Abstract
Regulatory T cells (Tregs) are crucial immune cells for tissue repair and regeneration. However, their potential as a cell-based regenerative therapy is not yet fully understood. Here, we show that local delivery of exogenous Tregs into injured mouse bone, muscle, and skin greatly enhances tissue healing. Mechanistically, exogenous Tregs rapidly adopt an injury-specific phenotype in response to the damaged tissue microenvironment, upregulating genes involved in immunomodulation and tissue healing. We demonstrate that exogenous Tregs exert their regenerative effect by directly and indirectly modulating monocytes/macrophages (Mo/MΦ) in injured tissues, promoting their switch to an anti-inflammatory and pro-healing state via factors such as interleukin (IL)-10. Validating the key role of IL-10 in exogenous Treg-mediated repair and regeneration, the pro-healing capacity of these cells is lost when Il10 is knocked out. Additionally, exogenous Tregs reduce neutrophil and cytotoxic T cell accumulation and IFN-γ production in damaged tissues, further dampening the pro-inflammatory Mo/MΦ phenotype. Highlighting the potential of this approach, we demonstrate that allogeneic and human Tregs also promote tissue healing. Together, this study establishes exogenous Tregs as a possible universal cell-based therapy for regenerative medicine and provides key mechanistic insights that could be harnessed to develop immune cell-based therapies to enhance tissue healing.
Subject terms: Regenerative medicine, Regulatory T cells, Monocytes and macrophages, Interleukins
Regulatory T cells (Tregs) are known for suppressing inflammatory processes, but their full capacity for tissue regeneration is yet to be harnessed. Here, the authors demonstrate the efficiency of Tregs in facilitating tissue healing in mouse models of bone, muscle, and skin injury, with monocytes/macrophages and interleukin-10 playing a key mechanistic role in the process.
Introduction
To create effective regenerative medicine therapies, it is critical to capitalise on the main players that regulate tissue repair and regeneration. Thus, given that numerous immune cell types orchestrate all stages of the tissue healing process following injury, manipulating the immune system to enhance tissue healing holds remarkable potential1. While immune cells initially drive a pro-inflammatory response, the transition into an anti-inflammatory and pro-healing immune phase is essential for tissue restoration2. Indeed, impairment of this transition, characterised by prolonged and sustained inflammation, disrupts tissue healing and leads to poor functional recovery1,2. To date, regenerative approaches aimed at modulating the immune system have predominantly focussed on utilising cytokines, biomaterials, or stem/progenitor cells, with immunomodulatory capabilities3. While these strategies have shown some promising results in various regenerative medicine contexts4, they often exert their effects via the modulation of endogenous immune cells. Therefore, a cell-based approach consisting of administering immune cells with pro-regenerative abilities could prove effective, especially when the delivered cell type is a master regulator of both the immune system and tissue healing. In this regard, regulatory T cells (Tregs) are very attractive candidates, due to their ability to modulate other immune cells and concurrently stimulate tissue healing via the secretion of effector molecules or through cell–cell interactions5–7. Indeed, endogenous Tregs that accumulate within injured tissues have been shown to play a key role in facilitating repair or regeneration in multiple tissues and organs, such as muscle8,9, skin10, heart11,12, central nervous system13, and lung14. Moreover, systemic Treg administration in mice has been shown to improve cardiac repair post-myocardial infarction15,16 and bone remodelling in osteogenesis imperfecta17. Thus, we investigated the potential of using local Treg administration into injured tissue as a regenerative medicine approach. Specifically, we selected acute injury models in mouse bone, muscle, and skin, which represent diverse tissue healing scenarios. We show that administration of exogenous Tregs into injured tissue greatly enhances tissue healing. The delivered Tregs rapidly adopt an injury-specific phenotype and express factors like interleukin (IL)-10 that trigger monocytes/macrophages (Mo/MΦ) to transition into an anti-inflammatory and pro-healing state. In addition, Tregs reduce neutrophil and cytotoxic T cell accumulation as well as IFN-γ production which facilitates the Mo/MΦ phenotypic switch. This study identifies exogenous Tregs as a potential universal cell-based therapy for tissue regeneration and reveals important mechanisms that could be leveraged to develop regenerative therapies based on immune cell delivery.
Results
Local Treg delivery facilitates bone, muscle, and skin tissue healing
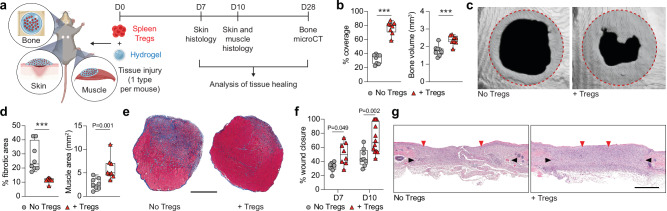
To investigate the extent to which local Treg administration promoted tissue healing, we chose three acute injury models: critical-size cranial defects18 for bone regeneration, volumetric muscle loss19 for muscle regeneration, and full-thickness dorsal excisional skin wounds19 for skin repair. We first verified the critical importance of Tregs in these tissue healing models, using the Foxp3DTR/GFP mouse. In this mouse line, the diphtheria toxin receptor (DTR) is expressed under the control of the Foxp3 promoter, enabling inducible Treg depletion upon diphtheria toxin (DT) administration20. Foxp3DTR/GFP and wildtype C57BL6/J control mice with bone, muscle, or skin injuries were treated with DT, leading to effective ablation of Tregs in Foxp3DTR/GFP mice (Supplementary Fig. 1a–f). Treg absence resulted in a significant impairment of tissue healing across all three tissues. This was evident through reduced defect coverage and bone volume in cranial injuries, diminished muscle regeneration accompanied by increased muscle fibrosis in volumetric muscle loss defects and compromised wound closure in full-thickness dorsal skin wounds (Supplementary Fig. 1g–l). We next investigated whether tissue healing could, conversely, be enhanced by rapidly augmenting Treg numbers via their local delivery into injured tissues of wildtype mice. For a more robust and reliable evaluation of tissue healing in wildtype mice, we increased the severity of the volumetric muscle loss injury to ensure that defects would not fully regenerate without therapeutic intervention21 and further delayed skin wound closure by applying splinting rings to limit wound contraction22. For bone, the cranial defect model was already severe enough to prevent regeneration without intervention18. Tregs were sorted from spleen (Supplementary Fig. 2a), and 2 × 105 cells were administrated locally to freshly performed injuries via a fibrin hydrogel directly polymerised in tissue defects (Fig. 1a). Fibrin hydrogels provided a suitable delivery system as fibrin is highly biocompatible and is commonly used for cell delivery in these models18,19. Remarkably, we found that local Treg delivery significantly improved bone, muscle, and skin tissue healing, compared to the control group of fibrin hydrogel without Tregs. This was demonstrated by enhanced cranial defect coverage and bone volume (Fig. 1b, c), reduced fibrotic area, increased muscle tissue and increased muscle fibre size (Fig. 1d, e; Supplementary Fig. 3a), and accelerated skin wound closure (Fig. 1f, g). In contrast, treatment with CD4+ conventional T cells (Tconvs), which were sorted as CD4+GFP− T cells from Foxp3DTR/GFP mice (Supplementary Fig. 2a) and delivered in the same manner, failed to demonstrate any significant improvement in tissue healing for all three injury types (Supplementary Figs. 3b, 4). Thus, our results provided strong evidence for the therapeutic potential of local Treg administration to promote healing of multiple tissue types.
Fig. 1. Local delivery of exogenous Tregs promotes healing of injured mouse tissues.
a Critical-size cranial defects, quadriceps volumetric muscle loss defect or full-thickness dorsal skin wounds were performed in wildtype C57BL6/J mice and treated with a fibrin hydrogel only, or hydrogel containing exogenous spleen Tregs. Tissue healing was assessed at different time points for each tissue. b Bone regeneration evaluated by microCT analysis of cranial defects expressed as defect coverage and new bone volume at D28 post-injury (n = 8 defects). c Representative cranial reconstructions. The original defect area is shaded with a dashed red outline. d Muscle regeneration represented by the percentage of fibrotic area and muscle area measured by histomorphometric analysis of tissue sections at D10 post-injury (n = 8 defects). e Representative muscle histology of a transverse section of the rectus femoris stained with Masson’s trichrome at D10 post-injury. Muscle tissue is stained in red, and the fibrotic area is in blue. Scale bar = 1 mm. f Percentage wound closure at D7 and D10 post-injury evaluated by histomorphometric analysis of tissue sections (n = 8 wounds for D7, n = 8 wounds for D10 ‘No Tregs’ and n = 12 wounds for D10 ‘+ Tregs’). g Representative histology of skin tissue stained with haematoxylin and eosin at D10 post-injury. Black arrows indicate wound edges and red arrows indicate tips of epithelium tongue. The epithelium (if any) is stained in purple, underneath which the granulation tissue is stained in pink–violet, with dark purple granulocyte nuclei. Scale bar = 1 mm. Data are plotted in box plots showing the median (central line) and IQR (bounds) with whiskers extending to the minimum and maximum values. Two-tailed unpaired Student’s t-test was used in (b), two-tailed Mann–Whitney U test for non-parametric data was used in (d). Two-way ANOVA with Bonferroni post hoc test was used in (f) for multiple comparisons. P values are indicated; ***P ≤ 0.001. a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Delivered Tregs adopt an injury-specific transcriptomic profile
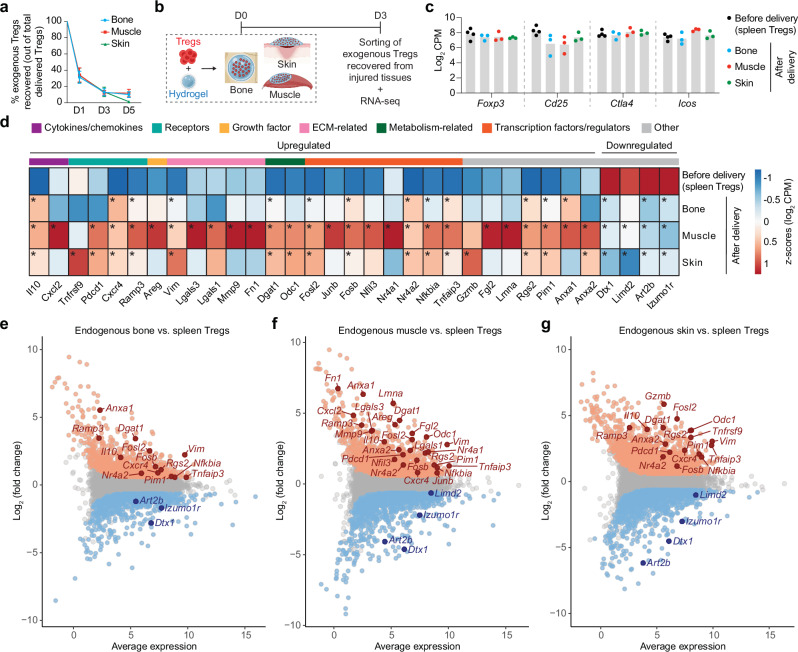
To understand the mechanisms behind exogenous Treg-mediated tissue healing, we evaluated the local retention and fate of the administered Tregs. To accurately detect exogenous Tregs post-delivery, we used Tregs from Foxp3IRES-mRFP mice (Supplementary Fig. 2b). These mice express the bright red fluorescent protein (RFP), facilitating detection using flow cytometry (Supplementary Fig. 5a). First, we found that RFP+ Tregs could be clearly detected for up to 5 days post-delivery in all three tissues, with a retention of around 30% after 1 day, followed by a gradual decline (Fig. 2a; Supplementary Fig. 5b, c). Importantly, no RFP+ Tregs were detected in the surrounding lymph nodes or spleen (Supplementary Fig. 5d).
Fig. 2. Exogenous Tregs delivered locally in injured tissues adopt an expression profile similar to endogenous Tregs.
Critical-size cranial defects, quadriceps volumetric muscle loss defect or full-thickness dorsal skin wounds were performed in wildtype C57BL6/J mice, and treated with a fibrin hydrogel only, or hydrogel containing exogenous spleen Tregs. a Percentage of delivered (exogenous) Tregs remaining in injured bone, muscle, and skin, on D1, D3, and D5 post-delivery (data are mean ± SD, n = 3 mice/time point for bone, n = 5 mice/time point for muscle and skin). b On D3 post-delivery, exogenous Tregs were sorted from injured tissues for RNA sequencing. c Log2 CPM values of genes classically expressed by Tregs in exogenous spleen Tregs before delivery (n = 4 mice) and exogenous Tregs recovered from injured tissues at D3 post-delivery (n = 3 mice/tissue). d Heat map depicting standardised expression values of selected differentially expressed genes (DEGs) in exogenous Tregs before delivery (n = 4 mice) and exogenous Tregs recovered from injured tissues at D3 post-delivery (n = 3 mice/tissue; average of replicates is shown). Genes marked with * are significantly up- or downregulated. Colour key above the heat map denotes the functional category of the genes. Endogenous Tregs were sorted from injured bone, muscle, and skin, on D7 post-injury for RNA sequencing. MA plots depict selected DEGs (FDR adjusted p value < 0.05) in endogenous tissue Tregs sorted from injured bone (e), muscle (f), and skin (g), compared to spleen Tregs sorted from uninjured (healthy) mice (n = 3 replicates/tissue, each comes from a pool of >2 mice). Genes marked in orange and blue represent significantly up- and downregulated genes respectively (FDR adjusted p value < 0.05). DEGs common between D7 endogenous Tregs and D3 exogenous Tregs from (d) are labelled on the MA plot for each tissue. b Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Next, we sought to explore the molecular mechanisms by which exogenous Tregs promoted tissue healing. Since previous studies have revealed a distinctive transcriptomic profile of endogenous Tregs accumulating within injured tissues8,12, we investigated the transcriptional changes occurring within the exogenous Tregs in our three injury models. To achieve this, we performed mini-bulk RNA-seq on the recovered RFP+ Tregs sorted from injured tissues at 3 days post-delivery and compared them to the sorted RFP+ spleen Tregs before delivery (Fig. 2b). RNA-seq analysis of these recovered exogenous Tregs showed that they retained expression of classic Treg markers such as Foxp3, Cd25, Ctla4 and Icos, confirming their Treg identity (Fig. 2c). Furthermore, analysis of differentially expressed genes (DEGs) (FDR < 0.05, fold change > |1.5|) in exogenous Tregs recovered at D3, compared to the Tregs before delivery, showed an upregulation of genes known to be involved in immunomodulation and in tissue healing (Supplementary Fig. 6a–c). Comparing the overlap of the upregulated and downregulated DEGs between all three tissues, it was observed that several genes were shared between bone, muscle, and skin (32 commonly upregulated and 161 commonly downregulated), while several others were unique to each tissue (Supplementary Fig. 6d). Among the commonly upregulated DEGs was the cytokine Il1023, which stood out as a key anti-inflammatory factor, known to be involved in macrophage modulation (Fig. 2d; Supplementary Fig. 6e). Additionally, we observed an upregulation of other anti-inflammatory mediators (e.g. Anxa124, Anxa225, Lgals126, Lgals327), elements of the TNFRSF-NF-kB signalling axis28 including Tnfrsf9, Nfkbia, Tnfaip3, and Pim1, and modulators of G-protein-coupled receptor signalling such as Rgs2 and Ramp326. Several genes that are characteristic of an effector Treg phenotype26 (e.g. Pdcd1, Gzmb, Fgl2), or are indicative of early non-lymphoid tissue (NLT) Treg cell adaptation (e.g. Junb29, Nr4a129, Nr4a230, Nfil326, and Dgat126) also showed increased expression in recovered Tregs, similar to other studies that have described NLT-specific adaptations of Tregs compared to their lymphoid counterparts26,29. Other genes upregulated in recovered Tregs included the growth factor amphiregulin (Areg) that plays an important role in improving muscle regeneration in vivo8, the transcription factor Fosl2, which has been reported to support wound healing31, as well as chemokine receptors (e.g. Cxcr426). Moreover, recovered Tregs showed a significant downregulation of some genes (Fig. 2d) like Izumo1r, which is known to be upregulated in Tregs from lymphoid organs but is downregulated in tissue Tregs32, as well as Dtx1 and Art2b, which are reportedly downregulated upon T cell activation33,34.
