Abstract
Redefinition of UAG, UAA and UGA to specify a standard amino acid occurs in response to recoding signals present in a minority of mRNAs. This ‘read-through’ is in competition with termination and is utilized for gene expression. One of the recoding signals known to stimulate read-through is a hexanucleotide sequence of the form CARYYA 3′ adjacent to the stop codon. The present work finds that of the 91 unique viral sequences annotated as read-through, 90% had one of six of the 64 possible codons immediately 3′ of the read-through stop codon. The relative efficiency of these read-through contexts in mammalian tissue culture cells has been determined using a dual luciferase fusion reporter. The relative importance of the identity of several individual nucleotides in the different hexanucleotides is complex.
INTRODUCTION
Standard decoding is enriched, in probably all organisms, by special signals, often called recoding signals, embedded in a subset of mRNAs. One facet of recoding is the redefinition of stop codons in certain sequence contexts to specify an amino acid. The same stop codon in the great majority of other contexts in the same cell retains the standard function of specifying termination. Thus, this redefinition is distinct from global reassignment that occurs in certain organelles and in some organisms. Where UGA is redefined to specify the 21st amino acid selenocysteine (1), the identity of the amino acid specified is the important feature. In other cases, in response to different recoding signals, the important consequence is that a proportion of the ribosomes continue translation beyond the stop codon in the same reading frame, i.e. they read through the stop codon. Though unimportant in itself, the most common amino acid encoded in the read-through of UGA is tryptophan and of UAG is glutamine. Read-through utilized for gene expression is typically 100–1000-fold above the error rate for sensing stop codons even though commonly only 1–10% of ribosomes read through such redefined stop codons. Read-through is utilized to synthesize a proportion of extended proteins that may have additional functions to that of the standard product. Synthesis of the extended product may be regulatory (2), or perhaps ribosome movement 3′ of the leaky stop codon may itself have regulatory significance for mRNA structure (3). UAA is often less efficiently read through than UAG, and UGA is more ‘leaky’ than UAG.
A recoding signal for read-through can be located many hundreds of nucleotides 3′ of the redefined stop codon as discovered by Miller and colleagues in their studies on barley yellow dwarf virus (4). It can also be an elaborate pseudoknot 3′ of the leaky stop codon as in the synthesis of the murine leukemia virus gag–pol precursor (5–7). However, in the original case of the RNA phage Q beta coat protein read-through (8,9) or in the case of Sindbis virus recoding (10) only the identity of the nucleotide 3′ adjacent to the UGA utilized is thought to be important. Release factors recognize the triplet stop codon and adjacent nucleotides; in particular, they recognize the 3′ adjacent nucleotide. In read-through cases the 3′ adjacent nucleotide is not one favored for recognition by the release factor (11). While the identity of the following two nucleotides has diminishing importance for release factor recognition, subsequent bases are not known to have any direct effect on that recognition. Nevertheless, for read-through to synthesize a replicase component of tobacco mosaic virus (TMV), the identity of six nucleotides 3′ adjacent to the stop codon, in the form CAR-YYA, is important (12–14).
The recent enormous increase in sequence information prompted us to assess nucleotide preferences in the vicinity of read-through stop codons. We have concentrated on read-through in viral expression since the great majority of cases currently known are in viral decoding. The importance of the different nucleotides within the contexts found were tested in a dual luciferase fusion reporter designed for this purpose (15). The leaky terminator and surrounding context are placed between a Renilla luciferase reporter, which provides a measure of termination, and a Photinus luciferase reporter, which provides a measure of read-through product.
MATERIALS AND METHODS
Collection and examination of viral sequences
Using the Taxonomy browser at the National Center for Biotechnology Information (NCBI) web site (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/), several representative nucleotide sequences from each currently accepted International Committee on Taxonomy of Viruses genus were examined to determine whether any members of that genus contained a read-through stop codon. In addition, an Entrez (http://www.ncbi.nlm.nih.gov/Entrez/) keyword search was performed for the terms ‘read-through’ or ‘transl_except’ and the resulting hits were further examined.
