Abstract
tmRNA acts to rescue stalled bacterial ribosomes while encoding a peptide tag added trans-translationally to the nascent peptide, targeting it for proteolysis. The permuted gene structure found in a group of cyanobacteria is shown to produce a two-piece mature tmRNA, as had been observed previously for the independently permuted gene of α-proteobacteria. The pieces have been mapped onto the gene sequence and aligned for the permuted cyanobacterial tmRNA sequences, including four novel sequences. Structural probing and base pair co-variations support a secondary structure model in which two pairings in the tRNA-like domain hold the two pieces together, and the coding piece bearing the tag reading frame additionally contains a single transient pseudoknot and three other stem–loops. This represents a dramatic reduction in pseudoknot number from the five present in one-piece cyanobacterial tmRNA.
INTRODUCTION
With both tRNA- and mRNA-like properties, transfer-message RNA (tm, SsrA or 10Sa RNA) solves problems that arise when bacterial ribosomes stall during translation (1,2). It is alanylated at the CCA tail of its tRNA-like domain prior to entering the stalled ribosome. That alanyl moiety is incorporated into the nascent protein, then translation resumes on a reading frame within the tmRNA and the ribosome is freed at the stop codon. The tmRNA-encoded peptide serves as a tag that promotes degradation of the tagged protein. tmRNA interacts specifically with several macromolecules, including SmpB (small protein B) (3), EF-Tu (4), ribosomal protein S1 (5), phosphoribosyl pyrophosphate synthase (6), RNase R and YfbG (7), and with the tRNA that decodes the resume codon (8).
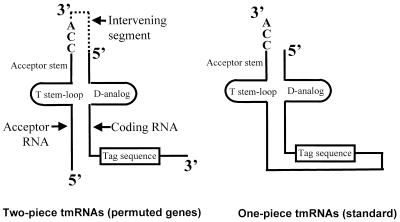
Standard tmRNA is found as a single RNA chain (Fig. 1, right). Although much longer than tRNA, it is likewise processed from a precursor containing extra RNA at either end, and probably by the same suite of processing enzymes (9–11). The tmRNA gene had been difficult to find in the α-proteobacteria (12,13) until it was realized that the gene is in a circularly permuted form, i.e. the segment normally at the 3′ end of tmRNA genes is instead found upstream of the segment normally at the 5′ end (14). Permutation of the tmRNA precursor places the two sites of tRNA-like processing at the borders of an internal intervening segment. Correspondingly, mature tmRNA was found in two pieces in an α-proteobacterium, with loss of the intervening segment (Fig. 1, left).
Figure 1.
Structural relationship between one-piece and two-piece tmRNAs. In the two-piece tmRNA the structure contains two distinct RNA molecules, the acceptor RNA and the coding RNA (14); the processed portion of the precursor is denoted by dotted lines.
Permuted tmRNA genes have also been found in a subgroup of cyanobacteria; the gene permutation event of the cyanobacteria was apparently independent of that of the α-proteobacteria. Here we show that mature tmRNA is found in two pieces in one of these cyanobacteria. In standard tmRNA, the tag reading frame is flanked by multiple pseudoknots in a looped RNA domain that emerges from the tRNA-like domain. It has been hypothesized that topological problems arise from the unusual situation that occurs with standard tmRNA, i.e. translating a looped mRNA; strain may develop in the loop during tag translation, but could be relieved by temporary opening of one of the pseudoknots (14). The two-piece construction breaks the reading frame loop, in both α-proteobacteria and cyanobacteria, offering a second alternative to the hypothetical problem. This might predict a concomitant loss of pseudoknots in the two-piece tmRNAs. We present structural probing data and phylogenetic analyses that produce a secondary structure model containing only one pseudoknot that is only partially stable in solution, as compared with five pseudoknots in one-piece tmRNAs from cyanobacteria.
MATERIALS AND METHODS
Bacterial growth
Synechococcus PCC6307 stock was purchased from the American Type Culture Collection and Synechococcus PCC7009 and PCC6904 stocks from the Pasteur Culture Collection. They were cultured in BG-11 medium under green light with CO2 bubbling, with the kind help of David Kehoe (Indiana University).
RNAs for mapping
Total RNA was prepared from Synechococcus PCC6307 cells (330 mg wet wt) using TRI reagent (Molecular Research Center), with a yield of 122 µg. Marker RNA (226-nt 3′-elongated version of the PCC6307 tmRNA coding piece) was transcribed with T7 RNA polymerase from a template DNA prepared by PCR from a plasmid containing the PCC6307 tmRNA gene.
