Abstract
Plasmid pIP501 encoded transcriptional repressor CopR is one of the two regulators of plasmid copy number. Previous data suggested that CopR is a HTH protein belonging to a family of 578 HTH proteins (termed HTH 3-family). Only a very limited number of proteins in this family, among them λ c1 repressor, 434 c1 repressor and P22 c2 repressor, have been characterized in detail so far. Previously, a CopR structural model was built based on structural homologies to the 434 c1 and P22 c2 repressor and used to identify amino acids involved in DNA binding and dimerization. Site-directed mutagenesis in combination with electrophoretic mobility shift assay (EMSA), dimerization studies and circular dichroism (CD) measurements verified the model predictions. In this study we used hydroxyl radical footprinting and fluorescence resonance energy transfer (FRET) measurements to obtain detailed information about the structure of the DNA in the CopR–DNA complex. Our results show that the DNA is bent gently around the protein, comparable to the bending angle of 20–25° observed in the 434 c1 repressor–DNA complex and the λ c1 repressor–DNA complex. The shape of CopR dimers as determined by sedimentation velocity experiments is extended and accounts for the relatively large area of protection observed with hydroxyl radical footprinting.
INTRODUCTION
The local structure and deformability of the DNA as defined by the base sequence are important for specific interactions between protein and DNA. Sequence-specific binding of proteins to DNA is often accompanied by bending of the DNA. Analyzing the structure of 86 protein–DNA complexes and the conformational changes in the DNA induced by protein binding revealed 28 examples of major DNA bending, e.g. CAP and IHF, and 58 complexes with minor DNA bending (1). Among the latter are the extensively characterized transcriptional repressors λ cI and Cro, P22 c2 and 434 c1 and Cro. They belong, together with the transcriptional repressor CopR, to a family of 578 HTH proteins named HTH 3-family (Interpro release 3.2.2001, IPR001387).
The 10.6 kDa CopR protein is one of the two regulators of the copy number of plasmid pIP501 (2). It has a dual function: it acts as transcriptional repressor at the essential repR promoter pII by binding to a 44 bp inverted repeat upstream of and overlapping pII (3). Furthermore, it prevents convergent transcription from pII and pIII (the antisense promoter), thereby indirectly increasing transcription initiation from pIII (4). Two almost identical Cop proteins (95% sequence similarity) with the same functions are encoded by the related streptococcal plasmids pAMβ1 (5) and pSM19035 (6). CopR forms sequence specific contacts with two consecutive sites (I and II) within the major groove of the DNA that share the sequence motif 5′-CGTG (7). It binds the DNA only as a preformed dimer, and not as a monomer (8). Based on structural homologies of CopR with the P22 c2 repressor and the 434 c1 repressor a structural model of the CopR–DNA complex comprising the N-terminal 63 amino acids of CopR was built (9). Using site-directed mutagenesis we could essentially prove the model predictions (9,10). This approach and the algorithm developed by Dodd and Egan (11) led to the conclusion that CopR binds the DNA via an HTH motif. Only very few other members of the above mentioned HTH 3-family have been characterized in detail until now and even less information is available on the conformational changes induced in the DNA upon binding of the HTH proteins.
In this study we used fluorescence resonance energy transfer (FRET) measurements to determine the degree of DNA bending induced by CopR binding and compared the data obtained with the data available from λ c1 repressor, 434 c1 and 434 Cro repressors. Similar to these repressors, CopR induced a minor bend of 20–25°. Furthermore, using sedimentation velocity experiments, we found that the full length CopR has an extended shape, which corresponds well to the large area of protection observed in hydroxyl radical footprinting experiments presented here.
MATERIALS AND METHODS
Preparation of fluorescence-labeled DNA sequences
The following 5′ fluorescence labeled oligonucleotides containing the binding site of CopR were used for the FRET experiments: (i) 6-FAM-CCG TGT GAA TAA TGC ACG G and 5-TMRh-CCG TGC ATT ATT CAC ACG G and (ii) 6-FAM-CCC ATG ATT TCG TGT GAA TAA TGC ACG AAA TCG G and 5-ROX-CCG ATT TCG TGC ATT ATT CAC ACG AAA TCA TGG G (6-FAM, 6-carboxyfluorescein; 5-TMRh, 5-carboxytetramethylrhodamine; 5-ROX, 5-rhodamine-X). The oligonucleotides were purified and annealed as described previously (12,13).
The buffer of the double-stranded DNA samples was changed to the protein-binding buffer (150 mM NaCl, 20 mM Tris–HCl pH 8 and 1 mM EDTA) on a Sephadex G25-column. Absorption measurements were made in this solution from 240 to 650 nm to determine the concentration of the samples and to control the quantity of the labeling. The obtained values indicated 100% labeling.
