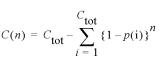Abstract
Although it has been recognized that PCR amplification of mixed templates may generate sequence artifacts, the mechanisms of their formation, frequency and potential elimination have not been fully elucidated. Here evidence is presented for heteroduplexes as a major source of artifacts in mixed-template PCR. Nearly equal proportions of homoduplexes and heteroduplexes were observed after co-amplifying 16S rDNA from three bacterial genomes and analyzing products by constant denaturing capillary electrophoresis (CDCE). Heteroduplexes became increasingly prevalent as primers became limiting and/or template diversity was increased. A model exploring the fate of cloned heteroduplexes during MutHLS-mediated mismatch repair in the Escherichia coli host demonstrates that the diversity of artifactual sequences increases exponentially with the number of both variable nucleotides and of original sequence variants. Our model illustrates how minimization of heteroduplex molecules before cloning may reduce artificial genetic diversity detected during sequence analysis by clone screening. Thus, we developed a method to eliminate heteroduplexes from mixed-template PCR products by subjecting them to ‘reconditioning PCR’, a low cycle number re-amplification of a 10-fold diluted mixed-template PCR product. This simple modification to the protocol may ensure that sequence richness encountered in clone libraries more closely reflects genetic diversity in the original sample.
INTRODUCTION
A severe problem in the analysis of genetic diversity by PCR-based methods is the potential generation of artifacts during the amplification of mixed templates using universal primers. Such artifacts have been recognized as arising from polymerase errors (1) and from the formation of chimeric molecules, the latter being formed when an incompletely extended PCR product acts as a primer on a heterologous sequence (2,3). However, recently heteroduplexes have also been implicated as sources of sequence artifacts (4–6). Here we focus on heteroduplex formation and its potential effect on genetic diversity, and we present a method to eliminate heteroduplexes from multi-template PCR products.
During the plateau phase of a mixed-template PCR, when decreasing primer:template ratios no longer favor primer annealing (7), cross-hybridization of heterologous sequences leads to the formation of heteroduplexes (8,9). In molecular diversity studies, such heteroduplexes can result in overestimation of the number of sequence variants in two ways. First, heteroduplexes can separate from homoduplexes during eletrophoretic migration due to conformational differences. Thus, heteroduplex bands or peaks, if sufficiently intense, can be interpreted as sequence variants. Separation is enhanced under denaturing conditions, such as during denaturant gradient gel electrophoresis (DGGE) or constant denaturant capillary electrophoresis (CDCE), two methods that fractionate DNA molecules by their dissociation–re-annealing kinetics for which melting temperature is a proxy. Secondly, when heteroduplexes are cloned, the host nick-directed mismatch repair system (MutHLS in Escherichia coli) can convert a heteroduplex into a single hybrid sequence by excision repair (10). As a result of the absence of methylation in the cloned insert, the repair enzymes cannot identify a parent strand and will independently choose either strand as a template for resynthesis of the complementary base (10–15). The repaired sequences are ‘mosaics’, i.e. composites of the two parent heterologs.
In this study we consider the formation, consequence and elimination of heteroduplexes in mixed-template PCR. First, we investigate heteroduplex formation by measuring its prevalence in multi-template PCR using CDCE. CDCE is uniquely suited for investigating heteroduplex formation between closely related sequences because of its high sensitivity in differentiating sequences diverging by as little as 1 single bp substitution within 100 bp (16). Secondly, we estimate the potential contribution of heteroduplexes to sequence diversity in clone libraries by modeling the MutHLS-directed mismatch repair of heteroduplex DNA during cloning. Thirdly, we present a quick and easy method to eliminate heteroduplexes from multi-template PCR products prior to cloning by subjecting the PCR products to ‘reconditioning PCR’, a dilution of amplification products into fresh reaction mixture followed by amplification for a low number of cycles.
