quantify sequence similarity, orthologue pairs were identified by reciprocal best BLAST between organism pairs and the full-length percent amino acid identity was calculated from the BLAST outputs, similar to the method used in108.
NUDT homology search
Whole genomes of free-living cyanobacterial relatives of the endosymbiont were curated along with available whole endosymbiont genomes (Table S1). These sequences were used as query for homology searches against the nuclear genomes of E. pelagica and E. clementina. Command line BLASTN with defaults, BLASTN using the custom settings previously validated for NUMT search81,109 (-reward 1 -penalty −1 -gapopen 7 -gapextend 2), minimap2 (-ax asm5 and -HK19 modes), and nucmer were all used to perform these homology searches71,110. As negative controls, the reversed sequence of the E. pelagica mitochondria and the E. coli genome were used. For all cyanobacterial and endosymbiont queries, BLASTN was the most sensitive and least stringent, identifying all homology regions identified by other programs. Contiguous regions of homology < 500bp in length were not considered, though most short alignments were < 100bp. The resulting > 500bp contiguous regions of homology were considered candidate NUDTs. Seven regions in total for E. clementina and none in E. pelagica. To verify that these alignments were not a result of misassembly, long-reads from nanopore sequencing of axenic cultures were aligned (minimap -ax map-ont) and the reads spanning the border of the insertion were counted and the depth compared to that of the contig. The read depth for the contig was calculated with samtools depth (considering only primary alignments to minimize skews from repetitive regions). To check for expression within the NUDTs, RNA-seq data from both polyA enrichment and rRNA depletion experiments was mapped as previously described and normalized with deeptools bamCoverage (--normalizeUsing BPM -p max -bs 100). Corresponding source regions from the endosymbiont and percent identities were pulled from the blast results. Using bedtools intersect, the source regions were overlapped with endosymbiont gene regions111. These coordinates were then mapped back to the nuclear region. Nuclear and diazoplast sequences corresponding to these identified gene fragment containing regions were aligned using EMBOSS Needle v6.6.0.0, which calculates percent identity112. The truncation was calculated by dividing the length of the gene fragment by the total length of the corresponding diazoplast gene. For both the nuclear and diazoplast genomes of E. clementina, GC content variation was analyzed in sliding windows of 5000bp with a step size of 1000bp using bedtools makewindows and bedtools nuc. All alignments were visualized with the Integrative Genomics Viewer113 (IGV) and plotted with circlize114.
Identification of Horizontal Gene Transfers
Diatom proteomes (Table S1) including the de novo predicted E. pelagica and E. clementina were used as input to a custom HGT pipeline adapted from115. In brief, the program uses diamond v2.0.14 to collect homologues from the NCBI NR database for each gene in an organism116. To best ensure representation of genes from a diverse range of taxa, three diamond runs were performed against different subsections of NR: Bacteria, the SAR supergroup, and the remainder of the database. These results are parsed so that, where possible, the final list of homologues for each gene consists of no more than 70% of any one kingdom and does not contain any hits to self (relevant for diatom proteins already in the NCBI NR). Proteins with under 10 identifiable homologues were excluded from further analysis. Proteins were aligned with mafft v7.525 (--auto) and poorly aligned regions trimmed with trimal (-automated1)117,118. The L-INS-i method in mafft was selected for most alignments. These alignments were used as inputs for generation of phylogenetic trees. For E. pelagica and E. clementina, IQ-Tree v2.2.0.3 and the inbuilt ModelFinder function was used with 1000 rounds of bootstrapping119–121. Because of runtime limitations, FastTree v2.1.1 was used to construct trees for all other diatoms122. The topology of these trees was parsed by PhySortR v1.0.8 (min.support = 0.7, min.prop.target = 0.7, clade.exclusivity = 0.9) to identify trees in which the diatom gene of interest is more closely related to bacterial homologues than to eukaryotic ones123. The results were parsed using custom scripts to remove genes with fewer than five bacterial taxa in the tree. PhySortR designates genes as All Exclusive, Exclusive, Non-Exclusive, or Negative based on the tree topology. We treated All Exclusive and Exclusive results as high confidence and Non-Exclusive as low confidence. In reality, HGT candidates with Non-Exclusive tree topology are a mix of ambiguous topology as well as likely real HGTs shared between the diatoms or other eukaryotes. The Non-Exclusive HGT candidates were further filtered based on Alienness score (AI)124. The alienness score was calculated with both the best prokaryote Evalue and with the best prokaryote Evalue after the first group of Bacillariophyta results, to account for HGTs that may be shared between diatom species. HGT candidates with positive AI scores were kept for subsequent analysis. Species of origin for HGT candidates was inferred using the taxonomic breakdown of the top blast result.
