Abstract
Hjc resolvase is an archaeal enzyme involved in homologous DNA recombination at the Holliday junction intermediate. However, the structure and the catalytic mechanism of the enzyme have not yet been identified. We performed database searching using the amino acid sequence of the enzyme from Pyrococcus furiosus as a query. We detected 59 amino acid sequences showing weak but significant sequence similarity to the Hjc resolvase. The detected sequences included DpnII, HaeII and Vsr endonuclease, which belong to the type II restriction endonuclease family. In addition, a highly conserved region was identified from a multiple alignment of the detected sequences, which was similar to an active site of the type II restriction endonucleases. We substituted three conserved amino acid residues in the highly conserved region of the Hjc resolvase with Ala residues. The amino acid replacements inactivated the enzyme. The experimental study, together with the results of the database searching, suggests that the Hjc resolvase is a distantly related member of the type II restriction endonuclease family. In addition, the results of our database searches suggested that the members of the RecB domain superfamily are evolutionarily related to the type II restriction endonuclease family.
INTRODUCTION
Homologous DNA recombination is a ubiquitous phenomenon found in every living organism and plays important roles in the generation of genetic diversity and the repair of DNA damage. Homologous DNA recombination occurs through the formation of a characteristic DNA structure called the Holliday junction. The molecular mechanism of homologous DNA recombination has been investigated and most of our knowledge about this process has been obtained from studies with Escherichia coli (reviewed in 1–7). Recently, however, information about recombination in Eukarya and, more recently, in Archaea has been accumulated. One focus of the progress in the field is the identification of the nucleases involved in the last stage of homologous DNA recombination, namely Holliday junction resolvase. Several enzymes in this category have been purified from various sources (reviewed in 8,9). RuvC, a junction resolvase derived from E.coli, is the most characterized enzyme to date. The tertiary structure of the enzyme has already been solved by an X-ray crystallographic study. This revealed that the enzyme shares a similar fold with retroviral integrase and RNase H (10,11). The other characterized enzymes include Cce1 from Saccharomyces cerevisiae mitochondria (12), Ydc2 from Schizosaccharomyces pombe mitochondria (13), RusA from lambdoid phage (14), T4 phage endonuclease VII (15) and T7 phage endonuclease I (16). The mitochondrial enzymes Cce1 and Ydc2 are similar in amino acid sequence (17). The other enzymes, however, do not show sequence similarity to each other. In other words, the resolvases involved in homologous DNA recombination are considered to have been replaced by non-orthologous enzymes in different organisms.
Recently, Komori et al. identified a junction resolvase from a hyperthermophilic archaeon, Pyrococcus furiosus (18). The enzyme, named Hjc (Holliday junction cleavage), has a function equivalent to that of eubacterial RuvC in homologous DNA recombination. It was the first report of an archaeal Holliday junction resolvase. They reported the nucleotide sequence of the gene encoding the Hjc resolvase and the length of the deduced amino acid sequence was 123 amino acid residues. Subsequently, Kvaratskhelia and White identified an enzyme equivalent to Hjc resolvase from a different archaeal organism, Sulfolobus solfataricus. In addition, they identified another junction resolvase activity from a cell extract of the same organism and named it Hje (Holliday junction endonuclease) (19,20). However, the Hje activity has not been purified to homogeneity and the corresponding gene has not yet been cloned, therefore, it is not known how structurally different the two resolvases are. It is now a very exciting issue to analyze the biochemical properties of the two activities in more detail to understand how they share roles in living cells.
In each of the archaeal genomes sequenced to date there is one ORF with high sequence similarity to that of P.furiosus Hjc. These ORF products are considered to be the counterparts of the Hjc resolvase. However, no clear sequence similarity between the archaeal Hjc and any other nuclease has been reported. Komori et al. tried to align the Hjc resolvase and E.coli RuvC sequences. However, the sequence similarity between them was too weak to infer the structure and function of Hjc resolvase (18). In order to obtain some clues about the structure, function and evolution of the archaeal junction resolvases, we performed database searches using the amino acid sequence of the P.furiosus Hjc resolvase as a query. The results of our database searches suggest that the archaeal Hjc resolvase is distantly related to the type II restriction endonucleases, which are widely distributed over eubacteria. At the same time, we introduced amino acid substitutions in the primary structure of the Hjc resolvase from P.furiosus and measured the changes in the various activities of the enzyme. The results of the experiments are consistent with predictions from database searches.
