Abstract
The survival of individual organisms facing stress is enhanced by the induction of a set of changes. As the intensity, duration and nature of stress is highly variable, the optimal response to stress may be unpredictable. To face such an uncertain future, it may be advantageous for a clonal population to increase its phenotypic heterogeneity (bet-hedging), ensuring that at least a subset of cells would survive the current stress. With current techniques, assessing the extent of this variability experimentally remains a challenge. Here, we use a bioinformatic approach to compare stress response genes with the rest of the genome for the presence of various kinds of repeated sequences, elements known to increase variability during the transfer of genetic information (i.e. during replication, but also during gene expression). We investigated the potential for illegitimate and homologous recombination of 296 Escherichia coli genes related to repair, recombination and physiological adaptations to different stresses. Although long repeats capable of engaging in homologous recombination are almost absent in stress response genes, we observed a significant high number of short close repeats capable of inducing phenotypic variability by slipped-mispair during DNA, RNA or protein synthesis.
INTRODUCTION
Stress is ubiquitous in nature. The survival of individual organisms faced by stress is facilitated by the induction of a set of changes. If the stress is of short duration, a physiological change may allow the organism to overcome stressful conditions. However, on a longer time scale, a genotypic modification may lead to a better adaptation to such stressful conditions. Stress can take many forms and in some environments cells may be faced with a combination of stressful changes. In addition to the intrinsic variability in nature, the intensity and duration of such a combination of stresses make it very unlikely that individual cells will attain adaptation for all general purposes. However, increasing phenotypic diversity among different cells may increase the chances that at least a subset of cells would survive among a large clonal population of bacteria facing such an uncertain future.
In the absence of stress, DNA replication is very faithful for copying normal DNA (10–9 errors per nucleotide). On the other hand, decoding this DNA into a functional protein is a more erroneous process, with estimates of 10–5 errors per nucleotide transcribed and 10–4 or more per amino acid incorporated (1). Naturally, errors due to slippage in repeated regions and to problems in protein folding are likely to increase the proportion of erroneous proteins. The accumulation of such errors leads to the production of >20% erroneous β-galactosidase enzymes under exponential growth phase (2). As errors in protein synthesis have been shown to accumulate under stress, and DNA mutations accumulate under a variety of stresses, it is not surprising that the number of aberrant proteins would increase under stressful conditions (3).
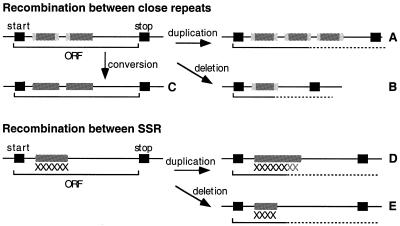
At the DNA level, repeated sequences are known to promote genetic variability in different ways (Fig. 1). Recombination between similar but not identical sequences may involve conversion or reciprocal strand exchange, thereby changing the DNA sequence. Recombination may also involve the duplication or deletion of regions of the chromosome, thereby introducing large changes. Homologous recombination between repeats is thought to be the basis of genetic variation in many bacteria (4). The minimal homology required for the action of RecA-mediated homologous recombination varies between 20 and 30 nt of strict identity in Escherichia coli (5) and in Bacillus subtilis (6). This coincides with the minimal significant length of large repeats in bacteria (25 nt in E.coli) (7).
Figure 1.
Scenarios of illegitimate recombination between close direct repeats and SSRs. Black boxes represent start and stop codons of the original gene, grey boxes represent strict repeats, and light grey boxes represent regions of weaker homology. Dashed lines indicate that deletions and duplication may induce frameshifts and therefore produce ORFs of very different length. (A and B) Duplication/deletion of the repeat and the region between occurrences. (C) The regions of non-strict similarity become similar after conversion. (D and E) Increase/decrease in the number of motifs of the SSR. Homologous recombination between long repeats closely follows the scenarios of (A), (B) and (C), except that large duplications are unstable and large deletions are strongly counter selected. Thus, conversions or reciprocal translocations are the most frequent outcome of homologous recombination between long distant repeats.
In addition, illegitimate recombination occurring by a RecA independent pathway may occur between repeats of ≥3 nt (8). The frequency of such recombination increases exponentially with length, and in E.coli becomes frequent for repeats >8 nt (9), although it also decreases exponentially with the distance between the occurrences (10). In consequence, long tandem repeats of small motifs [simple sequence repeats (SSRs)] are especially sensitive to this type of recombination in bacteria (11).
The effect of nucleotide repeats on the fidelity of gene transcription and translation is well documented. Because the RNA polymerase and the ribosomes are also prone to slippage, error rates increase on the regions of such motifs, with the formation of correspondingly erroneous mRNA and proteins. In these cases, recombination will transiently induce mutant phenotypes, as the DNA sequence is left unchanged, and only the protein is defective.
The physiological responses of E.coli to stress are probably among the best understood (12) due to decades of intensive research in the way these bacteria adapt to changes in temperature, osmolarity, pH and nutrients, or to the presence of toxic chemicals. To investigate the potential for recombination of E.coli stress genes, we compared them systematically with the rest of the genome for the presence of repeats. We selected a set of 296 stress response genes, including genes implicated in recombination and repair, but also physiological responses such as heat shock, cold shock or osmotic shock. Since intra-chromosomal recombination may proceed by either homologous recombination or illegitimate recombination, we have analysed the potential of both in the E.coli genome and in the stress response set of genes in particular. In every case, two types of controls were used: a first control uses statistics based on Markov chains to determine the significance of repeats, and a second biological control consists in comparing the stress subset with the remaining genes in order to identify potential differences. Finally, we compared the genes presenting an unusually high abundance of close repeats in the stress subset with orthologous genes in related bacteria.
MATERIALS AND METHODS
Data sets
Data on the complete bacterial genomes were taken from Entrez Genomes (http://www.ncbi.nlm.nih.gov). We used the annotations of E.coli K-12 version 53 (13), with nomenclature corrections and functional assignments included in Colibri (http://genolist.pasteur.fr/Colibri/) (14) and GeneProtEC (http://genprotec.mbl.edu/) (15).