Since the transcriptional changes observed in exogenous Tregs recovered from all three injured tissues suggested the acquisition of an effector-like phenotype, we decided to compare them to endogenous Tregs that naturally accumulate in sufficient numbers within 1-week post-injury in these tissues (Supplementary Fig. 7). Thus, we performed RNA-seq on Tregs isolated from these injured tissues at 7 days post-injury and compared them to spleen Tregs from uninjured mice (Supplementary Fig. 8a–c). Remarkably, the genes that were previously found to be differentially expressed in recovered exogenous Tregs, were also significantly up or downregulated (FDR < 0.05, fold change > |1.5|) in endogenous injured tissue Tregs when compared to healthy spleen Tregs (Fig. 2e–g).
To further explore the similarities between exogenous and endogenous Tregs, and to gain insights into the pathways by which exogenous Tregs likely promote tissue healing, we performed gene ontology (GO) analysis of the significantly upregulated DEGs (FDR < 0.05, fold change > |1.5|) that overlapped between exogenous and endogenous Tregs. This analysis revealed an enrichment of terms associated with tissue repair and regeneration, encompassing activities like cell morphogenesis, epithelial cell differentiation, muscle cell proliferation, and wound healing. Remarkably, immunomodulatory processes, including macrophage activation and mononuclear cell proliferation, were also enriched in the upregulated genes (Supplementary Fig. 8d). Indeed, some of these genes included factors that are well-known to promote macrophage polarisation from a pro-inflammatory to an anti-inflammatory and pro-healing state such as Il1023 and others including Anxa124, Fgl2 35, Lgals127, and Lgals327. The upregulation of some of these Treg-derived factors was also confirmed by RT-qPCR analysis of expression in endogenous and exogenous Tregs from muscle injury as an example, compared to healthy spleen Tregs (Supplementary Fig. 9). Altogether, we observed that exogenous Tregs adopted an “injury-specific” expression profile that was similar to endogenous Tregs, with exogenous Tregs likely modulating endogenous macrophages. Consequently, we decided to investigate the contribution of macrophages in exogenous Treg-mediated healing of bone, muscle, and skin injuries.
Delivered Tregs exert their pro-healing effects via macrophage modulation
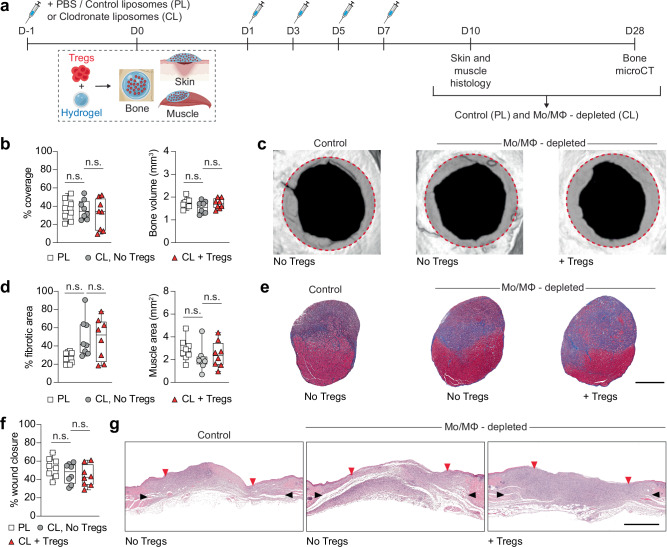
To determine the extent to which the therapeutic effect of Treg administration was dependent on macrophages, we tested the regenerative efficacy of exogenous Tregs in mice depleted of macrophages. We used clodronate liposomes (CL) to deplete Mo/MΦ, as this method is widely employed to transiently induce selective apoptosis in these cells36. Compared to mice that received control (PBS) liposomes, administration of CL led to >90% reduction in Mo/MΦ accumulating in injured tissues and spleen by D4 post-injury (Supplementary Fig. 10), without any significant increase in the expression of pro-inflammatory cytokines on D2 and D4 post-CL injection (Supplementary Fig. 11). Thus, Mo/MΦ were depleted via CL administration for a week and tissue healing outcomes were assessed post-Treg delivery (Fig. 3a). Strikingly, Mo/MΦ depletion abolished the therapeutic effect of Tregs, as demonstrated by the lack of significant improvement in bone, muscle, or skin healing when compared with control groups without exogenous Tregs (Fig. 3b–g; Supplementary Fig. 3c). This indicated that the Treg therapy was dependent on Mo/MΦ and suggested that exogenous Tregs exerted their pro-healing effects via Mo/MΦ, either directly or indirectly.
Fig. 3. Treg-mediated tissue healing depends on Mo/MΦ.
Wildtype C57BL6/J mice were depleted of Mo/MΦ using clodronate liposomes (CL), compared to control liposomes (PBS liposomes, PL), and tissue healing was assessed for critical-size cranial defects, quadriceps volumetric muscle loss defect or full-thickness dorsal skin wounds in response to Treg delivery. a Schematic of the macrophage depletion experiment. b–c Cranial regeneration assessed by microCT. Defect coverage and new bone volume at D28 post-injury in (b) (n = 8 defects). Representative cranial reconstructions in (c). The original defect area is shaded with a dashed red outline. d–e Muscle regeneration evaluated by histomorphometric analysis of tissue sections. Fibrotic area and muscle area at D10 post-injury in (d) (n = 8 defects). Representative histology of a transverse section of the rectus femoris stained with Masson’s trichrome at D10 post-injury in (e). Muscle tissue is stained in red, and the fibrotic area is in blue. Scale bar = 1 mm. f–g Skin wound closure measured by histomorphometric analysis of tissue sections. Wound closure at D10 post-injury in (f) (n = 8 wounds). Representative histology of haematoxylin and eosin staining at D10 post-injury in (g). Black arrows indicate wound edges and red arrows indicate tips of epithelium tongue. Scale bar = 1 mm. Data are plotted in box plots showing the median (central line) and IQR (bounds) with whiskers extending to the minimum and maximum values. One-way ANOVA with Bonferroni post hoc test was used in (b, d, f) for multiple comparisons; n.s.: non-significant. (PL: PBS/control liposomes, CL: clodronate liposomes, Mo/MΦ: monocytes/macrophages). a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
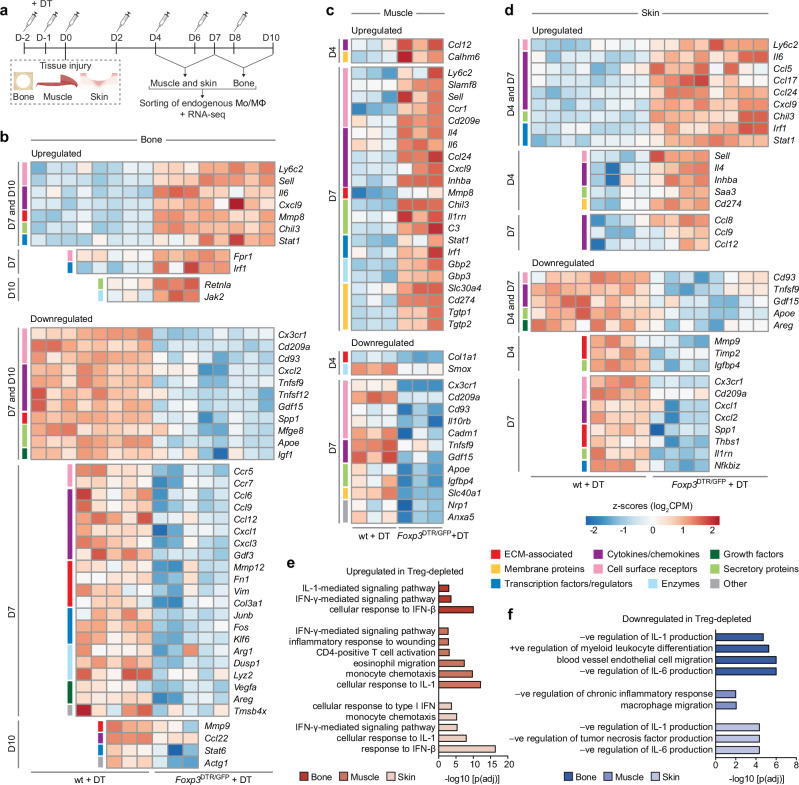
To further elucidate the mechanisms involved in exogenous Treg-mediated tissue healing, we sorted Mo/MΦ from Treg-treated tissues and controls on D4 and D7 post-injury (Supplementary Fig. 12). Although the total number of Mo/MΦ was not significantly different upon Treg treatment in all tissues (Supplementary Fig. 13a), we observed differences in the expression profile of the Mo/MΦ marker F4/80. Indeed, sterile inflammation is known to be accompanied by a rapid infiltration of pro-inflammatory Ly6C+/high, F4/80low Mo/MΦ into injured tissues, which gradually transition into an anti-inflammatory Ly6C−/low, F4/80high subset around 1-week post-injury37. Interestingly, we found that by D7 post-injury, Treg-treated tissues had a significantly lower proportion of the F4/80low subset (Supplementary Fig. 13b), which suggested an expedited transition towards an anti-inflammatory and pro-healing phase.
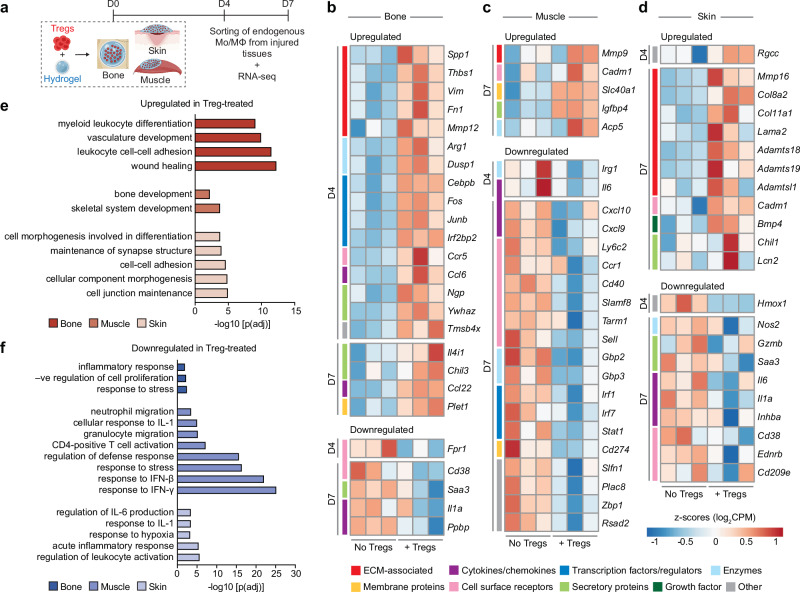
Next, we conducted RNA-seq analysis on Mo/MΦ at D4 and D7 post-injury to further explore their dynamic states (Fig. 4a). We found that the Mo/MΦ from bone, muscle, and skin displayed changes in different genes, with minimal overlap of DEGs between the three tissues, and across both time points (Supplementary Fig. 14). This was unlike our observations with Tregs, which had greater similarities between the three tissues, and is likely due to considerable plasticity in Mo/MΦ leading to greater tissue-specific diversity38. Nonetheless, the DEGs across all three tissues suggested the same functional trend. Focussing first on upregulated pro-healing DEGs (FDR < 0.05, fold change > |1.5|), Mo/MΦ from Treg-treated tissues significantly upregulated key anti-inflammatory genes, such as Arg139, Dusp140, Tmsb4x41, and Chil339 in bone (Fig. 4b), Slc40142 in muscle (Fig. 4c), and Chil143 in skin (Fig. 4d). Several genes linked to the ECM, known to be involved in minimising fibrosis and dampening inflammation, were also upregulated, such as Spp139, Thbs141, and Mmp1244,45 in bone (Fig. 4b), Mmp946 in muscle (Fig. 4c) and Adamtsl1 and Mmp1647 in skin (Fig. 4d). Furthermore, other factors that are expressed during anti-inflammatory “M2-like” macrophage polarisation were also upregulated, including transcription factors/regulators such as Cebpb48 and Irf2bp249, chemokines and their receptors such as Ccl650, Ccl2239 and Ccr551, secretory molecules such as Ngp52, Ywhaz53 and Il4i154 (Fig. 4b)43 and cell cycle regulator Rgcc55 (Fig. 4d).
Fig. 4. Mo/MΦ display a pro-healing transcriptomic profile upon Treg delivery.
a Wildtype C57BL6/J mice with bone, muscle or skin injuries were treated with fibrin hydrogel only or hydrogel containing exogenous Tregs. b–f Endogenous Mo/MΦ from injured tissues were sorted for RNA sequencing on D4 and D7 post-injury. Heat maps depicting standardised gene expression values of selected significantly upregulated and downregulated genes (FDR adjusted p value < 0.05) in Mo/MΦ from Treg-treated bone (b), muscle (c), and skin (d) injuries, compared to untreated controls (n = 3 mice/tissue per time point; individual replicates are shown). Colour key denotes the functional category of the genes. Gene ontology terms depicting enriched biological processes in significantly upregulated (e) and downregulated (f) genes in Treg-treated tissues, from both D4 and D7 macrophages combined (FDR < 0.01, adjusted by Benjamini–Hochberg correction). (Mo/MΦ: monocytes/ macrophages). a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Importantly, the upregulation of pro-healing and anti-inflammatory genes in the Treg-treated tissues was accompanied by the downregulation of genes associated with pro-inflammatory, “M1-like”, macrophages. This included receptors such as Ly6c2, Fpr1, Cd38, Cd40 and Sell56,57, secretory molecules such as Saa339, Gzmb58, and Irg159, and cytokine/chemokine genes such as Il1a, Il6, Cxcl9, Cxcl10 and Inhba39,56 (Fig. 4b–d), as well as pro-inflammatory interferon (IFN)-γ–stimulated genes, such as Stat160, Gbp361, Irf160, Cd27460 and Rsad256 in muscle, and Nos256 in skin (Fig. 4c, d).
