Abstract
We studied fluorescence intensity, polarization and lifetime of some commonly used fluorophores conjugated to oligodeoxyribonucleotides with different primary and secondary structures. We found that fluorescence intensity can increase or decrease upon hybridization of the labeled strand to its complement depending on the sequence and position of the fluorophore. Up to 10-fold quenching of the fluorescence upon hybridization was observed when the dye moiety was attached close to the 3′ end and the 3′-terminal base was either dG or dC. No quenching upon hybridization was observed when the dye was positioned within the same sequence context but close to the 5′ end. The presence of a dG overhang quenches the fluorescence less efficiently than a blunt end dG-dC or dC-dG base pair. When located internally in the double strand, the dG-dC base pair does not affect the fluorescence of the nearby dye. Guanosine in a single-stranded oligonucleotide quenches the fluorescence of nearby dye by <2-fold. Upon duplex formation, this quenching is eliminated and the fluorescence increases. This increase can only be detected when the fluorophore is located at least 6 nt from the terminal dG-dC base pair. The change of fluorescence polarization upon duplex formation inversely correlates with the change of intensity. Fluorescein conjugated to a single-stranded oligonucleotide or a duplex undergoes a bi-exponential decay with ∼4 and ∼1 ns lifetimes.
INTRODUCTION
Fluorescence techniques are used widely in nucleic acids research to study the structure of nucleic acids as well as their interaction with proteins (1). In addition, numerous methods for the detection and quantification of DNA and RNA based on fluorescence have been developed (2). That is why understanding the mechanisms of possible interactions between the fluorophores and nucleic acids is important. The initial work in this area used the free fluorescent dye molecules that change their intensity when bound to double-stranded nucleic acids through either intercalation or groove binding (3–7). It was shown that the fluorescent properties and the binding affinities of many of these dyes are dependent on the nucleic acid sequence (6,7). With the advent of automated DNA synthesis and phosphoramidite chemistry, it became possible to attach fluorophores covalently to an oligonucleotide of interest. In multiple biophysical and biochemical applications of fluorescent oligonucleotides it was observed that the fluorescence of some conjugated dyes was sensitive to the environment around the point of attachment (8–14). For example, the DNA base analogs 2-aminopurine and pteridine are partially quenched upon incorporation into double-stranded DNA. The degree of quenching depends on the fluorophore’s proximity to purines and on its position in the oligonucleotide (15–17). Likewise, dansyl, covalently linked to the C-5 of dU changes its fluorescence depending on structural changes in double-stranded DNA (18). Dye–nucleotide interactions were also reported for pyrene (19,20), tetramethylrhodamine (21,22) and the Cy dyes (23). The effect of individual nucleotides on the fluorescence of dyes has been studied in some detail for pyrene (24,25), coumarin (26), oxazine (13), Rhodamine 6G (27) and stilbene (28). In all of these cases it was proposed that photoinduced charge transfer between the dye and a nucleotide residue plays a crucial role in the process. Most of the dyes studied were quenched by guanosine, which was explained by the good electron donating properties of this nucleotide (26,29).
Hybridization of end-labeled single-stranded oligonucleotides to the complementary sequence can also quench the conjugated fluorophores. A number of researchers have reported that hybridization of an oligodeoxynucleotide labeled with fluorescein at the 5′ ends to the complementary sequence resulted in a decrease of fluorescence intensity (12,30,31). A similar effect has been shown for rhodamine (8) and BODIPY-Fl (32). In all these cases the quenching was attributed to the presence of a guanosine in the complementary strand in close proximity to the dye. An analogous quenching effect has been demonstrated for oligoribonucleotides labeled with fluorescein at the 3′ end upon binding to a ribozyme (14).
This paper describes the effects of primary and secondary structure of end-labeled and internally labeled oligodeoxyribonucleotides on the fluorescence properties of some conjugated dyes. Our data on fluorescence intensity, polarization and lifetime agree with the important role that guanosine plays as a quencher of fluorescence. However, this quenching effect also depends on secondary structure, local sequence and even the proximity of the 3′ end or the 5′ end of the oligonucleotide. Depending on these factors, the fluorescence of a labeled oligonucleotide can increase or decrease upon hybridization to the complementary sequence. The elucidation of the factors affecting the fluorescence has permitted us to modulate the fluorescence intensity of many conjugated dyes up to 10-fold, providing the basis for many interesting applications in nucleic acids research. The results obtained also contribute to the understanding of the mechanism of electron transfer in DNA, which is involved in a variety of oxidative processes (33,34).