Northern blot analysis
Total PCC6307 RNA (2.6 or 6.6 µg) or a 226-nt in vitro transcript RNA (2.8 pmol) was separated by electrophoresis in an 8% polyacrylamide–urea gel. A nylon membrane was blotted with the separated RNAs using a semi-dry blotter in 0.5× TBE and pre-hybridized in 4× SSC, 0.2× Denhardt’s, 1% SDS, 0.1 mg/ml salmon sperm DNA at 50°C. Either 5′-32P-end-labeled TGTCCGAGACACTGGTGTGG (acceptor probe) or GGAGAAACGAACGATGTTGT (coding probe) was added for hybridization at 50°C for 16 h. Blots were washed in 0.5× SSC and autoradiographed. Sizes of hybridizing RNAs were estimated using the dye bromophenol blue (whose mobility in this gel is equivalent to that of a 72-nt RNA) and the 226-nt transcript as standards.
Primer extension assay
5′-32P-labeled acceptor probe was annealed to 0.26 µg total PCC6307 RNA or to 0.8 pmol PCR product from the cloned PCC6307 tmRNA gene, by heating to 95°C and slow cooling. Reverse transcription of RNA was performed at 42°C for 50 min, then 70°C for 10 min. Dideoxynucleotide sequencing reactions were performed with the DNA template. An autoradiogram was taken after electrophoresis in a 12% polyacrylamide urea gel.
New sequences
tmRNA sequences from Prochlorococcus marinus MED4 and Synechococcus PCC6307 (originally reported under the name Cyanobium gracile) have been previously published (14). Sequences from Synechococcus WH8102 and P.marinus MIT9313 were identified in genomic data from the DOE Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html). Genomic DNAs were prepared from Synechococcus PCC7009 and PCC6904 by French pressure cell lysis and used for PCR with 5′-ACTAGGTGGTCCACACC and 5′-GGAAGGGCCCTTCCAGG, and the products sequenced directly after removing excess primers. Sequences were aligned manually. New sequences have been deposited in GenBank (accession numbers AY082654 and AY082655).
Probing reagents
All synthetic DNA oligonucleotides were synthesized by Genset (Paris, France). T7 RNA polymerase was prepared according to Wyatt et al. (15). The restriction enzyme EarI, alkaline phosphatase and T4 polynucleotide kinase were from New England Biolabs (Beverly, MA). AMV reverse transcriptase and T4 RNA ligase were from Gibco BRL Life Technologies (Cergy-Pontoise, France). RNases S1, V1, U2 and T1 were from Amersham-Pharmacia-Biotech (Orsay, France). Dimethylsulfate and N-cyclohexyl-N′-[2-(N-methyl-morpholino)-ethyl]-carbo-diimide-4-toluene sulfonate (CMCT) were from Aldrich (St Quentin-Fallavier, France). [γ-32P]ATP (3000 mCi/mmol) and [α-32P]pCp (3000 mCi/mmol) were from NEN (Paris, France).
RNAs for probing
DNA encoding a 260-nt P.marinus MED4 permuted pre-tmRNA was amplified by PCR from genomic DNA generously provided by Gabrielle Rocap and Penny Chisholm and cloned under T7 promoter control into a plasmid (p188A), verifying the expected sequence. The plasmid was linearized with EarI restriction nuclease before transcription, so that the RNA would end with its natural 3′-terminal sequence. In vitro transcription was performed as described (16), with electrophoresis on a denaturing gel to separate the transcribed RNA from unincorporated nucleotides and DNA fragments. The appropriate band was electroeluted and the RNA recovered by ethanol precipitation. Labeling at the 5′ end of the RNA was performed with [γ-32P]ATP and T4 polynucleotide kinase after dephosphorylation with alkaline phosphatase (17). Labeling at the 3′ end was carried out by ligation of [γ-32P]pCp using T4 RNA ligase. After labeling, the tmRNA was gel purified, eluted passively and ethanol precipitated.
Enzymatic digestions and chemical modifications
Enzymatic digestions and chemical modifications were performed on both 3′- and 5′-labeled tmRNA (30 000 c.p.m./reaction), supplemented with 1 µg total rRNA. Labeled RNA was heated to 75°C for 3 min and slowly cooled to room temperature. Enzymatic digestions (V1 at 2.5 × 10–6 U, S1 at 3 U, T1 at 0.1 U and U2 at 0.1 U) and chemical modifications (lead acetate at 1.3 mM and imidazole at 100 mM) of either 5′- or 3′-labeled RNA was as described (18). Cleavage and modification sites were detected by gel electrophoresis by direct identification with the statistical cleavage patterns of the RNA itself. Modification of N-3 atoms of cytosines and N-7 atoms of guanines by DMS or of N-1 atoms of adenosines and N-3 atoms of uridines by CMCT was done at 37°C as described (18). RNAs were ethanol precipitated, dried and counted and the RNA fragments separated by electrophoresis. The data were analyzed on a phosphorimager. The background present in control lanes was subtracted, but the quantitation of each fragment was scored manually.