The uniqueness of each sequence was determined using criteria that allowed different strains of the same virus to be included, but excluded any sequences with identical names or known aliases as determined by their NCBI taxonomic entries. The 82 nt long segment of sequence from the 19th nucleotide 5′ to the 60th nucleotide 3′ of the leaky stop codon was extracted from each identified distinct sequence. The segments were then compared to one another, and one sequence was excluded from any pair of sequences that shared >90% identity. Sequence controls were obtained by extracting the 82 nt region surrounding the in-frame, non-leaky stop codon downstream of each leaky stop codon.
The nucleotide triplet immediately 3′ of the stop codon (+1 triplet), was used to divide the sequences into groups. The non-randomness associated with each nucleotide position in each group was examined by chi-square (χ2) analysis. The secondary structure 3′ of the stop codon was examined using mFold (16,17). The folding was simulated at 37°C, and limitations were set so that no binding was allowed to occur with the stop codon or the sequence 5′ of it.
Assessing the ability of the sequence groups to signal read-through using p2luc
The dual luciferase reporter vector p2luc (DDBJ/EMBL/GenBank accession number AF043450) (15) was constructed with one of seventeen 18 bp sequences, representing groups 1, 2, 3 and 4 of the identified read-through groups, inserted between the BamHI and SalI restriction sites. The insert of each construct was synthesized as a pair of complementary oligonucleotides, such that the coding strand sequence read 5′-GATCC-CCC-AAA-WWW-XXX-XXX-CAG-3′, and the non-coding strand sequence read 5′-TCGAC-CTG-YYY-YYY-ZZZ-TTT-GGG-3′. GATCC and TCGAC were the complementary sticky ends of the BamHI and SalI sites respectively. The WWW was either TAG or TGA for the test sequences or CAG or TGG for the control sequences and the Zs represent the complement of the corresponding Ws. The Xs represent the +1 and +2 triplet nucleotide positions and the Ys represent the complement of the corresponding X. Each pair of oligonucleotides was then annealed and separately ligated into the digested p2luc vector. The raw results in relative light units (RLU), available at http://opbs.okstate.edu/Virevol/lucdat.html, were converted to %read-through (RT) using the formula RT = (RLU of test Photinus luc/RLU of test Renilla luc)/(RLU of control Photinus luc/RLU of control Renilla luc).
RESULTS
Collection and examination of viral sequences
Of the viral genera examined, 23 had read-through stop codons reported in their genome annotations. These 23 genera yielded 157 individual sequences for screening. The uniqueness of each sequence was determined as described in Materials and Methods, eliminating 66 sequences and leaving 91 unique sequences to be analyzed.
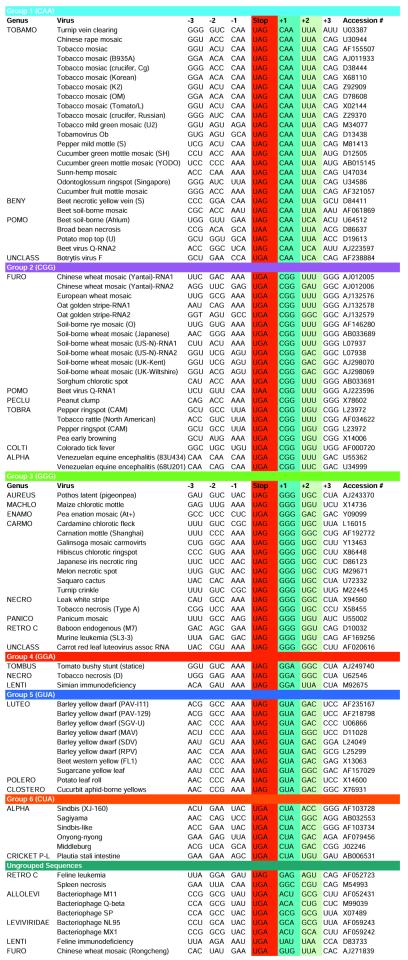
Since the nucleotides immediately 3′ of the leaky stop codon were previously implicated in read-through (4,5,10,12,18–22), the sequences were first categorized according to these nucleotides. It was found that 6 of the 64 possible nucleotide triplets accounted for 90% of the triplets in the +1 position of the read-through sequences, while being present in only two of the non-read-through control sequences. In contrast, the six most frequent triplets found in the +1 position of the non-read-through control groups only accounted for 35.1%. Each one of the six triplets, CAA, CGG, GGG, GGA, GUA and CUA, formed the basis of a sequence group into which all sequences containing that +1 triplet were placed (Table 1). For the purposes of discussion we have considered the nucleotides in sets of three, even though the influence of the nucleotides probably occurs at the nucleotide level rather than the codon level.