Diazoplast Isolation
E. clementina cells were harvested by scraping, then washed twice in CSI-N growth medium by centrifugation at 2,000xg, and re-suspensed in spheroid body isolation buffer (50 mM HEPES pH 8.0, 330 mM D-sorbitol, 2 mM EDTA NaOH pH 8.0, 1 mM MgCl2). Cells were then placed in a bath sonicator for 10 minutes followed by 3 low pressure cycles (500 psi) and by 5 high pressure cycles (2,000 psi) in an EmulsiFlex-C5 Homogenizer (Avestin) or until most cells appeared lysed under a microscope. After a 1-minute spin at 100xg to pellet the unbroken cells and broken frustules, the supernatant was collected and centrifuged at 3,000xg for 5 minutes to concentrate the diazoplasts and other organelles to a volume of 3–4 mL. This fraction was then split equally, and each half was laid on a discontinuous Percoll gradient. 89% Percoll, 10% 10xPBS, and 1% 1M HEPES pH 8.0 was diluted with SIB to generate the gradient, which consisted of 2 mL 90%, 3 mL 70%, 3 mL 60%, 3 mL 50%. The gradient was centrifuged for 20 minutes at 12,000xg, 4° using a Beckman Optima L-90K ultracentrifuge with SW-41 rotor.
The boundaries between the 60% and 70% layers and the 70% and 90% layers were collected, counted, and checked for purity via light microscopy. They were then diluted 1:6 in SIB Buffer and centrifuged at 2,000xg for 2–3 minutes to collect diazoplasts, which were resuspended in 200 μL Extraction Buffer (100mM Tris-HCl, pH8.0, 2% (wt/vol) SDS, 5mM EGTA, 10mM EDTA, 1mM PMSF, 2x protease inhibitor (1 tablet each of cOmplete™ Protease Inhibitor Cocktail, catalog number 4693116001 and Pierce™ Protease Inhibitor tablet, EDTA free, catalog number A32965)). During optimization, enrichment was assessed by Western blot for NifDK on both the diazoplast and whole cell extracts.
Protein Extraction, Preparation, and LCMS/MS
We generated whole cell lysate by homogenizing with a bead beater at 3000 strokes per minute for 3 minutes with 1 mm glass beads (BioSpec Products catalog number 11079110) or until most cells appeared lysed under a microscope. Diazoplasts were lysed similarly using 0.5 mm beads (BioSpec Products catalog number 11079105). Beads were pelleted at 100xg for 1 minute and the supernatant was removed; the beads were washed twice with 50 μL extraction buffer each by vortexing and spinning. These fractions were then added to the supernatant for a total of 300 μL, followed by an equal volume of cold Tris-buffered phenol (pH 7.5–7.9). This solution was vortexed for 1 minute, centrifuged at 18,000 × g for 15 minutes at 4°° C. The upper phase was discarded, then extracted with an equal volume of cold 50mM Tris-HCl, pH8.0. The phenol phase was extracted with Tris-HCl a total of three times, followed by addition of 0.1 M ammonium acetate in methanol and overnight incubation at −80° C. Samples were then transferred to new tubes and centrifuged at 18,00 × g for 20 minutes at 4° C. The supernatant was discarded and the pellet was washed once in 0.1 M ammonium acetate in methanol and twice in 1 mL cold methanol by centrifugation for 5 minutes at 18,00 × g at 4° C, followed by a final short spin and removal of trace methanol. The pellet was then resuspended in 150 μL resuspension buffer (6M Guandine-HCl in 25mM NH4HCO3 pH8.0). Each sample was then reduced with TCEP at a final concentration of 2 μM (Thermo Scientific catalog number 20490) for 1 hour at 56° C, alkylated with iodoacetamide (Thermo Scientific catalog number 90034) at a final concentration of 10 mM for 1 hour at ambient temperature, and then diluted with 3 volumes of 25mM NH4HCO3. Sequencing grade modified trypsin (Promega catalog number V5111) was added at a ratio of 1:50 followed by overnight incubation at 37° C, then repeated the next morning, followed by quenching the reaction by adding formic acid to a final concentration of 1%. Each sample was then loaded onto a C18 cartridge (Sep-pak waters catalog number WAT054960), activated with 80% acetonitrile and 0.1% formic acid. The flow-through was loaded a total of three times, followed by five washes with 1 mL 0.1% formic acid. The samples were then eluted with 200 μL of 80% acetonitrile 1% formic acid and the flow-through re-loaded a total of three times.