RESULTS
Two-piece tmRNA from a permuted cyanobacterial gene
The permuted gene of α-proteobacteria produces mature tmRNA in two pieces; that containing the CCA tail is termed the acceptor piece and that containing the tag reading frame is termed the coding piece. To determine whether the permuted tmRNA gene of a subgroup of cyanobacteria likewise produces a two-piece tmRNA, a northern blot of total Synechococcus PCC6307 RNA was probed separately for the two suspected pieces (Fig. 2). A probe for the suspected acceptor piece of tmRNA hybridized specifically to a single PCC6307 RNA species; by comparing its migration to that of standards, its size was estimated at 57 nt. A probe for the suspected coding piece also hybridized specifically to a single PCC6307 RNA species, clearly distinct from the RNA revealed by the other probe; its size was estimated at 198 nt. The discrepancy in signal intensity for the two RNAs could be a simple artifact of unequal efficiency of the two probes or, conceivably, indicates that the acceptor piece is more abundant in the cell than the coding piece. We would expect a 1:1 stoichiometry of the two pieces in the active tmRNA (see below) and also that either free RNA piece would be rapidly degraded, but features of the free acceptor piece such as alanylation might protect it from degradation.
Figure 2.
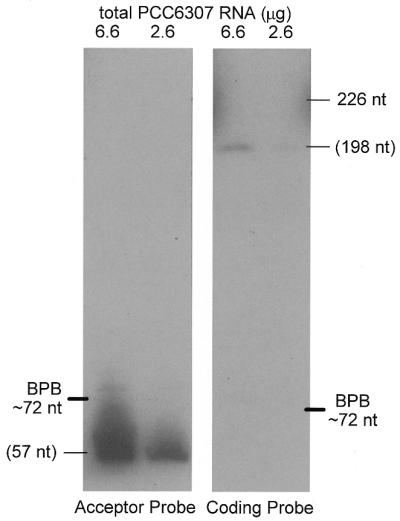
Two tmRNA pieces. Northern blot analysis of PCC6307 RNA using separate probes for the two tmRNA pieces. Sizes in parentheses were estimated by calibration with standards.
The sequences at the 3′ end of the acceptor piece and the 5′ end of the coding piece, which combine to form the tRNA-like domain, can be predicted with confidence by reference to other tmRNAs and to tRNAAla. These end sequences together with the size estimates for the two tmRNA pieces and the gene sequence from PCC6307 allow prediction of the sequence of each tmRNA piece. Primer extension with the acceptor probe was used to map directly the 5′ end of the acceptor piece RNA and thereby assess the accuracy of the size estimate from northern analysis. The 5′ end of the acceptor piece mapped precisely to the adenosine predicted from the size estimate (Fig. 3). This inspires confidence in the accuracy of the estimate for the coding piece, although with its larger size, an error of ±4 nt would not be unreasonable.
Figure 3.
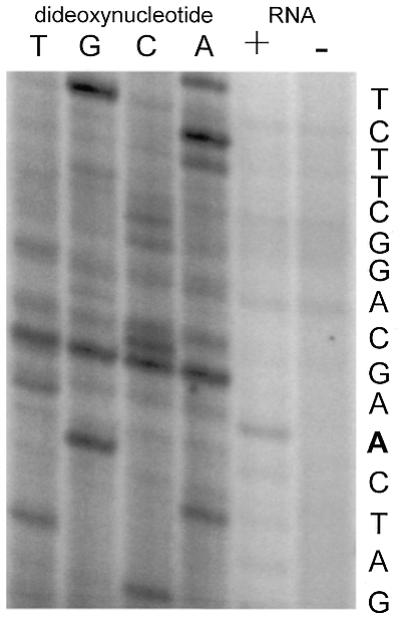
Determination of the acceptor piece 5′ end. Primer extension in the presence, but not absence, of total PCC6307 RNA produced a product that maps the 5′ end of tmRNA acceptor piece RNA to the adenosine highlighted in the gene sequence at right.