Table 1. Sequence groups based on nucleotides 3 adjacent to read-through stop codons.
Sequences in the +2 triplet position were also non-randomly distributed with the five most frequent accounting for 70.3%. These triplets were distributed among all the +1 groups. However, for five of the six +1 groups, there was a +2 triplet that was mostly non-random for each +1 triplet sequence: CAA-UUA (88.0%), CGG-UUU (55.0%), GGG-UGC (52.9%), GGA-GGC (66.3%) and GUA-GAC (80.0%). The sixth group, CUA, did not show any common +2 triplet sequence.
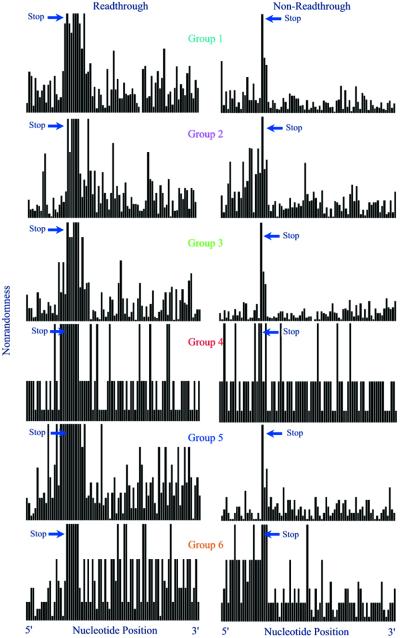
Examination of sequence variability within the groups using χ2 analysis (Fig. 1) showed that both the +1 and, to a lesser extent, +2 triplets in the read-through groups were much more non-random than their counterparts in the corresponding control groups. Read-through groups 4, 5 and 6 contain other nucleotides that have χ2 values as high as, or nearly as high as, their respective nucleotides in the +1 triplet. For group 4, the high level of background in both the read-through and control groups can be accounted for by the fact that this group only contains three viral sequences. For group 5, which consists mostly of luteoviruses, the other highly non-random nucleotides are expected because of the CCN-NNN repeat that has been shown to be important in signaling read-through for luteoviruses (4). This repeat is present to a limited extent in the other group 5 sequences, and in six of the nine ungrouped sequences, suggesting that this repeat may also play a role in signaling read-through in other genera. In group 6, the equality of other χ2 values with that of the +1 triplet is due to the higher level of sequence identity among the coding regions of the read-through proteins of the alpha viruses, as compared to members of other groups. This is evident by comparing the χ2 values 3′ of the leaky terminator in the read-through group and the χ2 values 5′ of the read-through protein stop codon in the control group with those 3′ of the control group.
Figure 1.
Examination of non-randomness in read-through groups. Each data set represents the 82 nt region surrounding either the leaky (left column) or non-leaky (right column) stop codon for each of the six +1 triplet groups. For each data set, the x-axis represents the individual nucleotide position from the –19th to the +60th relative to the stop codon, and the y-axis is a measure of the non-randomness associated with each nucleotide position in the form of a c2 value.