Spectroscopic studies
Absorption and fluorescence measurements were performed on a Specord M500 (Zeiss, Germany) and a SLM 48000S instrument (SLM Aminco, Urbana, IL). Fluorescence spectra were corrected for lamp fluctuation and polarization artifacts were avoided by using ‘magic angle’ conditions. The fluorescence samples were excited at 490 and 560 nm and the emission spectra were collected from 500 to 650 nm for the FRET spectra and from 570 to 650 nm for the acceptor spectra, respectively. 400 µl of ∼100 nM DNA were titrated with CopR to a final protein concentration of ∼2 µM and incubated for 10 min for each titration step. All measurements were carried out at 15°C.
Fluorescence anisotropies r were calculated from fluorescence intensity measurements by a vertical excitation polarizer with vertical (F││) and horizontal (F⊥) emission polarizers according to
with the experimental correction factor (14).
Fluorescence resonance energy transfer
The efficiency of FRET E from a fluorescence donor to an acceptor is related to the donor–acceptor distance R.
R0 is the Förster distance at which the energy transfer efficiency is 50%, calculated from
where J is the spectral overlap integral of the dyes, ΦD the quantum yield of the donor, n the refraction index of the medium and κ2 the orientation of the transition dipole moments (15–17). For a rapid randomization of the relative donor–acceptor orientation κ2 is two-thirds. The low anisotropy of the donor fluorescein implies that this is a good approximation for this study. Under the given buffer condition the Förster distance is 50 Å and 60 Å for 6-FAM/5-TMRh and 6-FAM/5-ROX, respectively (12,18,19).
The FRET efficiency E was determined by measuring the intensity of the sensitized emission of the acceptor normalized to the fluorescence of the acceptor alone (20).
= E × d+ × (ɛD,490/ɛA,560) + (ɛA,490/ɛA,560)
5
(ratio)A is linearly dependent on the efficiency of energy transfer, E. It normalizes the measured sensitized FRET signal for the concentration, for the quantum yield of the acceptor and for any errors in percentage of acceptor labeling. F(λem,490), F(λem,560) and FD(λem,490) are the fluorescence spectra of donor and acceptor labeled and donor-only labeled DNA samples excited at the given wavelength of 490 or 560 nm, respectively. ɛD and ɛA are the molar absorption coefficients of the donor and acceptor at the given wavelength. d+ is the fraction of donor labeled molecules, calculated from the absorption spectrum of the doubly labeled DNA molecule.
Electrophoretic mobility shift assay (EMSA) with fluorescence labeled operator DNA
For EMSA different concentrations of CopR were incubated for 15 min at 20°C with doubly labeled DNA (200 nM) in 150 mM NaCl, 20 mM Tris–HCl pH 8 and 1 mM EDTA. The samples were loaded on a native 8% polyacryamide gel and electrophoresed in 1× TBE-buffer. The gels were directly scanned using a FluorImager 595 (Molecular Dynamics). The DNA was excited with an argon laser at 488 nm and the fluorescence of the fluorescein detected in a range from 515 to 545 nm.
Preparation of radioactively labeled DNA fragments
Synthetic oligonucleotides were 5′ end-labeled with [γ-32P]ATP and purified from 8% denaturing polyacrylamide gels. Pairwise combinations of labeled/unlabeled oligonucleotides were annealed (68°C for 1 h, slow cooling overnight), resulting in a double-stranded DNA fragment containing the wild-type operator sequence. The oligonucleotides carry four G or C residues at their ends to facilitate correct annealing and to promote additional stability.
EMSA for competition experiments
Synthetic oligonucleotides (for sequences see Table 1) were annealed (68°C for 1 h, slow cooling down overnight). The concentration of the double-stranded DNA was determined based on absorption measurements. For the competition assay the radioactively labeled DNA fragment Ks61 was incubated with CopR in the presence/absence of unlabeled competitor DNA in 0.5× TBE. Complex formation was analyzed by electrophoretic mobility shift assay. Electrophoresis was performed in 8% polyacrylamide gels in 0.5× TBE.
Table 1. Sequences of the DNA fragments used in this work.
Underlined, protected region determined by hydroxyl radical footprinting; italics, artificial nucleotides introduced at the ends of the sequences to facilitate annealing.
Hydroxyl radical footprinting and gel electrophoresis
Hydroxyl radical treatment was performed essentially as described (21). A typical reaction mix contained 7.5 ng (0.2 pmol) radiolabeled DNA fragment, ∼80 pmol protein, 5 mM phosphate buffer pH 7.5, 150 mM NaCl and 5 µg herring sperm DNA in a final volume of 200 µl. After 15 min incubation at room temperature for equilibration sodium ascorbate, ammonium iron sulfate, EDTA and hydrogen peroxide were added with the final concentrations being 10 mM, 10 µM, 20 µM and 0.03% (v/v), respectively. After incubation at room temperature for 2 min the cleavage reaction was quenched by adding 24 µl of thiourea and EDTA (final concentrations 10 mM and 2 mM, respectively). The reaction mixtures were treated with phenol/chloroform to remove the protein, ethanol precipitated and subsequently dissolved in loading buffer containing 99% formamide.