MATERIALS AND METHODS
PCR amplification
PCR primers were designed to amplify a variable 114 bp region of the 16S rDNA from the genomic DNA of members of the genus Vibrio. The forward primer was modified for CDCE analysis with a 54 bp GC-rich sequence at the 5′ end attached to a fluorescein isothiocyanate molecule for fluorescence detection (17). The standard reaction mixture contained 1× Taq2000 enzyme and reagents (Stratagene, La Jolla, CA), 0.1 µM of each primer and 200 µM of each dNTP. The genomic DNA from three species, Vibrio cholera, Vibrio parahaemolyticus and Vibrio vulnificus, was used as template for amplification, either individually or combined as a two- or three-species template mixture to final reaction concentrations of 1.5–3 ng/µl. Amplification was performed on a Stratagene Robocycler and consisted of a 3 min denaturing step at 95°C followed by 30 cycles of 1.5 min at 95°C, 1 min at 50°C and 2 min at 72°C. To recondition the PCR product, the amplified reaction was diluted 10-fold into fresh reaction mixture of the same composition and cycled three times using the parameters specified above. The size and quality of resulting PCR products was confirmed by electrophoresis through a 1% agarose gel.
CDCE analysis of PCR products
Amplification products were separated and identified by CDCE as described by Khrapko et al. (17). The PCR products were diluted 10-fold into Milli-Q water and electro-injected into a 75 µm inner diameter glass capillary filled with a replaceable 6 × 106 molecular weight linear polyacrylamide gel matrix (Scientific Polymers, Ontario, NY) by applying a 2 µA current to a 20 µl sample volume for 40 s. Samples were then run at a current of 10 µA and a denaturing temperature of 78.0°C. Data were recorded as a time series of fluorescence signal. Peak areas, indicating relative DNA concentrations, were analyzed with AcqKnowledge™ 2.1 software (Biopac Systems, Santa Barbara, CA). Spectra of single-template reactions were used as standards for peak identification in mixed-template amplifications.
Model design and rationale
A model was created using Matlab 6.0 to simulate the effect of MutHLS-directed mismatch repair of heteroduplex DNA in a system initially defined by N sequence variants. First, we calculated the number of different heteroduplexes (HET) that could be produced by the pairwise combination of complementary heterologs in the system: HET = N × (N – 1). Next, the potential richness of sequences (N′) after cloning and mismatch repair of heteroduplexes was calculated as the number of unique sequences generated by combinatorial swapping of mismatch sites between heterologs. In the simplest case of a two-sequence system, the number of mosaic sequences that can be generated by mismatch repair is N′ = 2m, where m is the number of divergent sites between the sequences. To investigate template systems of higher complexity (N > 2), mosaics were generated for each potential heteroduplex combination, and the number of unique sequence types was summed to indicate N′ for the original N sequence system.
The frequencies of sequence variants after simulated mismatch repair of a clone library were determined for a system of three sequences containing three mismatched sites. Two cases were considered: first, where repair occurs independently for each mismatch, or second, where initiation of repair is restricted to nick sites 3′ or 5′ to the cloned heteroduplex insert, allowing co-repair of adjacent mismatches to the same template. Mosaic sequences were generated by manually stepping through the series of events required to repair the three mismatches, as mediated by different potential orientations of nick sites and MutS-mismatch binding. The probability of a given series of repair events was calculated based on the simplifying assumptions that all heteroduplexes are equally abundant among the cloned inserts, that all cloned heteroduplexes are repaired to the homoduplex state, and that all mismatches are repaired with the same efficiency in any orientation of the MutHLS system. The relative frequency of observing each sequence variant was determined by summing the probabilities of each series of repair events yielding that sequence. We considered conditions where heteroduplexes constitute 50, 5 and 0.1% of the cloned sequence inserts. Finally, theoretical sampling curves were calculated from the predicted mosaic frequencies using the formula for the average number of classes observed in a sample (18).
RESULTS
Heteroduplex formation
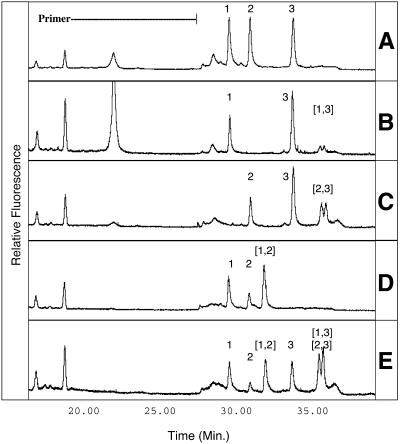
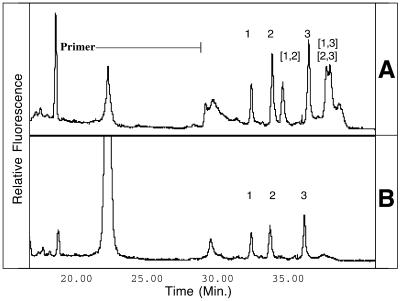
DNA from V.cholera, V.parahaemolyticus and V.vulnificus were individually amplified by PCR and subjected to combined CDCE analysis, yielding a single diagnostic peak for each species (Fig. 1A). The formation of heteroduplexes in mixed-template PCRs was indicated by the presence of additional peaks in CDCE spectra of co-amplified DNA templates (Fig. 1B–E). The double-stranded nature of the peaks was confirmed by denaturing a sample for 10 min prior to analysis. This treatment resulted in elimination of both the homoduplex and heteroduplex peaks and the formation of a single characteristic peak attributed to migration of single-stranded DNA (data not shown).