The terminal dipeptide has been implicated in translation termination efficiency (19,23,24). The chemical characteristics of the penultimate amino acid have been shown to influence the efficiency of termination, with basic residues yielding more efficient read-through in Saccharomyces cerevisiae (19), and acidic and hydrophobic residues giving higher read-through in Escherichia coli (23). Greater read-through efficiency is also associated with a higher likelihood of the ultimate amino acid participating in the formation of α-helices or β-sheets in E.coli (24). We examined the properties of the terminal dipeptides in both our sample and control groups and found no bias in the chemical characteristics of the penultimate amino acid, or in the α-helical or β-sheet propensities of the ultimate amino acid, suggesting that these 5′ signals are not utilized to facilitate viral read-through. However, comparison of the 5′ sequences revealed that there was a preference for adenine in the penultimate and ultimate nucleotide positions, accounting for 76 and 71% respectively of the total nucleotides in those positions compared to only 28 and 25% respectively in the non-read-through control groups.
The secondary structure 3′ of the stop codon was examined for stem–loop or pseudoknot structures reported to be important for efficient read-through in some viruses. Although some of the sequences examined displayed feasible structures, no consistent significant similarities were found with either the reported structures or with each other.
Construct analysis using luciferase reporter system
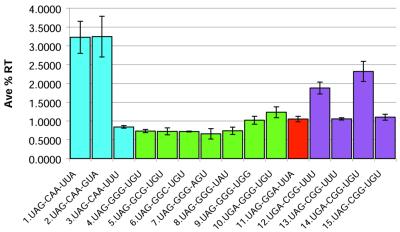
The test construct inserts were designed either to be exact replicas of the +1 and +2 triplets of the sequence groups, as was the case for constructs 1, 4, 11, 12 and 14 (groups 1, 3, 4, 2 and 2 respectively), or derivatives of those sequences with single nucleotide substitutions, as was the case for the remaining 10 constructs. Each of the 15 constructs inserted into the p2luc vector was tested for its ability to facilitate read-through expression of Photinus luciferase. All the constructs produced values greater than the low background level for spontaneous read-through for this vector (15). The raw output of the assays was converted to average %RT (Fig. 2). The sequence for construct 1 was taken directly from group 1 (CAA-UUA) and constructs 2 and 3 each make a single substitution in that sequence (CAA-GUA and CAA-UUU respectively). While the U to G substitution at position +4 had little effect on read-through, the A to U substitution at position +6 significantly reduced the level of read-through observed. These results are consistent with the results obtained by Skuzeski et al. examining the TMV read-through context in vivo (12) and the in vitro work of Zerfass and Beier (14).
Figure 2.
Read-through facilitated by p2luc constructs. Each of the 15 sequences listed on the x-axis were tested for their ability to facilitate read-through of Photinus luciferase using the fusion vector p2luc. Sequences 1, 4, 11, 12 and 14 represent exactly the +1 and +2 triplet sequences from groups 1, 3, 4, 2 and 2 respectively. The other 10 sequences were either substitutions of a single nucleotide position (sequences 2, 3 and 5–9) or a substitution of the stop codon (sequences 10, 13 and 15).
Constructs 4–10 were all based on group 3 (GGG). None of the single nucleotide substitutions made in the +1 and +2 triplets in constructs 5–8 had any significant effect on read-through levels, however in construct 9, where the +6 nucleotide U was substituted with a G, read-through increased significantly. In construct 10, where the UAG stop codon was changed to a UGA, the read-through rose ∼1.7-fold. A difference in read-through between identical constructs with different stop codons also occurred in constructs 12–14, which are based on Group 2 (CGG); both the CGG-UUU and CGG-UGU constructs showed significantly higher levels of read-through when placed downstream of a UGA terminator compared to their UAG counterparts. Groups 5–6 were not tested.
When comparing the constructs that represent the non-substituted read-through group sequences, there also appeared to be variation in the percentage of read-through each sequence group facilitated. Group 1 showed the highest levels (3.2% ± 0.4), followed by both group 2 constructs, themselves displaying variation (2.3% ± 0.3 and 1.9% ± 0.2). Group 4 had the third highest read-through levels (1.06% ± 0.08), and group 3 had the lowest of the groups tested (0.74% ± 0.05).