Prior to loading onto a 15% denaturating gel the samples were heat denatured (2 min at 90°C). After electrophoresis at 1200 V for 3 h the gels were transferred onto a filter paper and dried. The band pattern was visualized using a Fuji PhosphorImager.
In each lane the intensities of six bands well outside the footprint area were summed and subsequently used to obtain a normalization factor necessary for the elimination of loading errors in the different lanes. The normalized band intensities of the protein free control DNA were then subtracted from the corresponding band intensities of the lanes containing the footprint.
Preparation of proteins
Because glycerol is known to act as a scavenger of free hydroxyl radicals, Bacillus subtilis crude cell extracts containing CopR were prepared freshly prior to each use as described (3) with the exception that no glycerol was added. The purified His6-CopR used for the FRET experiments and for the analytical ultracentrifugation was obtained as described (8).
Ultracentrifugation
In order to get information about the shape of CopR dimers, sedimentation velocity experiments were carried out using an analytical ultracentrifuge XL-I (Beckman-Coulter, Palo Alto, CA). The data were analyzed by means of our computer program lamm (22). Since CopR in solution forms a monomer–dimer equilibrium which is characterized by an equilibrium dissociation constant of 1.5 µM (8) the experiments were performed in a larger concentration range. The sedimentation coefficients were fitted by the mentioned KDimer value and the relationship
s2 = 1.586 × s1 6
Modeling of CopR dimers
Based on the molecular mass (M) and the partial specific volume (![]() ), the dry volume (V) of the protein was calculated according to
), the dry volume (V) of the protein was calculated according to
with NA being the Avogadro number. The volume of the dimeric CopR was represented by 28 equal spheres supplied by a hydration shell and characterized by their Cartesian coordinates. For the models with different arrangements of the beads theoretical sedimentation coefficients were calculated using the program hydro (23) and adapted to the experimental one.
RESULTS
Hydroxyl radical footprinting
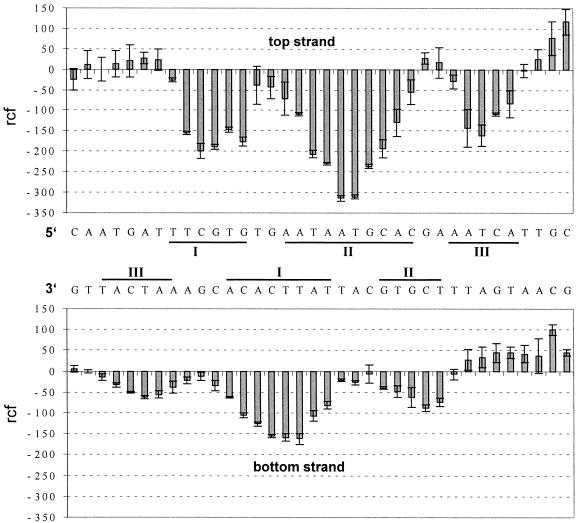
Hydroxyl radical cleavage has been used to obtain structural information about the CopR–DNA complex. Since a hydroxyl radical is roughly the size of a water molecule, the footprints reflect the solvent accessibility of the DNA in a complex. Therefore, hydroxyl radicals represent a very sensitive probe to acquire conformational details about the DNA in a protein–DNA complex (24). Figure 1 shows typical results of the hydroxyl radical cleavage in the absence and presence of CopR for both the top and the bottom strand, respectively. The corresponding histograms derived from at least five independent experiments are displayed in Figure 2. Three distinct areas of protection are visible on the top and on the bottom strand, labeled I, II and III, respectively. The protection at site III on the bottom strand is weaker compared to the other protection sites and clearly visible only in the histogram. The footprint pattern of the bottom strand is shifted by 4–5 nt in the 3′ direction compared with the top strand. The centers of the three footprints showing the lowest cutting frequency on each strand are separated by 10–11 bp and thus are precisely in phase with the helical repeat of the DNA. This observation indicates that the contacted sites are located on the same face of the DNA. Protection of the DNA backbone positions in the central footprint is more pronounced than in the flanking footprints. The hydroxyl radical footprints are symmetrically arranged about the pseudo-dyad axis of the operator sequence and reflect the 2-fold symmetry of the binding CopR-dimer.
Figure 1.
Hydroxyl radical footprinting. G>A, Maxam–Gilbert sequencing ladder; control, protein-free DNA; bound, DNA incubated with CopR prior to hydroxyl radical treatment. Protection sites I, II and III are indicated based on the results of the histograms (see Fig. 2).
Figure 2.
Histogram of the hydroxyl radical footprinting. Upper panel, top strand; lower panel, bottom strand. The relative cutting frequency (arbitrary units) of the bound DNA compared to the unbound control is shown. The bars represent average values from four and six experiments for the top strand and the bottom strand, respectively. The error bars show the standard deviation. Protection sites I, II and II are indicated.