MATERIALS AND METHODS
Oligodeoxyribonucleotides
All oligodeoxyribonucleotide synthesis reagents were purchased from Glen Research, except for wash acetonitrile, which was purchased from Fisher Scientific, and standard CPG, which was obtained from Applied Biosystems, Inc. (ABI). Fluorescent oligodeoxyribonucleotides were synthesized on a Perceptive Biosystems Expedite DNA synthesizer through direct incorporation of the fluorescein phosphoramidite to the 5′ end, fluorescein CPG to the 3′ end and C5-fluorescein-dT phosphoramidite internally. Other dyes were incorporated post-synthetically. A two-step procedure included the coupling of the amino modifier C6 T phosphoramidite {phosphoramidite 5-[N-(trifluoroacetylaminohexyl)-3-acrylimido]-2′-deoxyuridine phosphoramidite} during synthesis and post-synthetic modification with N-hydroxy succinimidyl ester of a fluorescent dye from Molecular Probes (35). Reverse phase HPLC analysis and purification of oligonucleotides was done using a Waters Alliance HPLC connected to a computer equipped with the Millennium software package (Version 3.1).
Melting curves of oligonucleotides
The fluorescence intensity, lifetime and polarization were measured for 200 nM fluorescent oligonucleotides in 20 mM Tris–HCl pH 8.4, 50 mM KCl, 2 mM MgCl2, and the concentration of unlabeled complementary oligonucleotide used for duplex formation was 1 µM, unless specified. Melting curves of fluorescent oligonucleotides and duplexes were measured on ABI PRISM 7700 in 50 µl of the above buffer using the following protocol: 25°C for 2 min, 95°C for 2 min, then decreasing the temperature to 25°C in 2°C per 15 s increments, incubation at 25°C for 2 min, then increasing the temperature to 95°C in 2°C per 15 s increments. For accurate comparison of the fluorescence of single-stranded oligonucleotides versus corresponding duplexes at room temperature, melting curves were normalized at 93–95°C. At this temperature duplexes are completely melted and the fluorescence of the single-stranded oligonucleotide became equal to the fluorescence of the corresponding duplex. This normalization procedure permitted to eliminate well-to-well variations in the fluorescence readings and directly compare the fluorescence of single-stranded and double-stranded oligonucleotides at 25°C.
For some experiments labeled oligonucleotides were hybridized to the complementary oligonucleotides with the protruding 5′ ends. In order to create the blunt-end duplex, the complex was incubated for 15 min at 37°C with 1 U of Taq DNA polymerase (Invitrogen) and 200 nM corresponding dNTP in 50 µl of 20 mM Tris–HCl pH 8.4, 50 mM KCl, 2 mM MgCl2. Melting of the created duplexes was performed as above.
PCR
PCR product of IL4 cDNA 133 bp fragment was synthesized with d(GAGTTGACCGTAACAGACATCTT) as a forward primer and d(CCTTCTCATGGTGGCTGTAG) as a reverse primer, where the T marks the position where the fluorescein is attached. In another experiment, the linear labeled primer was replaced with a labeled hairpin oligonucleotide d(CTACAGTCCTTCTCATGGTGGCTGTAG), where the underlined sequence is complementary to the 3′ end of the primer. A 50 µl PCR mixture contained 200 nM of each primer, 106 copies of cloned IL4 cDNA, 200 µM each dATP, dGTP, dCTP and TTP, 2 mM MgCl2, 20 mM Tris–HCl pH 8.4, 50 mM KCl and 1 U Platinum™ Taq DNA polymerase. Reactions were incubated at 25°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s and 72°C for 30 s.
RESULTS
Effect of terminal base pair on the fluorescence of conjugated fluorescein
We determined how the identity of the 5′-terminal base affects the fluorescence of 5′-labeled oligonucleotide upon duplex formation. Four oligonucleotides labeled at the 5′ end with 6-carboxyfluorescein were synthesized. Their sequences were identical, d(NNTTCTCATGGTGGCTGTAGAAC), except for the two bases at the 5′ end, which were d(AA), TT, d(CC) or d(GG). These labeled oligonucleotides were hybridized to an excess of unlabeled complementary sequences of the same length, melting curves were measured on an ABI PRISM 7700 and the normalized fluorescence at 25°C was calculated for single-stranded oligonucleotides and duplexes as described in the Materials and Methods. The fluorescence signal did not change upon hybridization when the oligonucleotide contains d(AA) or TT at the 5′ end. Conversely, there was an ∼40% decrease in fluorescence upon hybridization when the fluorescent oligonucleotide contained d(CC) at the 5′ end, and about a 30% decrease when the fluorescent oligonucleotide contained d(GG) at the 5′ end.