Peptide concentration was determined using Pierce™ Quantitative Colorimetric Peptide Assay (Thermo Fisher catalog number 23275). 1 μg of peptides from each sample was loaded on either on a Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher) (1 replicate) or an Eclipse Tribrid mass spectrometer (Thermo Fisher) (2 replicates), equipped with an Easy LC 1200 UPLC liquid chromatography system (Thermo Fisher). Peptides were first trapped using a trapping column (Acclaim PepMap 100 C18 HPLC, 75 μm particle size, 2 cm bed length), then separated using analytical column AUR3–25075C18, 25CM Aurora Series Emitter Column (25 cm × 75 μm, 1.7μm C18) (IonOpticks). The flow rate was 300 nL/min, and a 120-min gradient was used. Peptides were eluted by a gradient from 3 to 28 % solvent B (80 % acetonitrile, 0.1 % formic acid) over 106 min and from 28 to 44 % solvent B over 15 min, followed by a short wash (9 min) at 90 % solvent B. The Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer was configured as follows: Precursor scan was from mass-to-charge ratio (m/z) 375 to 1600 (resolution 120,000; AGC 3.0E6, maximum injection time 100ms) and top 20 most intense multiply charged precursors were selected for fragmentation (resolution 15,000, AGC 5E4, maximum injection time 60ms, isolation window 1.0 m/z, minimum AGC target 1.2e3, intensity threshold 2.0 e4, include charge state = 2–8). Peptides were fragmented with higher-energy collision dissociation (HCD) with normalized collision energy (NCE) 27. Dynamic exclusion was enabled for 24s. The Orbitrap Eclipse Tribrid mass spectrometer was configured as follows: Precursor scan was from mass-to-charge ratio (m/z) 375 to 1600 (resolution 120,000; AGC 200,000, maximum injection time 50ms, Normalized AGC target 50%, RF lens(%) 30) and the most intense multiply charged precursors were selected for fragmentation (resolution 15,000, AGC 5E4, maximum injection time 22ms, isolation window 1.4 m/z, normalized AGC target 100%, include charge state=2–8, cycle time 3 s). Peptides were fragmented with higher-energy collision dissociation (HCD) with normalized collision energy (NCE) 27. Dynamic exclusion was enabled for 30s.
Proteomics Data Analysis
Maxquant version 2.5.0 was used for proteomics database searches, using default parameters with the following changes: label-free and iBAQ quantification, matched between runs were enabled125. For identifications, peptides were searched against the Epithemia clementina reference host and diazoplast proteomes. The proteingroups.txt file output from MaxQuant was analyzed using Perseus version 2.0.10.0126. iBAQ values were imported and filtered to remove potential contaminants, reverse hits, and those only identified by site. Only proteins identified by two more unique peptides and with a minimum of 5% sequence coverage were included in further analysis. The iBAQ values were then log(2) transformed for normality, proteins with two or more non-valid values were removed, and missing values were imputed from a downshifted normal distribution of the total matrix (width 0.3 standard deviations, down shift 1.8 standard deviations). A two-sided students T-test using the significance analysis of microarrays method (s0=0.1, false discovery rate 0.05, 250 randomizations) was used to determine the enrichment of host-encoded proteins in the diazoplast.
Immunoblot
Whole cell and isolated diazoplast lysates were prepared as described above. Protein concentration was determined using Pierce™ BCA Protein Assay Kit (Thermo Fisher catalog number 23227). 0.5 μg of protein from each sample was diluted in lithium dodecyl sulfate buffer with 100 mM DTT and loaded onto a NuPage™ Bis-Tris gels 4–12% acrylamide (Thermo Fisher catalog number NP0321BOX) in MES Buffer, and separated by electrophoresis, using Precision Plus Protein All Blue Standard Precision Plus Protein™ Standards (BioRad catalog number 1610373). Proteins were then transferred into a nitrocellulose membrane using Bio-rad Transblot Turbo, followed by blocking in LiCOR blocking buffer (0.1% Casein, 0.2x PBS, 0.01% sodium azide) for 1 hour at room-temperature. The membrane was then incubated for two hours at room temperature with primary antibodies anti-NifDK (polyclonal goat at 1:5000 dilution, kindly provided by Dr. Dennis Dean from Virginia Tech, US) to detect nitrogenase and anti-PsbA (1:10,000 dilution rabbit from AgriSera AB, Vanas, Sweden) as an internal loading control. Antibodies were diluted in a solution of 50% TBST and 50% LiCOR blocking buffer. The membrane was then washed three times with TBST and incubated with LiCOR secondary antibodies (IRDye 800CW) for 1 hour (goat α-rabbit for PsbA and donkey α-goat for NifDK). After two rinses with TBST and one with PBS, the blot was imaged using an infra-red LiCOR imager. Intensity of the signal was quantified using Image Studio Lite software v5.2.
Supplementary Material
Table S1. NCBI Accessions and access links for all species used in this study
Table S2. RepeatMasker output tables for Epithemia
Table S3. Gene codes and putative identity of Epithemia-specific horizontally transferred genes.
Table S4. Processed and unprocessed proteomics data
Data S1. Integrative genomics viewer screenshots of genomic context, expression, and read support of all NUDTs
Acknowledgements
We thank Chriz Schvarcz and Kelsey McBeain for the generous sharing of E. pelagica cultures. We thank Dr. Devaki Bhaya and Dr. Arthur Grossman and members of their labs for their support and feedback on the project. We are grateful to Scott Miller and Heidi Abresch for their Epithemia expertise and discussion. Our thanks to Andres Reyes for mass spectrometry technical training. We thank Daniel S. Rokhsar, Jonathan Zehr, Kendra Turk-Kubo, Andy Alverson, Elizabeth Ruck, Paolo Carnevali, Dmitri Petrov, and Cedric Feschotte for expert advice during the project. Anti-NifDK polyclonal antibodies were kindly provided by Dennis Dean. E.Y. is a Chan Zuckerberg Biohub – San Francisco Investigator and supported by Burroughs Wellcome Fund. S.F. was partially funded by NIH training grant (T32GM007276). M.S.O. was partially funded by NIH training grant (5T32AI007328-32). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the National Institutes of Health. S.-L.X is funded by the National Institutes of Health grant S10OD030441 and by the Carnegie Endowment Fund to the Carnegie Mass Spectrometry Facility.