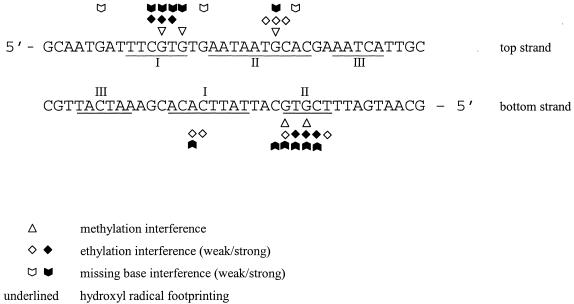
Figure 3 shows a comparison of the results of the hydroxyl radical with the data of the interference experiments taken from Steinmetzer and Brantl (7). Two sets of footprints, I and II (top strand) as well as the symmetry-related footprints I and II at the bottom strand correspond to the two binding half sites I and II identified previously by chemical interference experiments (7). However, backbone contacts within footprint I of the bottom strand are found to be significantly larger when compared with the respective site identified by interference experiments. The outer footprints III of both strands cover the sequence 5′ AATCA. No methylation-, ethylation- and missing base-interference signals were detected previously for this sequence (7). The lack of methylation- and missing base-interference signals for the footprints III of both strands can be explained by assuming that at these positions no base-specific contacts are formed between protein and DNA.
Figure 3.
Comparison of the footprinting data obtained in this study for the top and the bottom strands with data from ethylation-, methylation- and missing base-interference experiments (7). Protection sites I, II and III are underlined. Dark symbols indicate strong interference signals, light symbols represent weak interference signals.
However, there seems to be a contradiction between the lack of ethylation-interference signals and the observed protection for the sites III of both strands. CopR is still able to bind the DNA even in the presence of ethyl groups, indicating a relative high flexibility of the protein–DNA complex at these outer contact sites. This is in accordance with our observation that CopR is able to bind to a minimal operator of 17 bp length where both normally contacted sites III are absent. Another explanation arises from the possibility of an increased conformational flexibility of the sequence 5′-AATCA-3′ due to the CA step (25). Therefore, structural alterations of DNA conformation in this region—caused by binding of CopR—might be together with direct contacts jointly responsible for the decreased cutting frequency of hydroxyl radicals. Altogether, CopR protects a region of 29 bp, corresponding to 10 nm, which is surprisingly large for a small protein like CopR, a dimer of 21 kDa.
Competition assay
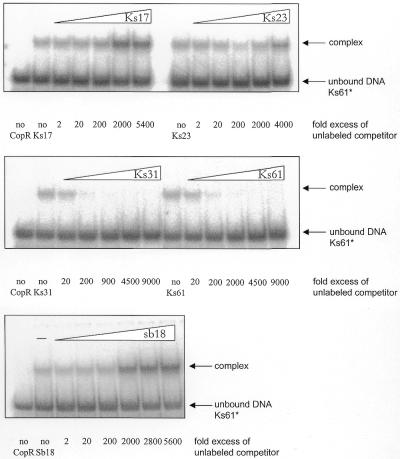
To evaluate the importance of the outer CopR–DNA contacts identified by hydroxyl radical footprinting for complex formation we performed competition assays with a radioactively labeled 61 bp fragment (Ks61, also used in footprinting experiments) and unlabeled DNA fragments containing either only the 17, 23 or 27 central base pairs (Ks17, Ks23, Ks31, respectively) of the operator sequence or the complete sequence (Ks61, see Table 1). The results are shown in Figure 4. As can be seen, the short 17 and 23 bp fragments containing only the central contact sites were unable to compete with the labeled 61 bp fragment whereas fragments Ks31 and Ks61 also containing the outer base pair contact sites efficiently displaced the radioactively labeled 61 bp fragment from the complex. This result indicates an additional stabilization of the complex by these outer base pairs.
Figure 4.
Competition assay of the labeled full length CopR target Ks61 with unlabeled DNA fragments containing the operator sequence (upper and middle panel) or an unrelated sequence as control (lower panel). The positions of unbound DNA and of the complex are indicated. The ration between the labeled fragment Ks61 and the unlabeled competitor is indicated on the bottom of the respective autoradiograms.
In the competition assays with the 17 and the 23 bp fragments an increased complex formation at higher concentrations of unlabeled competitor was observed. To further examine this unexpected effect, we used a completely different DNA fragment of similar length as a competitor (Fig. 4). The result shows also an increased complex formation due to an increased competitor concentration. Hence, the observed effect is due rather to a non-specific activation of complex formation than the result of specific interactions between protein and DNA.
FRET measurements
To determine conformational changes of the DNA by binding CopR, FRET experiments were performed. To maximize the effect of a change of the FRET efficiency by binding of CopR the DNA should be as small as possible. On the other hand, the DNA must be long enough to avoid interaction between the dyes and the protein. Therefore, based on the existing model of the CopR–DNA complex (9) we chose a 19 bp DNA containing the 17 bp minimal CopR binding site, two terminal CG pairs for stability and labeled the 5′ ends either with fluorescein or rhodamine (F19, see Table 1).