MATERIALS AND METHODS
Computational analysis of Hjc resolvase
We performed database searches with the computer program PSI-BLAST (21), using the amino acid sequence of Hjc resolvase as a query. The database searches were done at the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/blast/psiblast.cgi ). A multiple alignment was constructed according to the output of the PSI-BLAST search. The gap positions were slightly modified to increase the similarity.
Site-specific mutagenesis and measurement of the activity
Three residues of P.furiosus Hjc, D33, E46 and K48, were substituted with Ala by PCR-mediated mutagenesis using the Quick Change™ Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The NdeI–SacI fragment of pFUHJ2 (18), which contains the entire region of the hjc gene, was inserted into the pTV119 vector to reduce the size of the PCR template plasmid and the reactions were carried out according to the manufacturer’s instructions with some modifications. Mutagenized plasmids were selected by nucleotide sequencing and the NdeI–SacI fragments were returned to the pET21a vector. The nucleotide sequence of the entire region of the hjc gene was confirmed using a DNA sequencer (ABI Prism 310 Genetic Analyzer; PE Applied Biosystems, Foster City, CA). Expression and purification of the gene products were performed as in the case of the wild-type Hjc described earlier (18). Purified proteins were subjected to an endonuclease assay using a 32P-labeled synthetic Holliday junction as the substrate, as described (17).
RESULTS
PSI-BLAST search
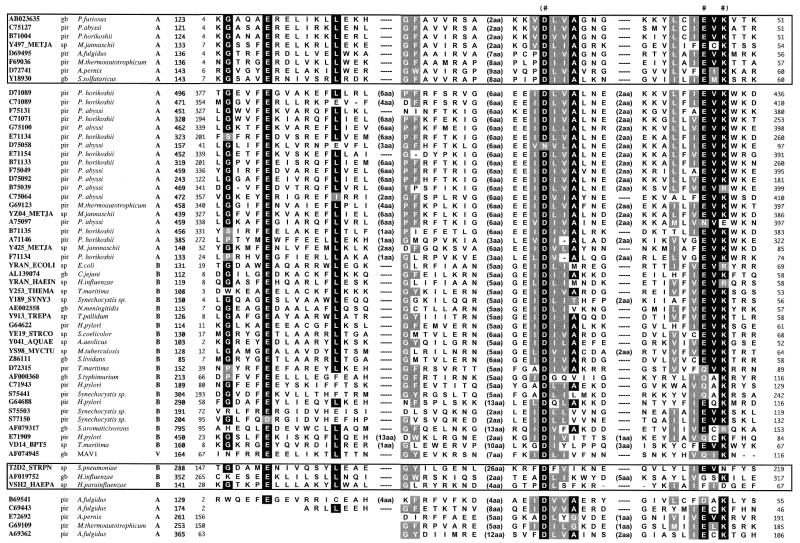
From the sequence homology search in the database using P.furiosus Hjc as a query we found 59 amino acid sequences that showed sequence similarity to the N-terminal half of Hjc resolvase, which is ∼60 amino acid residues in length. A multiple alignment of the detected sequences was constructed according to the output of the database search (Fig. 1). Of the detected 59 sequences, eight were the Hjc resolvase from P.furiosus itself and counterparts from other archaea. There were 48 sequences that were ORF products derived from eubacteria and archaea whose functions are unknown. The functions of the remaining three sequences have already been identified; these were DpnII (T2D2_STRPN), HaeII (AF019752) and Vsr endonuclease (VSH2_HAEPA). DpnII and HaeII are eubacterial enzymes and function as type II restriction endonucleases (reviewed in 22–24). The Vsr endonuclease is involved in the repair of TG mismatched base pairs (25,26). However, a recent X-ray crystallographic study revealed that Vsr endonuclease is a distant relative of the type II restriction endonuclease family (27). The sequence alignment included a conserved pattern, D-X6∼14-(E/Q)-X-(K/R) (see the region indicated by parentheses in Fig. 1), which is similar to the motif sequence of the type II restriction endonucleases. The type II restriction endonucleases share the sequence motif D-X6∼30-D/E-X-K, where X indicates any residue, although the corresponding residues of DpnII and HaeII deviate slightly from this motif. A structure comparison revealed that the corresponding region in Vsr endonuclease has the sequence pattern D-X10-F-X-H (27), which deviates from the motif sequence of the type II restriction endonucleases.