Phylogenetic analysis of two-piece cyanobacterial tmRNA
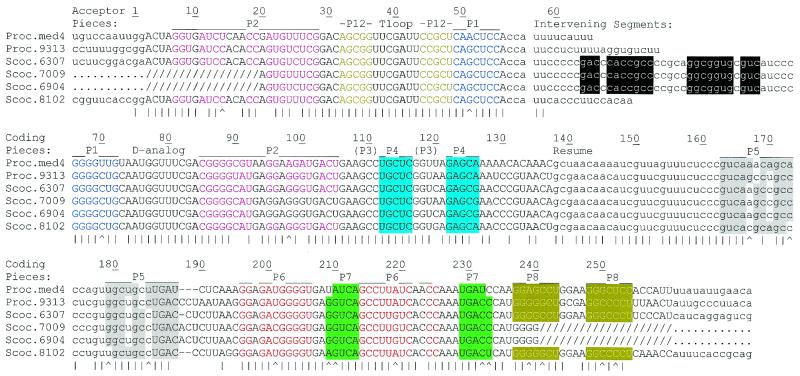
Previously, permuted tmRNA genes from only two cyanobacteria were known. This number has now increased to six; two more have been found in genome project databases and we generated two partial sequences by PCR from genomic DNAs. Based on our results for PCC6307 tmRNA we can expect that the permuted tmRNA genes of other cyanobacteria are in two pieces and we can predict the sequences of the pieces. For the predicted pieces, sequences from all six species are readily aligned, with a gap required at only one site (Fig. 4). The tag reading frame was readily identified in each coding piece, conforming to consensus features (19). Sequence variation is low: 188 of 252 mature positions (75%) are identical among the set (asterisks in Fig. 4). The sequences from PCC6307, PCC7009 and PCC6904 form an especially tight cluster. Homology among permuted cyanobacterial tmRNA genes decays at the flanks of the processed sequences and in the intervening segment.
Figure 4.
Alignment of two-piece cyanobacterial tmRNAs. Intervening and 5′ and 3′ flanking segments, tag reading frames and the CCA tails expected to be added after transcription are in lower case. Slashes show segments of unverified sequence where PCR was primed. Vertical lines mark positions conserved among all sequences and carats mark positions where proposed pairings (color coding) exhibit Watson–Crick base pair co-variation. Pairings are numbered according to an earlier proposal (14), but modified to reflect probing data (see below) that revealed extensions of P5 and P6 and that P3 does not form. Pm, P.marinus; Sc, Synechococcus. Numbering is for the pre-tmRNA transcript used for probing experiments.
A tentative secondary structure model had been proposed previously for the two-piece cyanobacterial tmRNA (14), guided by the structure of standard tmRNA. Despite the low sequence variation in the dataset, six of the nine pairings in that model receive support from positions with Watson–Crick base pair co-variation: one such position occurs in each of P1, P2 and P6 and two occur in each of P5, P7 and P8 (shown in Fig. 4 with extensions to P5 and P6 that were revealed by probing, as below). P1 and P2 hold the two tmRNA pieces together and P6 and P7 form a pseudoknot (PK2) downstream of the reading frame. Another pseudoknot upstream of the reading frame had been proposed in the earlier model, corresponding to the PK1 found in other tmRNAs and shown to be important in Escherichia coli (20). Neither of the stems proposed for PK1 (P3 and P4) receive phylogenetic support. P3 would be especially weak, usually comprising only 3 bp, one of which is a G:U pair, and without the benefit of stabilization by coaxial stacking on P2 that is possible for typical tmRNAs. In fact, the probing data show that P3, and therefore PK1, does not form in solution (see below).
