Abstract
There is a lack of information on how individual microsatellite loci differ with respect to their mutation properties. Such variation will have an important bearing on our understanding of the ubiquitous occurrence of simple repeat sequences in eukaryotic genomes and on deriving proper mutation models that can be incorporated into genetic distance estimates. We genotyped ∼100 families of the bird barn swallow (Hirundo rustica) for two hypervariable (heterozygosity >95%) microsatellite markers: HrU6, an (AAAG)n tetranucleotide repeat, and HrU10, an (AAGAG)n pentanucleotide repeat. A total of 27 germline mutation events were documented, corresponding to mutation rates of 0.57% (HrU6) and 1.56% (HrU10). The mutation rate increased with allele size, at ∼0.1% per repeat unit over the observed range of allele sizes (∼10–100 repeat units). Single repeat unit changes dominated, with 21/27 mutations representing the gain or loss of one repeat unit. There was no clear difference in the number of gains versus losses nor was there an effect of allele size on the magnitude or direction of mutation. Unexpectedly, the mutation rate of females (maternally transmitted mutations) was 2.5–5 times higher than that of males. Contrasting these observations with mutation data from other microsatellite loci reveals differences not only in the mutation rate, but also in the magnitude, direction and effect of sex on mutation. Thus, microsatellite mutation and evolution may be viewed as a dynamic and variable process.
INTRODUCTION
To be able to understand the ubiquitous occurrence of microsatellite DNA it is important to have a detailed picture of the instability of these simple sequence repeats. Moreover, elucidating the background of the extreme repeat expansion displayed by trinucleotide loci associated with human disease (1,2) and properly using microsatellites in evolutionary studies (3) require a profound understanding of the mutation process. Several approaches and model systems have been used for the characterisation of microsatellite mutations, e.g. yeast or bacterial plasmids with inserted microsatellite sequences (4,5), somatic mutations in transformed cell lines (6), comparative studies of evolutionarily accumulated mutations at orthologous loci in related species (7) and modelling based on population data (8). However, the information gained from most approaches is based on indirect observations only and might not always mirror the true in vivo mutation process in the germline (3). Direct records of new length variants identified in comparisons between parents and offspring in pedigrees may therefore be considered the most unambiguous way of analysing microsatellite mutation patterns (9). Only a limited number of such studies are available.
Recently, two surveys of microsatellite mutations identified in human pedigrees have revealed a general picture of the mutation process in the human genome (10,11). These studies suggest distinct mutation biases with respect to magnitude, direction, allele size, repeat type and sex. However, as they are based on data from only a few mutation events per locus there is still a lack of detailed information on if and, in that case, how individual microsatellite loci differ with respect to mutation properties. Clarifying this would be important for our understanding of the evolutionary dynamics and also the possible function of this ubiquitous class of repetitive DNA.
In this study we have analysed the mutational properties of two hypermutable (germline mutation rates of ∼1%) microsatellite loci, HrU6 and HrU10, in the passerine bird barn swallow (Hirundo rustica). HrU6 is a tetranucleotide repeat locus consisting of an (AAAG)n motif, while HrU10 is a pentanucleotide repeat with an (AAGAG)n motif. We followed the segregation of these two markers through 1000–2000 meioses in pedigrees generally consisting of mother, father and several chicks. The identification of a large number of de novo mutations allowed inferences on the mutational spectrum to be made separately for each locus.
MATERIALS AND METHODS
Samples and DNA extraction
Bird samples were collected at seven sites in Italy and four sites in Spain. Barn swallows breed in colonies of up to a few hundred individuals (12) and at each site almost all breeding individuals as well as their offspring were captured and a small blood sample was collected. Parentage testing using microsatellites was done as part of an ongoing ecological study of the social behaviour of barn swallows (13). Blood samples were kept frozen until a small amount was thawed and genomic DNA was extracted by either a Chelex-based method according to Walsh et al. (14) or by standard phenol/chloroform extraction.
Microsatellite markers
All individuals were genotyped for the microsatellite markers HrU6 (15) and HrU9 (16). In addition, all Spanish swallows were genotyped for HrU10 (16), while all Italian birds were genotyped for HrU5 (15). These markers are well suited for parentage testing since they display extreme polymorphism (see further below).
Whenever a mutation was suspected from non-congruence between parental and offspring genotypes, additional markers were genotyped to confirm parentage: HrU5 (for Spanish birds), HrU10 (for Italian birds), Pdo5 (17), POCC6 (18) and Mcy_4 (19) (both populations). Mutations found at HrU9 have been reported previously (20).