Figure 1.
CDCE separation of heterologous PCR products (A) combined after amplification and (B–E) co-amplified. Peak 1, V.vulnificus; peak 2, V.parahaemolyticus; peak 3, V.cholera. Heteroduplex peaks are denoted by bracketed numbers corresponding to the identities of the single-stranded heterologs. The indicated primer is a 20mer with a 54mer GC-rich clamp. Relative fluorescent intensities are indicated on the y-axis. (A) Peak identification standard, three single-template PCR products combined after amplification. (B–D) Co-amplification products of two-species template mixtures. (E) Three-species template mixture, heteroduplexes (2,3) and (1,3) co-migrate.
CDCE spectra of co-amplified DNA from V.cholera (peak 3) and either V.parahaemolyticus (peak 2) or V.vulnificus (peak 1) both revealed one homoduplex peak for each species and two heteroduplex peaks (Fig. 1B and C). The spectrum of co-amplified V.parahaemolyticus (peak 2) and V.vulnificus (peak 1), which diverge by 1 single bp substitution, showed only a single heteroduplex peak, presumably due to the overlap of peak areas from the two expected heteroduplexes (Fig. 1D). Similar proportions of heteroduplexes and homoduplexes were observed in mixed-template PCR products when the primer to product peak area ratio of less than 1 indicated the reactions had become primer limited (Fig. 1C–E). In contrast, when excess primer was present at the end of amplification, the homoduplex peaks were much larger than the heteroduplex peaks (Fig. 1B). The co-amplification product of the three-species template mixture, which indicated primer depletion, gave an even higher proportion of PCR product in the heteroduplex form, yielding the most intense signal of all peaks (Fig. 1E).
Heteroduplex contribution to genetic diversity
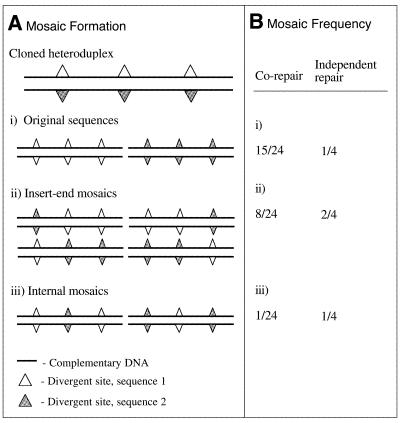
To illustrate how the cloning of heteroduplexes could contribute to the sequence diversity in clone libraries we modeled the MutHLS-directed mismatch repair of heteroduplex DNA during cloning. Our model demonstrates that the potential diversity of artifactual sequences in a clone library increases both with the number of sequence variants present in the original PCR and with the number of variable nucleotides. For simplicity, a case is considered where each heteroduplex contains variable sites at the same positions in the DNA sequence. For an initial template mixture of two sequences containing three mismatched nucleotides (m = 3), mismatch repair after cloning can yield eight sequence variants (Fig. 2). When the PCR template richness is increased to contain three or four sequences, fully divergent at the same three mismatch positions, 21 and 40 sequence variants can be generated, respectively. When a three-sequence system contains 10 fully divergent nucleotide positions, the potential cloned sequence richness increases to 3069 sequence variants (Table 1).
Figure 2.