Figure 1.
A multiple alignment of the amino acid sequences of the Hjc resolvases and their relatives. The first and second columns indicate the ID code and the corresponding database for each sequence. The third column indicates the source. The one letter characters, A, B and V, in the fourth column refer to the source: archaea, eubacteria or bacteriophage. The fifth column includes the total length of the sequence data. The sixth column indicates the residue number of the left-most residue of an aligned sequence. An aligned sequence is shown in the seventh column. An integer in parentheses in an aligned sequence indicates the number of residues that are not shown in the figure. The last column includes the residue number of the right-most residue of the aligned sequence. The N-terminal regions of the bottom four sequences are blank, because they did not show sequence similarity in this region to the remaining ORF products. When >70% of a site is occupied by an identical residue, the residue is indicated by a reversed character, and a residue physicochemically similar to the conserved residue is indicated by shadowing. Likewise, physicochemically similar residues at a site are shadowed when >70% of the site is occupied by physicochemically similar residues. A blank at a site for the bottom four ORF products was counted as a replacement with a physicochemically different residue. The conserved pattern similar to the motif of the restriction endonuclease family is indicated by parentheses over the alignment and three conserved residues in the pattern are indicated by #. There are two frames in the alignment. The upper frame includes the archaeal Hjc resolvases and their putative counterparts, while the three eubacterial enzymes with known functions are indicated in the lower frame.
Mutational analysis
To investigate if the amino acid residues are conserved in the predicted motif described above, we made three mutant Hjc proteins, D33A, E46A and K48A, by site-directed mutagenesis. The substituted residues were present at the alignment sites indicated by # in Figure 1. All of the mutant proteins produced in E.coli BL21(DE3) cells exhibited the same purification behavior as the wild-type proteins and similar amounts of homogeneous proteins were obtained from each recombinant E.coli strain (data not shown). The purified proteins were assayed for cleavage of the Holliday junction DNA. As shown in Figure 2A, no cleaved band was detected in the reactions using the mutant proteins under conditions in which the substrate DNA was almost completely cleaved by wild-type Hjc. The relative activities of the three mutant Hjc proteins were <1% of that of wild-type Hjc. The gel retardation assay showed that all of the mutant Hjc proteins can bind to the Holliday junction DNA with the same affinity as that of wild-type Hjc (Fig. 2B). These results support the idea that the three residues D33, E46 and K48 participate directly in catalysis by Hjc.
Figure 2.
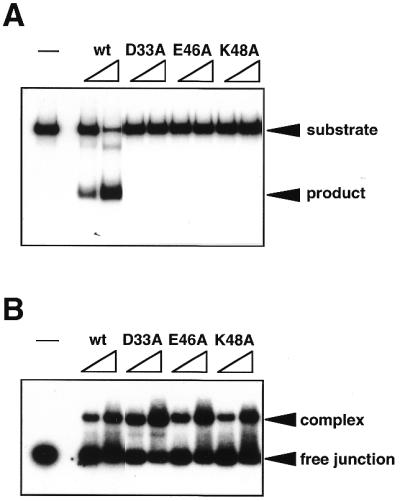
Cleavage and binding activities of wild-type and mutant Hjc proteins. (A) Cleavage assay using a 32P-labeled synthetic Holliday junction (100 nM) was done in the standard reaction buffer, as described in Materials and Methods. Each protein was added to the reaction to a final concentration of 2 or 5 nM. The reaction products were separated by PAGE and the results were visualized by autoradiography. (B) Binding activity to the junction was analyzed by a gel retardation assay. A 32P-labeled synthetic Holliday junction (10 nM) was incubated with Hjc proteins (20 or 50 nM). The electrophoretic profile was analyzed by autoradiography. In both panels lane – at the left side indicates the reaction without protein.