Footnotes
Declaration of interest
The authors declare no competing interest.
Declaration of generative AI and AI-assisted technologies
During the preparation of this work, the author(s) used GPT-4o mini and Claude 3.5 Sonnet for assistance with minor bioinformatic troubleshooting. The authors reviewed and edited all AI outputs and take full responsibility for the content of this publication.
REFERENCES
- 1.Archibald J.M. (2015). Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 25, R911–R921. 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 2.Martin W., and Herrmann R.G. (1998). Gene Transfer from Organelles to the Nucleus: How Much, What Happens, and Why? Plant Physiol. 118, 9–17. 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeling P.J., McCutcheon J.P., and Doolittle W.F. (2015). Symbiosis becoming permanent: Survival of the luckiest. Proc. Natl. Acad. Sci. 112, 10101–10103. 10.1073/pnas.1513346112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T., and Lee J.J. (1985). Protozoa as Hosts for Endosymbioses and the Conversion of Symbionts into Organelles,. J. Protozool. 32, 376–379. 10.1111/j.1550-7408.1985.tb04031.x. [DOI] [Google Scholar]
- 5.Marin B., Nowack M., E.C., and Melkonian M. (2005). A Plastid in the Making: Evidence for a Second Primary Endosymbiosis. Protist 156, 425–432. 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama T., and Ishida K. (2009). Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 19, R284–R285. 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 7.Nowack E.C.M., and Grossman A.R. (2012). Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc. Natl. Acad. Sci. 109, 5340–5345. 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer A., Poschmann G., Mühlich C., Valadez-Cano C., Hänsch S., Hüren V., Rensing S.A., Stühler K., and Nowack E.C.M. (2017). Massive Protein Import into the Early-Evolutionary-Stage Photosynthetic Organelle of the Amoeba Paulinella chromatophora. Curr. Biol. 27, 2763–2773.e5. 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Coale T.H., Loconte V., Turk-Kubo K.A., Vanslembrouck B., Mak W.K.E., Cheung S., Ekman A., Chen J.-H., Hagino K., Takano Y., et al. (2024). Nitrogen-fixing organelle in a marine alga. Science 384, 217–222. 10.1126/science.adk1075. [DOI] [PubMed] [Google Scholar]
- 10.Zakharova A., Tashyreva D., Butenko A., Morales J., Saura A., Svobodová M., Poschmann G., Nandipati S., Zakharova A., Noyvert D., et al. (2023). A neo-functionalized homolog of host transmembrane protein controls localization of bacterial endosymbionts in the trypanosomatid Novymonas esmeraldas. Curr. Biol. 33, 2690–2701.e5. 10.1016/j.cub.2023.04.060. [DOI] [PubMed] [Google Scholar]
- 11.McCutcheon J.P., Boyd B.M., and Dale C. (2019). The Life of an Insect Endosymbiont from the Cradle to the Grave. Curr. Biol. 29, R485–R495. 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Nowack E.C.M., Price D.C., Bhattacharya D., Singer A., Melkonian M., and Grossman A.R. (2016). Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc. Natl. Acad. Sci. 113, 12214–12219. 10.1073/pnas.1608016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponce-Toledo R.I., López-García P., and Moreira D. (2019). Horizontal and endosymbiotic gene transfer in early plastid evolution. New Phytol. 224, 618–624. 10.1111/nph.15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eastman K.E., Pendleton A.L., Shaikh M.A., Suttiyut T., Ogas R., Tomko P., Gavelis G., Widhalm J.R., and Wisecaver J.H. (2023). A reference genome for the long-term kleptoplast-retaining sea slug Elysia crispata morphotype clarki. G3 GenesGenomesGenetics 13, jkad234. 10.1093/g3journal/jkad234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartaxana P., Trampe E., Kühl M., and Cruz S. (2017). Kleptoplast photosynthesis is nutritionally relevant in the sea slug Elysia viridis. Sci. Rep. 7, 7714. 10.1038/s41598-017-08002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hehenberger E., Gast R.J., and Keeling P.J. (2019). A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc. Natl. Acad. Sci. 116, 17934–17942. 10.1073/pnas.1910121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkum A.W.D., Lockhart P.J., and Howe C.J. (2007). Shopping for plastids. Trends Plant Sci. 12, 189–195. 10.1016/j.tplants.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Keeling P.J. (2013). The Number, Speed, and Impact of Plastid Endosymbioses in Eukaryotic Evolution. Annu. Rev. Plant Biol. 64, 583–607. 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 19.Sibbald S.J., and Archibald J.M. (2020). Genomic Insights into Plastid Evolution. Genome Biol. Evol. 12, 978–990. 10.1093/gbe/evaa096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfitzer E. (1871). Untersuchungen über Bau und Entwicklung der Bacillariaceen (Diatomaceen) (A. Marcus).