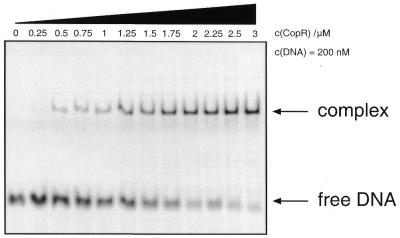
To visualize binding of CopR to the fluorescence labeled 19 bp DNA fragment an EMSA was performed. Doubly labeled DNA at 200 nM concentration was incubated with increasing amounts of CopR and the unbound DNA was separated from the DNA–protein complex on a native 8% polyacryamide gel (Fig. 5). The results obtained agree with EMSAs performed previously with radioactively labeled DNA (7) demonstrating that the introduced fluorescence label did not interfere with CopR–DNA complex formation.
Figure 5.
EMSA of CopR binding to the 19 bp DNA fragment F19 used in the FRET studies. 200 nM doubly labeled DNA samples were incubated with an increasing amount of proteins and separated on 8% native polyacrylamide gels.
Subsequently, the binding of CopR to DNA was analyzed in solution by measuring the anisotropy of both dyes, the donor FAM and the acceptor TMRh. In both cases the anisotropy increased by adding CopR indicating a binding of CopR to the 19 bp DNA fragment. The anisotropy of the donor increased from 0.08 to 0.10 for the donor-only sample and from 0.09 to 0.12 for the donor–acceptor sample, respectively. The anisotropy of the acceptor increased from 0.26 to 0.28. The volume of the nucleoprotein complex is larger than the volume of the free DNA resulting in a lower rotational rate of the dyes covalently attached to the DNA and, therefore, in an increased anisotropy (14). This slight increase of the anisotropy is due to an increased volume of the DNA by binding a CopR dimer and indicates that there is no direct interaction of the bound protein with the fluorescent dyes. However, the fluorescence intensity of the donor was quenched by the protein while the fluorescence intensity of the acceptor was unchanged. In donor-only samples the fluorescence intensity decreased to 70% and in donor–acceptor samples to 72% by adding CopR (Fig. 6).
Figure 6.
FRET efficiency versus CopR concentration. With increasing CopR concentration the FRET efficiency of the doubly labeled 19 bp DNA fragment containing the minimum operator sequence decreases by ∼3%. The insert shows the quenching of the donor fluorescein by increasing protein concentration. The donor intensity in a donor-only sample decreases by ∼30% while the quenching in the fluorescein and rhodamine labeled sample is ∼2–3% less corresponding with the decreasing FRET efficiency in the complex. While the donor fluorescence quantum yield has a direct influence on the Forster distance R0 (see Materials and Methods) and therefore an influence on the measured FRET efficiencies, this quenching effect has to be taken into account in calculating the change of the dye-to-dye distance in the free and bound DNA. Thus, the distance between the dyes attached to the DNA helix ends decreases from 6.4 ± 0.1 nm (R0 = 5.0 nm) in the free DNA to 6.2 ± 0.1 nm (R0 = 4.7 nm) in the complex with CopR indicating a slight bend of the DNA in the complex. The dissociation constant of the CopR–DNA complex in solution determined from the FRET efficiency and the quenching data was 1.6 ± 0.9 nM.
Binding of CopR to the DNA led to a decreased FRET efficiency. The energy transfer decreased from 0.19 ± 0.01 for free DNA to 0.16 ± 0.01 for the complexed DNA (Fig. 6). This corresponds to the observed quenching of the donor in the donor-only and the donor–acceptor sample by binding CopR. Due to a decreased energy transfer in the doubly labeled CopR–DNA complex the donor fluorescence intensity increases slightly resulting in a lower quenching than in the donor-only sample. Both the FRET efficiency decreased and the donor fluorescence intensity increased by ∼3%.
The dye-to-dye distances can be calculated from the FRET efficiencies. Under given buffer conditions the Förster distance R0 for the dye pair FAM/TMRh is 5.0 nm (see Materials and Methods) resulting in a dye-to-dye distance of 6.4 ± 0.1 nm in the free, unbound operator DNA. Due to a quenching of the donor fluorescence intensity to 70% in the complex, the Förster distance R0 decreased to 4.7 nm. This yielded a donor–acceptor distance of 6.2 ± 0.1 nm in the complex. Consequently, the dye-to-dye distance decreased by ∼0.2 nm when the DNA was complexed by CopR corresponding to a small DNA bending of ∼20–25°.
The decrease of FRET efficiency by an increasing CopR concentration was fitted with a two-step binding model published previously (8) to determine the binding constant of CopR to the operator DNA in solution (Fig. 6). In the first step, CopR monomers form a homodimer, and in the second step CopR dimers bind to the DNA.