PCR conditions
Primer sequences were as described in our original reports. For HrU6, HrU9 and HrU10 one primer was either fluorescently or radioactively labelled while for the other markers fluorescently labelled dCTP (Perkin-Elmer) was incorporated during PCR amplification. The 10 µl PCR reactions contained 1 µl of DNA, 5 pmol each primer, 1× PCR Buffer II (Perkin-Elmer), 1.75 mM MgCl2, 0.2 mM each dNTP (Amersham) and 0.5 U AmpliTaq (Perkin-Elmer). The thermal profile included an initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, annealing temperature as in the original reference for 40 s and 72°C for 40 s. The profile was ended by a 5 min prolonged extension step. For the Italian material a modified protocol was used (15).
Scoring and interpretation
PCR products were separated by electrophoresis in 6% polyacrylamide gels. Radioactively labelled PCR products were run together with a ladder of alleles of known sizes and were visualised by autoradiography. Fluorescently labelled products were run on an ABI377 instrument with the internal size standard GeneScan 500 (Perkin-Elmer). When testing for parentage, alleles scored with one of the two methods were only compared with alleles scored by the same method.
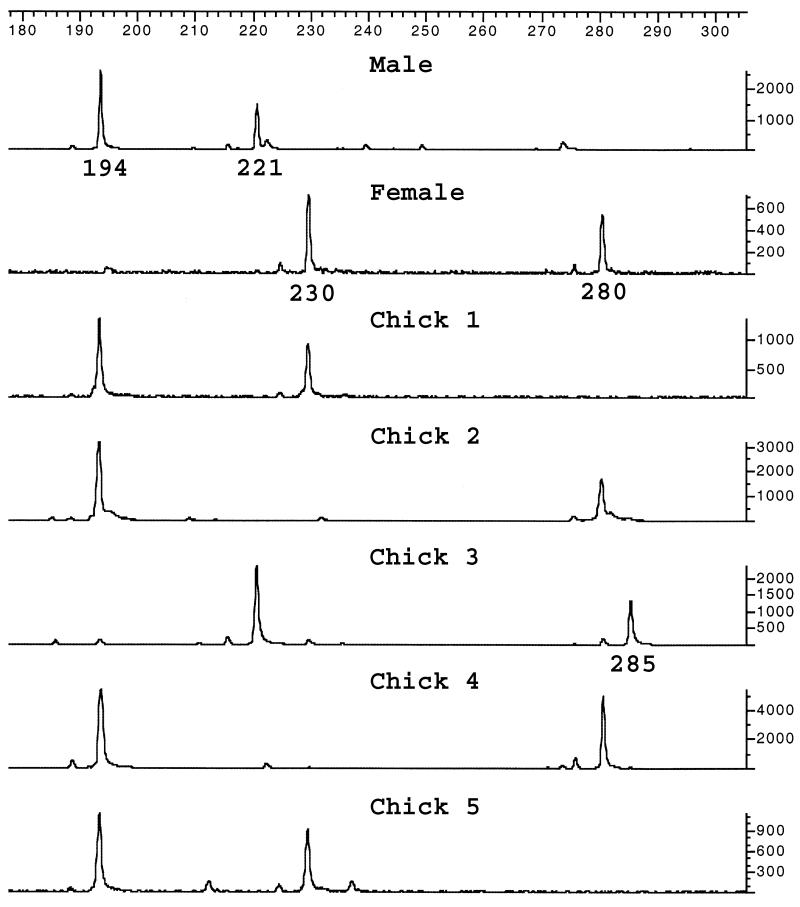
The barn swallow is a socially monogamous species in which extra-pair fertilisations occur (12). This means that non-congruence between presumed parent and offspring genotypes may either be due to the social father not being the biological father or to a germline mutation having arisen. To distinguish between these two possibilities we used the following criteria. When a chick’s genotype was non-congruent with one of the parent’s genotypes at all three loci initially scored we interpreted this as a case of extra-pair fertilisation. This seems like a plausible interpretation given the incidence of extra-pair fertilisation (>10%; 13) and the low probability of three independent mutation events at all loci (the likelihood of which is ∼0.013, i.e. 10–6). If one marker showed non-congruence while the two others were compatible with Mendelian inheritance (Fig. 1), we used the additional set of markers (see above) to elucidate whether the offspring was the product of an illegitimate mating or represented a de novo mutation event. In the Spanish material we also elucidated parentage with more markers when one marker showed congruence but two did not.
Figure 1.
Electropherograms showing a germline mutation event (chick 3) at the HrU10 pentanucleotide repeat locus.