Structural analysis of a permuted tmRNA precursor
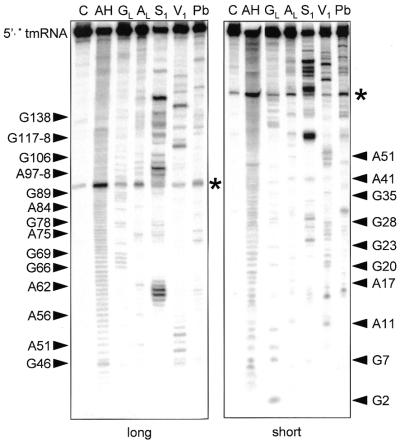
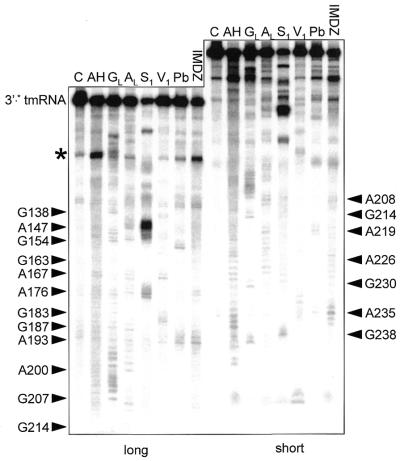
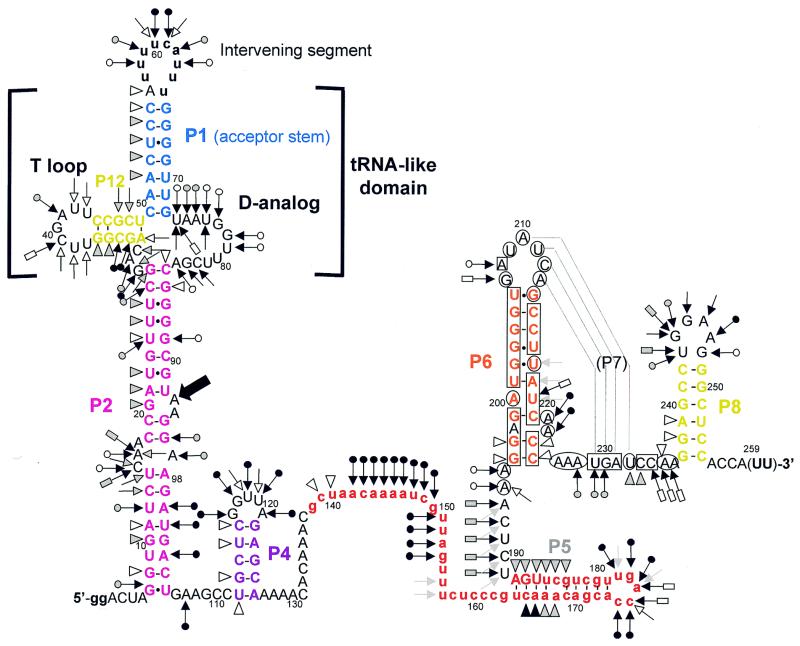
Sequence variation is too low to fully establish the secondary structure of the cyanobacterial two-piece tmRNA. We therefore employed structural probing, an approach that was instrumental in establishing the secondary structure model for standard tmRNA (18,21). For this study we reasoned that the one-piece permuted precursor form would be more manageable, yet still model well the relevant features of the two-piece mature form; the two forms differ only in whether the ends of the acceptor stem are free or connected by a short loop. A transcript of the P.marinus MED4 pre-tmRNA was end-labeled and its solution conformation was probed with chemicals and enzymes. RNase V1 cleaves double-stranded (ds)RNA or stacked nucleotides, while nuclease S1 and imidazole cleave single-stranded (ss)RNA. Lead acetate also cleaves ssRNA, but with sensitivity to subtle conformational changes of the RNA chain. The reactivity towards these probes was monitored for each nucleotide of the 258-nt-long synthetic pre-tmRNA. Nine independent experiments were performed (Figs 5 and 6 are representative). These data are summarized in Figure 7 on a secondary structure model that they, together with the phylogenetic analysis, support.
Figure 5.
Structural probing of the 5′ half of a two-piece tmRNA precursor transcript. Autoradiogram of cleavage products of 5′-labeled RNA by imidazole, lead and nucleases S1 and V1. Lanes C, incubation controls; lanes GL, RNase T1 hydrolysis ladder; lanes AL, RNase U2 hydrolysis ladder; lanes AH, alkaline hydrolysis ladder. The sequence is indexed at the sides. Asterisks show a strong spontaneous degradation site.
Figure 6.
Structural probing of the 3′-half. Autoradiogram of cleavage products of 3′-labeled RNA with indications as in Figure 5.
Figure 7.
Secondary structure model of the P.marinus MED4 pre-tmRNA, based on probing and phylogenetic data, showing the probing results. Triangles are V1 cuts; arrows capped by a circle are S1 cuts; uncapped arrows are lead acetate cuts; arrows capped by a rectangle are imidazole-induced cleavages. Intensities of cuts and cleavages are proportional to the darkness of the symbols: open, stippled and filled for weak, medium and strong cuts, respectively. A consistently observed degradation site (Fig. 5) is indicated by a bold arrow. Circled nucleotides are accessible to DMS or CMCT, squared nucleotides are protected. Structural domains P1, P2, P6, P8 and P12 are color coded as in Figure 4. The 3′-terminal nucleotides in parentheses are not present in the synthetic, but the precise length and 3′-terminal sequence of the coding piece is unknown. Lower case represents 5′ nucleotides added to improve in vitro transcription, the intervening segment and the tag reading frame (in red).