We then carried out functional enrichment analysis for each tissue using the differentially expressed Mo/MΦ genes (FDR < 0.05, fold change > |1.5|) combined from both time points (Fig. 4e, f). Enriched GO biological processes within the upregulated genes unveiled activities linked to a pro-healing phenotype, such as myeloid differentiation, wound healing, bone development, and morphogenesis (Fig. 4e). Correspondingly, downregulated genes showed an enrichment in GO biological processes associated with a pro-inflammatory phenotype, including inflammatory response, granulocyte migration, and cellular response to IFN-γ and interleukin (IL)-1 (Fig. 4f). The genes that contributed to these biological processes were visualised in gene network plots with each node representing a GO term (Supplementary Fig. 15).
Treg-treated mice show contrasting Mo/MΦ phenotypes to Treg-depleted mice
To gain deeper mechanistic insights into how treatment with exogenous Tregs modulated Mo/MΦ phenotypes, we conducted RNA-seq analysis on Mo/MΦ isolated from Treg-depleted mice, as Treg ablation mimics the opposite scenario to Treg treatment. Mo/MΦ were collected at two time points post-injury, based on the peak of Mo/MΦ accumulation in each tissue upon Treg ablation (D7 and D10 for bone, and D4 and D7 for muscle and skin) (Fig. 5a). We first observed that the total number of Mo/MΦ was significantly higher in Treg-depleted tissues at all time points tested (Supplementary Fig. 16a). Second, Mo/MΦ had an F4/80 expression profile that was opposite to that seen with Treg-treated tissues. The F4/80low subset, which typically represents pro-inflammatory Ly6Chigh Mo/MΦ37 in the initial healing phase, was significantly higher upon Treg depletion at both time points post-injury (Supplementary Fig. 16b). Concurrent with this finding, RNA-seq analysis of Mo/MΦ in Treg-depleted mice showed higher Ly6c expression and decreased Cx3cr1 expression across all three tissues, indicating an increased presence of the pro-inflammatory Ly6Chigh, CX3CR1low subset39 in the absence of Tregs (Fig. 5b–d).
Fig. 5. Mo/MΦ from Treg-depleted mice show an increased pro-inflammatory transcriptional signature.
a Wildtype (wt) C57BL6/J and Foxp3DTR/GFP mice were treated with diphtheria toxin (DT) to deplete Tregs in Foxp3DTR/GFP mice, and injuries were performed in bone (critical-size cranial defects), muscle (quadriceps volumetric muscle loss defect) or skin (full-thickness dorsal skin wounds). b–f Mo/MΦ were sorted from the injured tissues at two different time points per tissue for RNA sequencing. Heat maps depicting standardised gene expression values of selected significantly upregulated and downregulated DEGs (FDR adjusted p value < 0.05) in Mo/MΦ from Treg-depleted bone (b), muscle (c) and skin (d) injuries, compared to the wildtype controls, including DEGs that were both shared and unique, across both time points per tissue (n = 3–5 mice/tissue per time point; individual replicates are shown). Colour key on the left of the heat map denotes the functional category of the genes. Gene ontology terms depicting enriched biological processes in the significantly upregulated (e) and downregulated (f) genes in the Treg-depleted tissues, from both time points combined (FDR < 0.01, adjusted by Benjamini–Hochberg correction). (Mo/MΦ: monocytes/macrophages). a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Further supporting this, we found that the gene expression profile of Mo/MΦ from Treg-depleted mice were inversely correlated to that observed with Treg-treated mice (Fig. 5b–d). For instance, some pro-repair genes previously observed to be upregulated in Treg-treated mice were downregulated in Treg-depleted mice, such as Spp1, Thbs1, Mmp9, Slc40a1, and Tmsb4x. Similarly, several downregulated pro-inflammatory genes in Treg-treated mice were upregulated in Treg-depleted mice such as Ly6c2, Fpr1, Il6, Sell, Inhba, and various IFN-γ response genes like Stat1, Gbp3, Irf1, and Cd274, among others (Fig. 5b–d). The contrasting effect in Mo/MΦ inflammatory states between Treg-depleted and Treg-treated mice was also evident in the GO analyses (Fig. 5e–f; Supplementary Fig. 17), as the terms cellular response to IFN-γ, IFN-β, and IL-1 were enriched in the upregulated Mo/MΦ genes in the absence of Tregs in all tissues (Fig. 5e), while they were amongst the downregulated biological processes following Treg treatment. Moreover, downregulated genes in Treg-depleted mice exhibited enrichment in processes such as positive regulation of myeloid leucocyte differentiation (Fig. 5f), indicating a decrease in Mo/MΦ differentiation or polarisation, an effect that was again opposite to that observed in Treg-treated mice.
Delivered Tregs modulate other pro-inflammatory immune cells
To gain a more comprehensive understanding of the immune microenvironment after Treg administration, we assessed neutrophil, CD4+ T cell, and CD8+ T cell accumulation kinetics in Treg-treated tissues and controls using flow cytometry (Supplementary Figs. 18, 19a). Interestingly, all Treg-treated tissues showed a significantly lower number of neutrophils 3 days post-injury (Supplementary Fig. 19b). Regarding T cells, although CD4+ T cell levels were similar between Treg-treated and control conditions (Supplementary Fig. 19c), the number of CD8+ T cells was significantly decreased upon Treg treatment in all tissues (Supplementary Fig. 19d). Since we found that Treg treatment dampened CD8+ T cell accumulation, but the regenerative activity of the exogenous Tregs was ultimately dependent on macrophages, we investigated whether injured tissue CD8+ T cells directly influenced Mo/MΦ, using cells from muscle injury as a representative example. Mo/MΦ sorted from injured muscle, co-cultured with decreasing concentrations of CD8+ T cells sorted from the same tissue, displayed a steady reduction in the proportion of pro-inflammatory Ly6Chigh Mo/MΦ (Supplementary Fig. 20).
Additionally, T cells, along with natural killer (NK) cells, have previously been shown to produce increased levels of the pro-inflammatory cytokine IFN-γ upon Treg depletion10,60. Thus, we investigated whether this was impacted upon Treg administration in all three injury models. Indeed, we found that Treg delivery led to a reduction in the percentage and number of IFN-γ-producing T cells and NK cells in the injured tissues (Supplementary Fig. 21), which, in turn, is known to affect the composition and phenotype of Mo/MΦ during tissue healing10,60.
The therapeutic effect of exogenous Tregs depends on IL-10 expression
RNA-seq analysis showed that exogenous Tregs reduced Mo/MΦ response to IFN-γ and IL-1, which are well-known to induce a pro-inflammatory phenotype in Mo/MΦ56. Thus, we tested whether some of the factors expressed by exogenous Tregs could directly inhibit the response of Mo/MΦ to these pro-inflammatory cytokines in vitro. For this, we first used an in vitro culture system that mimicked differentiating monocytes within the injured tissue microenvironment, in the presence of pro-inflammatory cytokines (Supplementary Fig. 22a). We selected factors that have previously been shown to affect Mo/MΦ phenotype, such as IL-1023, Areg62, annexin A1 (ANXA1)24, annexin A2 (ANXA2)25, galectin-1 (Gal-1)26, galectin-3 (Gal-3)27 and fibrinogen-like protein 2 (FGL2)35, and assessed their ability to influence Mo/MΦ Ly6C expression. Interestingly, IL-10 was the only factor that led to a significant reduction in the proportion of Ly6C+ Mo/MΦ, while none of the other factors demonstrated this effect (Supplementary Fig. 22b). Next, we tested whether IL-10 could demonstrate a similar effect on Mo/MΦ sorted from injured tissues, and cultured ex vivo. As a model, we cultured Mo/MΦ sorted from injured muscle with or without pro-inflammatory cytokines IFN-γ and IL-1β, in the presence or absence of IL-10 (Supplementary Fig. 22c). We found that treatment with IL-10 strongly reduced the proportion of Ly6Chigh cells in injured tissue Mo/MΦ, with or without exposure to pro-inflammatory cytokines (Supplementary Fig. 22d). This is consistent with previous research in which IL-10 is known to limit inflammatory Mo/MΦ activation23. Thus, we reasoned that this cytokine likely plays an important role in mediating the immunomodulatory effect of exogenous Tregs on Mo/MΦ.
We further validated the increased expression of IL-10 in exogenous Tregs recovered on D3 post-delivery, compared to Tregs before delivery, using flow cytometry. Exogenous Tregs recovered from all three injured tissues showed a significantly higher expression of IL-10, which was not observed in exogenous Tregs recovered from uninjured tissue, on D3 post-delivery (Supplementary Fig. 23). This finding demonstrated that the increased IL-10 expression was truly reflective of exogenous Treg adaptation to the damaged microenvironment, rather than a mere response to a non-lymphoid tissue environment. It also suggested that IL-10 could be one of the main factors secreted by exogenous Tregs, responsible for inhibiting IFN-γ and IL-1–induced responses in injured tissue Mo/MΦ. Consequently, we decided to specifically investigate the extent to which exogenous Tregs relied on IL-10 expression to promote tissue healing. To evaluate this, we sorted CD25+ Tregs from Il10 knockout (KO) mice (Supplementary Fig. 2c) and delivered them locally into injured bone, muscle, and skin tissues via fibrin gels (Fig. 6a). Strikingly, we observed that Il10-deficient Tregs could not promote a significant improvement in any of the tissue healing parameters tested in bone, muscle, and skin injuries (Fig. 6b–g; Supplementary Fig. 3d). This emphasised the importance of IL-10 expression in exogenous Tregs for mediating their pro-healing effect post-injury.
Fig. 6. Treg-mediated acceleration of tissue healing depends on their production of IL-10.
a Critical-size cranial defects, quadriceps volumetric muscle loss defect or full-thickness dorsal skin wounds were performed in wildtype C57BL6/J mice and treated with a fibrin hydrogel only, or hydrogel containing exogenous IL10-deficient (Il10−/−) spleen Tregs. Tissue healing was assessed at different time points for each tissue. b Bone regeneration evaluated by microCT analysis of cranial defects expressed as defect coverage and new bone volume at D28 post-injury (n = 8 defects). c Representative cranial reconstructions. The original defect area is shaded with a dashed red outline. d Muscle regeneration represented by the percentage of fibrotic area and muscle area measured by histomorphometric analysis of tissue sections at D10 post-injury (n = 8 defects). e Representative muscle histology of a transverse section of the rectus femoris stained with Masson’s trichrome at D10 post-injury. Muscle tissue is stained in red, and the fibrotic area is in blue. Scale bar = 1 mm. f Percentage wound closure at D10 post-injury evaluated by histomorphometric analysis of tissue sections (n = 8 wounds). g Representative histology of skin tissue stained with haematoxylin and eosin at D10 post-injury. Black arrows indicate wound edges and red arrows indicate tips of epithelium tongue. The epithelium (if any) is stained in purple, underneath which the granulation tissue is stained in pink–violet, with dark purple granulocyte nuclei. Scale bar = 1 mm. Data are plotted in box plots showing the median (central line) and IQR (bounds) with whiskers extending to the minimum and maximum values. Two-tailed unpaired Student’s t-test was used in (b, d, f). n.s.: non-significant. a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Local Treg delivery of allogeneic and human Tregs accelerates tissue healing
The delivery of syngeneic Tregs represents autologous Treg administration in clinical settings. Since this may be challenging to achieve in a timely manner post-injury, we sought to evaluate the efficacy of allogeneic Tregs in our mouse injury models. Thus, we delivered spleen Tregs sorted from mice on a C57BL6/J background, into injured bone, muscle, and skin of BALB/c mice, representing allogeneic Treg delivery (Fig. 7a). Additionally, we assessed the ability of human Tregs to promote regeneration as a proof-of-concept for clinical translation. To achieve this, we delivered human Tregs isolated from donor peripheral blood mononuclear cells, which were expanded in vitro and tested for purity63,64 (Supplementary Fig. 24a), into injured tissues of BALB/c mice, representing xenogeneic Treg delivery (Fig. 7a). Strikingly, we found that both allogeneic and xenogeneic Treg administration led to a significant increase in bone and muscle regeneration (Fig. 7b–e; Supplementary Fig. 3e) and skin wound closure (Fig. 7f, g), demonstrating a regenerative capacity similar to syngeneic Tregs. We then tested the retention of the delivered allogeneic and xenogeneic Tregs by flow cytometry, using the bone injury model. Like syngeneic Tregs, we found that both allogeneic and xenogeneic Tregs could be recovered for up to 5 days post-delivery (Supplementary Fig. 24b–e). In fact, we observed a significantly higher recovery with delivered human Tregs at D3 and D5, compared to freshly sorted allogeneic Tregs (Supplementary Fig. 24b). This could have resulted from in vitro expansion of human Tregs, which may have conferred upon them improved stability following in vivo administration65. Indeed, we observed a similar trend upon delivering in vitro expanded murine syngeneic Tregs, which showed a remarkably higher recovery, with an average retention of 80% on D5 post-delivery (Supplementary Fig. 25).