Whenever a mutation was suspected both parents and offspring were independently reamplified and scored to eliminate genotyping error. Mutations were assumed to have occurred in the parental allele which required the fewest number of repeat unit changes to explain the observed length difference. It cannot be formally proven that this was consistently the case, but it is a commonly applied and reasonable assumption (20–23). One mutation could not be assigned to a particular parent since both parents carried the allele that presumably gave rise to the mutation. It should be noted that due to the high heterozygosity of the markers used in this study, all parents in which a mutation was inferred were heterozygous for the locus in question. This excludes the possibility of a segregating null allele in an apparently homozygous parent being incorrectly interpreted as a mutation to the same allele size as transmitted by the other parent. HrU6 and HrU10 possessed alleles that did not differ by multiples of the length of the respective repeat motif, indicating length polymorphism in the sequence adjacent to the main repeat or in the primary repeat structure (as was also suggested by the sequences of the original clones: HrU6, accession no. X84091; HrU10, accession no. X97562). For the analysis of the effect of allele size on mutation rate we therefore binned alleles into 4 (HrU6) and 5 bp (HrU10) size classes.
RESULTS
Ninety-two Spanish and 120 Italian barn swallow families were genotyped for two hypervariable microsatellite loci, HrU6 and HrU10, for the purpose of parentage testing. The numbers of observed alleles were 78 (HrU6) and 66 (HrU10) and the observed heterozygosities were 0.965 and 0.971, respectively. As a consequence of their extreme polymorphism, both loci contained unusually long alleles with a mean number of 29.5 repeat units at HrU6 (range 5–94) and 26.6 at HrU10 (15–91).
During the genotyping process HrU6 and HrU10 were found to be hypermutable. A total of 27 germline mutations were recorded, 12 at HrU6 and 15 at HrU10 (Table 1). The HrU6 mutations were scored among a total of 2093 meioses, corresponding to mutation rates of 0.57% (95% confidence interval 0.25–0.90%). For HrU10 960 meioses were scored and the mutation rate was 1.56% (0.78–2.35%).
Table 1. Characteristics of 27 germline microsatellite mutations from the barn swallow HrU6 and HrU10 loci.
| Marker | Parental sex | Direction and magnitude | Allele size (bp) | Estimated number of repeat units |
|---|---|---|---|---|
| HrU6 | Female | –1 | 335 | 53 |
| Female | –1 | 343 | 55 | |
| Female | –2 | 347 | 56 | |
| Female | –3 | 459 | 84 | |
| Female | –2 | 257 | 33 | |
| Female | +1 | 381 | 64 | |
| Female | +2 | 381 | 64 | |
| Female | +1 | 381 | 64 | |
| Female | –1 | 373 | 62 | |
| Female | +1 | 273 | 37 | |
| Male | +1 | 279 | 39 | |
| Male | +1 | 375 | 63 | |
| HrU10 | Female | +1 | 269 | 32 |
| Female | +1 | 235 | 26 | |
| Female | –1 | 235 | 26 | |
| Female | –1 | 366 | 52 | |
| Female | –2 | 232 | 25 | |
| Female | –1 | 232 | 25 | |
| Female | +1 | 225 | 24 | |
| Female | –1 | 264 | 31 | |
| Female | +1 | 280 | 35 | |
| Female | –1 | 278 | 34 | |
| Female | +1 | 231 | 25 | |
| Female | –1 | 269 | 32 | |
| Male | +1 | 265 | 32 | |
| Male | –2 | 234 | 25 | |
| a | +1 | 231 | 25 |
aParental origin of mutation could not be inferred.
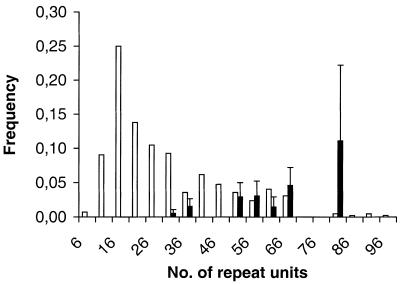
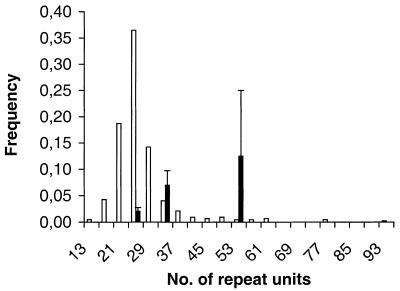
The size distribution of the mutating alleles was shifted towards longer alleles compared to the total allele frequency distribution (Figs 2 and 3). The mean length of mutating alleles at HrU6 was 346 bp, while it was 231 bp for non-mutating alleles (z = –4.564, P = 0.0001, Mann–Whitney U-test). For HrU10 the corresponding means were 256 and 242 bp, respectively (z = –2.300, P < 0.05).