2 CopR ↔ (CopR)2 + DNA ↔ [(CopR)2DNA]
Kdimer Kcomplex
The dissociation constant Kcomplex of the CopR–DNA complex with a 19 bp DNA fragment (the minimum operator size) was ∼1.6 ± 0.9 nM in solution and is higher than 0.4 ± 0.13 nM obtained for the complex with the full length operator DNA by EMSA with radioactively labeled DNA (the length of the fragment used was 61 bp) (8). This observation is in accordance with the results of the competition experiments but when comparing the two values for Kcomplex the different experimental conditions, i.e. DNA concentration (0.1 nM for radioactively labeled DNA, 200 nM for fluorescence labeled DNA), different protein preparations, different electrophoresis conditions (with versus without NaCl in the electrophoresis buffer, 4°C versus room temperature), have to be taken into account.
The hydroxyl radical footprinting experiments described above showed that CopR contacted 29 instead of just 17 bp determined with the interference experiments (7). However, the DNA fragment used for the above-described FRET experiments contained only the minimum size of the operator. To answer the question whether the presence of the outer base pairs corresponding to footprints c1 and a2 would increase the bending angle of the DNA in the complex, we repeated the FRET measurements with a 34 bp DNA fragment containing the whole 29 bp operator sequence. Since the end-to-end distance of the unbound 34 bp DNA fragment was ∼115 Å and out of range to detect with the dye-pair FAM/TMRh, we labeled the DNA helix ends with FAM and rhodamine-X, respectively. For this pair a larger Förster distance of ∼6.0 nm was described (19). Due to the large dye-to-dye distance in the free 34 bp DNA of ∼11.5 nm (this corresponds to an energy transfer efficiency of 0.02) only a large distance change (>1.5 nm) and therefore a DNA bending angle >40–50° results in a measurable increase of FRET.
The binding of CopR to the 34 bp DNA construct was confirmed by EMSA and anisotropy measurements (data not shown). However, no significant increase of the FRET efficiency was observed, indicating that the outer base pairs contacted by CopR did not increase the bending angle beyond 40–50° (data not shown). Thus, the center/s of the bend seems to be located within the central 19 bp of the operator sequence as described for the 434 repressor complex (26,27).
The obtained conformational change of the DNA in the CopR–DNA complex is in good agreement with the data obtained for the DNA bending in the 434 c1 repressor–DNA complex, where X-ray crystallography revealed a bending angle between 20 and 25° (26,27). Therefore, the DNA conformation in both, the 434 c1 repressor–DNA complex and in the CopR–DNA complex seems to be similar.
Analytical ultracentrifugation
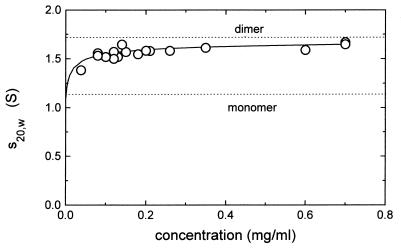
The existing model of the CopR dimer includes the amino acids 1–63 and has a length of ∼5 nm without a hydration shell. To obtain information about the in vivo shape of the full length CopR-dimers, sedimentation velocity experiments were performed. The results are shown in Figure 7. From these experiments a sedimentation coefficient for CopR of 1.71 (S) was estimated. Based on this value as well as on the molecular mass of dimeric CopR (23.8 kDa) and the partial specific volume of the protein a diffusion coefficient for dimeric CopR of 6.5 × 10–7 cm2/s was derived. Using these data and the procedure as given in Materials and Methods a model of the dissolved dimeric CopR with the maximal length of 8.4 nm was estimated for the fully hydrated protein. This is in good agreement with our results obtained in the footprinting experiments showing outer DNA contact sites and a DNA protection over ∼10 nm.
Figure 7.
Plot of sedimentation coefficients dependent on the CopR concentration. The solid line is the best fit based on the equilibrium constant KD = 1.5 µM. Whereas the sedimentation coefficient of the monomeric protein was estimated to be 1.08 (S), for the dimeric protein a sedimentation coefficient of 1.71 (S) was obtained.
DISCUSSION
A combination of hydroxyl radical footprinting and FRET measurements was used to determine the DNA conformation in the CopR–DNA complex. Furthermore, to resolve the shape of CopR sedimentation, velocity experiments were performed.
The footprints of CopR obtained by hydroxyl radical footprinting covered 29 bp and showed three distinct areas of protection for each strand and are similar to the results observed for λ-repressor, for the B.subtilis phage Φ105 repressor and for the phage Mu C protein (24,28,29).
One of the sites identified, namely site III of the bottom strand, was weaker than the other protection sites indicating that the interaction between CopR and the DNA had a slightly asymmetrical character. This is in agreement with previous observations (7) and also reflects the imperfect symmetry of the operator sequence. The area of protection was significantly larger than the area determined by the combination of methylation, ethylation and missing base interference experiments, where the distance between the outermost contacts was 17 bp (7). Protection sites I and II corresponded to the contacted sites I and II identified previously whereas the two outer contact sites III were not detected by interference experiments. The results of the competition assays showed that DNA fragments, containing only the minimal operator sequence of 17 bp length, were unable to compete with a 61 bp long DNA fragment containing the outer contact sites. Even a more than thousand-fold excess of the shorter fragments Ks17 and Ks23 containing the minimal recognition sequence was not sufficient to displace the full length CopR target. This indicated that CopR indeed made additional contacts outside of the 17-bp region. But these contacts were not essential for CopR binding as shown in the former interference experiments (7) and the FRET experiments presented in this study.