Double-strand-specific cuts from A51 to C55 suggest that the acceptor stem P1 forms in solution. The 5′ strand of P1 was susceptible to RNase V1, whereas its 3′ side was not, perhaps because of the local environment of P1 within the RNA structure. The intervening segment was heavily cleaved by probes specific for ssRNA. A base pair between A56 and U65 is possible (V1 cut at A56, lack of single-strand-specific cuts at U65), which would vanish when the intervening segment was excised. Lead, imidazole and nuclease S1 cleavages are consistent with both the D-analog region and the 7-nt T-loop being mostly single stranded in solution. Parts of P12, the equivalent of the T-stem, are folded (V1 cuts at G35 and G36), but surprisingly two lead cleavages and two nuclease S1 cuts were observed there, suggesting that pairs in P12 are disrupted. Between P2 and P12 the three joining nucleotides G29–C31 are apparently highly exposed within the folded RNA; they were heavily cleaved by nuclease S1 and lead.
Several RNase V1 cuts from G8 to U26 support the long stem P2. However C15–A17 and A97 were cut by lead, imidazole and S1, suggesting that P2 is disrupted by a 4-nt internal bulge. U12–U14 were also cleaved by single-strand-specific probes, probably because of the nearby C15–A17 bulge. U24 in P2 was cleaved by both single- and double-strand-specific probes, suggesting that parts of P2 breathe in solution, as in one-piece E.coli tmRNA (18). Strong degradation was consistently observed at the bond between U92 and A93; unusual instability of the UA dinucleotide has been noted in other RNAs (22). The V1 cut at G20 suggests that this nucleotide is stacked between its neighbors, as for G10, which may also hydrogen bond with G102. Parts of P2 (C27–G28 and A100–A103) within the permuted pre-tmRNA are unstable and their conformation is poorly defined in solution, as for one-piece tmRNAs (18).
In one-piece tmRNA from E.coli, P2 is followed by a functional pseudoknot (PK1; 20) made of two stems, P3 and P4; correlates of these stems had been proposed for the permuted cyanobacterial tmRNA (14). Our data support P4, in that U112–G113 and U115–C116 are cleaved by the double-strand-specific probe V1. However, probing does not support P3, which also has no phylogenetic support; instead, the heavy S1 cleavage of positions that would form P3 (A107 and G117–U120) suggests that P4 is an isolated stem–loop. Note that this failure to detect P3 occurred in its most favorable case; it would be 1 bp longer in P.marinus MED4 than for any of the other known two-piece tmRNA sequences.
The 36-nt stretch downstream of P4, containing the resume codon, shows many single-strand-specific cleavages. A cluster of double-strand-specific cuts at C165–A168 and G183–A188 supports the existence of helix P5, in a longer form than had been previously noticed. Nucleotides C174–U178 were cleaved by single-strand-specific probes, suggesting that P5 is capped by a 5-nt loop. The U189–A195 segment was cut by single-strand-specific probes.
Between G196 and A236, where phylogenetic analysis suggests the pseudoknot PK2, there were very few cuts by V1, S1 or lead. To increase the coverage of this region, DMS and CMCT were used as probes, detecting modification by reverse transcription with a labeled DNA primer complementary to the 3′ end of the RNA. Reverse transcription of the RNA from its 3′ end paused at positions C174–A176. Together these reagents modify accessible Watson–Crick faces of all four bases, A and C by DMS, and G and U by CMCT. Several segments through this region were modified by neither DMS nor CMCT, consistent with the presence of P6. RNase V1 cleavage and protection from DMS provide evidence for an extension of P6 by two pairs, G196:C225 and G197:C224, that had not been detected by phylogenetic analysis since it is separated from the rest of P6 and does not vary. Within P6, A222 and A223 appear to bulge, since they were cleaved by S1 and modified by DMS. Susceptibility of G207–A213 to single-strand-specific reagents suggests that it exists as a 7-nt loop at the end of P6. However, this loop was not as heavily cleaved by single-strand-specific probes as were the other loops in the RNA (at the intervening segment, the D-analog and capping P4 and P5). Phylogenetic analysis predicted that part of this loop (A210–A213) would form P7 by pairing with U229–U232, a segment whose conformation is ambiguous: for example, U229 and G230 were cut by S1 yet not modified by CMCT, and U232 was cut by V1 yet modified by CMCT. The light susceptibility of A210–A213 to single-strand-specific probes and the conformational ambiguity of U229–U232 may indicate that P7 forms only transiently in the presence of 20 mM MgCl2, such that the probing data reflect a mixture of two conformations in solution. It should be noted that P7 would be 1 bp shorter in P.marinus MED4 than in any of the other two-piece tmRNAs. The instability of PK2 may not be surprising considering how short (3 nt) the strand crossing the minor groove of P6 would be; the bulge at the end of P6 may be important in allowing P7 to form.