Figure 2.
Allele frequency distributions at the HrU6 locus. Alleles have been binned in five repeat unit (20 bp) intervals. Open bars represent frequencies of non-mutating alleles while filled bars represent mutating alleles. Error bars indicate the standard error of mutation rate estimates.
Figure 3.
As in Figure 2 but for the HrU10 locus. Alleles binned in four repeat unit (20 bp) intervals.
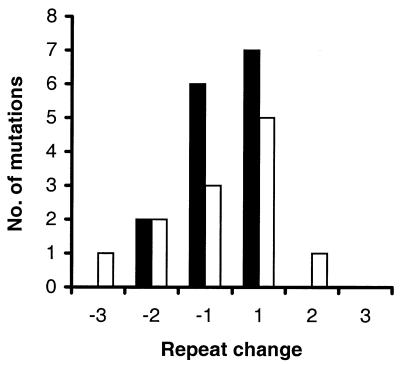
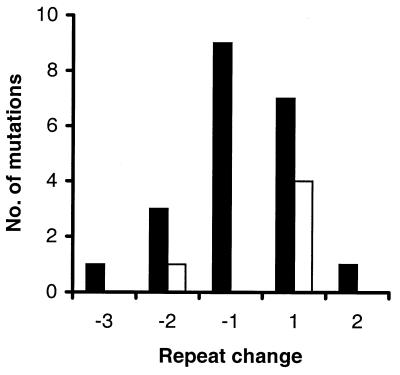
Single repeat unit changes dominated, with 8 out of 12 (HrU6, 67%) and 13 out of 15 (HrU10, 87%) representing gain or loss of one unit. There was no difference in the number of gains versus losses (Fig. 4), neither at HrU6 (6/6) nor at HrU10 (7/8). The magnitude of gains and losses was independent of allele size (magnitude regressed on allele size: HrU6, r2 = 0.011, P = 0.374; HrU10, r2 = 0.013, P = 0.341), thus not indicating a tendency for long alleles to decrease in size (see Discussion).
Figure 4.
Magnitude and direction of germline mutations detected at the HrU6 (open bars) and HrU10 (filled bars) loci.
There was an excess of maternally derived mutations for both loci. At HrU10 12 mutations arose in 530 female meioses (rate = 2.3%), while two mutations were revealed among 430 male meioses (0.47%, χ2 = 4.17, df = 1, P < 0.05). At HrU6 the corresponding figures were nine mutations in 1094 female meioses (0.82%) and three mutations in 999 male meioses (0.30%, χ2 = 1.67, df = 1, P = 0.196). Although not supported by strong statistical significance, these data suggest a female-biased mutation rate by a factor of 2.5–5 at these loci. There was no apparent difference in the distribution of gains and losses in relation to sex (Fig. 5; HrU6, P = 0.091; HrU10, P = 0.692, Fisher’s exact test).
Figure 5.
Magnitude and direction for HrU6 and HrU10 mutations in relation to sex. Filled bars, maternally derived mutations; open bars, paternally derived mutations.
DISCUSSION
An important impetus for this study was to address to what extent the mutation process may differ between individual microsatellite loci. Heterogeneity in the microsatellite mutation process is indicated by the failure of distance estimates based on simple mutation models to correctly date evolutionary divergences (see 24 and references therein). In the following we shall discuss this issue by contrasting our data on 27 de novo germline mutations at the barn swallow HrU6 and HrU10 loci to (i) the overall mutation pattern for microsatellites in the human genome (10,11), (ii) data from individual loci in human and other organisms including, in particular, (iii) a third hypermutable locus in barn swallows, HrU9 (20,25).
Mutation rate estimates are required in many models of DNA sequence evolution, yet this proves to be one of the most difficult parameters to estimate for evolutionary genetic analysis. Direct observations, by pedigree analysis, of mutations at human microsatellite loci have revealed estimates of mean mutation rates of 3 × 10–3–6 × 10–4 (26). Most of these studies have concerned a limited number of loci. Moreover, as studies reporting microsatellite mutations tend to deal with loci where mutations are relatively common, it is possible that the sets of loci used for analysis have been biased towards the upper end of the genomic distribution of mutation rates. Genethon’s extensive genotyping of >5000 microsatellite loci in the human genome, which might be considered a more representative data set in this respect, has suggested a lower mean genomic mutation rate, of ∼10–4 (27,28). However, even in this case the data set may be biased as it is based on highly polymorphic markers only. In the light of this it is clear that HrU6 and HrU10 are unusually mutable markers, the rate being close to the percentage level. Although an even higher rate (3–4%) has been reported for HrU9, an (AAAG)n barn swallow tetranucleotide repeat locus (20,25), repeat instability is not a species-specific trait for swallows, as other microsatellites have been documented with stable Mendelian inheritance in this species (15). Nor do bird microsatellites in general seem more unstable than human markers (29). Our data therefore add to a dynamic view of microsatellite evolution by demonstrating significant heterogeneity in the locus-specific mutation rate. As a consequence, using mean mutation rate estimates in the interpretation of microsatellite data can potentially flaw evolutionary analyses. This may be particularly problematic when the number of markers is low.