To determine the global shape of the DNA in complex with CopR FRET, experiments with two DNA fragments were performed: a 19-bp fragment containing only the minimal operator sequence plus two terminal base pairs for stability and a 34-bp fragment also including the outer contact sites. Both oligonucleotides were 5′ end-labeled with fluorescein as the donor and tetramethylrhodamine or rhodamine-X as the acceptor. Induced by the binding of CopR the dye-to-dye distance in the 19-bp fragment decreased by 0.2 nm. This reduction of the DNA end-to-end distance can be induced by bending and/or by un- or over-winding of the DNA upon protein binding. Taking into account the positions of the dyes at the DNA helix ends [rhodamine stacks on top of the helix end like an additional base pair while fluorescein points away from the DNA (13,30)] this decrease is consistent with the existing model of the CopR–DNA complex showing a bending angle of 20–25°. This model considers the DNA bending as well as an over-twisting of the DNA as described for the 434 repressor–DNA complex (31). For the larger DNA fragment containing the full length binding sequence no significant increase in the FRET efficiency was observed. These results indicate that the center of the DNA bending is inside the minimal operator sequence and that the additional outer base contacts did not increase the DNA bending beyond 40–50°. Neither of the two outer binding sites adds more than 10–15° to the overall bend.
The total size of the contacted area in the CopR–DNA complex consisting of three footprints on each strand was 29 bp, corresponding to a distance of ∼10 nm. CopR binds the DNA as a dimer (8). Analytical ultracentrifugation to determine the size and shape of CopR dimers revealed that CopR dimers had an extended shape with a size of 8.4 nm for the fully hydrated protein. Therefore, only a slight bending of the DNA is required to enable CopR to make additional contacts outside of the 17 bp recognition site that strengthen the protein–DNA interaction. A slight bend was in accordance with the fact that no pronounced hypersensitive sites of hydroxyl radical cleavage were observed indicating that no drastic distortion of the DNA backbone was caused by CopR binding. Similar to the conserved recognition sequence of the 434 repressor, the CopR operator contains two TG steps 11 bp apart that may serve as bending points by providing the flexibility required for the conformational changes of the DNA (32).
Because the current model of CopR consists only of amino acids 1–63 (9) and, therefore, represents only two-thirds of the protein, it was not sufficient to explain all determined contacts between protein and DNA. Analytical ultracentrifugation revealed that the full length protein dimers had a length of 8.4 nm and are considerably longer than CopR dimers lacking the C-terminus in our current model. It seems plausible that the C-terminal portion of CopR not included in the model formed the observed outer DNA contacts. Additionally, the observation that a truncated CopR lacking the 27 C-terminal amino acids (CopΔ27) has a slightly increased equilibrium dissociation constant KD of 3.8 nM for the protein–DNA complex (33) also indicated that additional contacts are formed between amino acids of the full length CopR C-terminus and the DNA backbone.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Birch-Hirschfeld (Institute of Virology, University Jena) for synthesizing the oligonucleotides used for the footprinting experiments and competition assays. This work was supported by grant BR 1552/4-3 of the Deutsche Forschungsgemeinschaft to S.B.
REFERENCES
- 1.Dickerson R.E. (1998) DNA bending. The prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brantl S. and Behnke,D. (1992) Copy-number control of the streptococcal plasmid pIP501 occurs at three levels. Nucleic Acids Res., 20, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brantl S. (1994) The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol. Microbiol., 14, 473–483. [DOI] [PubMed] [Google Scholar]
- 4.Brantl S. and Wagner,E.G.H. (1997) Dual function of the copR gene product of plasmid pIP501. J. Bacteriol., 179, 7016–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swinfield T.-J., Oultram,J.D., Thompson,E.E., Brehm,J.K. and Minton,N.P. (1990) Physical characterization of the replication region of the Streptococcus faecalis plasmid pAMβ1. Gene, 87, 79–90. [PubMed] [Google Scholar]
- 6.Ceglowski P. and Alonso,J.C. (1994) Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orfη-copS region. Gene, 145, 33–39. [DOI] [PubMed] [Google Scholar]
- 7.Steinmetzer K. and Brantl,S. (1997) Plasmid pIP501 encoded transcriptional repressor CopR binds asymmetrically at two consecutive major grooves of the DNA. J. Mol. Biol., 269, 684–693. [DOI] [PubMed] [Google Scholar]
- 8.Steinmetzer K., Behlke,J. and Brantl,S. (1998) Plasmid pIP501 encoded transcriptional repressor CopR binds to its target DNA as a dimer. J. Mol. Biol., 283, 595–603. [DOI] [PubMed] [Google Scholar]
- 9.Steinmetzer K., Hillisch,A., Behlke,J. and Brantl,S. (2000) Transcriptional repressor CopR: Structure model based localization of the DNA binding motif. Proteins, 38, 393–406. [PubMed] [Google Scholar]
- 10.Steinmetzer K., Hillisch,A., Behlke,J. and Brantl,S. (2000) Transcriptional repressor CopR: Amino acids involved in forming the dimeric interphase. Proteins, 39, 408–416. [DOI] [PubMed] [Google Scholar]
- 11.Dodd I.B. and Egan,J.B. (1990) Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res., 18, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz M., Hillisch,A., Payet,D., Buttinelli,M., Travers,A. and Diekmann,S. (1999) DNA bending induced by high mobility group proteins studied by fluorescence resonance energy transfer. Biochemistry, 38, 12150–12158. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz M., Hillisch,A., Goodman,S.D. and Diekmann,S. (1999) Global structure similarities of intact and nicked DNA complexed with IHF measured in solution by fluorescence resonance energy transfer. Nucleic Acids Res., 27, 4619–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakowicz J.R. (1983) Principles of Fluorescence Spectroscopy. Plenum Press, New York.