DISCUSSION
The degree to which the identity of the six nucleotides 3′ of viral read-through stop codons is restricted is remarkable. The sequences examined are from RNA-containing viruses in which mutation rates are notoriously high and different sequence combinations are undoubtedly frequently tested. Given the larger than triplet recognition in the release process (25), one would have expected the sequence 3′ of ‘tight’ (non-read-through) stop codons to be more restricted than leaky stop codons but this is not so, as the level of non-randomness associated with this position in the control groups is low (Fig. 1). However, with the read-through stop codons, restriction extends to the sixth following nucleotide and is even pronounced at this position. While other recoding signals are known to be operative in some of the available sequences analyzed (4,6,10,12), and are likely involved, though unrecognized, in others, it is clear from the statistical and experimental analysis performed that the 3′ hexanucleotide sequence is a major influence on read-through.
Stop codon recognition occurs in the ribosomal A-site. Stacking of the 3′ adjacent base has an influence on codon interactions in the A-site and influences both termination (26–28) and frameshifting (29). How the identity of up to six nucleotides affects read-through is less clear. In one case of frameshifting, that for yeast Ty3, there is provocative evidence that a local 3′ effect, which extends to 13 bases, is due to mRNA pairing with rRNA in the pre A-site (30).
An influence of specific rRNA segments on in-frame stop codon recognition has been proposed based on several experiments. These experiments suggest that in prokaryotes there is an interaction involving the stop codon and C1054 of helix 34 of the 16S rRNA (31). There is also evidence that a similar interaction is at work in eukaryotes because of the conservation of C1054 in yeast 18S rRNA (32). Also, the strength of the stop codon’s interaction in this trio may be in part due to the nucleotide sequence surrounding the stop codon (33). This is supported by site-directed crosslinking experiments showing that nucleotide positions +1 (34) and +4 to +6 (35) can be crosslinked to release factor 2.
Read-through signals
The number of nucleotides that appear to be necessary for the signaling of read-through vary. Group 1 adhered to the CAR-YYA formula found to be important for the in vivo read-through of the UAG of TMV (12) and the in vitro read-through of UAG, UAA and UGA in a TMV-specific context (14). Group 1 is also in agreement with the -CA(A/G)N(U/C/G)A- consensus sequence found to facilitate read-through in S.cerevisiae (36). The essential nucleotides in the spacer region between the stop codon and the beginning of the pseudoknot in MuLV (37) are mostly conserved in all of the members of group 3. The luteovirus proximal signaling sequence CCN-NNN is known to be necessary for read-through, whereas the +1 and +2 triplets appear to have no importance (4). This luteovirus signal appears to various extents in every group 5 sequence, as well as appearing in six of the nine ungrouped sequences. Therefore, perhaps group 5 should be redefined using the CCN-NNN repeat as the criterion instead of the +1 triplet, leaving only three ungrouped sequences.
The type of stop codon appears to be a determinant for the +1 triplet groups, as almost all the groups are stop codon specific, the only exceptions being broad bean necrosis virus and botrytis virus F in group 1, beet virus Q in group 2 and pea enation mosaic virus in group 3. Except for broad bean necrosis virus in group 1 and beet virus Q in group 2, the UAA stop codon does not appear in any of the sequences examined, which is consistent with UAA(A/G) being one of the most preferred termination sequences in eukaryotes (25). However, the context for UAG read-through in MuLV has been shown to work with UAA and UGA in vivo and in vitro (22). The TMV UAG read-through context also appears to function for both UAA and UGA in vitro (14). And the UGA read-through context of Sindbis virus can facilitate read-through for UAA and UAG as well (38). These data suggest that the stop codon dependence of the sequence groups may be the result of some other factor and not a necessity of the +1 triplet sequence.
The relative abundance of adenine in the penultimate and ultimate nucleotide positions relative to the leaky terminator suggests that the 5′ context may also play a role in signaling read-through. However, previous studies report conflicting evidence as to the influence of adenine in these positions raising questions, at least at the nucleotide level, about the importance of these positions (39,40).