DISCUSSION
We have identified the archaeal Hjc resolvases as distant relatives of the type II restriction endonucleases. The top eight sequences in Figure 1 are the Hjc resolvases from P.furiosus (18) and S.solfataricus (19) and putative counterparts from other archaea. They were derived from both Euryarchaeota and Crenarchaeota. The Hjc resolvase shares three biochemical characteristics with the type II restriction endonucleases. The first characteristic is a similarity in the requirement for a metal cation. Hjc resolvase requires Mg2+ ion(s) for catalytic activity (28). Likewise, the Mg2+ ion is essential for the catalytic activity of both type II restriction endonucleases and Vsr endonuclease. In the type II restriction endonucleases three conserved residues in the motif described above are directly or indirectly involved in Mg2+ ion binding. In contrast, only the first invariant Asp is involved in Mg2+ ion binding in Vsr endonuclease. Type II restriction endonucleases, including Vsr endonuclease, are known to require one or two Mg2+ ions per molecule (23,29). The number of Mg2+ ions in Hjc resolvase has not yet been determined. The residues indicated by # in Figure 1 correspond to the three residues involved in Mg2+ ion binding by the restriction endonuclease family. The second characteristic is the similarity in association of the molecules. The Hjc resolvase exists as a stable homodimer in solution (18) and, moreover, it binds to the Holliday junction as a dimer to exert its activity (28). Likewise, the type II restriction endonucleases form a homodimer or homotetramer, although the relative arrangement of the subunits in the complex is often different from endonuclease to endonuclease. In contrast, Vsr endonuclease functions as a monomer. Thirdly, both Hjc resolvase and the type II endonucleases show sequence specificity for the substrate. Hjc resolvase is a structure (Holliday junction)-specific endonuclease. However, it seems to have some sequence preference for cleavage (18; Komori et al., unpublished results). It is well known that type II restriction endonucleases show very strict sequence specificity for the cleavage point. Considering the sequence pattern and other biochemical characteristics, the features of Hjc resolvase seem to be closer to those of type II restriction endonucleases, rather than Vsr endonuclease.
The alignment shown in Figure 1 also includes 48 ORF products whose functions are unknown. Of the 48 ORF products, 25 were derived from archaea, while the remaining sequences were encoded by the genomes of eubacteria and a bacteriophage. G69019, an archaeal ORF product detected by our database searches, is classified as a member of the RecB domain superfamily (30). The RecB domain superfamily consists of the relatives of the C-terminal domains of RecB (E.coli) and AddA (B.subtilis), whose members are considered to be involved in DNA repair (30). The members of the RecB domain superfamily share a conserved segment similar to the motif of the type II restriction endonucleases (30). Recently, Wang et al. performed mutation studies to demonstrate that the conserved segment is involved in the nuclease activity of the RecB subunit (31). When we performed database searches with each of the detected sequences as a query, we were able to expand the members of the Hjc resolvase relatives to the members of the RecB domain superfamily and other members of the type II restriction endonucleases. To save space, however, we have only shown the results of database searches with Hjc resolvase as the query in Figure 1. The observations suggest that Hjc resolvase and its relatives form a diverse protein family together with members of the type II restriction enzyme family and the RecB domain superfamily and that the archaeal and eubacterial ORF products detected by our database searches may have endonuclease activities involved in homologous DNA recombination, DNA repair or restriction.