Fig. 7. Local delivery of allogeneic and xenogeneic Tregs promotes tissue healing.
a Skin, bone or muscle injuries were performed in BALB/c mice and treated with a fibrin hydrogel only, or hydrogel containing sorted spleen Tregs from C57BL6/J mice (allogeneic delivery), or hydrogel containing human Tregs isolated from peripheral blood and expanded in vitro (xenogeneic delivery). Tissue healing was assessed at different time points for each tissue. b Cranial regeneration at D28 post-injury evaluated by microCT and expressed as defect coverage and new bone volume (n = 8 defects). c Representative cranial reconstructions. The original defect area is shaded with a dashed red outline. d Muscle regeneration represented by the percentage of fibrotic area and muscle area measured by histomorphometric analysis at D10 post-injury (n = 8 defects). e Representative muscle histology of a transverse section of the rectus femoris stained with Masson’s trichrome at D10 post-injury. Muscle tissue is stained in red, and the fibrotic area is in blue. Scale bar = 1 mm. f Wound closure at D10 post-injury evaluated by histomorphometric analysis (n = 8 wounds). g Representative histology of skin tissue stained with haematoxylin and eosin at D10 post-injury. Black arrows indicate wound edges and red arrows indicate tips of epithelium tongue. The epithelium (if any) is stained in purple as a homogeneous keratinocyte layer on top of the wounds, underneath which the granulation tissue is stained in pink–violet, with dark purple granulocyte nuclei. Scale bar = 1 mm. Data are plotted in box plots showing the median (central line) and IQR (bounds) with whiskers extending to the minimum and maximum values. One-way ANOVA with Bonferroni post hoc test was used in (b, d: right) and a two-sided Kruskal–Wallis with Dunn’s post hoc test was used for non-parametric data in (d: left, f) for multiple comparisons. P values are indicated; ***P ≤ 0.001. (aTregs: allogeneic Tregs; hTregs: human Tregs). a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Discussion
The regulation of immune cells and immunomodulators is critical for successful tissue healing post-injury1,2. Thus, controlling immune components provides an attractive therapeutic approach to improve tissue repair outcomes. In particular, Tregs are known to play a pivotal role in modulating immune cells and are necessary for proper tissue repair and regeneration5,7. Tregs accumulate in injured tissues about 1-week post-injury, and act to suppress inflammation and stimulate regeneration via the secretion of effector proteins or cell–cell interactions5. Here, we first confirmed the crucial importance of Tregs in three injury models including critical-size cranial bone defects, volumetric muscle loss defects, and full-thickness skin wounds, by using the Foxp3DTR/GFP mouse, which allows inducible Treg ablation. As expected, Treg depletion severely impaired the tissue healing outcome in all tissues. Our results confirmed previous studies with mouse muscle8 and skin10 injury models, while providing additional evidence for the key role of Tregs after acute bone injury. Since Treg depletion disrupted the healing of all tissues tested, and endogenous Treg numbers typically peak during the late phase of tissue healing8,10, we explored the possibility of enhancing tissue healing by locally delivering exogenous Tregs to freshly injured tissues, via a fibrin hydrogel. Strikingly, this strategy significantly ameliorated healing in bone, muscle, and skin injury models, as shown by improved cranial defect coverage and bone volume, diminished fibrotic area in muscle accompanied by a higher muscle area and fibre size, and enhanced wound closure. The same was not observed upon local delivery of Tconvs to the injured tissues, thus emphasising the distinctive potential of Tregs as an effective immune cell therapeutic for regenerative medicine applications. Moreover, exogenous Tregs remained localised at the site of administration for up to 5 days post-delivery without significant recirculation into secondary lymphoid organs, while some Tregs might be diluted in surrounding tissues or may gradually undergo apoptosis in response to the injured tissue microenvironment66. Importantly, these exogenous Tregs delivered soon after injury rapidly responded to their microenvironment and adopted an “injury-specific” profile, akin to endogenous Tregs that naturally accumulate later. This was characterised by the upregulation of genes that are expressed by endogenous injured tissue Tregs, associated with the acquisition of an effector phenotype, and involved in immunomodulation and tissue healing. These included anti-inflammatory mediators that are known to influence Mo/MΦ, of which Il10 was one of the genes commonly upregulated in exogenous Tregs recovered from all three tissues. This cytokine has previously been shown to play a key role in promoting healing of several tissues such as muscle, skin, and heart23.
To further determine whether the pro-regenerative effect of Tregs was dependent on Mo/MΦ, we administered Tregs to mice that were depleted of Mo/MΦ using CL. Interestingly, the absence of Mo/MΦ abrogated the therapeutic effect of Tregs in all three models, demonstrating that exogenous Tregs likely mediated their effect via endogenous Mo/MΦ. Moreover, RNA-seq of Mo/MΦ sorted from injured bone, muscle, and skin showed an upregulation of genes associated with a pro-healing phenotype. For example, Mo/MΦ from Treg-treated bone showed a significantly higher expression of ECM-related genes such as Spp1 and Thbs1, which have been shown to dampen inflammatory responses and enhance tissue healing41,67–69, as well as the matrix metalloproteinase Mmp12, which has been shown to inhibit neutrophil infiltration and inflammation44,45. Similarly, Mo/MΦ in Treg-treated muscle upregulated Mmp9, which can reportedly minimise fibrosis and scar tissue formation46, promote inflammation resolution70, and is associated with a pro-healing macrophage subset21. In Treg-treated skin Mo/MΦ, upregulated ECM-associated genes included Adamtsl1 and Mmp16, with Adamtsl1 known to be expressed in later wound healing stages and Mmp16 contributing to wound healing through ECM protein degradation within granulation tissue47. Additionally, key anti-inflammatory genes like Arg139, Dusp140, Tmsb4x41, and Chil339 were upregulated in Mo/MΦ from Treg-treated bone, while Slc401 (ferroportin), which is required for muscle regeneration42, was upregulated in Mo/MΦ from Treg-treated muscle. Lastly, Chil1, which is highly expressed in pro-healing macrophages43, was upregulated in Mo/MΦ from Treg-treated skin. Furthermore, factors known to be induced upon monocyte-to-macrophage differentiation or that promote anti-inflammatory “M2-like” polarisation were also upregulated, such as Cebpb48, Irf2bp249, Ccl650, Ccl2239, Ngp52, Ywhaz53, and Il4i143,54 among others. Treg-treated tissue Mo/MΦ also displayed the concomitant downregulation of genes and pathways associated with a cellular response to pro-inflammatory cytokines like IFN-γ and IL-1. Taken together, Mo/MΦ in Treg-treated injuries adopted a more anti-inflammatory and pro-reparative phenotype. To further investigate the mechanisms by which Tregs modulated Mo/MΦ in these tissues, we performed RNA-seq analysis on Mo/MΦ sorted from Treg-depleted mice, which displayed an opposite transcriptional profile to Treg-treated mice, thereby supporting our observations. Contrary to Treg treatment, the absence of Tregs led to an increased proportion of the pro-inflammatory Ly6Chigh subset39 within Mo/MΦ and a downregulation of pro-repair gene expression (e.g. Spp1, Thbs1, Tmsb4x, etc.). This was accompanied by the upregulation of other pro-inflammatory genes showing an enrichment in the response to IFN-γ and IL-1, similar to other reports in skin10 and muscle60 injuries. Thus, the altered proportions of Mo/MΦ subsets in Treg-depleted mice demonstrated the influence of Tregs on inflammatory Mo/MΦ accumulation and/or their transition into anti-inflammatory Mo/MΦ, supporting our prior findings in Treg-treated mice. Overall, the combined analyses of Mo/MΦ in Treg-treated and Treg-depleted mice showcased that exogenous Tregs enhanced tissue repair and regeneration by fostering an anti-inflammatory and pro-healing switch in Mo/MΦ, primarily by inhibiting key inflammatory pathways and upregulating pro-healing factors. This aligns with previous studies on endogenous Tregs, that have demonstrated their influence on the phenotype and accumulation of macrophages in vitro71 and during wound healing10, muscle regeneration60, and cardiac repair11.
Changes in Mo/MΦ phenotype are likely to reflect the evolution of the immune response within Treg-treated tissues. In line with this, we observed that Treg treatment accelerated neutrophil clearance and reduced CD8+ T cell accumulation, both of which are important for the Mo/MΦ phenotypic switch72,73. The reduced number of neutrophils suggested an accelerated progression to the resolution phase, as neutrophils need to be rapidly cleared to induce a pro-resolution cascade72. Additionally, Tregs are known to suppress CD8+ T cell responses in various scenarios74, while elevated levels or prolonged presence of CD8+ T cells during tissue healing have been correlated with unfavourable tissue healing outcomes75. Conversely, the inhibition or depletion of CD8+ T cells has been demonstrated to enhance the healing process75,76. In fact, we showed that co-culture of Mo/MΦ from injured tissues, with decreasing concentrations of CD8+ T cells, reduced the proportion of the pro-inflammatory Ly6Chigh Mo/MΦ subset. This indicated that the reduced number of CD8+ T cells likely dampened the pro-inflammatory Mo/MΦ phenotype further, although the presence of more anti-inflammatory Mo/MΦ in the Treg-treated injuries could also in turn restrict CD8+ T cell accumulation77. Given that Treg delivery led to a reduction in CD8+ T cells, and previous studies have shown that T cells, along with NK cells, are the major producers of IFN-γ in Treg-depleted mice10,60, we analysed the levels of IFN-γ expression following Treg administration. Treg-treated tissues showed a reduction in IFN-γ–producing T cells (CD4+, CD8+) and NK cells, indicating that exogenous Tregs limited IFN-γ production, thereby controlling Mo/MΦ accumulation and phenotype10,60. This was consistent with the RNA-seq analysis of endogenous Mo/MΦ from Treg-treated mice that also showed a diminished cellular response to IFN-γ. Despite these significant changes in other immune cells, the profound impairment in Treg-mediated tissue healing upon Mo/MΦ depletion suggests that exogenous Treg treatment primarily promotes tissue healing by influencing Mo/MΦ.
To unveil the mechanism by which exogenous Tregs may exert a direct effect on Mo/MΦ, we tested whether some of the factors expressed by exogenous Tregs could directly impact Mo/MΦ phenotype, using an in vitro and an ex vivo culture system. We found IL-10 to be the only factor that could significantly reduce the proportion of Ly6C+ Mo/MΦ, both in the presence and absence of pro-inflammatory cytokines. Furthermore, delivering IL-10–deficient exogenous Tregs via fibrin gel into bone, muscle, and skin injuries failed to achieve any significant improvement in tissue healing, establishing that IL-10 played a key role in mediating the pro-healing effects of exogenous Tregs via Mo/MΦ modulation. In fact, Treg-derived IL-10 has been reported to be critical for restraining spontaneous inflammation in the colon, lung, and skin78, as well as for suppressing renal tissue injury in kidney disease79. Altogether, our findings support a model wherein exogenous Tregs rapidly adopt an injury-specific phenotype upon delivery into the injured tissue and affect endogenous Mo/MΦ by accelerating their transition to an anti-inflammatory and pro-repair phenotype, either directly through factors such as IL-10 or indirectly via reducing the number of pro-inflammatory cells and cytokines (Supplementary Fig. 26).
Finally, to provide a proof-of-concept for the clinical translation of this therapeutic approach, we also showed that allogeneic and human (xenogeneic) Tregs significantly ameliorated bone, muscle, and skin tissue healing. Indeed, both allogeneic and xenogeneic Tregs were detectable for up to 5 days post-administration, thus confirming their survival upon fibrin gel-mediated delivery. This was promising as there has been an increasing interest in Treg banking or off-the-shelf allogeneic Treg products, which would be particularly advantageous to improve consistency and accessibility80. Furthermore, since the administration of Tregs in a clinical setting would necessitate prior in vitro culture of Tregs, it was encouraging to note that the retention of delivered Tregs within the injured tissue increased dramatically when they were cultured in vitro, prior to administration, for both murine and human Tregs. This could be attributed to acquisition of more stability and survivability upon in vitro culture, leading to better viability and retention upon in vivo administration. Altogether this was promising, as future applications of this approach could potentially involve banking of in vitro expanded human donor Tregs, followed by allogeneic Treg transfer to patients.
Although this study focused on delivering exogenous Tregs immediately post-injury, future research is required to determine the maximum possible timeframe within which we can administer Tregs to damaged tissue to effectively facilitate healing. Undoubtedly, the presence of endogenous Tregs is crucial during the early stages following skin10 and muscle60 injury, and previous studies have shown that the delivery of exogenous Tregs either immediately or 1-day post-MI can promote tissue healing post-myocardial infarction81,82. Thus, we expect that augmenting Treg numbers locally as early as possible after tissue damage would likely provide the maximum benefit as a therapeutic strategy for tissue healing. However, Tregs are also important at a later stage of tissue healing, by expressing factors such as Areg which can stimulate progenitor cell proliferation8,9. Thus, it would be interesting to test whether exogenous Treg administration at a more advanced stage of healing can provide additional regenerative benefit, perhaps by supplementing the activities of endogenous Tregs that naturally accumulate at later time points post-injury8,10. Furthermore, since our data show that the acceleration of tissue healing by Tregs was mediated by their production of IL-10, future studies should explore the use of recombinant IL-10 as an alternative strategy to promote bone, muscle, and skin tissue healing. Indeed, IL-10 overexpression or sustained administration has been shown to improve cutaneous wound healing83 and muscle regeneration84, respectively.
In conclusion, while therapeutic strategies based on Treg delivery have mainly been explored for graft tolerance80, this study showed that local administration of exogenous Tregs is an effective approach to promote the repair and regeneration of multiple tissues. Exogenous Tregs act as a chief conductor orchestrating a rapid and effective healing response by modulating the accumulation and activity of other immune cells. Ultimately, the regenerative activity of exogenous Tregs relies on Mo/MΦ, although Mo/MΦ regulation by Tregs is possibly both direct and indirect. Factors expressed by exogenous Tregs, such as IL-10, exert a paracrine effect and facilitate a timely Mo/MΦ switch from a pro-inflammatory to an anti-inflammatory and pro-healing phenotype, essential for tissue healing. Lastly, we demonstrated the efficacy of this strategy with allogeneic or xenogeneic Treg delivery, highlighting the pro-healing activity of human Tregs and the potential of an allogeneic approach to develop off-the-shelf Treg-based therapies for regenerative medicine. Furthermore, this study provides important mechanistic insights into the pro-regenerative effects of exogenous Tregs, and the findings altogether could pave the way for developing regenerative strategies based on Treg delivery, either as standalone treatments or in conjunction with existing therapeutic approaches.
Methods
Mice
All animal studies were approved by the Monash Animal Research Platform Animal Ethics Committee, Monash University (approval number 17124). C57BL/6J and BALB/c-Asmu mice were purchased from the Monash Animal Research Platform or the Walter & Eliza Hall Institute of Medical Research. Foxp3GFP/DTR mice were purchased from The Jackson Laboratory (strain 016958) and bred in-house. Foxp3IRES-mRFP mice from The Jackson Laboratory (strain 008374) were received from A.V., Olivia Newton-John Cancer Research Institute and bred in-house. Il10−/− mice (B6.129P2-Il10tm1Cgn/J) were purchased from The Jackson Laboratory (strain 002251). Mice were bred in specific pathogen-free/SPF facilities, and all animals (both experimental and control) were co-housed in 12:12 light:dark light cycles at room temperatures ranging between 20 and 24 °C and humidities between 40 and 60%. Food and water were available ad libitum to the mice. Regular diet (BARASTOC) was provided. At any significant sign of distress or at the end of the experiments, mice were euthanized by cervical dislocation or CO2 asphyxiation.
Treg depletion by diphtheria toxin
Ten-week-old male Foxp3DTR/GFP and wildtype C57BL/6J mice received intraperitoneal injections of 20 ng/g diphtheria toxin, starting 2 days before injury and continuing daily until the injury day (day 0), then given every other day until day 8. Efficiency of Treg depletion was validated by assessing Foxp3+ Treg numbers within the CD4+ T cell population in blood, spleen, and injured tissues on day 7 post-injury using flow cytometry. To confirm Foxp3+ Treg depletion in Foxp3DTR/GFP mice used in sorting and regeneration experiments, tail vein blood was taken on day 7, or on the day of tissue harvest (if earlier than day 7), for flow cytometric analysis of CD4+, FOXP3+ Tregs.
Macrophage depletion by CL
Ten-week-old male wildtype C57BL/6J mice received intraperitoneal injections of clodronate encapsulated in liposomes or control PBS liposomes (200 μl of 5 mg/ml) (Liposoma B.V., CP-005-005) 1 day before injury. Injections continued every other day until day 7 post-injury to deplete macrophages. Efficiency of macrophage depletion was validated by assessing F4/80+ macrophage levels within the CD11b+ myeloid cell population in spleen using flow cytometry.