It is interesting to note that the individual groups are not host specific, since members of Furovirus, Coltivirus and Alphavirus all appear in group 2, and infect plants, bacteria and mammals, respectively. Host non-specificity combined with the small number of groups that are sufficient to accommodate all but three of the examined sequences suggests one of two conclusions: that either there are only a limited number of sequences that can signal read-through, and the members of each group co-evolved the same sequence; or, less likely given the diversity of the members within a group, that the members of a group came from a common ancestor that possessed the signaling sequence or a precursor to it.
Although we were not able to categorize three of the examined sequences, this is likely to be a consequence of the limited number of complete viral genomes containing read-through stop codons that have been sequenced to date. Identification of more read-through sequences may lead to the addition of other groups of conserved sequences, allowing the ungrouped sequences here to be categorized, revealing the total number of signaling sequence groups and how they facilitate read-through of a stop codon.
Read-through facilitated by the selected sequences in the p2luc
The varying amount of read-through observed with the different constructs suggests that some signals may play a larger role than others. The group 1 sequence CAA-UUA facilitates the highest amount of read-through of the tested sequences and reaches levels approaching the level of ∼5% reported by Skuzeski et al. for TMV (12). The constructs representing group 3, in contrast, show much lower levels than the ∼5% level reported by Jamjoom et al. for MuLV (41). However, both the pseudoknot and the spacer region are known to play a role in read-through signaling in MuLV (5,6), a member of group 3. So perhaps additional signaling sequences exist in the other members of group 3, and the CGG sequence is only part of a more complex signal. The presence of additional signals may also explain the presence of the CUA +1 sequence in group 5. As explained above, all the members of group 5 contain the CCN-NNN repeat known to be the proximal read-through signal in luteoviruses. Perhaps the CUA sequence increases the efficiency of the repeat.
Inspection of the luciferase assay results also reveals some differences in the importance of the individual nucleotide positions in the constructs. In all the constructs assayed, and in all but 4 of the 91 sequences examined, either a C or G is in the +1 nucleotide position. This supports evidence suggesting that the +1 position is critical for read-through in most systems, and that C or G in that position is important for efficient read-through to occur (10,18,42). In fact, C in the +1 nucleotide position is associated with all four constructs showing the highest levels of read-through.
The constructs that had substitutions in the +2 (construct 5), +3 (construct 6) and +4 (constructs 2 and 7) nucleotide position show no significant changes in the level of read-through facilitated, suggesting that these positions either have no role in signaling read-through or the nucleotides substituted are comparable to those they replaced. Previous work with the tobacco rattle virus (TRV) UGA context concluded that just a single nucleotide substitution was not sufficient to influence read-through (43). However, no substitutions to the +5 nucleotide were made in that study. Our results show that the +5 U to G substitution in TRV’s UGA significantly increases read-through in our system. In contrast, our data also indicate that the same substitution at the +5 position had no influence on either the group 2 UAG construct series or on the group 3 construct series, hinting that the importance of a position depends on the nature of the group and the stop codon. Alteration of the +6 position caused the most dramatic change in read-through of any subset tested. An A to U substitution in position +6 of group 1 decreased read-through 3.8-fold. In contrast, substituting a G for the +6 U in the group 3 construct series increased read-through significantly. The terminator also appeared to have an influence on read-through levels in COS cells. For both the group 3 and group 2 construct series, higher levels of read-through were displayed when comparing constructs with identical +1 and +2 sequences, but having a UGA terminator instead of a UAG. These findings hint at the complexity of read-through signaling and demonstrate the need for additional constructs to further explore the importance of each nucleotide position.
Since the contexts tested here were taken from groups that include viruses whose hosts are in different kingdoms, read-through signaling mechanisms may be universal. Indeed, in support of this view, the read-through analysis using luciferase reporter genes in mammalian COS cells is consistent with inferences derived primarily from viral sequences infecting plants. On the other hand, it may be possible that differences between the translation systems of the hosts would make the COS assay system used here a non-accurate representation of the performance of some of the contexts. Experiments are being designed to test the constructs used with COS cells in a plant based system.