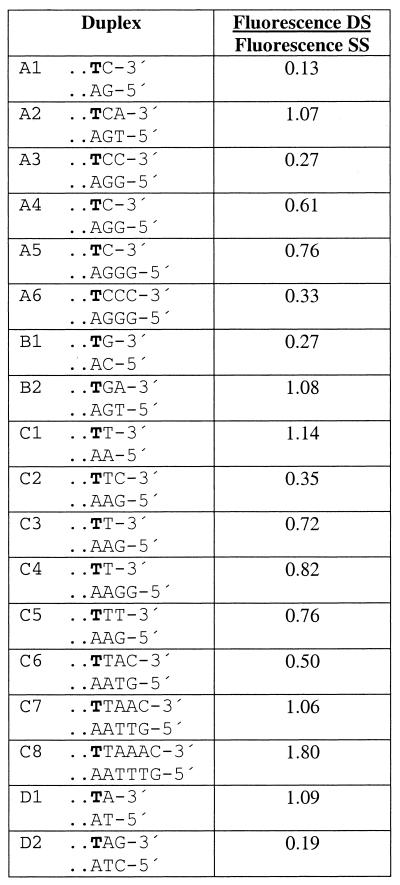
To examine the effect of the 3′-terminal base on fluorescence, a fluorescein was attached to a thymidine near the 3′ end of the oligonucleotide. Four labeled single-stranded oligonucleotides that differed only at the base on the 3′ end (dC, dG, T or dA) were synthesized (Table 1). Each oligonucleotide was hybridized to a variety of complementary strands of different lengths to form either a blunt-end duplex or a 5′ overhang. Based on 5′ overhangs, a number of duplexes with various blunt ends were generated by using appropriate dNTPs and DNA polymerase (see Materials and Methods). Some duplexes contained a mismatch between the 3′ end of the labeled strand and the 5′ end of the complementary oligonucleotide. The change in fluorescence upon duplex formation was calculated from the melting curves and presented in Table 1. A labeled oligonucleotide containing a 3′-terminal dC was quenched by 87% upon duplex formation (Table 1, A1). Addition of a single dA-T base pair to the end of the same duplex completely eliminated the quenching (A2). Replacing that dA-T base pair with another dC-dG base pair restored the quenching (A3). A 5′-dG overhang on the complementary unlabeled strand provides much less quenching than a dC-dG blunt-end base pair (A4). Adding a second dG to the complementary strand to give a two base overhang reduced the quenching effect even more (A5). Similar results were obtained when the labeled oligonucleotide ended with dG (Table 1, series B). Seventy-one percent quenching was observed in the duplex containing a dG-dC base pair at the end (B1). Extending the duplex by the addition of a dA-T base pair eliminated the quenching effect (B2). Conversely, an oligonucleotide containing a 3′-terminal T and a fluorescein on the penultimate base showed no decrease in fluorescence upon duplex formation (Table 1, C1). Extending this duplex by a single dG-dC base pair resulted in significant quenching of the fluorescence (duplex C2). Replacing the ultimate dG-dC base pair with a 5′-dG overhang or a terminal dG-T mismatch substantially reduced the quenching observed (duplexes C3–C5). A labeled oligonucleotide ending with adenosine (Table 1, series D) demonstrated results similar to the T-ended oligonucleotides. Only when the duplex was extended by the addition of a dG-dC base pair to the end was the fluorescence quenching observed (D2).
Table 1. Effect of the terminal structure of oligonucleotide duplexes on fluorescence.
d(CCGTACCTGGCTATCTGTGTN), where N is C in series A, G in series B, T in series C and A in series D were transformed into a number of different duplexes by the enzymatic reaction on the complementary sequences as described in the Materials and Methods. Positions labeled with fluorescein are marked in bold. Fluorescence intensity of the duplexes is presented relative to the single-stranded forms.
The results in Table 1 also show that the quenching of fluorescence by the terminal dG-dC or dC-dG base pair is dependent on the distance between the fluorophore and the blunt end of the duplex. When the fluorophore was positioned further from the 3′ end, the quenching upon hybridization decreased (Table 1, A1, A3 and A6; C2, C6–C8). When the fluorophore was at the sixth base from the 3′ end (C8), the quenching effect disappeared completely, and a strong enhancement of fluorescence was observed.
The effect of hybridization on fluorescence was compared for a fluorophore located on a base near the 3′ end versus the 5′ end. Two oligonucleotides of the same sequence were synthesized, so that their 3′ half was a mirror image of the 5′ half (Table 2). One of the oligonucleotides had fluorescein attached to the thymidine closest to the 5′ end, while the other oligonucleotide was labeled with fluorescein on the thymidine closest to the 3′ end. The 3′-terminal and 5′-terminal bases were dG. The fluorescence intensities per pmol of HPLC purified single-stranded oligonucleotides differed by <5%. The labeled oligonucleotides were hybridized to the unlabeled complementary sequence of the same size. The fluorescence intensity of the single-stranded and double-stranded structures was determined as described in the Materials and Methods and is shown as a ratio in Table 2. The oligonucleotide labeled close to the 3′ end showed substantial quenching upon hybridization, while the oligonucleotide labeled close to the 5′ end exhibited no decrease in fluorescence.
Table 2. Change of fluorescence upon duplex formation for oligonucleotides labeled with fluorescein internally close to the 5′ end or 3′ end.
Labeling positions are shown in bold.