Gene classification
Genes involved in adaptation or responses to stress were classified into 12 partially overlapping classes, according to standard bibliography (12). Functional assignments were updated taking into account the annotations of Colibri and GeneProtEC. We took into consideration the following 12 classes of stress response genes: response to atypical conditions, cold shock, detoxification, heat shock, response to molecular oxygen, osmotic stress, response to pH, recombination, repair, SOS, stationary phase (we removed genes related to energy metabolism) and stringent response. Overall, this set includes 296 genes, for a total of 4289 coding sequences in the genome. Although this classification may be punctually disputed, it was used because it reflects the work of many researchers of each domain (12). The list with the classification of these genes and their position in the sequence, as well as additional material for this paper, can be consulted at http://wwwabi.snv.jussieu.fr/~erocha/stress/.
Search for large repeats
The search for large strictly identical repeats was done using REPuter (16). We have searched for repeats with statistically significant lengths using a statistic of extremes that takes into account the composition in nucleotides and the length of the genome (17). Repeats >24 nt are statistically significant for the E.coli genome (P < 0.001) (7). We then computed the number of bases of such repeats present in each gene of E.coli—we call this the number of repeat bases per gene. This provides for a crude measure of the potential for homologous recombination of a given gene. Since genes have very different sizes, we compute a density of repeat bases per gene as the above quantity divided by the length of the gene. Note that this density can take values >1, since a single base can be in several relationships of similarity. A typical example of such a case is the set of rRNA genes of E.coli that is present in seven nearly identical copies.
Search for simple sequence repeats
Large SSRs. We searched for tandem repeats of motifs of 1–4 nt in length within genes, and in intergenic regions separately, since these two regions are compositionally quite different (18). We shall call X the motif of the SSR (e.g. CG in CGCGCG), n its multiplicity (e.g. 3 in CGCGCG) and L the cumulative length of the sequences (e.g. all genes or all intergenic regions). The probability of finding, by chance alone, a SSR of a motif X, with n motifs (Xn) anywhere in the set is given by: P = 1 – (1 – fXn)L where fX is the relative frequency of the motif X. We solved the above equation for all possible motifs X of 1–4 nt in length for P < 0.01. Through this approach we obtained the significant threshold for the length of SSR elements. We then searched for these exact SSR elements using standard pattern searching methods.
Density of SSRs. We also identified genes with high densities of small SSRs. We started by defining length thresholds for small SSRs. Considering that SSRs >3 nt have been shown to be engaged in illegitimate recombination in bacteria (8), we searched for mononucleotide runs of ≥5 nt (five motifs), for dinucleotide runs of ≥6 nt (three motifs), for trinucleotide runs of ≥6 nt (two motifs) and for tetranucleotide runs of ≥8 nt (two motifs). We computed the relative frequency of these small SSRs for each gene, separating SSRs with different motif sizes. The expected values were calculated using the observed number of such SSRs in 100 random sequences of equal length and equal frequencies of motifs (e.g. equal trinucleotide frequency for SSR of 3 nt motifs). Statistical significance of the difference between the observed and expected values were determined through the use of a Poisson distribution, with a mean estimated from the random experiments. These statistics are intended to compare the frequency of a word Xn given the frequency of words X. If there are more Xn than expected by chance alone, this suggests that the motif is over-represented and therefore a mutational or a selective force is probably at its origin. To compare different functional groups we performed Tukey–Kramer tests on the observed/expected ratio (19). Given that intergenic regions are quite small, this approach was only applied to genes.
Search for close repeats
Identification of close repeats. We used REPuter to identify repeats in regions defined by 500 nt upstream and downstream of each gene. In this case we imposed a length threshold of 9 nt, considered to be a conservative threshold for illegitimate recombination between close non-contiguous repeats. Since illegitimate recombination is rather sensitive to large distances between the occurrences of the repeats, we eliminated repeats whose occurrences were >1000 nt apart. We further eliminated repeats for which none of the occurrences was inside the gene. Using this methodology we obtained the number of repeats for each gene.
Statistical considerations. We made simulations to determine the empirical probability of finding a given number of close repeats. As stated above, each sequence was defined as the gene comprising the two 500 nt flanking regions. Therefore, we compared the observed number of close repeats in each sequence with the observed number in 1000 random sequences with the same size and composition in trinucleotides to take codon usage into account. The analysis of the random sequences provided an empirical distribution for the observed number of repeats. In the random experiments we did not shuffle both 500 nt flanking regions as these include coding and non-coding sequences, and because we were interested in the recombination potential of the gene in its genomic context.
Search for orthologues. Two genes were regarded as homologous if the proteins they code for are similar both in sequence and size. For this, we made pairwise comparisons of all proteins of all proteome pairs between E.coli and the set of free-living fully sequenced proteobacteria, filtering potential homologues using a threshold E-value in BlastP of 10–5 and a maximal difference of protein lengths of 20%. Subsequently, we aligned the sequences, using a variant of the classical dynamic programming algorithm for global alignment, where one counts 0-weight for gaps at both ends of the largest sequence, using the BLOSUM62 matrix (20). Finally, we retained reciprocal best hits with a similarity >50%. The following genomes of free-living proteobacteria were analysed: Caulobacter crescentus, E.coli 0157:H7, Haemophilus influenzae, Mesorhizobium loti, Neisseria meningitidis, Pasteurella multocida, Pseudomonas aeruginosa, Salmonella enterica Typhimurium, Sinorhizobium meliloti, Vibrio cholerae, Xyllela fastidiosa and Yersinia pestis.