At the 3′ end of the RNA, two V1 cuts were observed at G238 and A239, supporting P8. U243–G248 were cleaved by single-strand-specific probes, suggesting that they form a 6-nt loop capping P8.
DISCUSSION
Combining RNA structural mapping with phylogenetic sequence comparison is a powerful approach to RNA structure prediction. It proved helpful in elucidating the secondary structure of one-piece tmRNAs when only a few sequences were available (18). Our structural model differs in some respects from the standard model for one-piece tmRNA, especially within the coding piece. It should be noted that the structural analysis was performed on the precursor RNA and not on the mature RNA, hence, P1 and P2, which hold the two RNA pieces together, might be less stable when the intervening segment is removed. The secondary structure of the tRNA-like domain is remarkably conserved in both standard and permuted tmRNAs, but significant changes are observed in the RNA portions containing the tag reading frame; in the permuted pre-RNA from cyanobacteria the coding piece contains four stem–loops beyond P2, one of which can form a transient pseudoknot.
The most striking features of the two-piece cyanobacterial tmRNAs are the general loss of pseudoknots and the opening of the loop containing the reading frame. Loop opening may correct a topological problem of one-piece tmRNAs encountered during the unusual translation of a looped mRNA domain (14). One-piece tmRNAs may ameliorate this problem through the potential structural plasticity offered by their conserved pseudoknots. The value of these pseudoknots was questioned when in vitro activity was still observed when pseudoknots PK2–PK4 of E.coli tmRNA were either exchanged or substituted with ssRNAs (23); however, more aggressive deletion of two or more pseudoknots was not attempted. According to the hypothesis, both α-proteobacterial and cyanobacterial two-piece tmRNAs solve the topological problem by a similar strategy, by breaking the mRNA loop, therefore obviating the utility of pseudoknots. The dramatic reduction in pseudoknot number in the two-piece cyanobacterial tmRNA, from the five that occur in one-piece relatives (12) to one, supports this view. Another unusual feature is that the stop codon is inserted into the P5 helix, in contrast to E.coli tmRNA (18,24) and most of the known tmRNA gene sequences (25), where it occurs in the loop capping P5. A third unusual feature of the two-piece RNA from cyanobacteria is the susceptibility of P12 (the T-stem) to single-strand-specific probes.
Loss of the pseudoknot (PK1) upstream of the tag is a surprising feature of P.marinus MED4 tmRNA, because PK1 has been shown to be essential for E.coli tmRNA function in vitro (20). If general among the two-piece cyanobacterial tmRNAs, they may compensate in some way for the loss. However, as there has been no functional assessment of tmRNA activity for these cyanobacteria, it is conceivable that they simply suffer with less active tmRNA. Alternatively, PK1 may form in vivo in these mostly marine bacteria due to intracellular conditions of temperature, salt or cofactors that were not reproduced in our experimental conditions. Likewise PK2 could be more stable in vivo than the structural probing implies.
The vast majority of the tmRNA gene sequences exhibit strict conservation of two G residues within their D analogs that can potentially interact with the T loop, like the canonical tRNA tertiary interactions G18–ψ55 and G19–C56. The only known exceptions are from the two-piece tmRNA of the α-proteobacterium Caulobacter crescentus and a homolog from the Reclinomonas americana mitochondrion (14), suggesting that they have lost the capacity to form the putative interaction between their D and T analogs. Although no structural probing data are available for tmRNAs of the α-proteobacterial lineage, unusual sequence features have been noted in their tRNA-like domains, especially in their T loops. Thus, for unknown reasons, the tRNA-like domain in both lineages of two-piece tmRNA might generally be less constrained than that of one-piece tmRNAs.