Increase of fluorescence intensity of an internally conjugated fluorescein upon duplex formation
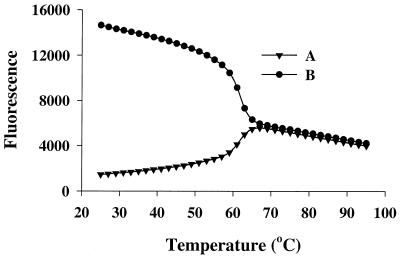
As shown in Table 1, when the fluorophore is positioned away from the 3′-terminal dG-dC base pair, the quenching became less pronounced and eventually an enhancement of fluorescence was observed (duplexes C7 and C8). To examine this further, two oligonucleotides were synthesized (Fig. 1). Oligonucleotide A, d(CCTTCTCATGGTGGCTGTAG), was labeled at the third base from the 3′-G. Oligonucleotide B, d(CCTTCTCATGGTGGCTGTAGAACT), was identical in sequence, except for an extra 4 nt added to the 3′ end. Thus, oligonucleotide B had a fluorescein six bases away from the 3′ end. Each oligonucleotide was hybridized to its complementary sequence and the melting curves of both duplexes are presented in Figure 1. The fluorescence intensity decreased upon hybridization when 2 nt separated the label and the terminal dG-dC base pair, but increased when the fluorophore was 6 nt away from the 3′ end.
Figure 1.
Melting curves of oligodeoxynucleotide duplexes labeled with fluorescein. (A) d(CCTTCTCATGGTGGCTGTAG) and (B) d(CCTTCTCATGGTGGCTGTAGAACT) were hybridized to an excess of the complementary oligonucleotide of the same size and melting curves were measured as described in the Materials and Methods. The labeling positions are shown in bold.
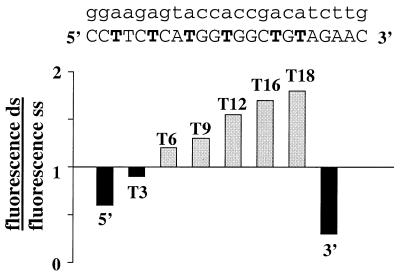
The increase of fluorescence upon hybridization is greatly dependent on the position of fluorophore within the oligonucleotide sequence (Fig. 2). Eight oligonucleotides that differed only in the position of the fluorescein were synthesized. The label was attached to the 5′ end, 3′ end or at one of six different internal thymidines. The ratios of the fluorescence intensity of the single-stranded and double-stranded oligonucleotides are presented in Figure 2. The fluorescence of the oligonucleotides labeled at the 5′ end, 3′ end or internal position T3 was quenched upon hybridization. However, for oligonucleotides labeled at internal positions starting with T6, an increase in fluorescence was observed. The further away the label was from the 5′ end, the larger was the increase in fluorescence upon hybridization.
Figure 2.
Change of the fluorescence intensity upon hybridization of oligodeoxynucleotides labeled with FAM at the 5′ end, 3′ end or internally. The internal labeling positions are shown in bold. The complementary strand is shown in lower case. The ratio between the fluorescence of the double-stranded and single-stranded labeled oligonucleotides was determined as described in the Materials and Methods.
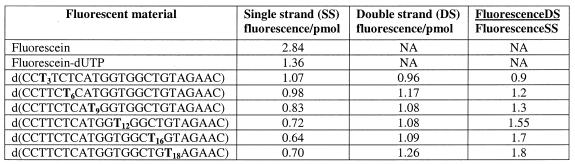
A possible explanation for the increase of fluorescence upon hybridization is that the fluorophore is quenched in the single strand and is dequenched as a result of duplex formation. If so, various labeled oligonucleotides that increase the fluorescence in duplex to a different degree should be quenched to a similar degree in the single-stranded form. We tested this hypothesis by measuring the fluorescence intensity per pmol for the single-stranded oligonucleotides labeled at different positions. The homogeneity of these oligonucleotides was confirmed by HPLC. As shown in Table 3, the specific fluorescence of free fluorescein is substantially higher than that of the dye-labeled mononucleotide, which in turn is higher than the fluorescence of the dye conjugated to oligonucleotides. Interestingly, the fluorescence/pmol of single-stranded oligonucleotides labeled at positions T3–T18 is inversely related to the fluorescence enhancement for the corresponding duplexes. For example, the oligonucleotides labeled at positions T16 and T18 are the least fluorescent in the single-stranded form, but resulted in the most fluorescence enhancement upon duplex formation. Therefore, hybridization appears to attenuate the quenching observed in the single-stranded oligonucleotides. To investigate the role of guanosine in the quenching of fluorescence in the single strand, two oligonucleotides were compared: d(CCTTCTCATGGTGGCTGTAGAAC) (T18, Table 3) and d(CCTTCTCATGGTGACTATAAAAC). Two dGs in the former oligonucleotides were replaced with two dAs in the latter one. No increase in fluorescence upon hybridization was observed with the latter oligonucleotide, indicating the important role of guanosine. The analysis of a large number of internally labeled oligodeoxyribonucleotides confirmed that the presence of at least one guanosine within 4 nt on either side of the label is required for the fluorescence to increase upon duplex formation (data not shown). In addition, we found no significant effect of 3′-terminal or 5′-terminal bases on the fluorescence increase when the label is located at least 6 nt from either end.