Cranial defect model
Male mice aged 10–12 weeks were anaesthetised using isoflurane and administered subcutaneous buprenorphine analgesia (0.1 mg/kg). The cranial region was shaved to reveal the calvarial bone area. An incision along the midline of the skin exposed the skull, with retraction of soft tissues to uncover the calvarial bone. Utilising a micromotor drill, two craniotomy defects (4.5 mm in diameter) were created on each side of the sagittal suture line, ensuring no dura perforation. After rinsing with saline, defects were either covered with a fibrin matrix for Treg delivery or 20 μl of bovine thrombin (10 U/ml) (Sigma-Aldrich) to prevent bleeding. The skin was then closed with staples.
MicroCT
At day 28 after the bone injury, mice were euthanised, and their skulls were extracted. Removal of surrounding muscle, soft tissue, and brain exposed the skull, which was fixed in 10% formalin for 24 h at room temperature and later transferred to 70% ethanol. A microCT 40 (Scanco Medical AG) scanner with 55 kVp energy and 145 ms intensity was used for medium resolution scans with a nominal 30 µm isotropic resolution. Images were filtered with a 3D Gaussian filter (sigma 1.2, support 1) after reconstruction. Bone was separated from the background using a global threshold of 22.4% of the maximum grey value. Subsequently, cylindrical masks were placed manually at the defect sites and imaged using the scanner software Image Processing Language (IPL) (Scanco Medical AG) or a NanoPET/CT scanner (Mediso) to calculate bone coverage and volume within these masks. Data analysis was performed on PMOD v3.3 biomedical image quantification software (PMOD Technologies). Standardised procedures for quantitative bone morphometry were employed to calculate bone volume, while coverage was determined on a dorso-ventral projection of the cylindrical area18.
Volumetric muscle loss model
Male mice aged 10–12 weeks were anaesthetised using isoflurane and given subcutaneous buprenorphine analgesia (0.1 mg/kg). The left hind limb was shaved, and a 1 cm unilateral incision exposed the underlying fascia. By retracting the surrounding tissue, the quadriceps muscle was brought out through the incision. Using a 3 mm biopsy punch (Kai Medical), volumetric muscle loss defects were created, or a 2.5 mm × 5 mm section of the quadriceps involving the rectus femoris muscle was excised. Muscle defects were either covered with a fibrin matrix for Treg delivery or left uncovered. The soft tissue was closed with sutures.
Histomorphometric analysis of muscle regeneration
At D10 after the injury, mice were euthanised, and the skinless hind limb was collected and fixed in 10% formalin for 24 h at room temperature. The injury site, encompassing the proximal and distal sections of the quadriceps muscle (including rectus femoris, vastus medialis, and vastus lateralis), was removed and embedded in paraffin. Muscles were sliced into 4 μm thick transverse sections at five depths, starting from the patella’s edge, passing the wound’s centre, up to the proximal end of the defect site. Sections were stained with Masson’s Trichrome and muscle regeneration was assessed by averaging the blue-stained fibrotic area’s percentage (normalised to total muscle area) and the remaining non-fibrotic muscle area across five tissue section depths, using Aperio ImageScope software (Leica Biosystems). To analyse muscle fibre area, the section with the largest number of centrally nucleated fibres, which represents regenerating muscle fibres9, from each sample was identified. For three representative regions in each of these sections, the cross-sectional area of individual muscle fibres was determined by manually tracing the fibre circumference in Fiji. Approximately 300 fibres were traced per sample to provide the mean cross-sectional area for each sample.
Skin wound healing model
Male mice aged 10–12 weeks were anaesthetised using isoflurane and given subcutaneous buprenorphine analgesia (0.1 mg/kg). Their backs were shaved, and two full-thickness excisional wounds were created using a 5 mm biopsy punch (Kai Medical). In the non-splinting model, wounds were covered with adhesive round spot plasters (22.5 mm, Livingstone International, Australia) held in place with 3 M Blenderm surgical tape. In the splinting wound healing model, each wound was splinted with an 8 mm nylon ring (M8 nylon washer, Zenith ITWProline) glued to the skin with Ultra-fast super glue (UHU). This prevented wound healing via local skin contraction, maintaining wound structure and creating a model closer to human re-epithelialisation and new tissue formation during wound healing85. Immediately after, wounds were covered with either a fibrin matrix for Treg delivery or adhesive round spot plasters secured using 3M Blenderm surgical tape. Bandages and rings were inspected every alternate day and replaced if they started to loosen.
Histomorphometric analysis of skin wound closure
At D7 and/or D10 post-injury, mice were euthanised and the dorsal skin including the wounded area was excised. Wounds were fixed in 10% neutral buffered formalin for 24 h at room temperature. Wounds were then harvested using an 8 mm biopsy punch (Kai Medical) around the original wound, embedded in paraffin, and sectioned at 4 μm until the centre of the wound was passed. Sections were stained with haematoxylin and eosin and re-epithelialisation was measured by histomorphometric analysis of tissue sections using Aperio ImageScope (Leica Biosystems). The wound’s centre was determined by measuring the distance of the gap between the edges of the panniculus carnosus muscle that is severed upon wound creation. Wound closure was calculated at the centre, as the ratio of epidermis closure to the length of the panniculus carnosus gap.
T-cell delivery into injured tissue via fibrin hydrogels
Mouse or human Tregs, or mouse CD4+ Tconvs were delivered by adding them to fibrin hydrogels immediately post-wound or post-defect creation. Fibrin hydrogels were directly applied to the site of the defect—over the dura within bone defects, within the muscle tissue gap, and within the punched skin area. Each defect received 40 µl of fibrin hydrogel containing 20 µl of RPMI 1640 with or without 200,000 sorted Tregs, mixed with a final concentration of fibrinogen (Enzyme Research Laboratories) at 10 mg/ml for bone, 8 mg/ml for muscle, or 6 mg/ml for skin, along with 2 U/ml of thrombin (Sigma-Aldrich), 5 mM CaCl2 and 25 µg/ml aprotinin (Roche) in HEPES buffer. GFP+ Tregs from Foxp3DTR/GFP mice were used for Treg delivery, except for recovery experiments where RFP+ Tregs from Foxp3IRES-mRFP mice were used to enhance exogenous Treg detection through the brighter expression of RFP.
Treg delivery into uninjured tissue via subcutaneous fibrin hydrogel implant
Male mice aged 10 weeks were anaesthetised using isoflurane and administered subcutaneous buprenorphine analgesia (0.1 mg/kg). Dorsal hair was shaved, and a 5 mm skin incision was made along the sagittal plane in the dorsothoracic region, 10 mm lateral to the spine on each side. A small pocket was formed in the subcutaneous space by blunt dissection to accommodate the fibrin gel for implant. For each implant, an 80 μl hydrogel was prepared containing 40 μl of RPMI 1640 (Thermo Fisher Scientific) with 400,000 sorted RFP+ Tregs, mixed with a final concentration of 10 mg/ml fibrinogen (Enzyme Research Laboratories), 2 U/ml thrombin (Sigma-Aldrich) and 5 mM CaCl2 and 25 µg/ml aprotinin (Roche) in HEPES buffer. Hydrogels were polymerised at the time of surgery and implanted into the subcutaneous pocket created, after which incision sites were closed using surgical sutures.
Human Treg isolation
Human CD4+CD25hiFoxp3+ naturally occurring Tregs were isolated from healthy volunteers63,64, with informed consent and approval from the Newcastle and North Tyneside Research Ethics Committee (UK, approval number 09/H0907/61). Naturally occurring Tregs, defined as CD4+CD25hiFOXP3+ cells were isolated by negative enrichment of CD4+ T cells (RosetteSep, STEMCELL Technologies) followed by CD25hi positive selection (RoboSep; STEMCELL Technologies). The isolated Tregs were activated and expanded in vitro for 1–2 weeks using Dynabeads Human CD3/CD28 T-cell expander beads (Thermo Fisher Scientific), cultured in RPMI 1640 with 10% heat-inactivated FBS. The expansion was supported by addition of high-dose IL-2 (500 IU/ mL, Roche) and 100 nM rapamycin (Sigma-Aldrich) in the culture medium. Over 95% of the expanded Tregs were Foxp3+. Their immunosuppressive function was verified through a mixed lymphocyte reaction with CD8+ T cells, wherein the expanded Tregs effectively suppressed CD8+ T cell survival, proliferation, and cytokine production63,64. Tregs were cryopreserved and displayed more than 90% viability according to the trypan blue exclusion assay and showed stable expression of FOXP3 (>98%) and HELIOS (>95%) post-thawing.
Leucocyte isolation from mouse spleen and lymph nodes
Spleens from euthanised mice were crushed between glass slides, and released cells were strained through a 70 μm cell strainer (Greiner) into complete media (RPMI 1640 with 10% heat-inactivated FBS). Lymph nodes were pressed through a 70 μm cell strainer (Greiner) using a syringe plunger, and cells were washed with complete media. After centrifugation at 500 × g for 10 min at 4 °C, cells were treated with ice-cold red blood cell (RBC) lysis buffer (0.15 M ammonium chloride, 10 mM potassium bicarbonate, and 1 mM EDTA in distilled water) for 2 min, washed, and centrifuged to create a leucocyte pellet for antibody staining.
Leucocyte isolation from injured bone, muscle, and skin tissues
Skin wounds were harvested using an 8 mm biopsy punch, muscle defects including the proximal and distal segments of the quadriceps were excised, and injured calvarial bone including the original craniotomy defects were cut out, after which harvested tissues were finely minced with scissors. Tissue mincing was followed by enzymatic digestion using 2 mg/ml Collagenase II (Thermo Fisher Scientific, #17101015) for muscle and bone, and Collagenase XI (Sigma-Aldrich, #C7657) for skin, supplemented with 100 µg/ml DNase I (Roche). Two 20-min digestions at 37 °C were conducted, and after each, the supernatant with released cells was collected and enzymes were neutralised with DMEM/F12 (for macrophage isolation) or RPMI 1640 (for Treg isolation) containing 10% heat-inactivated FBS and 5 mM EDTA. This mixture was kept chilled while the remaining undigested tissue underwent the second digestion. All released cells and digested tissue were passed through a 70 μm cell strainer (Greiner), with residual tissue further mashed using a syringe plunger. The resulting single-cell suspensions were centrifuged at 500 × g for 10 min at 4 °C, then treated with ice-cold RBC lysis buffer for 40 s on ice, washed, and centrifuged to yield a leucocyte pellet for antibody staining.
Flow cytometry staining for analysis of live cells
Cells were incubated with TruStain FcX™ anti-mouse CD16/32 antibodies (10 μg/ml; clone 93, BioLegend) diluted in flow cytometry (FACS) buffer (5% heat-inactivated FBS + 5 mM EDTA in PBS), for 20 min on ice, and subsequently incubated with cell surface antibodies in FACS buffer for 30 min on ice. Cell surface antibodies were added according to the target population. To sort mouse spleen Tregs for delivery into injured tissues via fibrin hydrogels, GFP+ or RFP+ Tregs were sorted from the spleens of female Foxp3DTR/GFP and Foxp3IRES-mRFP mice, respectively, using APC anti-mouse CD4 (BioLegend, Clone GK1.5, 2 µg/ml). Tregs from Il10−/− female mice spleens were sorted as CD4+ CD25+ Tregs using the following anti-mouse antibodies: APC anti-CD4 (BioLegend, Clone GK1.5, 2 µg/ml) and PE anti-CD25 (Biolegend, Clone PC61, 1 µg/ml). To sort endogenous injured tissue Tregs and healthy spleen Treg controls for bulk RNA sequencing, the following anti-mouse antibodies from BioLegend were used: PE/Cyanine7 anti-CD3 (clone 17A2, 2.5 µg/ml), APC anti-CD4 (clone GK1.5, 2 µg/ml), BV711 anti-CD11b (clone M1/70, 2 µg/ml), and BV711 anti-F4/80 (clone BM8, 4 µg/ml). To sort endogenous Mo/MΦ from injured tissues for mini-bulk RNA sequencing, the following anti-mouse antibodies from BioLegend were used: PerCP/Cyanine5.5 anti-Siglec-F (clone S17007L, 2 µg/ml), PerCP/Cyanine5.5 anti-Ly6G (clone 1A8, 2 µg/ml), APC-Fire750 anti-CD11b (clone M1/70, 2 µg/ml), and PE anti-F4/80 (clone BM8, 4 µg/ml). To detect exogenous RFP+ Tregs recovered from injured tissues post syngeneic or allogeneic Treg delivery, the following anti-mouse antibodies from BioLegend were used: PE/Cyanine7 anti-CD3 (clone 17A2, 2.5 µg/ml), APC anti-CD4 (clone GK1.5, 2 µg/ml), BV711 anti-CD11b (clone M1/70, 2 µg/ml), BV711 anti-F4/80 (clone BM8, 4 µg/ml), and BV711 anti-Ly6G (clone 1A8, 2 µg/ml). To detect exogenous human Tregs recovered from injured tissues post xenogeneic Treg delivery, the following anti-mouse antibodies from BioLegend were used: PE/Cyanine7 anti-CD3 (clone 17A2, 2.5 µg/ml), BV711 anti-CD11b (clone M1/70, 2 µg/ml), BV711 anti-F4/80 (clone BM8, 4 µg/ml) and BV711 anti-Ly6G (cone 1A8, 2 µg/ml), and the following anti-human antibodies that were a kind gift from Associate Professor Martin Davey from Professor Jamie Rossjohn’s lab (Monash University) were used: PE anti-CD3 (BioLegend, clone UCHT1, 2 µg/ml) and AF700 anti-CD4 (BioLegend, clone SK3, 2 µg/ml). Viability was detected by adding 1 µg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (BV421) diluted in FACS buffer, after washing out the cell surface antibodies, to exclude any DAPI-positive dead cells. For analysis of pro-inflammatory marker expression in the in vitro cultured Mo/MΦ, cells were first washed with PBS, stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation (Thermo Fisher Scientific, #L34957, 1:400) for 30 min on ice, then incubated with TruStain FcX anti-CD16/32 (10 μg/ml; clone 93, BioLegend) for 20 min on ice, followed by cell surface staining for 30 min on ice, with the following anti-mouse antibodies from BioLegend: BV711 anti-CD11b (clone M1/70, 2 µg/ml), PE anti-F4/80 (clone BM8, 4 µg/ml) and FITC anti-Ly6C (clone HK1.4, 2.5 µg/ml), diluted in FACS buffer. Stained cells were washed twice before sorting, or analysis using the BD LSR Fortessa X-20 flow cytometer (BD Biosciences). The acquired data were analysed using FlowJo v10 Software (BD Biosciences).