RESULTS
Distribution of large strict repeats in the genome
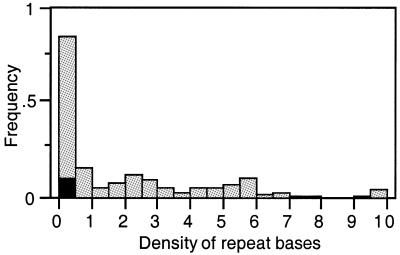
Large repeats are abundant in a small number of genes. The genome of E.coli contains a large number of repeats, half of which are inside genes (7). Dividing the sum of the lengths of these repeats by the number of genes gives an average of 177 nt repeats per gene. This suggests the existence of a large number of genes containing repeats capable of performing homologous recombination. However, the concentration of these repeats is not homogeneous since 92% of the genes do not contain any such repeat sequences. In fact, the majority of the large repeats of the E.coli genome are in rRNA operons, unknown function ORFs and genes related to mobile genetic elements (phages, plasmids and transposons). Two groups of genes are particularly over-represented. The rRNA operons have densities of repeat bases (i.e. number of repeats bases per nucleotide) ranging between 5.7 and 7, which is concordant with their number in the genome (seven complete operons). The 11 unknown function ORFs trs5_X, possess densities in the range 6.1–9.6.
Large repeats are rare in the stress response set. The gene presenting the largest density of repeats in the stress subset is ranked 230 (cspF) in the sorted list of the 347 genes containing large repeats (Fig. 2). Hence, all the genes of the stress subset including at least one repeat (20 out of the 347 genes) score a density of repeats smaller than the median. As a result, the analysis of genes containing repeats indicates that the stress response subset contains significantly fewer large repeats (P < 0.001, Wilcoxon test).
Figure 2.
Histogram of the cumulated length of repeats present in E.coli genes, divided by the length of genes (density of repeat bases). Only 8% of the genes contain repeats, and most of them contain densities of repeats smaller than 0.5. The repeats concerning the stress response genes are drawn in black (they are all in the first bin of the histogram).
Simple sequence repeats
Large SSRs are rare in E.coli K12. We searched for SSRs with motifs ranging in length from 1 to 4 nt in different sets of genes: stress response genes, other genes and the regulatory regions of these two classes of genes. We defined potential regulatory regions as the 100 bp regions preceding the genes. No significantly large SSRs with mononucleotide and dinucleotide motifs were identified among stress response genes. Also, we only found one trinucleotide repeat, CAT4 at gyrB, and one tetranucleotide repeat, CCAA3 at intA. In the regulatory regions of stress response genes we found a CGG5 and a CCAG3 near uvrC and ftsZ, respectively.
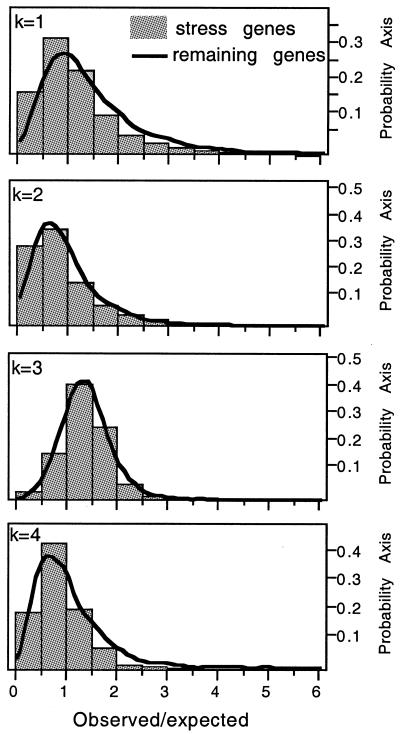
Large SSR densities are rare for dinucleotide and tetranucleotide motifs. Having observed that single large SSRs are rare in the genomic text, we have performed a complementary analysis to check if some genes contain large densities of smaller SSRs. SSRs of mononucleotides and trinucleotides are significantly over-represented in the genes of E.coli K12 (P < 0.001), however the median values of observed/expected densities are larger for trinucleotides (1.34) than for mononucleotides (1.15), and this difference is apparent from the frequency distribution curves (Fig. 3). SSRs with dinucleotide and tetranucleotide motifs are under-represented (P < 0.001), with median observed/expected values of 0.74 and 0.87, respectively.
Figure 3.
Distribution of observed/expected values for SSR densities for k-tuples from 1 to 4, in the stress subset (bars) and in remaining genes of the genome (lines). Expected values were calculated using the observed number of SSRs in 1000 random sequences of equal length and equal frequency of the motif (see Materials and Methods).
Large SSR densities are not over-represented in the stress response subset. We then tested if there were significant differences in terms of SSR densities in the stress response genes by comparison with the rest of the genome. As for the complete set of E.coli genes, we observed larger densities of trinucleotide SSRs, then mononucleotides, and finally tetranucleotides and dinucleotides (Fig. 3). The only significant difference identified in the stress response subset by comparison with the complete genome is a smaller over-representation of monunucleotide SSRs (P < 0.05). In our website we present the full list of genes presenting more significant over-representation of SSR density in the stress subset.
Analysis of occurrences of close repeats in the genome
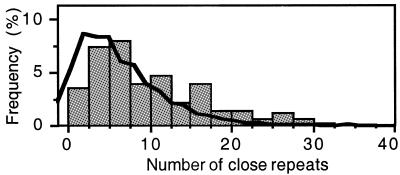
Over-representation of close repeats. We observed an average of 7.9 close direct repeats per gene of the E.coli genome (Fig. 4). Performing the same analysis on 1000 random sets of genes with the same composition in trinucleotides, we observed a significantly smaller average number of repeats (6.5 repeats per gene, P < 0.001, signed-rank test), indicating an over-representation of close repeats in the genes of E.coli. Classification of the codon usage of E.coli genes using factorial correspondence analysis into normal, highly expressed and horizontally transferred genes (21) reveals a significantly smaller number of repeats in the class of horizontally transferred genes (P < 0.001, Tukey–Kramer test). The observed number of close repeats is not significantly different from the expectation in the latter set (P > 0.05, signed-rank test). The other two sets of genes show significantly higher numbers of close repeats and the differences between them are not statistically significant (P > 0.05, Wilcoxon test). Within the functional classes defined in the E.coli sequencing paper (13), the class of ‘transcription, RNA processing and degradation’ genes presents an over-representation of close repeats by comparison with the remaining genes (P < 0.01, signed-rank test), followed by the classes of ‘DNA replication, recombination and repair’, ‘translation’ and ‘transport binding proteins’ at a smaller level of significance (P < 0.05).