- 21.Drum R.W., and Pankratz S. (1965). Fine structure of an unusual cytoplasmic inclusion in the diatom genus,Rhopalodia. Protoplasma 60, 141–149. 10.1007/BF01248136. [DOI] [Google Scholar]
- 22.DeYoe H.R., Lowe R.L., and Marks J.C. (1992). Effects of Nitrogen and Phosphorus on the Endosymbiont Load of Rhopalodia Gibba and Epithemia Turgida (bacillariophyceae). J. Phycol. 28, 773–777. 10.1111/j.0022-3646.1992.00773.x. [DOI] [Google Scholar]
- 23.Prechtl J., Kneip C., Lockhart P., Wenderoth K., and Maier U.-G. (2004). Intracellular Spheroid Bodies of Rhopalodia gibba Have Nitrogen-Fixing Apparatus of Cyanobacterial Origin. Mol. Biol. Evol. 21, 1477–1481. 10.1093/molbev/msh086. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama T., Kamikawa R., Tanifuji G., Kashiyama Y., Ohkouchi N., Archibald J.M., and Inagaki Y. (2014). Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci. 111, 11407–11412. 10.1073/pnas.1405222111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moulin S.L.Y., Frail S., Braukmann T., Doenier J., Steele-Ogus M., Marks J.C., Mills M.M., and Yeh E. (2024). The endosymbiont of Epithemia clementina is specialized for nitrogen fixation within a photosynthetic eukaryote. ISME Commun. 4, ycae055. 10.1093/ismeco/ycae055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruck E.C., Nakov T., Alverson A.J., and Theriot E.C. (2016). Phylogeny, ecology, morphological evolution, and reclassification of the diatom orders Surirellales and Rhopalodiales. Mol. Phylogenet. Evol. 103, 155–171. 10.1016/j.ympev.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Schvarcz C.R., Wilson S.T., Caffin M., Stancheva R., Li Q., Turk-Kubo K.A., White A.E., Karl D.M., Zehr J.P., and Steward G.F. (2022). Overlooked and widespread pennate diatom-diazotroph symbioses in the sea. Nat. Commun. 13, 799. 10.1038/s41467-022-28065-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster R.A., and Zehr J.P. (2006). Characterization of diatom–cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ. Microbiol. 8, 1913–1925. 10.1111/j.1462-2920.2006.01068.x. [DOI] [PubMed] [Google Scholar]
- 29.Benson M.E., Kociolek P.J., Spaulding S.A., and Smith D.M. (2012). Pre-Neogene non-marine diatom biochronology with new data from the late Eocene Florissant Formation of Colorado, USA. Stratigraphy 9, 121–152. [Google Scholar]
- 30.Abresch H., Bell T., and Miller S.R. (2024). Diurnal transcriptional variation is reduced in a nitrogen-fixing diatom endosymbiont. ISME J. 18, wrae064. 10.1093/ismejo/wrae064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama T., and Inagaki Y. (2017). Genomic divergence within non-photosynthetic cyanobacterial endosymbionts in rhopalodiacean diatoms. Sci. Rep. 7, 13075. 10.1038/s41598-017-13578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muñoz-Marín M. del C., Shilova I.N., Shi T., Farnelid H., Cabello A.M., and Zehr J.P. (2019). The Transcriptional Cycle Is Suited to Daytime N2 Fixation in the Unicellular Cyanobacterium “Candidatus Atelocyanobacterium thalassa” (UCYN-A). mBio 10, 10.1128/mbio.02495-18. 10.1128/mbio.02495-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landa M., Turk-Kubo K.A., Cornejo-Castillo F.M., Henke B.A., and Zehr J.P. (2021). Critical Role of Light in the Growth and Activity of the Marine N2-Fixing UCYN-A Symbiosis. Front. Microbiol. 12. 10.3389/fmicb.2021.666739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamakura S., Mann D.G., Nakamura N., and Sato S. (2021). Inheritance of spheroid body and plastid in the raphid diatom Epithemia (Bacillariophyta) during sexual reproduction. Phycologia 60, 265–273. 10.1080/00318884.2021.1909399. [DOI] [Google Scholar]
- 35.Moulin S.L.Y., Frail S., Doenier J., Braukmann T., and Yeh E. (2023). The endosymbiont of Epithemia clementina is specialized for nitrogen fixation within a photosynthetic eukaryote. Preprint at bioRxiv, 10.1101/2023.03.08.531752 10.1101/2023.03.08.531752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki S., Kawachi M., Tsukakoshi C., Nakamura A., Hagino K., Inouye I., and Ishida K. (2021). Unstable Relationship Between Braarudosphaera bigelowii (= Chrysochromulina parkeae) and Its Nitrogen-Fixing Endosymbiont. Front. Plant Sci. 12. 10.3389/fpls.2021.749895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schvarcz C.R., Stancheva R., Turk-Kubo K.A., Wilson S.T., Zehr J.P., Edwards K.F., Steward G.F., Archibald J.M., Oatley G., Sinclair E., et al. (2024). The genome sequences of the marine diatom Epithemia pelagica strain UHM3201 (Schvarcz, Stancheva & Steward, 2022) and its nitrogen-fixing, endosymbiotic cyanobacterium. Wellcome Open Res. 9, 232. 10.12688/wellcomeopenres.21534.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S., Suleski M., Craig J.M., Kasprowicz A.E., Sanderford M., Li M., Stecher G., and Hedges S.B. (2022). TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 39, msac174. 10.1093/molbev/msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kooistra W.H.C.F., Gersonde R., Medlin L.K., and Mann D.G. (2007). The Origin and Evolution of the Diatoms: Their Adaptation to a Planktonic Existence. In Evolution of Primary Producers in the Sea, Falkowski P. G. and Knoll A. H., eds. (Academic Press; ), pp. 207–249. 10.1016/B978-012370518-1/50012-6. [DOI] [Google Scholar]