Flow cytometry staining for analysis of fixed cells
For detection of neutrophils, CD4+/CD8+ T cells, Mo/MΦ, and Tregs at different time points in mouse injured tissues, spleen or blood, cells were fixed prior to analysis on a flow cytometer. Samples that required fixation were first stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation (Thermo Fisher Scientific, #L34957, 1:400) for 30 min on ice, followed by incubation with TruStain FcX anti-CD16/32 (10 μg/ml; clone 93, BioLegend) for 20 min on ice. Subsequently, cell surface staining was performed for 30 min on ice, with the following anti-mouse antibodies from BioLegend: myeloid cells were stained with BV711 anti-CD11b (clone M1/70, 2 µg/ml), BV421 anti-Ly6G (clone 1A8, 2 µg/ml), and PE anti-F4/80 (clone BM8, 4 µg/ml); lymphocytes were stained with PE/Cyanine7 anti-CD3 (clone 17A2, 2.5 µg/ml), APC anti-CD4 (clone GK1.5, 2 µg/ml) and BV421 anti-CD8a (clone 53-6.7, 2 µg/ml). For intracellular staining of Foxp3, cells were fixed with the eBioscience Fixation/Permeabilization diluent and concentrate (Thermo Fisher Scientific, #00-5223-56, #00-5123-43), then stained with FITC anti-mouse Foxp3 antibody (Tonbo Biosciences, Clone 3G3, 6.6 µg/ml) in 0.5% saponin diluted in FACS buffer, for 45 min at 4 °C. For intracellular cytokine staining of IL-10, cells were fixed with Intracellular Staining Fixation Buffer (BioLegend, #420801) for 20 min at room temperature in the dark, followed by permeabilisation with 1X Intracellular Staining Permeabilization Wash Buffer (BioLegend, #421002) according to the manufacturer’s instructions. Fixed cells were then incubated with the APC anti-mouse IL-10 antibody (BioLegend, Clone JES5-16E3, 2 µg/ml) diluted in 1X Intracellular Staining Permeabilization Wash Buffer (BioLegend), for 30 min at 4 °C. For analysis of human Treg purity, cell surface staining was performed with FITC anti-human CD4 (eBioscience, clone OKT4, 2.5 µg/ml), followed by intracellular staining with PE anti-human Foxp3 antibody (eBioscience, Clone 236A/E7, 3 µg/ml) and PE-Cy7 anti-human Helios antibody (Biolegend, Clone 22F6, 2.5 µg/ml), in 0.5% saponin diluted in FACS buffer, for 45 min at 4 °C. Stained cells were washed twice before analysis using the BD LSR Fortessa X-20 flow cytometer (BD Biosciences). The acquired data were analysed using FlowJo v10 Software (BD Biosciences).
Isolation and stimulation of leucocytes for intracellular cytokine staining
For samples that required intracellular cytokine staining with IFN-γ, the CD45+ fraction was first enriched using CD45 Microbeads (1:10, Miltenyi Biotec, # 130-052-301) according to the manufacturer’s instructions. Briefly, after RBC lysis, the CD45 beads were incubated with the cell suspension for 15 min at 4 °C. After incubation, the cells were washed by adding 5 ml of FACS buffer (PBS + 5% HI-FBS) and centrifuging at 500 × g for 10 min at 4 °C. The supernatant was discarded, and the cell pellet containing CD45 beads was resuspended in 2 ml of FACS buffer. These CD45+ cells containing lymphocytes were purified by positive selection using a MACS Separation LS Column (#130-042-401, Miltenyi Biotec) and MidiMACS Separator (#130-042-302, Miltenyi Biotec) according to the manufacturer’s instructions. Briefly, the LS Column was placed on the MidiMACS separator and 2 ml of cell suspension containing CD45 beads was added into the LS Column. Following this, the column was washed three times with 3 ml of FACS buffer each time. The magnetically labelled fractions that were retained in the LS Column were then removed from the MidiMACS Separator by adding 5 mL of FACS buffer into the LS Column and flushing out the CD45+ cell suspension with a plunger. The cell suspension was centrifuged at 500 × g for 10 min at 4 °C, supernatant discarded, and the cells were resuspended in RPMI 1640 + 10% FBS-HI containing a PMA & ionomycin cell stimulation cocktail (Tonbo Biosciences, # TNB-4970-UL400) in the presence of 1 µg/ml Monensin solution (Australian Biosearch, #420701). The stimulation was performed for 4 h at 37 °C, after which the cells were washed with FACS buffer, then with PBS, and stained with LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation (Thermo Fisher Scientific, #L34957, 1:500) to detect cell viability. This was followed by blocking and surface staining steps with the following anti-mouse cell surface antibodies: FITC anti-CD11b (BioLegend, Clone M1/70, 2 µg/ml), FITC anti-F4/80 (BioLegend, Clone BM8, 4 µg/ml), BV711 anti-CD3 (BioLegend, Clone 17A2, 2.5 µg/ml), BV605 anti-CD4 (BioLegend, Clone GK1.5, 2 µg/ml), APC anti-CD8a (BioLegend, Clone 53-6.7, 2 µg/ml), and BV421 anti-NK-1.1 (BioLegend, Clone PK136, 2.5 µg/ml). Cell surface antibody staining was followed by fixation with Intracellular Staining Fixation Buffer (BioLegend, #420801) for 30 min at 4 °C and subsequent permeabilization with 1X Intracellular Staining Permeabilization Wash Buffer (BioLegend, #421002) according to the manufacturer’s instructions. Then intracellular cytokine staining was performed with PE anti-mouse IFN-γ (BioLegend, Clone XMG1.2, 2.5 µg/ml) diluted in the 1X Intracellular Staining Permeabilization Wash Buffer (BioLegend), for 30 min at 4 °C. Stained cells were washed twice before analysis using the BD LSR Fortessa X-20 flow cytometer (BD Biosciences). The acquired data were analysed using FlowJo v10 Software (BD Biosciences).
Cell sorting from mouse spleen and injured tissues
Sorting of immune cells was performed on the BD Influx Cell Sorter (BD Biosciences) through a 70 µm nozzle for spleen, and a 100 µm nozzle for endogenous Tregs and Mo/MΦ from injured bone, muscle or skin tissues, which were sorted into DMEM/F12 (for macrophages) or RPMI 1640 (for Tregs) supplemented with 10% heat-inactivated FBS. Sorting of exogenous Tregs was performed on the BD FACSAria Fusion flow cytometer (BD Biosciences) through a 70 µm nozzle. Triplicate wells (of 50 cells/well) were sorted per mouse for each of the three tissues (bone, muscle, and skin). The cells were directly sorted into a chilled 384-well PCR plate (Greiner, 785290), containing 1.2 μl of primer/lysis mix in each well [20 nM indexed polydT primer (custom made, IDT), 1:6,000,000 dilution of ERCC RNA spike-in mix (Ambion, 4456740), 1 mM dNTPs (NEB - N0446S), 1.2 units SUPERaseIN Rnase Inhibitor (Thermo Fisher, AM2696), 0.2% Triton X-100 solution (100 ml, Sigma-Aldrich, 93443), DEPC water (Thermo Fisher, AM9920)]. Sorted plates were sealed, centrifuged for 1 min at 3000 rpm and immediately frozen upside down at −80 °C until further processing for mini-bulk RNA sequencing.
RNA extraction and quantification
Total RNA was isolated from the sorted endogenous Mo/MΦ, as well as endogenous Tregs (from healthy spleen and injured tissues) using the RNeasy Plus Micro Kit (Qiagen, 74034) according to the manufacturer’s instructions. RNA concentration was measured using a Qubit Fluorometer (Invitrogen) and RNA quality was assessed using the Bioanalyzer 2100 system (Agilent Technologies).
Mini-bulk RNA sequencing
Mini-bulk RNA sequencing was performed for exogenous Tregs and endogenous Mo/MΦ from Treg-treated mice (and controls), using a modified CEL-Seq2 protocol86. For the recovered exogenous Tregs, mini-bulk RNA sequencing was performed on 27 wells of sorted samples in a 384-well plate (3 tissues, 3 mice/tissue, each mouse sorted as 3 technical replicates), containing 50 cells/well. For the endogenous Mo/MΦ from injured tissue, 1.1 μl of purified RNA (6 ng/ μl) from each sample was added to a well of a 384-well PCR plate (Greiner, 785290) containing 20 nM of an air-dried indexed polydT primer (custom made, IDT). First strand cDNA synthesis was performed by using a reverse transcription reaction mix (Invitrogen, 18064-014) according to the manufacturer’s instructions. The recovered Treg samples were then pooled into 1.5 ml Eppendorf DNA LoBind microcentrifuge tubes, while the individual Mo/MΦ samples were transferred separately to other 1.5 ml tubes. These samples were then treated with Exonuclease I (20 U/μl, NEB, M0293L) for 30 min at 37 °C, followed by 80 °C for 10 min, followed by a 1.2X bead clean-up. The eluted cDNA was then used for second strand synthesis performed with the NEBNext Second Strand Synthesis Module (NEB, E6111S) in a final reaction volume of 20 μl to generate double-stranded cDNA followed by a 1.2X bead clean-up. All DNA purification and size selection steps were done using the NucleoMag NGS Clean-up and Size select magnetic beads (Macherey-Nagel, 7449970.5), according to the manufacturer’s instructions. In vitro transcription (IVT) was then performed on the eluted cDNA using the T7 enzyme (MEGAscript T7 Transcription Kit, Ambion, AM1334) incubated at 37 °C for 13 h, after which left-over primers were removed using the ExoSAP-IT For PCR Product Clean-Up kit (Affymetrix, 78200). The sample was then subjected to RNA fragmentation by chemical heat incubation with RNA fragmentation reagents (Ambion AM8740) and subsequent purification using 1.8X RNAClean XP beads (Beckman Coulter, A63987) according to the manufacturer’s instructions. The fragmented RNA was transcribed into cDNA using 5′-tagged-random-hexamer primers (GCCTTGGCACCCGAGAATTCCANNNNNN) introducing a partial Illumina adapter as also described in the CEL-Seq2 protocol86 to perform the 5′-tagged-random-hexamer reverse transcription (ranhexRT). PCR was performed using half of the ranhexRT sample as a template [1X KAPA HiFi HotStart ReadyMix (KapaBiosystems, KK2602), 400 nM each primer]. Then the final PCR-amplified library was submitted to two consecutive 1X bead clean-up steps and then used for sequencing on the Illumina NextSeq 2000 instrument (50 cycle kit) including 5% PhiX in the sequencing run.
Analysis of mini-bulk RNA sequencing data
Mini-bulk reads for both sequencing experiments were mapped to the Mus musculus GRCm38.p6 genome and ERCC spike-in sequences using the Subread aligner (v2.10.5)87 and assigned to genes using scPipe (v1.18.0)88 with GENCODE M25 primary assembly annotation. Gene-based annotations were obtained from the “EnsDb.Mmusculus.v79” R/Bioconductor package89, which was used to map the GENCODE/Ensembl gene IDs to gene symbols and to other info (e.g. gene biotype, chromosome location). Gene counts were exported as a matrix by scPipe with UMI-aware counting. For Treg samples, technical replicates were aggregated by summing the UMI counts, to generate pseudobulked data resulting in three biological replicates for each of the four tissues (D0 spleen, and D3 recovered bone, muscle, and skin). The UMI-deduplicated read count files containing this Treg pseudobulked data (combining all technical replicates for each sample into one) was uploaded to the Degust web tool90 for further processing. For the endogenous Mo/MΦ samples, since there were no technical replicates, the original UMI-deduplicated read count files were uploaded to Degust. Degust performs normalisation using trimmed mean of M values (TMM)91, and differential gene expression analysis was conducted using limma/voom92 for the exogenous Tregs and the quasi-likelihood functionality of edgeR93 for the endogenous Mo/MΦ. Genes with a false-discovery rate (FDR) adjusted p value < 0.05 were considered differentially expressed (DE). The heat map was generated using the pheatmap function from the pheatmap package (version 1.0.12)94, along with the RColorBrewer package (version 1.1-3)95 in R (version 4.2.2). Functional enrichment analysis for Gene Ontology Biological Processes was performed for the differentially upregulated genes using gProfiler96 using the Benjamini–Hochberg FDR method for multiple testing correction (with the significance threshold of p < 0.01). Gene-concept network plots were made using the cnetplot function under the Enrichplot package (version 1.22.0)97 and the ClusterProfiler package (version 3.0.4)98 to visualise the network of differentially expressed genes contributing to each gene ontology biological process term.
Bulk RNA sequencing
Bulk RNA sequencing was performed for endogenous Tregs from injured tissues, and healthy spleen controls, as well as for Mo/MΦ from Treg-depleted mice. For Tregs, RNA quantity/quality assessment, library preparation and sequencing were performed at the Medical Genomics Facility, Monash Health Translation Precinct (MHTP) and at Micromon Genomics (Monash University). Libraries for endogenous Tregs were constructed using the Tecan/Nugen Ovation Solo Library prep kit, starting with 1 ng of input total RNA. For Mo/MΦ, RNA quantity/quality assessment, library preparation and sequencing were performed at MHTP. Libraries were constructed starting with 20 ng of input total RNA. Individual first strand synthesis was performed to add the Illumina P7 primer containing an 8 bp sample index and 10 bp unique molecular identifier (UMI) and standard i7/R2 primer during the initial polyA priming. Samples were processed as 3 pools, which were amplified using P7 and a template switching oligonucleotide that had been added to the 5′ end of RNA. Three different i5 indexes were used to distinguish the 3 sample pools. The Illumina P5 was added by tagmentation by Nextera transposase during amplification. Libraries were quantified and quality assessed by Qubit, Bioanalyzer and RT-qPCR, and one equimolar pool was prepared based upon size-corrected RT-qPCR quantitation. This library pool was loaded for on-board denaturation and clustering into one lane of the Illumina NextSeq 2000 platform for sequencing (at MHTP) using the P3 cycle kit, to generate 60 bp single-end 3′ end reads, with a depth of minimum 15 million reads/sample, or into one lane of the MGITech MGISEQ2000RS hardware (at Micromon), to generate 100 bp paired-end reads, with a minimum depth of 30 million reads/sample.
Analysis of bulk RNA sequencing data
The sequenced raw FASTQ files for the endogenous Tregs were analysed with the RNASik pipeline99 using STAR aligner100 with Mus musculus GRCm38 reference. For the macrophages, the analysis was performed using the nfcore/rnaseq 3.3 pipeline (forward-stranded), using STAR aligner100 with the Mus musculus GRCm39 reference, including “with_umi” flag for UMI-deduplication. Reads were quantified using featureCounts101 producing the raw genes count matrix and various quality control metrics. Raw counts (forward-stranded reads) were then analysed with Degust web tool90 which performs normalisation using trimmed mean of M values (TMM)91 and differential expression analysis using limma/voom92. Samples were batch corrected to account for variation between two different sequencing facilities across replicates. This was done within Degust by adding a covariate to the linear model for each sequencing batch which reduced the noise and improved the power of the test. Genes with an FDR adjusted p value < 0.05 were considered differentially expressed (DE). The MA plots and Volcano plots were generated using GGPlot2102 in R (version 4.2.2). Venn diagrams were generated using InteractiVenn103.