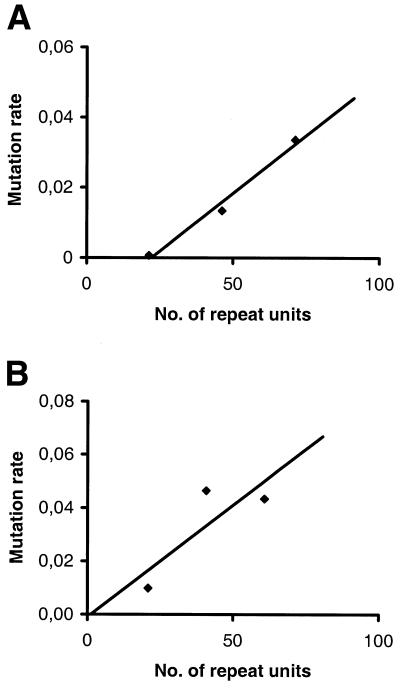
Previous studies of both individual loci (16,20,30) and of pooled data from the human genome (10) have demonstrated a positive relationship between microsatellite repeat length (allele size) and mutation rate. A similar trend was found at HrU6 and HrU10. Regressing mutation rate on allele size for three major allele size classes (10–30, 30–50 and 50+ repeat units) roughly suggests that the mutation rate increases by ∼0.1% per repeat unit over this size range (Fig. 6). If correct, the mutation rate would thus differ by about one order of magnitude between the shortest and the longest alleles at HrU6 and HrU10 (Fig. 6).
Figure 6.
Estimated rates of mutation at (A) HrU6 and (B) HrU10 in relation to allele size. Alleles have been grouped in three size classes: 10–30, 30–50 and 50+ repeat units.
There was no propensity for mutations at HrU6 or HrU10 to either lead to an increase or decrease in repeat length. In contrast, data from individual loci in several species, including the HrU9 locus in barn swallows (20,25), and pooled data on dinucleotide repeats from the human genome (10,31) show directionality in the mutation process, with an excess of insertions over deletions. However, Xu et al. (11) found no such directionality when analysing mutations at human tetranucleotide repeat loci. A microsatellite locus may ‘grow’ if processes promoting its expansion dominate over processes acting in the opposite direction. Not invoking selection, growth may either be an active process due to truly biased mutation or simply represent stochastic variation between loci. Under both scenarios it is likely that some loci will eventually reach appreciable lengths unless there is some mechanism counteracting further expansion. For instance, an upper limit of allele size has been incorporated in some models of microsatellite mutation (32,33), although it is not clear what biological mechanism could account for such a constraint. Alternatively, it has been indicated by several studies that long alleles are more prone to decrease in size (10,11,20,34,35), a situation that would obviously counteract evolution into huge arrays. The absence of such a trend for HrU6 and HrU10 could potentially be related to the fact that they are unusually long.
Point mutations causing interruptions in perfect repeat arrays are likely to abridge the mutation rate to new length variants (compare the positive relationship between repeat length with mutation rate discussed above). A plausible model to explain why most microsatellites do not attain appreciable lengths therefore posits that the equilibrium distribution of microsatellite repeat lengths is reflected in a balance between the point mutation rate and the rate of replication slippage (36). According to this view, it is possible that if the point mutation rate is sufficiently low or the slippage rate is sufficiently high, some loci may evolve into huge arrays. By this reasoning the extensive repeat lengths displayed by HrU6 and HrU10 can potentially be explained by a high slippage mutation rate, as we have documented, even if there is no directionality in the mutation process. Alternatively, the rate of point mutation could be low. It is interesting in this respect that a negative relationship between the substitution rate in flanking regions and microsatellite length has recently been demonstrated for orthologous microsatellites in mouse and rat (37).