With the mechanism of translation termination only partially understood, it is difficult to determine with certainty what role the sequence groups described here play in altering that mechanism. The sequences in these groups could influence the binding equilibrium of either release factor or amino-acyl-tRNA, through direct contact or indirectly through interactions with the ribosome. It is unlikely that these groups have no role because of their high level of non-randomness, their conspicuous under-representation in the control groups and the ability of the plant viral sequences to facilitate read-through even using an animal based assay system.
While this manuscript was in final preparation, another classification scheme for read-through stop codon contexts was published by Beier and Grimm (44). This classification differs from the one presented here in that the sequences are divided into three types. Type I represents plant viruses containing the CAA-UYA consensus sequence of TMV (12), similar to our group 1. Type II contains both plant and animal viruses that have either a CGG or CUA +1 triplet 3′ of a UGA terminator, where we have each of these triplets separated into different groups. Type III is based on the linear, purine-rich octanucleotide found in the spacer region of MuLV.
Acknowledgments
ACKNOWLEDGEMENTS
This work was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported under OKL01789 from that station and GM48152 from the National Institutes of Health. We thank Lorin Petros and Xiufen Li for help with the luciferase experiments.
REFERENCES
- 1.Low S.C. and Berry,M.J. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci., 21, 203–208. [PubMed] [Google Scholar]
- 2.Robinson D.N. and Cooley,L. (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development, 124, 1405–1417. [DOI] [PubMed] [Google Scholar]
- 3.Atkins J.F., Weiss,R.B. and Gesteland,R.F. (1990) Ribosome gymnastics–degree of difficulty 9.5, style 10.0. Cell, 62, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C.M., Dinesh-Kumar,S.P. and Miller,W.A. (1996) Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol., 70, 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y.X., Yuan,H., Rein,A. and Levin,J.G. (1992) Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol., 66, 5127–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills N.M., Gesteland,R.F. and Atkins,J.F. (1991) Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA, 88, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ten Dam E.B., Pleij,C.W. and Bosch,L. (1990) RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes, 4, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofstetter H., Monstein,H.J. and Weissmann,C. (1974) The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta, 374, 238–251. [DOI] [PubMed] [Google Scholar]
- 9.Weiner A.M. and Weber,K. (1973) A single UGA codon functions as a natural termination signal in the coliphage Q beta coat protein cistron. J. Mol. Biol., 80, 837–855. [DOI] [PubMed] [Google Scholar]
- 10.Li G. and Rice,C.M. (1993) The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol., 67, 5062–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tate W.P. and Mannering,S.A. (1996) Three, four or more: the translational stop signal at length. Mol. Microbiol., 21, 213–219. [DOI] [PubMed] [Google Scholar]
- 12.Skuzeski J.M., Nichols,L.M., Gesteland,R.F. and Atkins,J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- 13.Stahl G., Bidou,L., Rousset,J.P. and Cassan,M. (1995) Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res., 23, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerfass K. and Beier,H. (1992) Pseudouridine in the anticodon G psi A of plant cytoplasmic tRNA(Tyr) is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res., 20, 5911–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grentzmann G., Ingram,J.A., Kelly,P.J., Gesteland,R.F. and Atkins,J.F. (1998) A dual-luciferase reporter system for studying recoding signals. RNA, 4, 479–486. [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 17.Zuker M., Mathews,D.H. and Turner,D.H. (1999) In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43.