Processing of the internal loop of permuted pre-tmRNA to form the tRNA-like ends need not differ from processing of one-piece tmRNA or tRNA. However, two-piece tmRNA could have as many as two additional processing steps, which occur at the 5′ and 3′ flanks of the precursor through unknown mechanisms. Alternatively, the ends in question could be the unprocessed start and termination sites of transcription. In α-proteobacteria a candidate –10 promoter hexamer is always found 6 or 7 nt upstream of the mature 5′ end, suggesting that the precursor 5′ end remains unprocessed; similar conservation is not found upstream of the permuted cyanobacterial genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Séan Turner (Indiana University) for sharing unpublished phylogenetic data that led to the choice of species for tmRNA sequencing, David Kehoe for help with growing cyanobacteria and Gabrielle Rocap and Penny Chisholm for the gift of P.marinus MED4 genomic DNA. This work was supported by a Human Frontier Science Program research grant (RG0291/2000-M100), a research grant entitled ‘Recherche Fondamentale en Microbiologie et Maladies Infectieuses’ (Institut Pasteur) and an ‘Action Concertée Incitative Jeunes Chercheurs 2000’ from the French Ministry of Research to B.F. and by National Institutes of Health grant GM59881 to K.P.W.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +33 2 23 23 48 51; Fax: +33 2 23 23 44 56; Email: brice.felden@univ-rennes1.fr AY082654 and AY082655
REFERENCES
- 1.Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- 2.Gillet R. and Felden,B. (2001) Emerging views on tmRNA mediated protein tagging and ribosome rescue. Mol. Microbiol., 42, 879–885. [DOI] [PubMed] [Google Scholar]
- 3.Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudinger-Thirion J., Giege,R. and Felden,B. (1999) Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA, 5, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wower I.K., Zwieb,C.W., Guven,S.A. and Wower,J. (2000) Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J., 19, 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ando H., Kitabatake,M. and Inokuchi,H. (1996) 10Sa RNA complements the temperature-sensitive phenotype caused by a mutation in the phosphoribosyl pyrophosphate synthetase (prs) gene in Escherichia coli. Genes Genet. Syst., 71, 47–50. [DOI] [PubMed] [Google Scholar]
- 7.Karzai A.W. and Sauer,R.T. (2001) Protein factors associated with the Ssra-SmpB tagging and ribosome rescue complex. Proc. Natl Acad. Sci. USA, 98, 3040–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillet R. and Felden,B. (2001) Transfer RNAAla recognizes transfer-messenger RNA with specificity; a functional complex prior to entering the ribosome? EMBO J., 20, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin-Chao S., Chia-Li,W. and Yi-Tzu,L. (1999) RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl Acad. Sci. USA, 96, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Pandit,S. and Deutscher,M.P. (1998) 3′Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komine Y., Kitabatake,M., Yokogawa,T., Nishikawa,K. and Inokuchi,H. (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams K.P. (1998) The tmRNA website. Nucleic Acid Struct., 26, 16163–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felden B., Gesteland,R.F. and Atkins,J.F. (1999) Eubacterial tmRNAs: everywhere except the alpha-proteobacteria? Biochim. Biophys. Acta, 1446, 145–148. [DOI] [PubMed] [Google Scholar]
- 14.Keiler K.C., Shapiro,L. and Williams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl Acad. Sci. USA, 37, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt J.R., Chastain,M. and Puglisi,J.D. (1991) Synthesis and purification of large amounts of RNA oligonucleotides. BioTechniques, 11, 764–769. [PubMed] [Google Scholar]
- 16.Felden B., Florentz,C., Giege,R. and Westhof,E. (1994) Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol., 235, 508–531. [DOI] [PubMed] [Google Scholar]
- 17.Silberklang M., Gillum,A.M. and RajBhandary,U.L. (1977) The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res., 4, 4091–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felden B., Himeno,H., Muto,A., McCutcheon,J.P., Atkins,J.F. and Gesteland,R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–104. [PMC free article] [PubMed] [Google Scholar]
- 19.Williams K.P., Martindale,K.A. and Bartel,D.P. (1999) Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J., 18, 5423–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nameki N., Felden,B., Atkins,J.F., Gesteland,R.F., Himeno,H. and Muto,A. (1999) Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J. Mol. Biol., 286, 733–744. [DOI] [PubMed] [Google Scholar]
- 21.Hickerson R.P., Watkins-Sims,C.D., Burrows,C.J., Atkins,J.F., Gesteland,R.F. and Felden,B. (1998) A nickel complex cleaves uridine in folded RNA structures: application to E. coli tmRNA and related engineered molecules. J. Mol. Biol., 279, 577–587. [DOI] [PubMed] [Google Scholar]
- 22.Kierzek R. (1992) Nonenzymatic hydrolysis of oligoribonucleotides. Nucleic Acids Res., 20, 5079–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nameki N., Tadaki,T., Himeno,H. and Muto,A. (2000) Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett., 470, 345–349. [DOI] [PubMed] [Google Scholar]
- 24.Williams K.P. and Bartel,D.P. (1996) Phylogenetic analysis of tmRNA secondary structure. RNA, 2, 1306. [PMC free article] [PubMed] [Google Scholar]
- 25.Williams K.P. (2002) The tmRNA website: invasion by an intron. Nucleic Acids Res., 30, 179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]