- 40.Bowler C., Allen A.E., Badger J.H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R.P., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 41.Vancaester E., Depuydt T., Osuna-Cruz C.M., and Vandepoele K. (2020). Comprehensive and Functional Analysis of Horizontal Gene Transfer Events in Diatoms. Mol. Biol. Evol. 37, 3243–3257. 10.1093/molbev/msaa182. [DOI] [PubMed] [Google Scholar]
- 42.Van Etten J., and Bhattacharya D. (2020). Horizontal Gene Transfer in Eukaryotes: Not if, but How Much? Trends Genet. 36, 915–925. 10.1016/j.tig.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Mitra R., Li X., Kapusta A., Mayhew D., Mitra R.D., Feschotte C., and Craig N.L. (2013). Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc. Natl. Acad. Sci. 110, 234–239. 10.1073/pnas.1217548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González-Pech R.A., Stephens T.G., Chen Y., Mohamed A.R., Cheng Y., Shah S., Dougan K.E., Fortuin M.D.A., Lagorce R., Burt D.W., et al. (2021). Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium. BMC Biol. 19, 73. 10.1186/s12915-021-00994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dougan K.E., Bellantuono A.J., Kahlke T., Abbriano R.M., Chen Y., Shah S., Granados-Cifuentes C., van Oppen M.J.H., Bhattacharya D., Suggett D.J., et al. (2024). Whole-genome duplication in an algal symbiont bolsters coral heat tolerance. Sci. Adv. 10, eadn2218. 10.1126/sciadv.adn2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryłka K., Alverson A.J., Pickering R.A., Richoz S., and Conley D.J. (2023). Uncertainties surrounding the oldest fossil record of diatoms. Sci. Rep. 13, 8047. 10.1038/s41598-023-35078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez J.V., Yuhki N., Masuda R., Modi W., and O’Brien S.J. (1994). Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J. Mol. Evol. 39, 174–190. 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- 48.Timmis J.N., Ayliffe M.A., Huang C.Y., and Martin W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 49.Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theissen U., and Martin W. (2006). The difference between organelles and endosymbionts. Curr. Biol. 16, R1016–R1017. 10.1016/j.cub.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 51.Nowack E.C.M. (2014). Paulinella chromatophora – rethinking the transition from endosymbiont to organelle. Acta Soc. Bot. Pol. 83, 387–397. 10.5586/asbp.2014.049. [DOI] [Google Scholar]
- 52.Keeling P.J. (2024). Horizontal gene transfer in eukaryotes: aligning theory with data. Nat. Rev. Genet. 25, 416–430. 10.1038/s41576-023-00688-5. [DOI] [PubMed] [Google Scholar]
- 53.Muller H.J. (1964). The relation of recombination to mutational advance. Mutat. Res. 106, 2–9. 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 54.Allen J.M., Light J.E., Perotti M.A., Braig H.R., and Reed D.L. (2009). Mutational Meltdown in Primary Endosymbionts: Selection Limits Muller’s Ratchet. PLOS ONE 4, e4969. 10.1371/journal.pone.0004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tyra H.M., Linka M., Weber A.P., and Bhattacharya D. (2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8, R212. 10.1186/gb-2007-8-10-r212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foster R.A., Kuypers M.M.M., Vagner T., Paerl R.W., Musat N., and Zehr J.P. (2011). Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J. 5, 1484–1493. 10.1038/ismej.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tschitschko B., Esti M., Philippi M., Kidane A.T., Littmann S., Kitzinger K., Speth D.R., Li S., Kraberg A., Tienken D., et al. (2024). Rhizobia–diatom symbiosis fixes missing nitrogen in the ocean. Nature 630, 899–904. 10.1038/s41586-024-07495-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie R.J. (2013). The ammonia transport, retention and futile cycling problem in cyanobacteria. Microb. Ecol. 65, 180–196. 10.1007/s00248-012-0111-1. [DOI] [PubMed] [Google Scholar]
- 59.Keeling P.J., and Archibald J.M. (2008). Organelle Evolution: What’s in a Name? Curr. Biol. 18, R345–R347. 10.1016/j.cub.2008.02.065. [DOI] [PubMed] [Google Scholar]
- 60.Liu F., Fernie A.R., and Zhang Y. (2024). Can a nitrogen-fixing organelle be engineered within plants? Trends Plant Sci. 10.1016/j.tplants.2024.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Elhai J. (2023). Engineering of crop plants to facilitate bottom-up innovation: A possible role for broad host-range nitroplasts and neoplasts. Preprint at OSF, 10.31219/osf.io/ny2rc 10.31219/osf.io/ny2rc. [DOI] [Google Scholar]
- 62.Bombar D., Heller P., Sanchez-Baracaldo P., Carter B.J., and Zehr J.P. (2014). Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. ISME J. 8, 2530–2542. 10.1038/ismej.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCutcheon J.P., and Moran N.A. (2012). Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26. 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 64.Stein-Taylor J.R. and Phycological Society of America (1973). Handbook of Phycological Methods: Culture methods and growth measurements (University Press Cambridge; [England: ]). [Google Scholar]
- 65.Workman Rachel, Timp Winston, Fedak Renee, Kilburn Duncan, Hao Stephanie, and Liu Kelvin (2018). High Molecular Weight DNA Extraction from Recalcitrant Plant Species for Third Generation Sequencing. Protoc. Exch. 10.1038/protex.2018.059. [DOI] [Google Scholar]