Classes and frequencies of mosaic sequences possible from mismatch repair of heteroduplex containing three mismatches. (A) Mismatch repair of unmethylated heteroduplexes containing three mismatches can generate three classes of mosaic sequences: (i) restoration of an original sequence, (ii) creation of mosaics with distinct domains at the 3′ and 5′ ends and (iii) creation of mosaics with related insert termini but internal hybridism. (B) Relative frequencies of the mosaic classes produced by MutHLS repair of one heteroduplex containing three mismatches are indicated for independent repair of each mismatch or for co-repair of adjacent mismatches where repair initiates only at nick sites flanking the heteroduplex insert.
Table 1. Potential sequence richness in cloned PCR products after mismatch repair of heteroduplexes.
| Original sequence richness (N) | Unique heteroduplexes in final PCR product (HET) | Shared mismatch sites (m) | Potential cloned sequence richness (N′) |
|---|---|---|---|
| 2 |
2 |
3 |
8 |
| 3 |
6 |
3 |
21 |
| 3 |
6 |
4 |
40 |
| 3 |
6 |
10 |
3069 |
| 4 | 12 | 10 | 6136 |
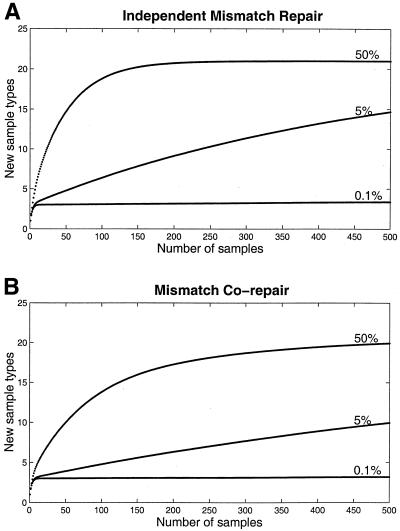
To illustrate how the proportion of cloned heteroduplex inserts influences the number of mosaic sequences detected while sampling a clone library, we considered a test case of three divergent sequences containing three mismatched sites. We estimated the frequency and distribution of mosaics generated by mismatch repair when heteroduplexes constitute 50, 5 or 0.1% of the cloned inserts, assuming either independent repair of each mismatch (Fig. 3A), or allowing co-repair of adjacent mismatches (Fig. 3B). For example, assuming independent repair, if 5% of the inserts are heteroduplexes, analysis of 500 clones in the clone library should reveal, on average, approximately 15 sequence types. However, if the heteroduplex insert fraction is reduced to 1 in 1000 inserts, sampling to 500 yields the three original templates with a low, but non-zero, probability of observing several low-frequency artifacts. When repair proceeds independently for each mismatch, the relative frequencies of mosaic sequences observed in the clone library is the highest relative to the original sequences (Fig. 3A). When co-repair of adjacent mismatches is possible, the frequency at which mosaics are detected decreases (Fig. 3B) as restoration of a parent sequence is the predominant class of repair products (Fig. 2B).
Figure 3.
Mean number of classes observed in n samples. Curves calculated by iteration of the formula described by Ewens (18)
where C(n) is the average number of sequence variants observed at sample n, Ctot is the total number of sequence variants in the population after cloning and mismatch repair, and p(i) is the probability of observing the ith sequence variant in the population. When 50, 5 and 0.1% of cloned inserts are heteroduplexes, mosaic sequences are detected at decreasing frequencies (A) via independent repair of adjacent mismatches by the MutHLS system, and (B) allowing co-repair of adjacent mismatches.
Heteroduplex elimination
As illustrated by our relative frequency model, reducing proportions of cloned heteroduplex inserts should dramatically reduce the frequency of mosaic sequences detected while screening genetic diversity. Because of the potentially high contribution of heteroduplexes to sequence richness in cloned PCR products, we investigated the possibility of eliminating heteroduplexes from multi-template PCR products prior to cloning. CDCE spectra of co-amplified PCR products before and after the reconditioning PCR were compared to detect elimination of the heteroduplex peaks in the mixed-template samples. The genomic DNA of V.cholera, V.parahaemolyticus and V.vulnificus was co-amplified (Fig. 4A), then diluted 10-fold and subjected to a three-cycle ‘reconditioning PCR’. This procedure resulted in the absence of artifactual peaks in CDCE spectra of mixed-template reactions suggesting at least 50-fold reduction of heteroduplex abundance based on the background detection limit of the method (<1%; Fig. 4B).
Figure 4.