In vitro culture of Mo/MΦ and cytotoxic T cells isolated from injured muscle
Mo/MΦ and CD8+ T cells were sorted from injured muscle at D3 post-injury, and were centrifuged at 500 × g for 10 min at 4 °C. Both cell types were resuspended in an appropriate volume of RPMI 1640 + 5% heat-inactivated FBS, with 100 units/ml penicillin/streptomycin and 10 ng/ml M-CSF for subsequent seeding. For CD8+ T cell and Mo/MΦ co-culture, 10,000 Mo/MΦ were added to ultra-low binding 96-well plates, along with either no CD8+ T cells, or with 1000, or 2000 CD8+ T cells, representing a CD8+ T cells:Mo/MΦ ratio of 1:10 (low) and 1:5 (high), respectively. The suspension of CD8+ T cells and Mo/MΦ was cultured at 37 °C with 5% CO2 for 48 h before collecting them for subsequent staining steps.
In vitro culture of bone marrow-derived monocytes or injured muscle-derived Mo/MΦ
For treatment of monocytes or injured tissue Mo/MΦ with different Treg-derived factors, 1 × 105 primary monocytes or 20,000 sorted Mo/MΦ were added per well of a 96-well plate. Primary monocytes were isolated using the EasySep Mouse Monocyte Isolation Kit (STEMCELL Technologies), according to the manufacturer’s instructions, and cultured in DMEM/F12 (Thermo Fisher Scientific) with 10% FBS, 10 ng/ml M-CSF (PeproTech) and 100 units/ml penicillin/streptomycin, for 3 days, with or without 0.1 ng/ml IFN-γ (STEMCELL Technologies, #78021), and/or 1 ng/ml IL-1β (PeproTech, #211-11B), in the presence of one of the Treg-derived factors: recombinant mouse IL-10 (10 ng/ml, PeproTech, #210-10-10), recombinant human ANXA1 (500 ng/ml, R&D systems, #3770-AN-050), recombinant human ANXA2 (500 ng/ml, R&D systems, #9409-AN-050), recombinant mouse FGL-2 (375 ng/ml, R&D systems, #5257-FL), recombinant mouse Areg (500 ng/ml, PeproTech, #315-36), recombinant mouse Gal-1 (500 ng/ml, R&D Systems, #1245-GA), or recombinant mouse Gal-3 (500 ng/ml, R&D Systems, #1197-GA). Monocytes were cultured at 37 °C with 5% CO2 for 3 days, while sorted Mo/MΦ were cultured for 48 h before detaching them with TrypLE (Thermo Fisher Scientific) containing 2 mM EDTA and resuspending them in PBS for subsequent staining steps.
ELISAs for cytokines in injured muscle
Homogenised muscle tissue was incubated for 30 min on ice in T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific) using 10 ml/g of tissue containing 1 tablet of protease inhibitor (Roche)/7 ml. Samples were then centrifuged at 10,000 × g for 5 min and supernatants were stored at −80 °C. Total protein concentration was measured with a Bradford assay (Millipore) and cytokine/chemokine concentrations were detected using ELISA kits from R&D Systems: Mouse IL-1 beta/IL-1F2 DuoSet ELISA (#IL-1B-DY401-05); Mouse CCL2/JE/MCP-1 DuoSet ELISA (#DY479-05), Mouse IFNgamma DuoSet ELISA (#IFN-DY485-05); Mouse TNF-alpha DuoSet ELISA (#DY410-05); and Mouse IL-6 DuoSet ELISA (#DY406-05).
Immunofluorescence of injured tissue cryosections
To process the tissues for immunostaining, tissues were briefly washed in PBS, and fixed in 4% paraformaldehyde for 4 h, washed with PBS, cryoprotected overnight in 30% sucrose, and embedded in optimum cutting temperature (O.C.T) compound for cryo-sectioning (10 μm thickness). Sections were stored at −20 °C. Prior to processing, sections were briefly thawed at room temperature for 15 min and washed with PBS to prepare the slides for immunostaining. For detection of endogenous Tregs and myeloid cells, permeabilization was first performed with PBS containing 0.2% Triton X-100 for 4 min, followed by rinsing with PBS-T (PBS with 0.05% tween-20). Then blocking was performed with PBS containing 1% bovine serum albumin (BSA) and 10% normal goat serum (NGS) for 1 h at room temperature. Sudan Black B solution (0.1% in 70% ethanol, Sigma, #199664) was applied to tissue sections for 10 min to reduce tissue autofluorescence. Sections were stained with primary anti-mouse antibodies: rat anti-mouse F4/80 (Thermo Fisher Scientific, #14-4801-82, 5 μg/ml) and rat anti-human/mouse Foxp3 (PCH101) (Thermo Fisher Scientific, #14-4776-82, 5 μg/ml), added in 1% normal goat serum in 0.1% BSA in PBS overnight at 4 °C. After two washes of 5 min in PBS-T, sections were incubated for 2 h in a secondary antibody: goat anti-rat Alexa Fluor-488 (Thermo Fisher Scientific, #A-11006, 2 µg/ml) in 1% normal goat serum in 0.1% BSA in PBS at room temperature. For detection of exogenous Tregs, permeabilization, blocking and staining steps were skipped. Nuclei were stained with 0.1 mg/ml of DAPI. Sections were imaged on a Leica DMi8 fluorescent microscope and processed using Fiji.
In vitro culture and expansion of murine Tregs
CD4+, GFP+ Tregs sorted from spleens of Foxp3DTR/GFP mice were cultured in vitro using Dynabeads Mouse T-Activator CD3/CD28 for T-Cell Expansion and Activation (Thermo Fisher Scientific, #11452D) according to the manufacturer’s instructions. Briefly, Tregs were plated in 96-well ultra-low attachment plates at a density of 1.5 × 106 cells/ml with Dynabeads at a bead-to-cell ratio of 2:1 and cultured for 1 week in RPMI 1640 + 10% heat-inactivated FBS, with 100 units/ml penicillin/streptomycin and recombinant mouse IL-2 (2000 U/ml). On day 7, cells were detached from the plate by washing with PBS, centrifuged at 500 × g for 10 min, and used for in vivo delivery via fibrin gels. Over 95% of the expanded Tregs were FOXP3+.
RT-qPCR on sorted exogenous and endogenous Tregs
Exogenous Tregs were sorted on day 3 post-delivery, while endogenous Tregs were sorted on day 7 post-injury, for RT-qPCR using Taqman Fast Advanced Cells-to-CT (Thermo Fisher Scientific), according to the manufacturer’s instructions. Briefly, 2000 cells/subset were used as input, and lysed in 50 μl of Lysis solution containing DNase I, for 5 min at room temperature, followed by incubation with 5 μl Stop solution for 2 min at room temperature. Reverse transcription was performed using the Fast Advanced RT Buffer and RT Enzyme Mix, followed by qPCR using the Fast Advanced Master Mix and the following Taqman Gene Expression Assays (Thermo Fisher Scientific) for mouse genes: Areg (Mm01354339_m1), Il10 (Mm00439614_m1), Anxa1 (Mm00440225_m1), Lgals3 (Mm00802901_m1), Fgl2 (Mm00433327_m1), Gapdh (Mm99999915_g1), and Mrpl32 (Mm00777741_sH). Gapdh and Mrpl32 were used as housekeeping genes during data analysis.
Statistics & reproducibility
Statistical analyses were performed using GraphPad Prism 9 statistical software (GraphPad, USA). Data were tested for normality using the Shapiro–Wilk test. For normally distributed data, significant differences were calculated with Student’s t-test for pairwise comparisons, or by analysis of variance (ANOVA) for three or more groups, followed by Bonferroni post hoc test when performing multiple comparisons between groups. For non-normally distributed data, a Mann–Whitney U test was performed for pairwise comparisons, while a Kruskal–Wallis test was performed for three or more groups followed by the Dunn’s post hoc test for multiple comparisons. P < 0.05 was considered statistically significant. P values are indicated and the symbol *** indicates P values ≤ 0.001, while n.s. indicates not significant. No statistical method was used to predetermine sample size. No data were excluded from study reporting. For all in vivo experiments, mice were preselected based on age and then randomly assigned to treatment groups. Histological analyses of the bone and muscle tissues were blinded.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the staff at the Monash Animal Research Platform for technical services and colony maintenance, Dr. Michael de Veer at Monash Biomedical Imaging and Dr. Celeste Piotto for help with microCT scanning of some calvarial bone samples, Dr. Casey J. A. Anttila and Dr. Tracey M. Baldwin from The Walter and Eliza Hall Institute Flow Cytometry Facility and Advanced Genomics Facility for sorting and processing of mini-bulk RNA sequencing samples, Micromon Genomics (Monash University) for sequencing of endogenous skin Treg samples, Anh Vo at the Medical Genomics Facility in Monash Health Translation Precinct (MHTP) for assistance with assessing the RNA quantity and quality prior to sequencing, and the Monash Bioinformatics Platform (Prof. David Powell, Dr. Deanna Deveson Lucas, Dr. Andrew Perry and Ms Adele Barugahare) for assistance with processing and analysis of RNA sequencing data. We acknowledge the use of equipment and technical assistance of the Monash Histology Platform, Department of Anatomy and Developmental Biology, Monash University. We also thank Dr. Ziad Julier and Dr. Alon M. Douek for experimental advice, and Ms. Monika Mohenska for Bioinformatics support. Biorender.com was used to create some illustrations in the figures. This work was funded in part by the National Health and Medical Research Council (APP1140229 and APP1176213) to M.M.M., the Viertel Charitable Foundation Senior Medical Researcher Fellowship to M.M.M., and the Osaka University International Joint Research Promotion Program to S.A. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
Author contributions
M.M.M. conceptualised the initial study. B.N., J.L.T., Y.K.A., Y.-Z.L., J.M.D.L., N.H., and M.M.M. designed and conducted the experiments and interpreted the results. S.L. assisted with animal surgeries and histology. A.J.P. established the mouse colonies at Monash and the Treg depletion protocol. X.-N.W. provided the human Tregs. D.A.-Z. performed protocol development and optimisation, data generation, and study design of the mini-bulk RNA-sequencing. P.F.H. performed study design, data processing, and QC of the mini-bulk RNA sequencing data. T.W. designed and performed the bulk RNA sequencing strategy. G.A.K. and R.M. performed some of the microCT analyses. A.V. and S.A. gave conceptual insights and helped with experimental design and data interpretation. B.N. and M.M.M. wrote the initial draft and made the figures. B.N., J.L.T., Y.K.A., J.M.D.L., A.V., S.A., and M.M.M. edited the manuscript.
Peer review
Peer review information
Nature Communications thanks Qizhi Tang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data associated with this study are present in the main text or the Supplementary Materials. RNA sequencing data generated for this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession code GSE228871 for bulk RNA-seq of endogenous Tregs, GSE230173 for mini-bulk RNA-seq of monocytes/macrophages from Treg-treated mice, GSE230177 for mini-bulk RNA-seq of recovered exogenous Tregs and GSE268828 for bulk RNA-seq of macrophages from Treg-depleted mice. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51353-2.