Table 3. Fluorescence intensity per pmol of fluorescein, fluorescein-dUTP and oligonucleotides labeled with fluorescein at different positions.
Internal labeling positions are shown in bold.
Effect of primary and secondary structure of oligonucleotide on fluorescence, polarization and lifetime of the conjugated fluorescein
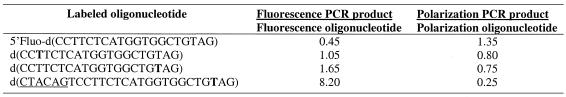
Fluorescence intensity and polarization of oligonucleotides labeled with fluorescein at positions T3, T18 and at the 5′ end were compared with the intensity and polarization of the 133 bp double-stranded PCR products derived from the use of these oligonucleotides as primers (see Materials and Methods). In addition to linear oligonucleotides, a hairpin oligonucleotide labeled at position T18 was also examined along with the corresponding PCR product. To form a blunt-end hairpin, 6 nt were added to the 5′ end of the linear oligonucleotide. The fluorescence intensity and polarization of the single-stranded and double-stranded structures are shown in Table 4. For the fluorescein attached to the 5′ position of oligonucleotide, the fluorescence intensity decreased upon the formation of the double-stranded structure and the polarization increased. The opposite effect was observed for the internally labeled oligonucleotides. In this case, the increased fluorescence intensity was accompanied by decreased polarization. The internally labeled hairpin oligonucleotide exhibited the largest change in both fluorescence intensity and polarization upon the formation of the PCR product.
Table 4. Change of fluorescence intensity and polarization of fluorescein-labeled oligonucleotides upon incorporation into the PCR product.
Internal labeling positions are shown in bold. Fluorescence intensity and polarization were measured on a Polarion (Tecan) according to vendor protocol.
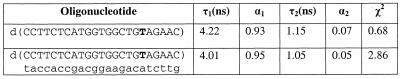
The fluorescence lifetimes were determined for the fluorescein labeled single-stranded oligonucleotide and its duplex that demonstrated 1.8-fold difference in fluorescence intensity (T18, Table 3). Measurements were performed at 25°C in the solution described in the Materials and Methods in frequency domain mode using a SPEX spectrofluorometer. Data were analyzed with DataMax software (Table 5). A biexponential decay model provided the best fit to the data for both the single-stranded and double-stranded oligonucleotides with decay components of ∼4 and ∼1 ns. The lifetime of the primary component was similar to the lifetime of free fluorescein and its fractional amplitude is much higher than that of the short lifetime component. No significant difference in lifetime parameters between the single-stranded and double-stranded structures was discovered. However, the χ2 values indicate that other multiexponential decay models may provide a better fit to the data and resolve a more complex mixture of the decay components.
Table 5. Fluorescence lifetimes of single-stranded and double-stranded fluorescein-labeled oligonucleotides.
Internal labeling positions are shown in bold.
Effect of primary and secondary structure of oligonucleotides on the fluorescence of dyes other than fluorescein
All of the data presented above were obtained with oligonucleotides containing fluorescein conjugated through the C5 position of thymidine, or attached to the terminus by a short alkyl linker. In addition to fluorescein, a variety of other fluorophores were examined. Similar to fluorescein, JOE, HEX, TET, Alexa 594, ROX, MAX and TAMRA are quenched at the proximity of terminal dG-dC and dC-dG base pairs and enhance their fluorescence upon duplex formation when located internally at least 6 nt away from the ends of oligonucleotide. Certain other dyes, such as Texas Red, BODIPY TR and the Cy3 and Cy5 dyes did not follow this pattern (data not shown).
DISCUSSION
We synthesized a large number of labeled oligodeoxyribonucleotides of different sequences to systematically investigate the effect of their primary and secondary structure on the properties of some commonly used fluorescent dyes. Fluorescence intensity may decrease or increase upon duplex formation depending on the sequence of the oligonucleotide and the position of the fluorophore. Oligodeoxyribonucleotides containing fluorescein conjugated to the 5′-terminal dC or dG decrease their fluorescence by ∼40% upon duplex formation. No quenching is observed when the labeled oligodeoxynucleotide ends in dA or T. The quenching of fluorescence upon duplex formation for an oligodeoxynucleotide ending in dC was reported previously and may be explained by the quenching effect of guanosine located on the complementary strand in the proximity of the dye (8,12,30–32). The quenching properties of guanosine can be attributed to its electron donating ability, which permits the charge transfer between the nucleobase and a nearby dye (26,29). However, in our experiments, fluorescence was also quenched when the fluorescent oligonucleotide containing dG at its 5′ end was hybridized to its dC containing complement. This result cannot be explained by a scheme where a guanosine opposing the fluorophore in the complementary strand is acting as a quencher of fluorescence. Rather, the terminal dG-dC and dC-dG base pairs are responsible for the fluorescence quenching.