- 66.Ranallo-Benavidez T.R., Jaron K.S., and Schatz M.C. (2020). GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432. 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marçais G., and Kingsford C. (2011). A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770. 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Coster W., D’Hert S., Schultz D.T., Cruts M., and Van Broeckhoven C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrew S. (2019). Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 70.Hu J., Wang Z., Sun Z., Hu B., Ayoola A.O., Liang F., Li J., Sandoval J.R., Cooper D.N., Ye K., et al. (2024). NextDenovo: an efficient error correction and accurate assembly tool for noisy long reads. Genome Biol. 25, 107. 10.1186/s13059-024-03252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv. [Google Scholar]
- 73.Vaser R., Sović I., Nagarajan N., and Šikić M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zimin A.V., Puiu D., Luo M.-C., Zhu T., Koren S., Marçais G., Yorke J.A., Dvořák J., and Salzberg S.L. (2017). Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 27, 787–792. 10.1101/gr.213405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zimin A.V., and Salzberg S.L. (2020). The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLOS Comput. Biol. 16, e1007981. 10.1371/journal.pcbi.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laetsch D.R., and Blaxter M.L. (2017). BlobTools: Interrogation of genome assemblies. Preprint at F1000Research, 10.12688/f1000research.12232.1 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- 77.Mikheenko A., Prjibelski A., Saveliev V., Antipov D., and Gurevich A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150. 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rhie A., Walenz B.P., Koren S., and Phillippy A.M. (2020). Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 245. 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manni M., Berkeley M.R., Seppey M., Simão F.A., and Zdobnov E.M. (2021). BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 38, 4647–4654. 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., and Smit A.F. (2020). RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. 117, 9451–9457. 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T.L. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26. 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kriventseva E.V., Kuznetsov D., Tegenfeldt F., Manni M., Dias R., Simão F.A., and Zdobnov E.M. (2019). OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47, D807–D811. 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stanke M., Diekhans M., Baertsch R., and Haussler D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 85.Stanke M., Schöffmann O., Morgenstern B., and Waack S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62. 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gabriel L., Hoff K.J., Brůna T., Borodovsky M., and Stanke M. (2021). TSEBRA: transcript selector for BRAKER. BMC Bioinformatics 22, 566. 10.1186/s12859-021-04482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brůna T., Hoff K.J., Lomsadze A., Stanke M., and Borodovsky M. (2021). BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics Bioinforma. 3, lqaa108. 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brůna T., Lomsadze A., and Borodovsky M. (2020). GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics Bioinforma. 2, lqaa026. 10.1093/nargab/lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., and Borodovsky M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506. 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwata H., and Gotoh O. (2012). Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40, e161. 10.1093/nar/gks708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gotoh O., Morita M., and Nelson D.R. (2014). Assessment and refinement of eukaryotic gene structure prediction with gene-structure-aware multiple protein sequence alignment. BMC Bioinformatics 15, 189. 10.1186/1471-2105-15-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S., Zhou Y., Chen Y., and Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim D., Paggi J.M., Park C., Bennett C., and Salzberg S.L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pertea G., and Pertea M. (2020). GFF Utilities: GffRead and GffCompare. Preprint at F1000Research, 10.12688/f1000research.23297.1 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dainat J. (2022). AGAT: Another Gff Analysis Toolkit to handle annotations in any GTF/GFF format. Version v1.0.0 (Zenodo). 10.5281/zenodo.11106497 10.5281/zenodo.11106497. [DOI] [Google Scholar]