- 15.Clegg R.M. (1996) In Wang,X.F. and Herman,B. (eds), Fluorescence Imaging Spectroscopy and Microscopy. John Wiley & Sons, New York, NY, pp. 179–252.
- 16.Förster T. (1948) Intermolecular energy migration and fluorescence. Ann. Phys., 2, 55–75.
- 17.Van der Meer B.S., Coker,G. and Chen,S.-Y.S. (1994) Resonance Energy Transfer-Theory and Data. VCH, New York, NY.
- 18.Stühmeier F., Welch,J.B., Murchie,A.I., Lilley,D.M. and Clegg,R.M. (1997) Global structure of three-way DNA junctions with and without additional unpaired bases: a fluorescence resonance energy transfer analysis. Biochemistry, 36, 13530–13538. [DOI] [PubMed] [Google Scholar]
- 19.Toth K., Sauermann,V. and Langowski,J. (1998) DNA curvature in solution measured by fluorescence resonance energy transfer. Biochemistry, 37, 8173–8179. [DOI] [PubMed] [Google Scholar]
- 20.Clegg R.M. (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol., 211, 353–388. [DOI] [PubMed] [Google Scholar]
- 21.Zorbas H., Rogge,L., Meisterernst,M. and Winnacker,E.L. (1989) Hydroxyl radical footprints reveal novel structural features around the NFI binding site in adenovirus DNA. Nucleic Acids Res., 17, 7735–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behlke J. and Ristau,O. (1997) Molecular mass determination by sedimentation velocity experiments and direct fitting of the concentration profiles. Biophys. J., 72, 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia de la Torre J.H., Navarro,S., Lopez Martinez,M.C., Diaz,F.G. and Lopez Cascales,J.J. (1994) HYDRO: a computer program for the prediction of hydrodynamic properties of macromolecules. Biophys. J., 67, 530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tullius T.D. and Dombrowski,B.A. (1986) Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to λ repressor. Proc. Natl Acad. Sci. USA, 83, 5469–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Packer M.J., Dauncey M.P. and Hunter C.A. (2000) Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol., 295, 71–83. [DOI] [PubMed] [Google Scholar]
- 26.Aggarwal A.K., Rodgers,D.W., Drottar,M., Ptashne,M. and Harrison,S.C. (1988) Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science, 242, 899–907. [DOI] [PubMed] [Google Scholar]
- 27.Shimon L.J. and Harrison,S.C. (1993) The phage 434 OR2/R1-69 complex at 2.5 Å resolution. J. Mol. Biol., 232, 826–838. [DOI] [PubMed] [Google Scholar]
- 28.Van Kaer L., Van Montagu,M. and Dhaese,P. (1989) Purification and in vitro DNA-binding specificity of Bacillus subtilis phage Φ105 repressor. J. Biol. Chem., 264, 14784–14791. [PubMed] [Google Scholar]
- 29.Ramesh V. and Nagaraja,V. (1996) Sequence-specific DNA binding of the phage Mu C protein: Footprinting analysis reveals altered DNA conformation upon protein binding. J. Mol. Biol., 260, 22–33. [DOI] [PubMed] [Google Scholar]
- 30.Norman D.G., Grainger,R.J., Uhrin,D. and Lilley,D.M. (2000) Location of cyanine-3 on double-stranded DNA: importance for fluorescence resonance energy transfer studies. Biochemistry, 39, 6317–6324. [DOI] [PubMed] [Google Scholar]
- 31.Koudelka G.B., Harbury,P., Harrison,S.C. and Ptashne,M. (1988) DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc. Natl Acad. Sci. USA, 85, 4633–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzou W.S. and Hwang,M.J. (1999) Modeling helix-turn-helix protein induced DNA bending with knowledge-based distance restraints. Biophys. J., 77, 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn K., Steinmetzer,K. and Brantl,S. (2000) Transcriptional repressor CopR: The structured acidic C terminus is important for protein stability. J. Mol. Biol., 300, 1021–1031. [DOI] [PubMed] [Google Scholar]