- 18.Kopelowitz J., Hampe,C., Goldman,R., Reches,M. and Engelberg-Kulka,H. (1992) Influence of codon context on UGA suppression and readthrough. J. Mol.Biol., 225, 261–269. [DOI] [PubMed] [Google Scholar]
- 19.Mottagui-Tabar S., Bjornsson,A. and Isaksson,L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valle R.P., Drugeon,G., Devignes-Morch,M.D., Legocki,A.B. and Haenni,A.L. (1992) Codon context effect in virus translational readthrough. A study in vitro of the determinants of TMV and Mo-MuLV amber suppression. FEBS Lett., 306, 133–139. [DOI] [PubMed] [Google Scholar]
- 21.Honigman A., Wolf,D., Yaish,S., Falk,H. and Panet,A. (1991) cis Acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology, 183, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y.X., Levin,J.G., Hatfield,D.L., Schaefer,T.S., Gorelick,R.J. and Rein,A. (1989) Suppression of UAA and UGA termination codons in mutant murine leukemia viruses. J. Virol., 63, 2870–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S.P. (1996) Functional interaction between release factor one and P-site peptidyl-tRNA on the ribosome. J. Mol. Biol., 261, 98–107. [DOI] [PubMed] [Google Scholar]
- 24.Tate W.P., Poole,E.S., Dalphin,M.E., Major,L.L., Crawford,D.J. and Mannering,S.A. (1996) The translational stop signal: codon with a context, or extended factor recognition element? Biochimie, 78, 945–952. [DOI] [PubMed] [Google Scholar]
- 25.Brown C.M., Stockwell,P.A., Trotman,C.N. and Tate,W.P. (1990) Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res., 18, 6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayer D. and Yarus,M. (1986) The context effect does not require a fourth base pair. Science, 231, 393–395. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen W.T. and Curran,J.F. (1991) Effects of the nucleotide 3′ to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J. Mol. Biol., 219, 231–241. [DOI] [PubMed] [Google Scholar]
- 28.Stormo G.D., Schneider,T.D. and Gold,L. (1986) Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res., 14, 6661–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertrand C., Prere,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (2002) Influence of the stacking potential of the base 3′ of tandem shift codons on –1 ribosomal frameshifting used for gene expression. RNA, 8, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., Stahl,G. and Farabaugh,P.J. (2001) Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA, 7, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prescott C., Krabben,L. and Nierhaus,K. (1991) Ribosomes containing the C1054-deletion mutation in E.coli 16S rRNA act as suppressors at all three nonsense codons. Nucleic Acids Res., 19, 5281–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernoff Y.O., Newnam,G.P. and Liebman,S.W. (1996) The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: genetic evidence in yeast. Proc. Natl Acad. Sci. USA, 93, 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter A.E., Turner,D.H., Kim,J., Lyttle,M.H., Muller,P., Mathews,D.H. and Zuker,M. (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA, 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arkov A.L. and Murgola,E.J. (1999) Ribosomal RNAs in translation termination: facts and hypotheses. Biochemistry, 64, 1354–1359. [PubMed] [Google Scholar]
- 35.Brown C.M. and Tate,W.P. (1994) Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem., 269, 33164–33170. [PubMed] [Google Scholar]
- 36.Namy O., Hatin,I. and Rousset,J.P. (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep., 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills N.M., Gesteland,R.F. and Atkins,J.F. (1994) Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J., 13, 4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G.P. and Rice,C.M. (1989) Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol., 63, 1326–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mottagui-Tabar S., Tuite,M.F. and Isaksson,L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S., Ryden-Aulin,M. and Isaksson,L.A. (1999) Interaction between a mutant release factor one and P-site peptidyl-tRNA is influenced by the identity of the two bases downstream of the stop codon UAG. FEBS Lett., 455, 355–358. [DOI] [PubMed] [Google Scholar]
- 41.Jamjoom G.A., Naso,R.B. and Arlinghaus,R.B. (1977) Further characterization of intracellular precursor polyproteins of Rauscher leukemia virus. Virology, 78, 11–34. [DOI] [PubMed] [Google Scholar]
- 42.Phillips-Jones M.K., Hill,L.S., Atkinson,J. and Martin,R. (1995) Context effects on misreading and suppression at UAG codons in human cells. Mol. Cell Biol., 15, 6593–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urban C., Zerfass,K., Fingerhut,C. and Beier,H. (1996) UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Res., 24, 3424–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beier H. and Grimm,M. (2001) Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res., 29, 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]