References
- 1.Julier, Z., Park, A. J., Briquez, P. S. & Martino, M. M. Promoting tissue regeneration by modulating the immune system. Acta Biomater.53, 13–28 (2017). 10.1016/j.actbio.2017.01.056 [DOI] [PubMed] [Google Scholar]
- 2.Forbes, S. J. & Rosenthal, N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat. Med.20, 857–869 (2014). 10.1038/nm.3653 [DOI] [PubMed] [Google Scholar]
- 3.Weber, E. W., Maus, M. V. & Mackall, C. L. The emerging landscape of immune cell therapies. Cell181, 46–62 (2020). 10.1016/j.cell.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legrand, J. M. D. & Martino, M. M. Growth factor and cytokine delivery systems for wound healing. Cold Spring Harb. Perspect. Biol.14, a041234 (2022). 10.1101/cshperspect.a041234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, J., Tan, J., Martino, M. M. & Lui, K. O. Regulatory T-cells: potential regulator of tissue repair and regeneration. Front. Immunol.9, 585 (2018). 10.3389/fimmu.2018.00585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, C. et al. Repair’ Treg cells in tissue injury. Cell. Physiol. Biochem.43, 2155–2169 (2017). 10.1159/000484295 [DOI] [PubMed] [Google Scholar]
- 7.Lei, H., Schmidt-Bleek, K., Dienelt, A., Reinke, P. & Volk, H. D. Regulatory T cell-mediated anti-inflammatory effects promote successful tissue repair in both indirect and direct manners. Front. Pharmacol.6, 184 (2015). 10.3389/fphar.2015.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell155, 1282–1295 (2013). 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuswanto, W. et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity44, 355–367 (2016). 10.1016/j.immuni.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nosbaum, A. et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J. Immunol.196, 2010–2014 (2016). 10.4049/jimmunol.1502139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weirather, J. et al. Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res.115, 55–67 (2014). 10.1161/CIRCRESAHA.115.303895 [DOI] [PubMed] [Google Scholar]
- 12.Xia, N. et al. A unique population of regulatory T cells in heart potentiates cardiac protection from myocardial infarction. Circulation142, 1956–1973 (2020). 10.1161/CIRCULATIONAHA.120.046789 [DOI] [PubMed] [Google Scholar]
- 13.Ito, M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature565, 246–250 (2019). 10.1038/s41586-018-0824-5 [DOI] [PubMed] [Google Scholar]
- 14.Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell162, 1078–1089 (2015). 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang, R. et al. CD4+FoxP3+CD73+ regulatory T cell promotes cardiac healing post-myocardial infarction. Theranostics12, 2707–2721 (2022). 10.7150/thno.68437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang, T.-T. et al. Regulatory T cells ameliorate cardiac remodeling after myocardial infarction. Basic Res. Cardiol.107, 232 (2011). 10.1007/s00395-011-0232-6 [DOI] [PubMed] [Google Scholar]
- 17.Kang, I.-H. et al. Quantitative increase in T regulatory cells enhances bone remodeling in osteogenesis imperfecta. iScience25, 104818 (2022). 10.1016/j.isci.2022.104818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julier, Z. et al. Enhancing the regenerative effectiveness of growth factors by local inhibition of interleukin-1 receptor signaling. Sci. Adv.6, eaba7602 (2020). 10.1126/sciadv.aba7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alshoubaki, Y. K. et al. A superior extracellular matrix binding motif to enhance the regenerative activity and safety of therapeutic proteins. NPJ Regen. Med.8, 25 (2023). 10.1038/s41536-023-00297-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol.8, 191–197 (2007). 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- 21.Ratnayake, D. et al. Macrophages provide a transient muscle stem cell niche via NAMPT secretion. Nature591, 281–287 (2021). 10.1038/s41586-021-03199-7 [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., Ge, J., Tredget, E. E. & Wu, Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc.8, 302–309 (2013). 10.1038/nprot.2013.002 [DOI] [PubMed] [Google Scholar]
- 23.King, A., Balaji, S., Le, L. D., Crombleholme, T. M. & Keswani, S. G. Regenerative wound healing: the role of interleukin-10. Adv. Wound Care3, 315–323 (2014). 10.1089/wound.2013.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur, S. et al. Annexin A1 drives macrophage skewing to accelerate muscle regeneration through AMPK activation. J. Clin. Investig.130, 1156–1167 (2020). 10.1172/JCI124635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bittel, D. C. et al. Annexin A2 mediates dysferlin accumulation and muscle cell membrane repair. Cells9, 1919 (2020). 10.3390/cells9091919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity50, 493–504.e497 (2019). 10.1016/j.immuni.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, F.-T. Regulatory roles of galectins in the immune response. Int. Arch. Allergy Immunol.136, 385–400 (2005). 10.1159/000084545 [DOI] [PubMed] [Google Scholar]
- 28.Vasanthakumar, A. et al. The TNF receptor superfamily-NF-κB axis is critical to maintain effector regulatory T cells in lymphoid and non-lymphoid tissues. Cell Rep.20, 2906–2920 (2017). 10.1016/j.celrep.2017.08.068 [DOI] [PubMed] [Google Scholar]
- 29.DiSpirito, J. R. et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol.3, eaat5861 (2018). 10.1126/sciimmunol.aat5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W. et al. Steroid nuclear receptor coactivator 2 controls immune tolerance by promoting induced Treg differentiation via up-regulating Nr4a2. Sci. Adv.8, eabn7662 (2022). 10.1126/sciadv.abn7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Povoleri, G. A. M. et al. Human retinoic acid–regulated CD161+ regulatory T cells support wound repair in intestinal mucosa. Nat. Immunol.19, 1403–1414 (2018). 10.1038/s41590-018-0230-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarin, P. et al. Treg cells require Izumo1R to regulate γδT cell-driven inflammation in the skin. Proc. Natl Acad. Sci. USA120, e2221255120 (2023). 10.1073/pnas.2221255120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, W. H. & Lai, M. Z. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell. Biol.25, 1367–1378 (2005). 10.1128/MCB.25.4.1367-1378.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahl, S. et al. Metalloprotease-mediated shedding of enzymatically active mouse ecto-ADP-ribosyltransferase ART2.2 upon T cell activation. J. Immunol.165, 4463–4469 (2000). 10.4049/jimmunol.165.8.4463 [DOI] [PubMed] [Google Scholar]
- 35.Hou, X. X., Wang, X. Q., Zhou, W. J. & Li, D. J. Regulatory T cells induce polarization of pro-repair macrophages by secreting sFGL2 into the endometriotic milieu. Commun. Biol.4, 499 (2021). 10.1038/s42003-021-02018-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summan, M. et al. Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. Am. J. Physiol. Regul. Integr. Comp. Physiol.290, R1488–R1495 (2006). 10.1152/ajpregu.00465.2005 [DOI] [PubMed] [Google Scholar]
- 37.Arnold, L. et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med.204, 1057–1069 (2007). 10.1084/jem.20070075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapouri-Moghaddam, A. et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol.233, 6425–6440 (2018). 10.1002/jcp.26429 [DOI] [PubMed] [Google Scholar]
- 39.Varga, T. et al. Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J. Immunol.196, 4771–4782 (2016). 10.4049/jimmunol.1502490 [DOI] [PubMed] [Google Scholar]
- 40.Korhonen, R. et al. Attenuation of the acute inflammatory response by dual specificity phosphatase 1 by inhibition of p38 MAP kinase. Mol. Immunol.48, 2059–2068 (2011). 10.1016/j.molimm.2011.06.439 [DOI] [PubMed] [Google Scholar]
- 41.Brady, R. D. et al. Thymosin β4 administration enhances fracture healing in mice. J. Orthop. Res.32, 1277–1282 (2014). 10.1002/jor.22686 [DOI] [PubMed] [Google Scholar]
- 42.Corna, G. et al. The repair of skeletal muscle requires iron recycling through macrophage ferroportin. J. Immunol.197, 1914–1925 (2016). 10.4049/jimmunol.1501417 [DOI] [PubMed] [Google Scholar]
- 43.Ye, T. et al. Cardiac overexpression of Chil1 improves wound healing to prevent cardiac rupture after myocardial infarction. J. Cardiovasc. Transl. Res.16, 608–623 (2023). [DOI] [PubMed]
- 44.Wynn, T. A. & Vannella, K. M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity44, 450–462 (2016). 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufour, A. et al. C-terminal truncation of IFN-γ inhibits proinflammatory macrophage responses and is deficient in autoimmune disease. Nat. Commun.9, 2416 (2018). 10.1038/s41467-018-04717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabrera, S. et al. Overexpression of MMP9 in macrophages attenuates pulmonary fibrosis induced by bleomycin. Int. J. Biochem. Cell Biol.39, 2324–2338 (2007). 10.1016/j.biocel.2007.06.022 [DOI] [PubMed] [Google Scholar]
- 47.Oh, S. Y., Lee, S. J., Jung, Y. H., Lee, H. J. & Han, H. J. Arachidonic acid promotes skin wound healing through induction of human MSC migration by MT3-MMP-mediated fibronectin degradation. Cell Death Dis.6, e1750–e1750 (2015). 10.1038/cddis.2015.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham, T.-H. et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood119, e161–e171 (2012). 10.1182/blood-2012-01-402453 [DOI] [PubMed] [Google Scholar]
- 49.Chen, H. H. et al. IRF2BP2 reduces macrophage inflammation and susceptibility to atherosclerosis. Circ. Res.117, 671–683 (2015). 10.1161/CIRCRESAHA.114.305777 [DOI] [PubMed] [Google Scholar]
- 50.Feng, X. et al. CCL6 promotes M2 polarization and inhibits macrophage autophagy by activating PI3-kinase/Akt signalling pathway during skin wound healing. Exp. Dermatol.32, 403–412 (2023). 10.1111/exd.14718 [DOI] [PubMed] [Google Scholar]
- 51.Kaufmann, A., Salentin, R., Gemsa, D. & Sprenger, H. Increase of CCR1 and CCR5 expression and enhanced functional response to MIP-1 alpha during differentiation of human monocytes to macrophages. J. Leukoc. Biol.69, 248–252 (2001). 10.1189/jlb.69.2.248 [DOI] [PubMed] [Google Scholar]
- 52.Liu, K. et al. Neutrophilic granule protein (NGP) attenuates lipopolysaccharide-induced inflammatory responses and enhances phagocytosis of bacteria by macrophages. Cytokine128, 155001 (2020). 10.1016/j.cyto.2020.155001 [DOI] [PubMed] [Google Scholar]
- 53.Kong, J. S. et al. Dynamic transcriptome analysis unveils key proresolving factors of chronic inflammatory arthritis. J. Clin. Investig.130, 3974–3986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue, Y. et al. IL4I1 is a novel regulator of M2 macrophage polarization that can inhibit T cell activation via L-tryptophan and arginine depletion and IL-10 production. PLoS ONE10, e0142979 (2015). 10.1371/journal.pone.0142979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, R., Zhang, G. & Chen, S. Y. Response gene to complement 32 protein promotes macrophage phagocytosis via activation of protein kinase C pathway. J. Biol. Chem.289, 22715–22722 (2014). 10.1074/jbc.M114.566653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jablonski, K. A. et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE10, e0145342 (2015). 10.1371/journal.pone.0145342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, H., Manivannan, A., Crane, I., Dawson, R. & Liversidge, J. Critical but divergent roles for CD62L and CD44 in directing blood monocyte trafficking in vivo during inflammation. Blood112, 1166–1174 (2008). 10.1182/blood-2007-06-098327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hiebert, P. R. & Granville, D. J. Granzyme B in injury, inflammation, and repair. Trends Mol. Med.18, 732–741 (2012). 10.1016/j.molmed.2012.09.009 [DOI] [PubMed] [Google Scholar]
- 59.Ganta, V. C. et al. A microRNA93–interferon regulatory factor-9–immunoresponsive gene-1–itaconic acid pathway modulates M2-like macrophage polarization to revascularize ischemic muscle. Circulation135, 2403–2425 (2017). 10.1161/CIRCULATIONAHA.116.025490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panduro, M., Benoist, C. & Mathis, D. T. Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc. Natl Acad. Sci. USA115, E2585–E2593 (2018). 10.1073/pnas.1800618115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tretina, K., Park, E. S., Maminska, A. & MacMicking, J. D. Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J. Exp. Med.216, 482–500 (2019). 10.1084/jem.20182031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin, R. M., Warunek, J. & Wohlfert, E. A. Therapeutic administration of IL-10 and amphiregulin alleviates chronic skeletal muscle inflammation and damage induced by infection. Immunohorizons2, 142–154 (2018). 10.4049/immunohorizons.1800024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, X. N. et al. Regulatory T-cell suppression of CD8+ T-cell-mediated graft-versus-host reaction requires their presence during priming. Transplantation88, 188–197 (2009). 10.1097/TP.0b013e3181ac14ce [DOI] [PubMed] [Google Scholar]
- 64.Mavin, E., Dickinson, A. & Wang, X. N. Do cryopreserved regulatory T cells retain their suppressive potency? Transplantation95, e68–e70 (2013). 10.1097/TP.0b013e318291175c [DOI] [PubMed] [Google Scholar]
- 65.Fraser, H. et al. A rapamycin-based GMP-compatible process for the isolation and expansion of regulatory T cells for clinical trials. Mol. Ther. Methods Clin. Dev.8, 198–209 (2018). 10.1016/j.omtm.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yolcu, E. S. et al. Apoptosis as a mechanism of T-regulatory cell homeostasis and suppression. Immunol. Cell Biol.86, 650–658 (2008). 10.1038/icb.2008.62 [DOI] [PubMed] [Google Scholar]
- 67.Uaesoontrachoon, K., Wasgewatte Wijesinghe, D. K., Mackie, E. J. & Pagel, C. N. Osteopontin deficiency delays inflammatory infiltration and the onset of muscle regeneration in a mouse model of muscle injury. Dis. Model. Mech.6, 197–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kyriakides, T. R. & Maclauchlan, S. The role of thrombospondins in wound healing, ischemia, and the foreign body reaction. J. Cell Commun. Signal.3, 215–225 (2009). 10.1007/s12079-009-0077-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao, C., Guo, H., Mi, Z., Grusby, M. J. & Kuo, P. C. Osteopontin induces ubiquitin-dependent degradation of STAT1 in RAW264.7 murine macrophages. J. Immunol.178, 1870–1881 (2007). 10.4049/jimmunol.178.3.1870 [DOI] [PubMed] [Google Scholar]
- 70.Greenlee, K. J. et al. Proteomic identification of in vivo substrates for matrix metalloproteinases 2 and 9 reveals a mechanism for resolution of inflammation. J. Immunol.177, 7312–7321 (2006). 10.4049/jimmunol.177.10.7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tiemessen, M. M. et al. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc. Natl Acad. Sci. USA104, 19446–19451 (2007). 10.1073/pnas.0706832104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, J. Neutrophils in tissue injury and repair. Cell Tissue Res.371, 531–539 (2018). 10.1007/s00441-017-2785-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishimura, S. et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med.15, 914–920 (2009). 10.1038/nm.1964 [DOI] [PubMed] [Google Scholar]
- 74.McNally, A., Hill, G. R., Sparwasser, T., Thomas, R. & Steptoe, R. J. CD4+CD25+ regulatory T cells control CD8+ T-cell effector differentiation by modulating IL-2 homeostasis. Proc. Natl Acad. Sci. USA108, 7529–7534 (2011). 10.1073/pnas.1103782108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reinke, S. et al. Terminally differentiated CD8+ T cells negatively affect bone regeneration in humans. Sci. Transl. Med.5, 177ra136 (2013). 10.1126/scitranslmed.3004754 [DOI] [PubMed] [Google Scholar]
- 76.Davis, P. A., Corless, D. J., Aspinall, R. & Wastell, C. Effect of CD4+ and CD8+ cell depletion on wound healing. Br. J. Surg.88, 298–304 (2001). 10.1046/j.1365-2168.2001.01665.x [DOI] [PubMed] [Google Scholar]
- 77.Vermare, A., Guérin, M. V., Peranzoni, E. & Bercovici, N. Dynamic CD8+ T cell cooperation with macrophages and monocytes for successful cancer immunotherapy. Cancers14, 3546 (2022). 10.3390/cancers14143546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity28, 546–558 (2008). 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- 79.Ostmann, A. et al. Regulatory T cell–derived IL-10 ameliorates crescentic GN. J. Am. Soc. Nephrol.24, 930–942 (2013). 10.1681/ASN.2012070684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raffin, C., Vo, L. T. & Bluestone, J. A. Treg cell-based therapies: challenges and perspectives. Nat. Rev. Immunol.20, 158–172 (2020). 10.1038/s41577-019-0232-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharir, R. et al. Experimental myocardial infarction induces altered regulatory T cell hemostasis, and adoptive transfer attenuates subsequent remodeling. PLoS ONE9, e113653 (2014). 10.1371/journal.pone.0113653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang, T. T. et al. Regulatory T cells ameliorate cardiac remodeling after myocardial infarction. Basic Res. Cardiol.107, 232 (2012). 10.1007/s00395-011-0232-6 [DOI] [PubMed] [Google Scholar]
- 83.Short, W. D. et al. IL-10 promotes endothelial progenitor cell infiltration and wound healing via STAT3. FASEB J.36, e22298 (2022). 10.1096/fj.201901024RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huynh, T. et al. Local IL-10 delivery modulates the immune response and enhances repair of volumetric muscle loss muscle injury. Sci. Rep.13, 1983 (2023). 10.1038/s41598-023-27981-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunn, L. et al. Murine model of wound healing. J. Vis. Exp.75, e50265 (2013). [DOI] [PMC free article] [PubMed]
- 86.Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol.17, 77 (2016). 10.1186/s13059-016-0938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res.41, e108 (2013). 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian, L. et al. scPipe: a flexible R/Bioconductor preprocessing pipeline for single-cell RNA-sequencing data. PLoS Comput. Biol.14, e1006361 (2018). 10.1371/journal.pcbi.1006361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rainer, J. EnsDb.Mmusculus.v79: Ensembl based annotation package. R package version 2.99.0 (2017).
- 90.Powell, D. R. Degust: interactive RNA-seq analysis. Zenodo (2019).
- 91.Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol.11, R25 (2010). 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol.15, R29 (2014). 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12 (2019).
- 95.Neuwirth, E. RColorBrewer: ColorBrewer Palettes. R package version 11–13 (2022).
- 96.Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res.47, W191–W198 (2019). 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu, G. Enrichplot: visualization of functional enrichment result. R package version 1.20.0 (2023).
- 98.Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsyganov, K., Perry, A., Archer, S. K., Powell, D. RNAsik: a pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw.3, 583 (2018).
- 100.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 102.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2016).
- 103.Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P. & Minghim, R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform.16, 169 (2015). 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the main text or the Supplementary Materials. RNA sequencing data generated for this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession code GSE228871 for bulk RNA-seq of endogenous Tregs, GSE230173 for mini-bulk RNA-seq of monocytes/macrophages from Treg-treated mice, GSE230177 for mini-bulk RNA-seq of recovered exogenous Tregs and GSE268828 for bulk RNA-seq of macrophages from Treg-depleted mice. Source data are provided with this paper.