Terminal dG-dC and dC-dG base pairs also quench the fluorophore located internally close to the 3′ end of the oligonucleotide. The efficiency of quenching of internal fluorescein is much higher than that for the 5′ label or 3′ label and can reach 10-fold. Terminal dG-T mismatch or 5′-dG overhangs result in much less pronounced quenching than the blunt-end dG-dC and dC-dG base pairs. Only a terminal dG-dC pair affects the fluorescence of duplex; when flanked with a dA-T pair, no quenching occurs. Our data are consistent with the proposed formation of the charge transfer complexes between some fluorophores and nucleobases (26,28,29). Guanosine, which has the highest electron donating ability of all the bases, plays a critical role in this process. However, our data show that the ability of guanosine to quench the fluorescence dramatically depends on whether it is involved in hydrogen bonding and whether it is located at the end of the double-stranded structure or internally.
We compared quenching upon hybridization for fluorescein located on the base within the same sequence close to the 5′ end versus the 3′ end. Interestingly, several-fold quenching upon duplex formation occurred only when the fluorescein was attached near the 3′ end. This result cannot be explained by the difference in distance between fluorescein and guanine of two structures. The distance between the CH3 of thymidine and the N7 of guanine calculated using the Macromodel molecular modeling program for the B-form helix, was found to be very similar in both cases: 8.275 Å for 5′-GAT and 9.351 Å for 3′-GAT. Other steric and electron donating properties of the double helix need to be explored to explain this phenomenon.
The distance between the terminal dG-dC base pair and the fluorophore is another important factor affecting the decrease of fluorescence upon hybridization. Quenching was only observed when the fluorophore was attached within 4 nt from the 3′ end. When the fluorescein is located five or more residues away from the 3′ end, hybridization results in increase of fluorescence instead of quenching. This increase of fluorescence does not depend on the identity of the terminal base pair, but instead depends on the presence of at least one guanosine within 4 nt on either side of the dye. Here we demonstrate that the mechanisms of this fluorescence increase upon hybridization are based on the quenching of the fluorophore in the single-stranded structure by a neighboring guanosine, and its subsequent dequenching upon duplex formation. A similar dequenching mechanism was suggested previously for end-labeled oligonucleotides (31). Although the role of guanosine in the quenching of a fluorophore attached to a single-stranded oligonucleotide is compelling, other possible factors should not be excluded from consideration, such as other nucleobases, the linker arm between the base and the dye, or solvent effects. Another factor that might be important is the conformational flexibility of the single strand, which is greater than the double strand, and may affect the fluorescence through a secondary structure within a single-stranded oligonucleotide.
To shed more light onto the mechanism of fluorescence changes, the fluorescence lifetime and polarization were measured for various single-stranded and double-stranded labeled oligonucleotides. To measure the fluorescence lifetime we chose the internally labeled linear oligonucleotide that increased its fluorescence intensity upon hybridization by 1.8-fold. Biexponential decay with two different lifetimes was observed for single-stranded and double-stranded structures, indicating the existence of the fluorophore in two different electronic states. This is consistent with previous work on oligonucleotides labeled with rhodamine, TAMRA and oxazine that demonstrated two or more conformations of the dye as it associated and disassociated from the nucleobases (13,21,22,27,36). The decreased lifetime of one of the decay components would support a dynamic rather than static quenching mechanism (37). On the other hand, there is no significant difference in lifetime parameters between the single-stranded and double-stranded fluorescent oligonucleotides, while their difference in fluorescent intensities is substantial. This indicates that complex formation between the fluorophore and a nucleobase, or a static quenching, plays an important role in decreasing the fluorescence intensity of the single-stranded oligonucleotide. In the duplex this complex formation is eliminated and the fluorescence intensity increases.
Our preliminary study of fluorescence polarization showed that the change of fluorescence polarization upon duplex formation inversely correlates with the change of intensity indicating a decreased mobility of the fluorophore in the quenched state. When the 5′-labeled oligonucleotide was incorporated into the PCR product its fluorescence intensity decreased and polarization increased. This increase may be attributed to the increased size of the PCR product and to the quenching of the fluorophore by the terminal G-C base pair. However, when fluorescein was located internally the opposite effect was observed, fluorescence intensity increased and polarization decreased upon the formation of the PCR product. This may be explained by the quenched state of the fluorophore in the unincorporated primer (especially hairpin), which is characterized by low mobility of the fluorescent moiety and high polarization. As a result, polarization of the primer is higher than the polarization of the PCR product despite of the higher size of the latter. Besides, the size of our PCR product is relatively small, only 133 bp, so its effect on polarization is not significant. The most significant change of both intensity and polarization happens when an internally labeled oligonucleotide in a hairpin conformation is incorporated into the PCR product. The more efficient quenching of the fluorophore in the hairpin primers compared with the linear ones, which causes low fluorescence intensity and high polarization of unincorporated primers may explain this.