- 97.Tanaka T., Maeda Y., Veluchamy A., Tanaka M., Abida H., Maréchal E., Bowler C., Muto M., Sunaga Y., Tanaka M., et al. (2015). Oil Accumulation by the Oleaginous Diatom Fistulifera solaris as Revealed by the Genome and Transcriptome. Plant Cell 27, 162–176. 10.1105/tpc.114.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hongo Y., Kimura K., Takaki Y., Yoshida Y., Baba S., Kobayashi G., Nagasaki K., Hano T., and Tomaru Y. (2021). The genome of the diatom Chaetoceros tenuissimus carries an ancient integrated fragment of an extant virus. Sci. Rep. 11, 22877. 10.1038/s41598-021-00565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Osuna-Cruz C.M., Bilcke G., Vancaester E., De Decker S., Bones A.M., Winge P., Poulsen N., Bulankova P., Verhelst B., Audoor S., et al. (2020). The Seminavis robusta genome provides insights into the evolutionary adaptations of benthic diatoms. Nat. Commun. 11, 3320. 10.1038/s41467-020-17191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zepernick B.N., Truchon A.R., Gann E.R., and Wilhelm S.W. (2022). Draft Genome Sequence of the Freshwater Diatom Fragilaria crotonensis SAG 28.96. Microbiol. Resour. Announc. 11, e00289–22. 10.1128/mra.00289-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paajanen P., Strauss J., van Oosterhout C., McMullan M., Clark M.D., and Mock T. (2017). Building a locally diploid genome and transcriptome of the diatom Fragilariopsis cylindrus. Sci. Data 4, 170149. 10.1038/sdata.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Armbrust E.V., Berges J.A., Bowler C., Green B.R., Martinez D., Putnam N.H., Zhou S., Allen A.E., Apt K.E., Bechner M., et al. (2004). The Genome of the Diatom Thalassiosira Pseudonana: Ecology, Evolution, and Metabolism. Science 306, 79–86. 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 103.Roberts W.R., Downey K.M., Ruck E.C., Traller J.C., and Alverson A.J. (2020). Improved Reference Genome for Cyclotella cryptica CCMP332, a Model for Cell Wall Morphogenesis, Salinity Adaptation, and Lipid Production in Diatoms (Bacillariophyta). G3 GenesGenomesGenetics 10, 2965–2974. 10.1534/g3.120.401408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ferrante M.I., Broccoli A., and Montresor M. (2023). The pennate diatom Pseudo-nitzschia multistriata as a model for diatom life cycles, from the laboratory to the sea. J. Phycol. 59, 637–643. 10.1111/jpy.13342. [DOI] [PubMed] [Google Scholar]
- 105.Lommer M., Specht M., Roy A.-S., Kraemer L., Andreson R., Gutowska M.A., Wolf J., Bergner S.V., Schilhabel M.B., Klostermeier U.C., et al. (2012). Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 13, R66. 10.1186/gb-2012-13-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Emms D.M., and Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Conway J.R., Lex A., and Gehlenborg N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rohwer R.R., Hamilton J.J., Newton R.J., and McMahon K.D. (2018). TaxAss: Leveraging a Custom Freshwater Database Achieves Fine-Scale Taxonomic Resolution. mSphere 3, 10.1128/msphere.00327-18. 10.1128/msphere.00327-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsuji J., Frith M.C., Tomii K., and Horton P. (2012). Mammalian NUMT insertion is non-random. Nucleic Acids Res. 40, 9073–9088. 10.1093/nar/gks424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., and Zimin A. (2018). MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14, e1005944. 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quinlan A.R., and Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rice P.M., Bleasby A.J., and Ison J.C. EMBOSS User’s Guide: Practical Bioinformatics with EMBOSS (Cambridge University Press; ). [Google Scholar]
- 113.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gu Z., Gu L., Eils R., Schlesner M., and Brors B. (2014). circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 115.Lhee D., Lee J., Ettahi K., Cho C.H., Ha J.-S., Chan Y.-F., Zelzion U., Stephens T.G., Price D.C., Gabr A., et al. (2020). Amoeba Genome Reveals Dominant Host Contribution to Plastid Endosymbiosis. Mol. Biol. Evol. 38, 344–357. 10.1093/molbev/msaa206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Buchfink B., Reuter K., and Drost H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Katoh K., and Standley D.M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Capella-Gutiérrez S., Silla-Martínez J.M., and Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., and Lanfear R. (2020). IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., and Jermiin L.S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., and Vinh L.S. (2018). UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 35, 518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Price M.N., Dehal P.S., and Arkin A.P. (2010). FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE 5, e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stephens T.G., Bhattacharya D., Ragan M.A., and Chan C.X. (2016). PhySortR: a fast, flexible tool for sorting phylogenetic trees in R. PeerJ 4, e2038. 10.7717/peerj.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rancurel C., Legrand L., and Danchin E.G.J. (2017). Alienness: Rapid Detection of Candidate Horizontal Gene Transfers across the Tree of Life. Genes 8, 248. 10.3390/genes8100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tyanova S., Temu T., and Cox J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 126.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. NCBI Accessions and access links for all species used in this study
Table S2. RepeatMasker output tables for Epithemia
Table S3. Gene codes and putative identity of Epithemia-specific horizontally transferred genes.
Table S4. Processed and unprocessed proteomics data
Data S1. Integrative genomics viewer screenshots of genomic context, expression, and read support of all NUDTs