Figure 4.
Distribution of the average number of close repeats in the stress subset (bars) and in remaining genes (lines).
Close repeats are longer than expected. Since longer repeats are expected to induce more frequent recombination events, we searched to determine if the observed close repeats are longer than expected. Statistical analysis corroborates this hypothesis since the longest repeats per gene are, on average, 10.5 bp whereas the average longest repeats in the random genes is 9.9 bp (P < 0.01, signed-rank test).
Close inverted repeats are rare. We have searched for the existence of inverted close repeats, but these elements were found to be significantly under-represented in the genes in comparison with either forward repeats or random sequences (P < 0.001, signed-rank test).
Distance between occurrences and induction of frameshifts. The computation of expected values of close repeats takes codon usage into account. Nevertheless, since the clustering of amino acids may originate repeats at the DNA level, we checked that repeats are indeed over-represented in all three reading frames. Also, if the distance between the two occurrences of a repeat is not a multiple of three, duplications or deletions will introduce a frameshift. Defining a frameshift as the difference between the codon positions of the first nucleotide of the two occurrences of a repeat, we observed a significant over-representation of close repeats in all types of frameshifts (from 0 to 2, P < 0.01, Wilcoxon tests), although repeats on the same reading frame are more over-represented (P < 0.01, Tukey–Kramer test).
Close repeats within the stress response set
Over-representation of close repeats in the set. We tested if over-representation is larger in the stress response genes by comparison with the other genes of the genome. The results show that the stress genes have, on average, 10.5 repeats per gene, which is significantly more than the 7.8 repeats per gene for the other E.coli genes (P < 0.001, Wilcoxon test). Therefore, close direct repeats are over-represented in the stress response set of genes when compared with random genes with the same composition in trinucleotides and when compared with the remaining genes of E.coli. This is also the case when one analyses the absolute number of repeats (Fig. 4). In terms of the induction of frameshifts by duplication or deletion of genetic material, the repeats of this set are over-represented in all potential reading frame frameshifts, just as the average E.coli genes. We then searched for functional classes of stress response genes that over-represent close repeats, by comparison with the set of all stress responses. Results indicate significant over-representation in genes related to recombination, pH and response to oxygen stress (P < 0.01, Tukey–Kramer test). However, some of the other categories contain few genes, which may render difficult the draw of meaningful comparative statistical tests.
Close repeats are larger than expected. We observed a median of 11 nt for the largest repeat in each gene, which is larger than the median found in the randomised genes (9.9 nt, P < 0.01, signed-rank test), and in the average E.coli gene (10.5 nt, P < 0.01, Wilcoxon test).
Close repeats in orthologues. We have searched for repeats in the orthologous genes of other completely sequenced proteobacteria (see Materials and Methods), partitioning the genes into those orthologous to the stress response set and to other genes (Table 1). For all the analysed genomes, a significantly larger number of close repeats is observed in the genes of the stress subset by comparison with the remaining genes. The differences between genomes can partly been explained by different sets of orthologues and by the very different nucleotide composition. Indeed, genomes with more biased G+C contents will tend to contain larger numbers of repeats.
Table 1. Orthologues of E.coli genes for the stress subset and for the remaining genes, identified with the stringent criteria defined in Materials and Methods.
| Stress | Others | Comparison | |||
|---|---|---|---|---|---|
| No. of orthologues | No. of repeats | No. of orthologues | No. of repeats | P < | |
| C.crescentus | 136 | 27.4 | 1110 | 21.7 | 0.002 |
| E.coli O157:H7 | 282 | 11.2 | 3382 | 7.9 | 0.001 |
| H.influenzae | 154 | 13.5 | 1073 | 11.1 | 0.003 |
| M.loti | 154 | 21.4 | 1378 | 17.1 | 0.005 |
| N.meningitidis | 117 | 17.4 | 943 | 12.7 | 0.001 |
| P.multocida | 169 | 11.2 | 1292 | 8.8 | 0.001 |
| P.aeruginosa | 196 | 26.8 | 1773 | 21.7 | 0.001 |
| S.enterica | 274 | 11.7 | 2789 | 9.0 | 0.001 |
| S.meliloti | 139 | 18.0 | 1151 | 13.9 | 0.003 |
| V.cholerae | 189 | 10.1 | 1675 | 7.1 | 0.001 |
| X.fastidiosa | 125 | 10.9 | 944 | 8.2 | 0.003 |
| Y.pestis | 216 | 8.7 | 2132 | 6.8 | 0.001 |
The table displays the average number of repeats in each subset of orthologs, and the comparison between the stress subset and the remaining genes in the genomes (Wilcoxon test).
Analysis of most repetitive genes of the stress subset
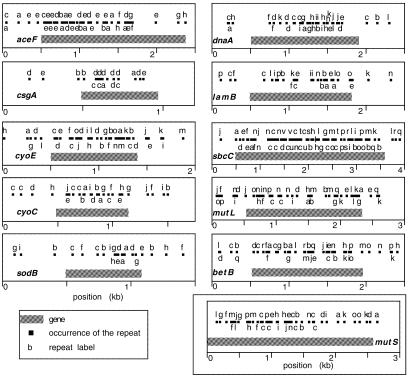
We extracted the genes over-representing close repeats in E.coli K12 (P < 0.05) and ranked them in terms of observed/expected values (see full tables at our web site). We then depicted the 10 first elements of this list, along with mutS, that, although less highly ranked (position 40), is the most well studied of mutator genes (Fig. 5). Results indicate rather different patterns. First, whereas some genes display repeats in a homogeneous way along most of the gene (e.g. mutL and sbcC), others possess regions with almost no repeats (e.g. edges of dnaA and end of aceF), and others strongly non-homogeneous distributions (e.g. sodB). Second, the effect of the context of the gene (its environment in the genome), varies in an important way between genes including a significant number of copies in these regions (e.g. cyoE) and others presenting very few (e.g. csgA and mutS). Third, whereas some genes present, almost exclusively, repeats in single copies (e.g. mutL, sbcC), others include many repeats in multiple copies (e.g. csgA and aceF).