Most of the data presented here were obtained with oligonucleotides containing fluorescein conjugated through the C5 position of thymidine. The choice of thymidine was based on its commercial availability, and investigation of oligonucleotides labeled at other bases is in progress. A number of other fluorophores, including JOE, HEX, TET, Alexa 594, ROX and TAMRA, change their fluorescence depending on the sequence and structure of the oligonucleotide similar to fluorescein. Some other dyes, such as Texas Red, some BODIPY dyes and the Cy dyes, did not exhibit this behavior, or else their pattern of modulation was different to that of fluorescein. Such differences may be ascribed to the different redox potential of these dyes. The measurement of the redox potentials of specific dyes may help to predict the effect of nucleotide sequences on their fluorescence.
The ability to change the fluorescence intensity of labeled oligonucleotides through their primary and secondary structures can provide a valuable tool for the detection of nucleic acids and to study the interaction of proteins with nucleic acids. One of the applications is the use of mono-labeled oligonucleotides as cost-effective fluorogenic PCR primers and hybridization probes. The use of fluorogenic primers in multiplex quantitative PCR is described elsewhere (38).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Malek Masoud (Invitrogen) for valuable help in producing fluorogenic primers and Prof. Robert Coleman for his assistance in oligonucleotide structure analysis using the Macromodel program.
REFERENCES
- 1.Millar D.P. (1996) Fluorescence studies of DNA and RNA structure and dynamics. Curr. Opin. Struct. Biol., 6, 322–326. [DOI] [PubMed] [Google Scholar]
- 2.Glazer A.N. and Mathies,R.A. (1997) Energy-transfer fluorescent reagents for DNA analyses. Curr. Opin. Biotech., 8, 94–102. [DOI] [PubMed] [Google Scholar]
- 3.LePecq J.-B. and Palotti,C. (1967) A fluorescent complex between ethidium bromide and nucleic acids. Physical–chemical characterization. J. Mol. Biol., 27, 87–106. [DOI] [PubMed] [Google Scholar]
- 4.Löber G. (1981) The fluorescence of dye–nucleic acids complexes. J. Lumin., 22, 221–265. [Google Scholar]
- 5.Dougherty G. and Pigram,W.J. (1982) Spectroscopic analysis of drug–nucleic acid interactions. CRC Crit. Rev. Biochem., 12, 103–132. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer C. and Wähnert,U. (1986) Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol., 47, 31–112. [DOI] [PubMed] [Google Scholar]
- 7.Loontiens F.G., Regenfuss,P., Zechel,A., Dumortier,L. and Clegg,R.M. (1990) Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A–T)] and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry, 29, 9029–9039. [DOI] [PubMed] [Google Scholar]
- 8.Cardullo R.A., Agrawal,S., Flores,C., Zamecnik,P.C. and Wolf,D.E. (1988) Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 85, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murchie A.I.H., Clegg,R.M., von Kitzing,E., Duckkett,D.R., Diekmann,S. and Lilley,D.M.J. (1989) Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature, 341, 763–766. [DOI] [PubMed] [Google Scholar]
- 10.Cooper J.P. and Hagerman,P.J. (1990) Analysis of fluorescence energy transfer in duplex and branched DNA molecules. Biochemistry, 29, 9261–9268. [DOI] [PubMed] [Google Scholar]
- 11.Clegg R.M., Murchie,A.I.H., Zechel,A., Carlberg,C., Diekmann,S. and Lilley,D.M.J. (1992) Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry, 31, 4846–4856. [DOI] [PubMed] [Google Scholar]
- 12.Lee S.P., Porter,D., Chirikjian,J.G., Knutson,J.R. and Han,M.K. (1994) A fluorometric assay for DNA cleavage reactions characterized with BamHI restriction endonuclease. Anal. Biochem., 220, 377–383. [DOI] [PubMed] [Google Scholar]
- 13.Sauer M., Drexhage,K.H., Lieberwirth,U., Muller,R., Nord,S. and Zander,C. (1998) Dynamics of the electron transfer reaction between an oxazine dye and DNA oligonucleotides monitored on the single-molecule level. Chem. Phys. Lett., 284, 153–163. [Google Scholar]
- 14.Walter N.G. and Burke,J.M. (1997) Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA, 3, 392–404. [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D., Evans,K.O. and Nordlund,T.M. (1994) Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry, 33, 9592–9599. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins M.E., Pfeiderer,W., Balis,F.M., Porter,D. and Knutson,J.R. (1997) Fluorescence properties of pteridine nucleoside analogs as monomers and incorporated into oligonucleotides. Anal. Biochem., 244, 86–95. [DOI] [PubMed] [Google Scholar]