REFERENCES
- 1.Holliday R. (1964) Genet. Res., 5, 282–304. [Google Scholar]
- 2.Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara A. and Ogawa,T. (1995) Trends Biochem. Sci., 20, 387–391. [DOI] [PubMed] [Google Scholar]
- 4.Shinagawa H. and Iwasaki,H. (1996) Trends Biochem. Sci., 21, 107–111. [PubMed] [Google Scholar]
- 5.Roca A.I. and Cox,M.M. (1997) Trends Biochem. Sci. Sci., 20, 387–391. [Google Scholar]
- 6.West S.C. (1997) Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 7.Kowalczykowski S.C. (2000) Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 8.Lilley D.M. (1995) In Lilley,D.M. (ed.), DNA–Protein: Structural Interactions. Oxford University Press, Oxford, UK, pp. 114–140.
- 9.White M.F., Giraud-Panis,M.-J.E., Pohler,J.R.G. and Lilley,D.M.J. (1997) J. Mol. Biol., 269, 647–664. [DOI] [PubMed] [Google Scholar]
- 10.Ariyoshi M., Vassylyev,D.G., Iwasaki,H., Nakamura,H., Shinagawa,H. and Morikawa,K. (1994) Cell, 78, 1063–1072. [DOI] [PubMed] [Google Scholar]
- 11.Saito A., Iwasaki,H., Ariyoshi,M., Morikawa,K. and Shinagawa,H. (1995) Proc. Natl Acad. Sci. USA, 92, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White M.F. and Lilley,D.M. (1996) J. Mol. Biol., 257, 330–341. [DOI] [PubMed] [Google Scholar]
- 13.Oram M., Keeley,A. and Tsaneva,I. (1998) Nucleic Acids Res., 26, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan S.N., Harris,L., Bolt,E.L., Whitby,M.C. and Lloyd,R.G. (1997) J. Biol. Chem., 272, 14873–14882. [DOI] [PubMed] [Google Scholar]
- 15.Raaijmakers H., Vix,O., Toro,I., Golz,S., Kemper,B. and Suck,D. (1999) EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson M.J., Pohler,J.R. and Lilley,D.M. (1999) Nucleic Acids Res., 27, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White M.F. and Lilley,D.M. (1997) Mol. Cell. Biol., 17, 6465–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komori K., Sakae,S., Shinagawa,H., Morikawa,K. and Ishino,Y. (1999) Proc. Natl Acad. Sci. USA, 96, 8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kvaratskhelia M. and White,M.F. (2000) J. Mol. Biol., 297, 923–932. [DOI] [PubMed] [Google Scholar]
- 20.Kvaratskhelia M. and White,M.F. (2000) J. Mol. Biol., 295, 193–202. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts R.J. and Halford,S.E. (1993) In Linn,S.M., Lloyd,R.S., Roberts,R.J. (eds), Nuclease. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 35–88.
- 23.Aggarwal A.K. (1995) Curr. Opin. Struct. Biol., 5, 11–19. [DOI] [PubMed] [Google Scholar]
- 24.Pingoud A. and Jeltsch,A. (1997) Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 25.Sohail A., Lieb,M., Dar,M. and Bhagwat,A.S. (1990) J. Bacteriol., 172, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennecke F., Kolmar,H., Brundl,K. and Fritz,H.J. (1991) Nature, 353, 776–778. [DOI] [PubMed] [Google Scholar]
- 27.Tsutakawa S.E., Muto,T., Kawate,T., Jingami,H., Kunishima,N., Ariyoshi,M., Kohda,D., Nakagawa,M. and Morikawa,K. (1999) Mol. Cell, 3, 621–628. [DOI] [PubMed] [Google Scholar]
- 28.Komori K., Sakae,S., Fujikane,R., Morikawa,K., Shinagawa,H. and Ishino,Y. (2000) Nucleic Acids Res., 28, 4544–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutakawa S.E., Jingami,H. and Morikawa,K. (1999) Cell, 99, 615–623. [DOI] [PubMed] [Google Scholar]
- 30.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Chen,R. and Julin,D.A. (2000) J. Biol. Chem., 275, 507–513. [DOI] [PubMed] [Google Scholar]