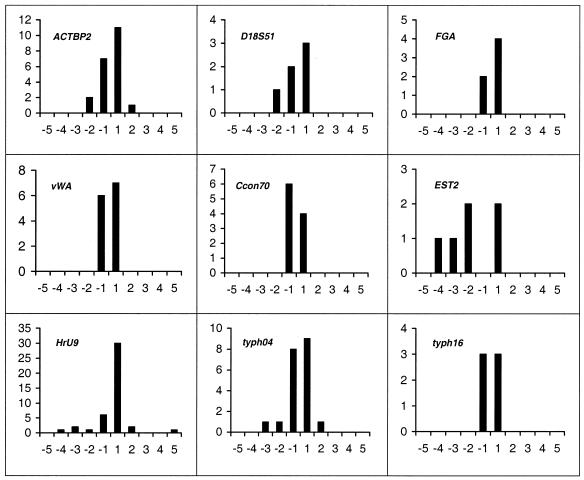
The magnitude of mutation at HrU6 and HrU10 favours a revised stepwise mutation model (4,38) where single step changes dominate but occasional multistep mutations do occur. To compare the mutational spectra at different microsatellite loci we extracted from the literature data for individual loci in different species where at least six germline mutations have been reported (Fig. 7). Generally, single step changes seem to dominate. However, at the Australian lizard EST2 locus six of eight mutations involved two or more repeat unit changes (χ2 = 14.85, df = 3, P < 0.05). Moreover, at the barn swallow HrU9 locus the spectrum is significantly different from that at the other loci (χ2 = 14.09, df = 3, P < 0.05), one repeat unit gains clearly outnumbering one repeat unit losses. It thus seems evident that a single mutation model cannot be applied to all microsatellite loci.
Figure 7.
Magnitude and direction of microsatellite mutations (n ≤ 6) from a range of species: human, Homo sapiens, ACTBP2, D18S51, FGA and vWA (10); ant, Camponotus consobrinus, Ccon70 (30); Australian lizard, Egernia stokesii, EST2 (23); barn swallow, Hirundo rustica, HrU9 (20); pipe fish, Syngnathus typhle, typh04 and typh16 (22).
An increasing body of evidence has indicated that the germline mutation rate of many vertebrates is male-biased (39–43). Tentatively, such a sex difference can be explained by the fact that more cell divisions precede the formation of male than female gametes and assuming that a significant proportion of nucleotide substitutions arise from replication errors. For instance, the number of human germ cell divisions has been estimated to be at least an order of magnitude higher in males than in females at average ages of reproduction (44).
Sex-specific mutation rates have mainly been analysed by indirect approaches, comparing the relative rates of sequence evolution on autosomes and sex chromosomes (39). The extraordinary germline mutation rate of microsatellites allows the effect of sex to be analysed by a direct approach, through pedigree analysis (10). Accepting replication-dependent slipped strand mispairing to be the prevailing source of microsatellite mutation, an excess of male germ cell divisions should be mirrored in a corresponding excess of paternally derived mutations. Thus, we unexpectedly found a 2.5–5-fold excess of maternally derived mutations for HrU6 and HrU10 (P < 0.05). If correct, this is unlikely to be explained by avian reproductive biology. Although there are no quantitative estimates available for passerine birds, the number of germ cell divisions should also be significantly higher in the male than female germline in these organisms, given the massive production of sperm (45) [for galliform birds Kahn and Quinn (45) estimated male germ cell divisions to be in excess by a factor of 3–5].
Evolutionary analyses of sex chromosome sequences have shown that the avian mutation rate is male-biased, including in passerine birds (41,45). Furthermore, for the barn swallow HrU9 locus we previously observed a 2-fold excess of male mutations (20). This is significantly different from the sex ratio at HrU6 (χ2 = 4.24, df = 1, P < 0.05) and HrU10 (χ2 = 8.48, df = 1, P < 0.01). We therefore conclude that HrU6 and HrU10 are unusual in terms of the relative contribution of mothers and fathers to germline mutation. To our knowledge, these data represent the first evidence that the effect of sex on the mutation rate varies between individual neutral microsatellite loci within a genome. Deviating sex-specific mutation rates have been reported for trinucleotide repeat expansion loci associated with human disease (1), although the cause of this variation is not known. Deviating sex-specific rates of point mutations have also been seen at human disease-causing loci (42). In the latter case, sex-specific methylation patterns can potentially account for such variation, e.g. at CpG sites. Methylation has not been implicated as playing a role in the process of microsatellite mutation but it cannot be completely excluded that methylation status affects the stability of the repeat array.
Acknowledgments
ACKNOWLEDGEMENTS
Nicola Saino and J. J. Cuervo are gratefully acknowledged for barn swallow samples. H.E. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg foundation. This study was supported by the Swedish Research Council for Agriculture and Forestry.