CDCE separation of three-template PCR amplification products before and after three-cycle reconditioning PCR. Peak identities are as indicated in Figure 1. (A) The peak areas of homoduplex and heteroduplex DNA indicate they are at similar proportions in the final PCR product (∼50% heteroduplexes). (B) Heteroduplex peaks are eliminated by a three-cycle reconditioning PCR such that their relative frequency is below the background detection limit (<1%), illustrating at least a 50-fold reduction in heteroduplex frequency.
DISCUSSION
The presence of heteroduplex molecules in PCR-amplified samples containing unknown genetic diversity complicates the interpretation of sequence diversity. The potential number of unique heteroduplexes formed in the last annealing step of a PCR will be greater than, or equal to, the number of original sequence types. Thus, the sequence richness inferred by methods such as DGGE and CDCE, where heteroduplexes appear at discrete migration distances, may be dramatically overestimated if heteroduplex formation is not considered. If heteroduplexes are cloned into a host capable of mismatch repair, the sequence richness present in the clone library (N′) may increase exponentially from the initial number of unique double-stranded sequences (N) to include the sum of unique mosaics potentially formed from each heteroduplex present in the PCR product. The resulting increase of apparent sequence richness may explain part of the difficulty in sampling the genetic diversity present in clone libraries to saturation. Heteroduplexes have also been implicated as a source of sequence artifacts in other diversity-screening techniques, including RFLP (19), and RAPD analysis (20).
The cloning and subsequent mismatch repair of unmethylated heteroduplexes has been well established (11,13–15) and the potential for mosaic formation recognized as problematic, although its contribution to apparent genetic diversity not fully characterized (5,6,21). Our CDCE results indicate that late-stage PCR heteroduplexes can constitute a significant fraction of the PCR product and that the highest proportion of DNA in heteroduplex form is obtained by increasing the template diversity in the amplification mixture. This behavior is expected as increasing the diversity of templates in the mixture will lead to an increased probability of reannealing with a complementary heterolog. Furthermore, our modeling results demonstrate that the potential richness of mosaics formed from cloning heteroduplexes increases exponentially as nucleotide divergence increases.
The relative frequency of each mosaic sequence generated by mismatch repair of cloned heteroduplexes will differ depending on the series and direction of repair events mediated by the MutHLS system. To initiate repair of heteroduplex DNA, MutS recognizes and binds to a mismatch site while MutH recognizes a neighboring exonuclease cleavage site d(GATC) and nicks an unmethylated DNA strand (10,12,22,23). MutL connects the activities of MutS and MutH by directing digestion of the DNA strand from the nick site to the MutS–mismatch complex (12,24) in either the 3′ or 5′ direction (14,25). As the 4 bp MutH exonuclease recognition sequences should occur at a rough average of every 256 bp, a 1000 bp heteroduplex insert will likely contain three or four MutH exonuclease sites, enabling independent repair of interspersed mismatches. However, the MutL-mediated interaction of DNA-bound MutS and MutH can occur over distances of several kilobases (12,25), allowing co-repair of multiple mismatches to the same template over the length of the insert (11). Depending on the actual sequence of the cloned insert, MutHSL-mediated repair will be an intermediate combination of independent repair and co-repair mechanisms, and could create the frequently observed chimeric sequences defined by distinct domains at the 5′ and 3′ ends.
The several studies that have investigated the diversity of sequence artifacts in clone libraries of mixed-template PCR amplified DNA suggest increases in apparent diversity that parallel our model of heteroduplex mismatch repair. Borriello and Krauter (21) demonstrated that MutS-mediated mismatch repair was responsible for a 3-fold increase in sequence richness during analysis of 20 cloned PCR inserts targeting a five-member gene family. Similarly, after DGGE analysis of 66 clones Speksnijder et al. (5) observed nine additional low-frequency sequences in a clone library constructed from PCR amplification of an original seven-template system. Our model for a three-template system (Fig. 3) suggests that when 5% of cloning inserts are heteroduplexes, mismatch repair can result in a doubling of apparent sequence richness after analysis of 80 samples and a 5-fold increase in apparent sequence richness after 500 samples. By reducing the heteroduplex insert fraction to 0.1%, sampling to 500 yields, on average, only the three original templates. Thus, if PCR-generated cloning inserts contain a substantial fraction of heteroduplex molecules, which are indistinguishable from homoduplex DNA by routine agarose gel fractionation, as suggested by Figure 1C–E, there is a high probability that a PCR/cloning artifact will be sampled during the analysis of cloned sequence richness. However, if the cloning of heteroduplexes is minimized, given the diversity of potential mosaic sequences generated by mismatch repair, it is unlikely that the same mosaic sequence will be encountered twice.