Figure 5.
Spatial distribution of close direct repeats in E.coli K12 top 10 genes + mutS. Black boxes represent repeats and grey boxes represent genes. Different occurrences of the same repeat are marked with the same letter.
Naturally the role of such repeats can differ significantly in consideration of their spatial distribution. Thus, AceF (pyruvate dehydrogenase) presents the highest observed/expected values of close repeats. It also presents significantly aggregated repeats (P < 0.05, r-scan). These repeats are aggregated in the early regions of the gene concerning the multiple domains for Lipoyl binding. One may then suppose the existence of important functional constrains associated with these multiple repeats, which are documented at the amino acid level in SwissProt (entry P06959). Repeats randomly distributed along the gene are much less likely to constitute multiple protein domains. Naturally, the >50% of repeats with occurrences that are not in the same reading frame are not likely to be caused by functional constraints on the protein structure.
Repeats in orthologous genes of other genomes
The existence of abundant close repeats in these genes may allow high levels of illegitimate recombination. However, the abundance of such repeats can be motivated by selection for recombination events or by functional constrains (e.g. repeated protein folding motifs). Hence, we compared the sequences of the highest-ranking E.coli K12 genes (and mutS) with their orthologues among 12 proteobacteria to analyse if the repeated regions in E.coli K12 are more or less conserved than the remaining regions of the genes (Table 2).
Table 2. Conservation of the repeated regions of the 10 (+ mutS) most biased genes of E.coli K12 comparing with other proteobacteria.
| No. of orthologues | Biased genes | DNA | Protein | |||
|---|---|---|---|---|---|---|
| – | + | – | + | |||
| aceF | 8 | 8 | 5 | 0 | 0 | 1 |
| betB | 10 | 2 | 0 | 0 | 0 | 0 |
| csgA | 2 | 2 | 0 | 0 | 0 | 0 |
| cyoC | 8 | 6 | 0 | 0 | 0 | 0 |
| cyoE | 11 | 4 | 0 | 1 | 0 | 0 |
| dnaA | 12 | 4 | 0 | 1 | 0 | 0 |
| lamB | 4 | 1 | 0 | 0 | 1 | 0 |
| mutL | 12 | 4 | 0 | 2 | 0 | 7 |
| mutS | 12 | 3 | 4 | 2 | 2 | 0 |
| sbcC | 5 | 5 | 1 | 0 | 0 | 1 |
| sodB | 12 | 2 | 0 | 0 | 0 | 0 |
The table displays the number of orthologues among these genomes, the number of such orthologues over-representing close direct repeats, and the relative conservation of the regions bearing repeats in the E.coli K12 gene. For the latter, we aligned the protein orthologues and analysed if the regions including repeats in E.coli K12 presented a higher number of mismatches in the alignment (χ2 test, P < 0.05). For the analyses at the DNA level, the protein alignments were back translated to DNA and the same procedure was applied. Columns under ‘–’ indicate the number of orthologues with significantly smaller conservation at the location of repeats in E.coli K12, ‘+’ indicates significantly higher conservation.
The most interesting result is that in 8 out of 11 genes, the regions bearing the repeats and the other regions of the genes have not diverged at a significantly different pace. This observation holds for the analysis at both the DNA and the protein level, suggesting that these regions are not ‘special’ in any way, except by the existence of repeats in the E.coli K12 gene, and eventually in the orthologues. In particular, the hypothesis that these are highly variable regions, such as immunodominant proteins in pathogens, does not hold. One is also tempted to rule out the hypothesis that these regions constitute duplicated amino acid motifs with relatively important functional constraints. An exemplary case is provided by sodB, whose repeated regions in E.coli K12 evolve at the rate of the remaining gene sequence, even though only two other genomes contain a sodB gene with more than the expected number of close direct repeats. Three genes constitute an exception to this trend: in some genomes the regions with repeats evolve faster in aceF and mutS, and slower in mutL. Among the 11 proteins, AceF is one of the most conserved, MutS is an intermediate statute and MutL is one of the least conserved (data not shown). The three genes are widespread among proteobacteria.
DISCUSSION
Are the repeats due to chance or functional constraints?
Even though repeats may be a source of instability in the transfer of genetic information, they could be present in different genes for very different reasons. Chance and functional constraints are the first candidates that come to mind. However, statistical tests and the conservation of the repeats in orthologues belonging to different species suggest that chance is not the most likely explanation for the existence of the majority of these repeats.
A well-known category of functional constraints is the one linked with protein structure; a repeated amino-acid motif in a protein could be involved in the folding or in the interaction with another molecule. However, >50% of close repeats are in different frames and, thus, not encoding the same amino-acid motifs, cannot be explained by such a constraint. Alternatively, such repeats could either be due to other functional or historical constraints not yet identified. If this were the case, it would be interesting to investigate such constraints further. However, this is not very likely, as the study of orthologues shows that these repeats are generally not more conserved than the rest of the gene in which they are found. Therefore, other explanations must be considered for the over-representation of close repeats. Before discussing them, we shall first comment on the presence of the different repeats.
Large repeats
A general comparative analysis of large repeats in bacterial genomes has been published elsewhere (7). Here, we searched the chromosome of E.coli K12 for large repeats able to perform intra-chromosomal homologous recombination in order to identify stress response genes that might endure sequence variation through this mechanism. Results indicate that repeats are concentrated in a few (less than 350) genes, of which only 20 belong to the stress response set. Moreover, all the genes of this set presented less repeats than the average. It seems, therefore, that intra-chromosomal recombination does not constitute a major mechanism for the evolution of these genes.
Simple sequence repeats
Though SSRs play an important role in the dynamics of eukaryotes, their presence in bacteria is rare, and mostly reduced to pathogenic organisms (for a review see ref. 11). Using a conservative length threshold, we found almost no large SSRs in the genome of E.coli, confirming previous observations (22,23). However, the absence of very large SSRs does not exclude slippage of small SSRs. In fact, we found a large number of genes containing high densities of SSRs. This kind of data is difficult to analyse, since with current knowledge we cannot predict the instability of such small SSRs from sequence alone. Nevertheless, these results can be a useful starting point for further experimental studies.