REFERENCES
- 1.Usdin K. and Grabczyk,E. (2000) DNA repeat expansions and human disease. Cell. Mol. Life Sci., 57, 914–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings C.J. and Zoghbi,H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 3.Golding D.B. and Schlötterer,C. (1999) Microsatellites Evolution and Applications. Oxford University Press, Oxford, UK.
- 4.Levison G. and Gutman,G.A. (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol., 4, 203–221. [DOI] [PubMed] [Google Scholar]
- 5.Strand M., Prolla,T.A., Liskay,R.M. and Petes,T.D. (1993) Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature, 365, 274–276. [DOI] [PubMed] [Google Scholar]
- 6.Talbot C.C. Jr, Avramopoulos,D., Gerken,S., Chakravarti,A., Armour,J.A., Matsunami,N., White,R. and Antonarakis,S.E. (1995) The tetranucleotide repeat polymorphism D21S1245 demonstrates hypermutability in germline and somatic cells. Hum. Mol. Genet., 4, 1193–1199. [DOI] [PubMed] [Google Scholar]
- 7.Schlotterer C., Amos,B. and Tautz,D. (1991) Conservation of polymorphic simple sequence loci in cetacean species. Nature, 354, 63–65. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty R., Kimmel,M., Stivers,D.N., Davison,L.J. and Deka,R. (1997) Relative mutation rates at di-, tri- and tetranucleotide microsatellite loci. Proc. Natl Acad. Sci. USA, 94, 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber J.L. and Wong,C. (1993) Mutation of human short tandem repeats. Hum. Mol. Genet., 2, 1123–1128. [DOI] [PubMed] [Google Scholar]
- 10.Ellegren H. (2000) Heterogeneous mutation processes in human microsatellite DNA sequences. Nature Genet., 24, 400–402. [DOI] [PubMed] [Google Scholar]
- 11.Xu X., Peng,M. and Fang,Z. (2000) The direction of microsatellite mutations is dependent upon allele length. Nature Genet., 24, 396–399. [DOI] [PubMed] [Google Scholar]
- 12.Møller A.P. (1994) Sexual Selection in the Barn Swallow. Oxford University Press, Oxford, UK.
- 13.Saino N., Primmer,C.R., Ellegren,H. and Moller,A.P. (1997) An experimental study of paternity and tail ornamentation in the barn swallow (Hirundo rustica). Evolution, 51, 562–570. [DOI] [PubMed] [Google Scholar]
- 14.Walsh P.S., Metzger,D.A. and Higuchi,R. (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques, 10, 506–513. [PubMed] [Google Scholar]
- 15.Primmer C.R., Moller,A.P. and Ellegren,H. (1995) Resolving genetic relationships with microsatellite markers: a parentage testing system for the swallow Hirundo rustica. Mol. Ecol., 4, 493–498. [DOI] [PubMed] [Google Scholar]
- 16.Primmer C.R., Moller,A.P. and Ellegren,H. (1996) New microsatellites from the pied flycatcher Ficedula hypoleuca and the swallow Hirundo rustica genomes. Hereditas, 124, 281–283. [DOI] [PubMed] [Google Scholar]
- 17.Griffith S.C., Stewart,I.R.K., Dawson,D., Owens,I.P.F. and Burke,T.A. (1999) Contrasting levels of extra-pair paternity in mainland and island populations of the house sparrow (Passer domesticus): is there an ‘island effect’? Biol. J. Linn. Soc. Lond., 68, 303–316. [Google Scholar]
- 18.Bensch S., Price,T. and Kohn,J. (1997) Isolation and characterization of microsatellite loci in a Phylloscopus warbler. Mol. Ecol., 6, 91–92. [DOI] [PubMed] [Google Scholar]
- 19.Double M.C., Dawson,D., Burke,T. and Cockburn,A. (1997) Finding the fathers in the least faithful bird: a microsatellite-based genotyping system for the superb fairy-wren Malurus cyaneus. Mol. Ecol., 6, 691–693. [Google Scholar]
- 20.Primmer C.R. and Ellegren,H. (1998) Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol., 15, 997–1008. [DOI] [PubMed] [Google Scholar]
- 21.Brinkmann B., Klintschar,M., Neuhuber,F., Huhne,J. and Rolf,B. (1998) Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet., 62, 1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones A.G., Rosenqvist,G., Berglund,A. and Avise,J.C. (1999) Clustered microsatellite mutations in the pipefish Syngnathus typhle. Genetics, 152, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner M.G., Bull,C.M., Cooper,S.J.B. and Duffield,G.A. (2000) Microsatellite mutations in litters of the Australian lizard Egernia stokesii. J. Evol. Biol., 13, 551–560. [Google Scholar]