- 17.Nordlund T.M., Andersson,S., Nilsson,L., Rigler,R., Graslund,A. and McLaughlin,L.W. (1989) Structure and dynamics of a fluorescent DNA oligomer containing the EcoRI recognition sequence: fluorescence, molecular dynamics and NMR studies. Biochemistry, 28, 9095–9103. [DOI] [PubMed] [Google Scholar]
- 18.Barawkar D.A. and Ganesh,K.N. (1995) Fluorescent d(CGCGAATTCGCG): characterization of major groove polarity and study of minor groove interactions through a major groove semantophore conjugate. Nucleic Acids Res., 23, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koenig P., Reines,S.A. and Cantor,C.R. (1977) Pyrene derivatives as fluorescent probes of conformation near the 3′ termini of polyribonucleotides. Biopolymers, 16, 2231–2242. [DOI] [PubMed] [Google Scholar]
- 20.Manoharan M., Tivel,K.L., Zhao,M., Nafisi,K. and Netzel,T.L. (1995) Base sequence dependence of emission lifetimes for DNA oligomers and duplexes covalently labeled with pyren: relative electron transfer quenching efficiencies of A, G, C and T nucleoside towards pyren. J. Phys. Chem., 99, 17461–17472. [Google Scholar]
- 21.Edman L., Mets,U. and Rigler,R. (1996) Conformational transitions monitored for single molecules in solution. Proc. Natl Acad. Sci. USA, 93, 6710–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggeling C., Fries,J.R., Brand,L., Gunther,R. and Seidel,C.A.M. (1998) Monitoring conformational dynamics of a single molecule by selective fluorescence spectroscopy. Proc. Natl Acad. Sci. USA, 95, 1556–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randolph J.B. and Waggoner,A.S. (1997) Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res., 25, 2923–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lianos P. and Georghiou,S. (1979) Complex formation between pyrene and the nucleotides GMP, CMP, TMP and AMP. Photochem. Photobiol., 29, 13–21. [Google Scholar]
- 25.Shafirovich V.Y., Courtney,S.H., Ya,N. and Geacintov,N.E. (1995) Proton-coupled photoinduced electron transfer, deuterium isotope effects and fluorescence quenching in noncovalent benzo[α]pyrenetetraol-nucleoside complexes in aqueous solutions. J. Am. Chem. Soc., 117, 4920–4929. [Google Scholar]
- 26.Seidel C.A.M., Schulz,A. and Sauer,M.H.M. (1996) Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem., 100, 5541–5553. [Google Scholar]
- 27.Widengren J., Dapprich,J. and Rigler,R. (1997) Fast interactions between Rh6G and dGTP in water studied by fluorescence correlation spectroscopy. Chem. Phys., 216, 417–426. [Google Scholar]
- 28.Lewis F.D., Letzinger,R.L. and Wasielewski,M.R. (2001) Dynamics of photoinduced charge transfer and hole transport in synthetic DNA hairpins. Acc. Chem. Res., 34, 159–170. [DOI] [PubMed] [Google Scholar]
- 29.Steenken S. and Jovanovic,V. (1997) How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc., 119, 617–618. [Google Scholar]
- 30.Knemeyer J.P., Marme,N. and Sauer,M. (2000) Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem., 72, 3717–3724. [DOI] [PubMed] [Google Scholar]
- 31.Crockett A.O. and Wittwer,C.T. (2001) Fluorescein-labeled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem., 290, 89–97. [DOI] [PubMed] [Google Scholar]
- 32.Kurata S., Kanagawa,T., Yamada,K., Torimura,M., Yokomaku,T., Kamagata,Y. and Kurane,R. (2001) Fluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY‚ FL-labeled probe or primer. Nucleic Acids Res., 29, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy C.J., Arkin,M.R., Jenkins,Y., Ghatlia,N.D., Bossman,S.H., Turro,N.J. and Barton,J.K. (1993) Long-range photoinduced electron transfer through a DNA helix. Science, 262, 1025–1029. [DOI] [PubMed] [Google Scholar]
- 34.Porath D., Bezryadin,A., de Vries,S. and Dekker,C. (2000) Direct measurement of electrical transport through DNA molecules. Nature, 403, 635–638. [DOI] [PubMed] [Google Scholar]
- 35.Ju J., Ruan,C., Fuller,C.W., Glazer,A.N. and Mathies,R.A. (1995) Fluorescence energy transfer dye-labeled primers for DNA sequencing and analysis. Proc. Natl Acad. Sci. USA, 92, 4347–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vamosi G., Gohlke,C. and Clegg,R.M. (1996) Fluorescence characteristics of 5-carboxytetramethylrhodamine linked covalently to the 5′ end of oligonucleotides: multiple conformers of single-stranded and double-stranded dye-DNA complexes. Biophys. J., 71, 972–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakowicz J.R. (1999) Principles of Fluorescence Spectroscopy, 2nd Edn. Kluver Academic/Plenum Publishers, New York, pp. 185–210.
- 38.Nazarenko I., Lowe,B., Darfler,M., Ikonomi,P., Schuster,D and Rashtchian,A. (2002) Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res., 30, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]