Among the six genes with higher observed/expected ratios of mononucleotide SSRs there are two genes that are known to induce mutator phenotypes: mutT and dam. MutT prevents mispair of 8-oxoG with template A during replication. Mutant alleles of mutT specifically increase AT to CG mutations by several thousand-fold. Inactivation and over-expression of Dam methyltransferase has also been associated with mutator phenotypes (24). mutY and vsr are among the genes with larger densities of dinucleotide SSRs. MutY removes A from 8-oxoG–A or G–A mispairs, and its inactivation significantly increases mutation rate. It is probably not a coincidence to find MutT and MutY among genes with large densities of SSRs. These genes are both involved in the repair of oxidised guanines and are among some of the most commonly found mutator phenotypes (25). Vsr codes for the very short patch repair protein involved in the correction of T–G mismatches. Tetranucleotide SSRs present several genes induced by DNA damage, such as dinJ and ruvC. It also contains genes coding for subunits of several proteins such as cyoC, sodC and sbcB. Since these genes, or genes coding for other subunits of their proteins, also contain large numbers of close repeats, we discuss them below.
Close repeats
Contrary to the results for the elements capable of enduring homologous recombination or SSR slippage, we have observed an important over-representation of close direct repeats in E.coli genes, and especially in the stress response. Unfortunately, most studies on the impact of illegitimate recombination have been focused on SSRs or on large palindromes. Our results suggest that the study of close direct repeats may provide important insights on the evolution of bacterial chromosomes, since the over-representation of close repeats may allow for frequent conversion, duplication or deletion of genetic material by illegitimate recombination. In this work, we have restricted our attention to repeats with occurrences closer than 1000 bp, since for larger distances recombination becomes rare. However, laboratory experiments have shown a significant number of recombination events for 8 nt repeats at a distance of 987 bp (26), for 18 nt repeats at 2313 bp (10), for 24 nt repeats at 1741 bp (27) and for 100 nt repeats up to 7000 bp (28).
Three different outcomes are possible from illegitimate recombination (Fig. 1). A conversion of the repeats region will lead to slightly modified proteins. A duplication of a part of a gene will probably render it temporarily inactive (by blocking transcription or by leading to a defective protein). However, large tandem repeats are unstable, and a wild-type genotype will be easily recovered by deletion. Deletions of genetic material are more difficult to revert, and eventually require recombination with foreign genetic information. However, mutator phenotypes have been shown to reduce the recombination barrier (29). A recent study identified in Pseudomonas putida a mutS gene with an important deletion that provided a mutator phenotype intermediate between mutS+ and mutS– (30). This indicates that partially amputated genes may fulfil their function, albeit less efficiently. Therefore, different modulations of the mutator phenotype may be provided by different outcomes of the recombination process.
Illegitimate recombination between inverted repeats is extremely rare for chromosomal repeats and produces dimers in plasmids (31). Furthermore, these inverted repeats could block replication, transcription or translation by forming hairpins. This may explain why close inverted repeats are avoided in most genes, including in the stress subset.
All genes of the cold shock subset revealed an over-representation of close direct repeats smaller than the average. On the other side, one of the classes that most over-represents close direct repeats (among the stress response genes) is the one related to recombination. In fact, in this class only 3 genes (ruvB, sbcB and recT) out of 19 show an over-representation of repeats smaller than the average stress gene.
Comparison with orthologous genes from other bacteria
We have made a preliminary analysis of the existence of repeats among orthologues of the E.coli stress subset in other proteobacteria. Generally, these genes are biased in terms of the number of close repeats in all proteobacterial genomes. However, some genes are systematically biased in all genomes (e.g. aceF and sbcC), whereas for others the bias is restricted to the bacterial species closer to E.coli (e.g. dnaA).
For example, the cyoC gene contains high numbers of close repeats and SSR in E.coli and over-represents close repeats in all analysed proteobacterial genomes. The regions with repeats are not more conserved than the remaining regions of the gene, but this is also one of the most conserved genes of the set. CyoC codes for the subunit III of the aa3-type cytochrome c oxidase, a component of the aerobic respiratory chain of E.coli. CyoC does not contain any of the redox centres and can be removed from the purified enzyme but has a function during biosynthesis of the enzyme. In the absence of the COIII gene, only a fraction of the oxidase is assembled into an enzyme with low but significant activity (32).
The data on the conservation of the repeated regions is therefore quite difficult to interpret, given the functional constrains, the rate of evolution of the gene, the potential advantage of such a mutant phenotype, and the ecology of each bacterial species.
Repeats in genes involved in transfer of genetic information
It is interesting to note that within the functional classes defined by Blattner et al. (13), the classes involved in the transfer of genetic information show the most over-representation of close repeats. Given this over-representation, the list of most biased genes may provide a first set of candidate genes for screening for activities involved in stress response and/or transfer of genetic information. Looking more specifically at genes that have been classified both among stress genes and among genes involved in the transfer of genetic information can be useful to understand the distribution of these repeats.
The observation that mutS and mutL are among the genes that show the most over-representation of close direct repeats is consistent with experimental evidence showing that deletions in these genes (and in particular in mutS) are a major source of mutator phenotypes in pathogenic and commensal strains of E.coli (33). Mutator phenotypes have also been associated with MMR mutants in P.aeruginosa (34), and we do find an over-representation of repeats in mutS and mutL. Some of these mutants were shown to be the result of small deletions by recombination between two 8 bp repeats in the mutS gene (35), which corroborates our analysis. Interestingly, the genomes for which mutS and mutL exhibit over-representation of close repeats are typically the same (P < 0.05). Neisseria meningitidis is the only exception, since its mutL over-represents repeats but not its mutS gene. However, over-representation in mutS would be accepted at a lower significance threshold (P < 0.075, instead of P < 0.05). These results suggest co-evolution of these genes in respect to the over-abundance of close repeats. However, contrary to mutS, the repeated regions in mutL are frequently more conserved than the remaining coding sequences (in DNA and protein), even though mutL is one of the least conserved among the analysed genes. This might suggest a selective pressure at the amino acid level as the basis of the existence of such repeats. The observation that 8 out of the 12 mutL orthologues do not over-represent close repeats renders this hypothesis less likely.