Heteroduplex elimination from multi-template PCR products prior to cloning has been attempted previously. Some heteroduplexes can be eliminated from PCR products by using a single-strand cleaving endonuclease to resolve internal single-strand loop structures (4,26). Single-stranded loop structures can cause heteroduplexes to migrate anomalously during gel electrophoresis forming additional bands on agarose gels (19,27), which can also be removed by gel purification of the desired PCR product (4). However, as our results indicate, when heterologous sequences are of the same length and diverge by base pair mismatches the resulting heteroduplexes can maintain a Watson–Crick configuration (28,29) and migrate with homoduplex DNA during agarose gel electrophoresis.
Our ‘reconditioning PCR’ method to eliminate heteroduplexes is based on the principle that formation of homoduplex DNA will be favored to the exclusion of heteroduplex DNA in the presence of excess primer. By restoring the initial primer concentrations during the ‘reconditioning PCR’ a denatured DNA molecule will have a higher probability of annealing with a primer than with a heterolog, leading to extension of the homoduplex. By the same principle, the formation of PCR chimeras from annealing and extension of heterologous DNA fragments can be reduced by optimizing the number of amplification cycles to maintain an excess of primer through the endpoint of the reaction. We found that a 10-fold dilution of amplification product, followed by three-cycle re-amplification, effectively removed heteroduplexes from our mixed-template amplifications as detected by CDCE. Higher initial dilutions (e.g. 100-fold) for reconditioning will create even more favorable conditions for homoduplex formation while maintaining product yield within an acceptable range for cloning (i.e. 0.1–50 ng). Both dilution and reconditioning cycle number can be adjusted for adaptation to different amplification protocols.
In summary, our experiments demonstrate that when the PCR is used to analyze the genetic diversity present in a community of DNA, heteroduplex DNA may represent a large fraction of the final reaction product. Our modeling results indicate that the cloning and mismatch repair of such heteroduplexes can create an explosion of sequence diversity in a clone library. Avoidance of primer limitation during amplification, followed by the low cycle number reconditioning PCR suggested here, quantitatively reduces the abundance of heteroduplexes in the final PCR product. Interpretation of sequence diversity represented at least twice in a clone library minimizes the potential of including low-frequency mosaic sequences in the analysis. These recommendations help ensure that the sequence richness observed during analysis reflects the genetic diversity present in the original PCR template.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Aoy Tomita-Mitchell, of People’s Genetics, Inc. (MA, USA) for help in primer design and CDCE analysis, Dr William G. Thilly of the Massachusetts Institute of Technology (MA, USA) for use of the CDCE machine, Weiming Zeng of the Massachusetts Institute of Technology for helpful discussion, and Dr Andy Solow of the Woods Hole Oceanographic Institute (Woods Hole, MA) for help with statistical analysis. This work was funded in part by MIT Sea Grant and by a National Science Foundation Graduate Research Fellowship to J.R.T.