- 24.Calabrese P.P., Durrett,R.T. and Aquadro,C.F. (2001) Dynamics of microsatellite divergence under stepwise mutation and proportional slippage/point mutation models. Genetics, 159, 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primmer C.R., Saino,N., Moller,A.P. and Ellegren,H. (1996) Directional evolution in germline microsatellite mutations. Nature Genet., 13, 391–393. [DOI] [PubMed] [Google Scholar]
- 26.Ellegren H. (2000) Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet., 16, 551–558. [DOI] [PubMed] [Google Scholar]
- 27.Weissenbach J., Gyapay,G., Dib,C., Vignal,A., Morissette,J., Millasseau,P., Vaysseix,G. and Lathrop,M. (1992) A second-generation linkage map of the human genome. Nature, 359, 794–801. [DOI] [PubMed] [Google Scholar]
- 28.Dib C., Faure,S., Fizames,C., Samson,D., Drouot,N., Vignal,A., Millasseau,P., Marc,S., Hazan,J., Seboun,E. et al. (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature, 380, 152–154. [DOI] [PubMed] [Google Scholar]
- 29.Neff B.D. and Gross,M.R. (2001) Microsatellite evolution in vertebrates: inference from AC dinucleotide repeats. Evolution, 55, 1717–1733. [DOI] [PubMed] [Google Scholar]
- 30.Crozier R.H., Kaufmann,B., Carew,M.E. and Crozier,Y.C. (1999) Mutability of microsatellites developed for the ant Camponotus consobrinus. Mol. Ecol., 8, 271–276. [DOI] [PubMed] [Google Scholar]
- 31.Amos W., Sawcer,S.J., Feakes,R.W. and Rubinsztein,D.C. (1996) Microsatellites show mutational bias and heterozygote instability. Nature Genet., 13, 390–391. [DOI] [PubMed] [Google Scholar]
- 32.Nauta M.J. and Weissing,F.J. (1996) Constraints on allele size at microsatellite loci: implications for genetic differentiation. Genetics, 143, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldman M.W., Bergman,A., Pollock,D.D. and Goldstein,D.B. (1997) Microsatellite genetic distances with range constraints: analytic description and problems of estimation. Genetics, 145, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harr B. and Schlotterer,C. (2000) Long microsatellite alleles in Drosophila melanogaster have a downward mutation bias and short persistence times, which cause their genome-wide underrepresentation. Genetics, 155, 1213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wierdl M., Dominska,M. and Petes,T.D. (1997) Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics, 146, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruglyak S., Durrett,R.T., Schug,M.D. and Aquadro,C.F. (1998) Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl Acad. Sci. USA, 95, 10774–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santibanez-Koref M.F., Gangeswaran,R. and Hancock,J.M. (2001) A relationship between lengths of microsatellites and nearby substitution rates in mammalian genomes. Mol. Biol. Evol., 18, 2119–2123. [DOI] [PubMed] [Google Scholar]
- 38.Di Rienzo A., Peterson,A.C., Garza,J.C., Valdes,A.M., Slatkin,M. and Freimer,N.B. (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc. Natl Acad. Sci. USA, 91, 3166–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyata T., Hayashida,H., Kuma,K., Mitsuyasu,K. and Yasunaga,T. (1987) Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harbor Symp. Quant. Biol., 52, 863–867. [DOI] [PubMed] [Google Scholar]
- 40.Shimmin L.C., Chang,B.H. and Li,W.H. (1993) Male-driven evolution of DNA sequences. Nature, 362, 745–747. [DOI] [PubMed] [Google Scholar]
- 41.Ellegren H. and Fridolfsson,A.K. (1997) Male-driven evolution of DNA sequences in birds. Nature Genet., 17, 182–184. [DOI] [PubMed] [Google Scholar]
- 42.Hurst L.D. and Ellegren,H. (1998) Sex biases in the mutation rate. Trends Genet., 14, 446–452. [DOI] [PubMed] [Google Scholar]
- 43.Haldane J.B.S. (1947) The rate of mutation of the gene for hemophilia and its segregation in males and females. Ann. Eugen., 13, 262–271. [DOI] [PubMed] [Google Scholar]
- 44.Vogel F. and Motulsky,A.G. (1997) Human Genetics: Problems and Approaches, 3rd Edn. Springer, Berlin, Germany.
- 45.Kahn N.W. and Quinn,T.W. (1999) Male-driven evolution among Eoaves? A test of the replicative division hypothesis in a heterogametic female (ZW) system. J. Mol. Evol., 49, 750–759. [DOI] [PubMed] [Google Scholar]