The mutT and mutY genes of E.coli, whose mutation induce mutagenesis but no hyper-recombination (36), revealed no significant over-representation of close repeats. However, they both reveal an abundant density of SSR. One might speculate that since their recovery by horizontal transfer is difficult, a mechanism relying on SSR slippage could be positively selected. Interestingly, all ‘minor’ components of the human DNA mismatch repair system contain mononucleotide microsatellites in their coding sequences (37).
The sbcC gene, which codes for a co-suppressor with sbcB of recB and recC mutations, is one of the 10 genes that most over-represents close direct repeats. sbcB does not over-represent close repeats but contains instead a high density of tetranucleotide SSRs, which might engage into illegitimate recombination. The SbcCD protein cleaves hairpin structures that halt the progress of the replication fork, allowing homologous recombination to restore DNA replication (38). Given these properties, a duplication or deletion in sbcB and a slippage in sbcC could be associated with a phenotype affecting DNA metabolism.
Among the genes related to oxygen response we found sodB to over-represent close repeats, sodA to over-represent mononucleotide SSRs and sodC to over-represent tetranucleotide SSRs. These genes code for the three different superoxide dismutases of E.coli and the coincidence of all having elements capable of engaging in illegitimate recombination, but of different type, is difficult to explain by random effects. Indeed mutator phenotypes are known to result from mutations in sodA and sodB (24).
The cases of the mut, sbc and sod genes suggest that SSR and close repeats may have equivalent or complementary roles in terms of inducing phenotypes affecting DNA metabolism.
CONCLUSION
The observation that stress genes together with genes involved in the transfer of genetic information (DNA, RNA or protein metabolism) contain more repeats that the rest of the genome merits some discussion. We have previously mentioned that chance or functional constraints on the protein level are not the most likely explanations for at least some of these repeats. This can seem surprising if one considers that the evolution of such essential house-keeping processes involved in the management of genetic information should be strongly constrained. Thus, repeats would have long been eliminated to minimise variance in phenotype if there is a simple evolved solution to maximise growth rate during the exponential phase.
However, if one does not consider the ‘feast’ lifestyle most encountered under laboratory conditions, but the various, often unpredictable, stresses that bacteria meet in nature, it may be interesting for bacteria to use another strategy: ‘bet-hedging’. Bet-hedging has long been known in several disciplines (e.g. economy or evolution; for a historical review see ref. 39), it can be seen as the classical ‘don’t put all your eggs in one basket’. Such a strategy is known to be useful when environments are risky. Thus, according to the bet-hedging strategy, polymorphic bacteria populations under stressful conditions could have some enhanced chances of survival.
A plethora of mechanisms allows bacterial populations to deal with common stresses, from the modification of their physiology (12) to the change of their genetic information (40). However, for novel or infrequent kinds of stress, requiring new solutions, a transient mutant phenotype may be positively selected. This could be achieved by slippage of SSR (as for contingency loci) or by illegitimate recombination between close repeats. Recombination at the DNA level would produce a defective gene inducing a mutant phenotype. Reversion of the slippage could proceed by point mutation for conversions, by deletion for duplications and by recombination with foreign DNA for deletions. Naturally, the exactness of this hypothesis will have to be tested using information on the sequence of mutant genes found in natural isolates. At the moment few data are available, but much of it points to local recombination events at the origin of transient mutator phenotypes.
The above reflects a more standard scenario of DNA deletion by illegitimate recombination between close repeats compatible with observations of deletions in mutator genes. However, an alternative hypothesis can be put forward. Repeats may mediate slippage by RNA polymerase or ribosome leading to amputated proteins. Such aberrant proteins would create mutant phenotypes, but without changes in the DNA sequence. If such slippage occurred among mutator genes bearing close repeats, one would obtain a transient mutator phenotype (41).
Because they could explain part of the versatility of micro-organisms, such bet-hedging scenarios will have to be validated experimentally, but because some of the phenotypes would be transient and present only in a subset of cells, they will require the development of new experimental protocols.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the following people for criticism, discussion and encouragement: Antoine Danchin, Julian Davies, Francisco Dionísio, Erick Denamur, Guillaume Lecointre, Miroslav Radman and anonymous referees.
REFERENCES
- 1.Kurland C.G. (1992) Translational accuracy and the fitness of bacteria. Annu. Rev. Genet., 26, 29–50. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen F. and Kurland,C.G. (1990) Processivity errors of gene expression in Escherichia coli. J. Mol. Biol., 215, 511–521. [DOI] [PubMed] [Google Scholar]
- 3.Ballesteros M., Fredriksson,A., Henriksson,J. and Nystrom,T. (2001) Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J., 20, 5280–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha E.P.C., Danchin,A. and Viari,A. (1999) Functional and evolutionary roles of long repeats in prokaryotes. Res. Microbiol., 150, 725–733. [DOI] [PubMed] [Google Scholar]
- 5.Watt V.M., Ingles,C.J., Urdea,M.S. and Rutter,W.J. (1985) Homology requirements for recombination in Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 4768–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majewski J. and Cohan,F.M. (1999) DNA sequence similarity requirements for interspecific recombination in bacillus. Genetics, 153, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocha E.P.C., Danchin,A. and Viari,A. (1999) Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol., 16, 1219–1230. [DOI] [PubMed] [Google Scholar]
- 8.Michel B. (1999) Illegitimate recombination in bacteria. In Charlebois,R.L. (ed.), Organization of the Prokaryotic Genome. ASM Press, Washington DC, pp. 129–150.