REFERENCES
- 1.Keohavong P. and Thilly,W.G. (1989) Fidelity of DNA-polymerases in DNA amplification. Proc. Natl Acad. Sci. USA, 86, 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang G.C.Y. and Wang,Y. (1996) The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology, 142, 1107–1114. [DOI] [PubMed] [Google Scholar]
- 3.Wang G.C.Y. and Wang,Y. (1997) Frequency of formation of chimeric molecules is a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol., 63, 4645–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu X.Y., Wu,L.Y., Huang,H.S., McDonel,P.E., Palumbo,A.V., Tiedje,J.M. and Zhou,J.Z. (2001) Evaluation of PCR-generated chimeras: mutations and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol., 67, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speksnijder A., Kowalchuk,G.A., De Jong,S., Kline,E., Stephen,J.R. and Laanbroek,H.J. (2001) Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol., 67, 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Wintzingerode F., Gobel,U.B. and Stackebrandt,E. (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev., 21, 213–229. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki M.T. and Giovannoni,S.J. (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol., 62, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen M.A. and Straus,N. (1993) Effect of PCR conditions on the formation of heteroduplex and single-stranded-DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl., 3, 186–194. [DOI] [PubMed] [Google Scholar]
- 9.Ruano G. and Kidd,K. (1992) Modeling of heteroduplex formation during PCR from mixtures of DNA templates. PCR Methods Appl., 2, 112–116. [DOI] [PubMed] [Google Scholar]
- 10.Modrich P. (1987) DNA mismatch correction. Annu. Rev. Biochem., 56, 435–466. [DOI] [PubMed] [Google Scholar]
- 11.Carraway M. and Marinus,M.G. (1993) Repair of heteroduplex DNA-molecules with multibase loops in Escherichia coli. J. Bacteriol., 175, 3972–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrich P. (1989) Methyl-directed DNA mismatch correction. J. Biol. Chem., 264, 6597–6600. [PubMed] [Google Scholar]
- 13.Learn B.A. and Grafstrom,R.H. (1989) Methyl-directed repair of frameshift heteroduplexes in cell-extracts from Escherichia coli. J. Bacteriol., 171, 6473–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahue R.S., Au,K.G. and Modrich,P. (1989) DNA mismatch correction in a defined system. Science, 245, 160–164. [DOI] [PubMed] [Google Scholar]
- 15.Parker B.O. and Marinus,M.G. (1992) Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khrapko K., Coller,H.A., Andre,P.C., Li,X.C., Hanekamp,J.S. and Thilly,W.G. (1997) Mitochondrial mutational spectra in human cells and tissues. Proc. Natl Acad. Sci. USA, 94, 13798–13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khrapko K., Hanekamp,J.S., Thilly,W.G., Belenkii,A., Foret,F. and Karger,B.L. (1994) Constant denaturant capillary electrophoresis (CDCE)—a high-resolution approach to mutational analysis. Nucleic Acids Res., 22, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewens W.J. (1972) The sampling theory of selectively neutral alleles. Theor. Popul. Biol., 3, 87–112. [DOI] [PubMed] [Google Scholar]
- 19.Jansen R. and Ledley,F.D. (1990) Disruption of phase during PCR amplification and cloning of heterozygous target sequences. Nucleic Acids Res., 18, 5153–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayliffe M.A., Lawrence,G.J., Ellis,J.G. and Pryor,A.J. (1994) Heteroduplex molecules formed between allelic sequences cause nonparental RAPD bands. Nucleic Acids Res., 22, 1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borriello F. and Krauter,K.S. (1990) Reactive site polymorphism in the murine protease inhibitor gene family is delineated using a modification of the PCR reaction (PCR+1). Nucleic Acids Res., 18, 5481–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahue R.S., Su,S.S. and Modrich,P. (1987) Requirement for d(GATC) sequences in Escherichia coli MutHLS mismatch correction. Proc. Natl Acad. Sci. USA, 84, 1482–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsh K.M., Lu,A.L., Clark,S. and Modrich,P. (1987) Isolation and characterization of the Escherichia coli MutH gene-product. J. Biol. Chem., 262, 15624–15629. [PubMed] [Google Scholar]
- 24.Sancar A. and Hearst,J.E. (1993) Molecular matchmakers. Science, 259, 1415–1420. [DOI] [PubMed] [Google Scholar]
- 25.Dao V. and Modrich,P. (1998) Mismatch-, MutS-, MutL- and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J. Biol. Chem., 273, 9202–9207. [DOI] [PubMed] [Google Scholar]
- 26.Lowell J.L. and Klein,D.A. (2000) Heteroduplex resolution using T7 endonuclease I in microbial community analyses. Biotechniques, 28, 676–678. [DOI] [PubMed] [Google Scholar]
- 27.Zorn A.M. and Krieg,P.A. (1991) PCR analysis of alternative splicing pathways—identification of artifacts generated by heteroduplex formation. Biotechniques, 11, 180–184. [PubMed] [Google Scholar]
- 28.Brown T., Kennard,O., Kneale,G. and Rabinovich,D. (1985) High-resolution structure of a DNA helix containing mismatched base-pairs. Nature, 315, 604–606. [DOI] [PubMed] [Google Scholar]
- 29.Hunter W.N., Brown,T., Anand,N.N. and Kennard,O. (1986) Structure of an adenine.cytosine base pair in DNA and its implications for mismatch repair. Nature, 320, 552–555. [DOI] [PubMed] [Google Scholar]