- 9.Pierce J.C., Kong,D. and Masker,W. (1991) The effect of the length of direct repeats and the presence of palindromes on deletion between directly repeated DNA sequences in bacteriophage T7. Nucleic Acids Res., 19, 3901–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chédin F., Dervyn,E., Ehrlich,S.D. and Noirot,P. (1994) Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol. Microbiol., 12, 561–569. [DOI] [PubMed] [Google Scholar]
- 11.van Belkum A., Scherer,S., van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neidhardt F., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds) (1996) Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn, 2 vol. ASM Press, Washington DC.
- 13.Blattner F.R., Plunkett,G.P.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1461. [DOI] [PubMed] [Google Scholar]
- 14.Moszer I., Glaser,P. and Danchin,A. (1995) Subtilist: a relational database for the Bacillus subtilis genome. Microbiology, 141, 261–268. [DOI] [PubMed] [Google Scholar]
- 15.Riley M. and Labedan,B. (1997) Protein evolution viewed through Escherichia coli protein sequences: introducing the notion of a structural segment of homology, the module. J. Mol. Biol., 268, 857–868. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz S. and Schleiermacher,C. (1999) REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics, 15, 426–427. [DOI] [PubMed] [Google Scholar]
- 17.Karlin S. and Ost,F. (1985) Maximal segmental match length among random sequences from a finite alphabet. In Cam,L.M.L. and Olshen,R.A. (eds), Proceedings of the Berkeley Conference in Honor of Jerzy Neyman and Jack Kiefer. Wadsworth, Inc., Belmont, CA, Vol. I, pp. 225–243.
- 18.Rocha E.P.C., Viari,A. and Danchin,A. (1998) Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res., 26, 2971–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zar J.H. (1996) Biostatistical Analysis. Prentice Hall, New Jersey.
- 20.Erickson B.W. and Sellers,P.H. (1983) Recognition of patterns in genetic sequences. In Sankoff,D. and Kruskal,J.B. (eds), Time Warps, String Edits and Macromolecules: the Theory and Practice of Sequence Comparison. Addison-Wesley, New Jersey, pp. 55–91.
- 21.Médigue C., Rouxel,T., Vigier,P., Henaut,A. and Danchin,A. (1991) Evidence for horizontal gene transfer in E.coli speciation. J. Mol. Biol., 222, 851–856. [DOI] [PubMed] [Google Scholar]
- 22.Field D. and Wills,C. (1998) Abundant microsatellite polymorphism in Saccharomyces cerevisiae and the different distributions of microsatellites in 8 prokaryotes and S.cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl Acad. Sci. USA, 95, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gur-Arie R., Cohen,C.J., Eitan,Y., Shelef,L., Hallerman,E.M. and Kashi,Y. (2000) Simple sequence repeats in Escherichia coli: abundance, distribution, composition and polymorphism. Genome Res., 10, 62–71. [PMC free article] [PubMed] [Google Scholar]
- 24.Horst J.-P., Wu,T.-H. and Marinus,M.G. (1999) Escherichia coli mutator genes. Trends Microbiol., 7, 29–36. [DOI] [PubMed] [Google Scholar]
- 25.Miller J.H. (1996) Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol., 50, 625–643. [DOI] [PubMed] [Google Scholar]
- 26.Albertini A.M., Hofer,M., Calos,M.P. and Miller,J.H. (1982) On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell, 29, 319–328. [DOI] [PubMed] [Google Scholar]
- 27.Singer B.S. and Westlye,J. (1988) Deletion formation in bacteriophage T4. J. Mol. Biol., 202, 233–243. [DOI] [PubMed] [Google Scholar]
- 28.Lovett S.T., Gluckman,T.J., Simon,P.J., Sutera,V.A. and Drapkin,P.T. (1994) Recombination between repeats in E.coli by a recA-independent, proximity-sensitive mechanism. Mol. Gen. Genet., 245, 294–300. [DOI] [PubMed] [Google Scholar]
- 29.Vulic M., Dionisio,F., Taddei,F. and Radman,M. (1997) Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl Acad. Sci. USA, 94, 9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurusu Y., Narita,T., Suzuki,M. and Watanabe,T. (2000) Genetic analysis of an incomplete mutS gene from Pseudomonas putida. J. Bacteriol., 182, 5278–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyu Y.L., Lin,C.-T. and Liu,L.F. (1999) Inversion/dimerization of plasmids mediated by inverted repeats. J. Mol. Biol., 285, 1485–1501. [DOI] [PubMed] [Google Scholar]
- 32.Haltia T., Saraste,M. and Wikstrom,M. (1991) Subunit III of cytochrome c oxidase is not involved in proton translocation: a site-directed mutagenesis study. EMBO J., 10, 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeClerc J.E., Li,B., Payne,W.L. and Cebula,T.A. (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science, 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- 34.Oliver A., Canton,R., Campo,P., Baquero,F. and Blazquez,J. (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288, 1251–1254. [DOI] [PubMed] [Google Scholar]
- 35.Oliver A., Baquero,F. and Blázquez,J. (2002) The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol., 43, 1641–1650. [DOI] [PubMed] [Google Scholar]
- 36.Denamur E., Lecointre,G., Darlu,P., Tenaillon,O., Acquaviva,C., Sayada,C., Sunjevaric,I., Rothstein,R., Elion,J., Taddei,F., Radman,M. and Matic,I. (2000) Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell, 103, 711–721. [DOI] [PubMed] [Google Scholar]
- 37.Chang D.K., Metzgar,D., Wills,C. and Boland,C.R. (2001) Microsatellites in the eukaryotic DNA mismatch repair genes as modulators of evolutionary mutation rate. Genome Res., 11, 1145–1146. [DOI] [PubMed] [Google Scholar]
- 38.Connelly J.C., Kirkham,L.A. and Leach,D.R. (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA, 95, 7969–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stearns S.C. (2000) Daniel Bernoulli (1738): evolution and economics under risk. J. Biosci., 25, 221–228. [DOI] [PubMed] [Google Scholar]
- 40.Arber W. (2000) Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev., 24, 1–7. [DOI] [PubMed] [Google Scholar]
- 41.Ninio J. (1991) Transient mutators: a semiquantitative analysis of the influence of translation and transcription errors in mutation rates. Genetics, 129, 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]