Abstract
Transcription initiation by the σ54 RNA polymerase requires specialised activators and their associated nucleoside triphosphate hydrolysis. To explore the roles of σ54 in initiation we used random mutagenesis of rpoN and an in vivo activity screen to isolate functionally altered σ54 proteins. Five defective mutants, each with a different single amino acid substitution, were obtained. Three failed in transcription after forming a closed complex. One such mutant mapped to regulatory Region I of σ54, the other two to Region III. The Region I mutant allowed transcription independently of activator and showed reduced activator-dependent σ54 isomerisation. The two Region III mutants displayed altered behaviour in a σ54 isomerisation assay and one failed to stably bind early melted DNA as the holoenzyme; they may contribute to a communication pathway linking changes in σ to open complex formation. Two further Region III mutants showed gross defects in overall DNA binding. For one, sufficient residual DNA binding activity remained to allow us to demonstrate that other activities were largely unaffected. Changes in DNA binding preferences and core polymerase-dependent properties were evident amongst the mutants.
INTRODUCTION
In bacteria σ factors are pivotal in establishing regulated and specific initiation of transcription. They bind to the core RNA polymerase and contribute promoter-specific recognition through their DNA binding activities. Additional functions, such as being targets for activator proteins and assisting the DNA strand separation required for RNA synthesis, underline their key roles in gene expression (1). Numerous σ factors are present in bacteria, belonging to two classes. The major class is the σ70 family of proteins; the minor class is represented by σ54 (2,3). The σ54 protein participates in a system of positive control that involves enhancer binding proteins that must hydrolyse a nucleoside triphosphate (NTP) in order to drive the σ54 holoenzyme closed complex to the open complex (4,5). The NTP hydrolysis and enhancer involvement suggest similarities to features of eukaryotic transcription initiation (6). In bacteria, σ54 functions to express genes associated with diverse activities in many different areas of metabolism (reviewed in 7).
The domain organisation and functions of σ54 have been explored previously using directed mutagenesis and biochemical characterisation of σ fragments. The domains clearly interact for the full function of σ54 and several distinct activities reside in discrete parts of σ54 (3,8–11). The N-terminal 50 residues (Region I) are required for activator responsiveness and also function to keep the holoenzyme in a transcriptionally silent state prior to activation (5,12–18). Region I directs σ54 to the DNA fork junction that is created when DNA melting starts next to the –12 GC element of the promoter (13,14,19,20). Region I and the C-terminal DNA-binding domain in Region III of σ54 interact (21). Some Region I and II phenotypes are shared and seem to have a common basis in directing fork junction binding and so restricting the conformation of the holoenzyme (22). Protein footprint studies have shown that core polymerase interacts with sequences in Region I and the DNA-binding domain (residues 329–477) suggesting that the interface of σ54 with the core is extensive and specialised (21,23–25). A major core binding determinant is located in Region III between residues 120 and 215 of Klebsiella pneumoniae σ54 (10).
Identification of σ54 sequences involved in interactions with promoter DNA, core polymerase and activator provides a basis for understanding the mechanism of transcription initiation. Both directed and random mutagenesis of σ54 have been used. (15–18,26–31). Here we have used random mutagenesis of the full K.pneumoniae rpoN gene, encoding σ54, to identify residues important for functioning of the σ54 holoenzyme in vivo. Subsequently, purified single amino acid substitution mutant proteins were assayed to place limits upon the functional defects in σ54. We identified single amino acids important for functioning of regulatory Region I, for interactions with the core polymerase and for activity of the DNA-binding domain. Mutants defective in transcription initiation but still able to direct closed complex formation were obtained. In addition to its primary core and DNA binding functions, it seems that Region III contributes to activities required for conversion of the closed complex to the open complex. Some of these activities are closely associated with DNA binding preferences and some phenotypes further suggest that the interface of σ54 with core polymerase is extensive.
MATERIALS AND METHODS
Mutagenesis
The low copy number plasmid pMM83 (26) encoding K.pneumoniae rpoN was transformed into the mutator strain Escherichia coli XL1 Red (Stratagene). Cells were grown to saturation in Luria broth twice or four times from a 10% inoculum at 37°C. Total growth time was 24 or 48 h. Randomly mutated pMM83 plasmid DNA was then isolated from each culture and mixed together.
Growth screen
Mutated DNA was transformed into UNF2792 (26), a K.pneumoniae rpoN mutant strain, selecting with 15 µg/ml chloramphenicol on Luria agar plates at 37°C. Transformants were picked into Luria broth in microtiter plate wells (200 µl) with 15 µg/ml chloramphenicol and grown at 37°C for 24 h. Subsequently, 5 µl of cells were spotted onto a M9 minimal medium plate (32) supplemented with 25 µg/ml histidine and either 0.5 or 1 mg/ml arginine to screen for the aut phenotype. After 24 h incubation at 37°C cells were scored for growth.
Immunoblots
Transformants displaying the aut phenotype were tested for σ54 protein levels using a polyclonal antibody to E.coli σ54 as described previously (33). Transformants were grown in Luria broth overnight, from a single colony inoculum. Cultures (5 ml) were centrifuged, then resuspended in water to a final volume of 50 µl. An aliquot of 10 µl of concentrated cells was lysed with 10 µl of 2× SDS sample buffer, heated at 95°C and 10 µl of each loaded on denaturing 7.5% SDS–PAGE minigels. Separated proteins were blotted onto PDVF membranes. Anti-σ54 (a gift from A. Ishihama; see also 34) and alkaline phosphatase-conjugated anti-rabbit IgG (Promega) antibodies were used for detection.
In vivo promoter activation assays
Klebsiella pneumoniae UNF2792 containing pMM83 derivatives was transformed with a K.pneumoniae nifH::lacZ translational fusion reporter plasmid pMB1 (35) or a derivative, pWVC88049 (36), with a higher affinity binding site for the holoenzyme. Cells grown in nutrient broth were used to inoculate 4 ml of NFDM (37) supplemented with histidine (125 µg/ml), 20% glucose as the carbon source and aspartic acid (100 µg/ml) as a poor nitrogen source. They were grown overnight at 30°C. In order to allow derepression of nifA synthesis and activity (22), replica cultures were grown in sealed bijoux. Subsequently, β-galactosidase activity was measured.
DNA sequencing
Small scale plasmid preparations were made using the Qiagen midi column method and DNA sequences obtained by the ABI prism big dye terminator cycle sequencing ready reaction method (Perkin Elmer) using primers distributed along the coding sequence and upstream of the natural promoter of rpoN. At least two independent sequencing reactions for each strand were performed.
Purification of σ54
Wild-type and mutant proteins were prepared as N-terminal His-tagged proteins expressed from pET28b(+) in E.coli B834(DE3) (Novagen). Refolding of the L200P, Q351R and S379F mutants was necessary, since these were found in inclusion bodies following overproduction, and a refolded wild-type protein was included for strict comparison. Refolding was from urea as described (8). Where required, mutants were further chromatographed on a heparin column to remove residual core RNA polymerase. Proteins were finally stored in 10 mM Tris–HCl, pH 8.0, 50% (v/v) glycerol, 0.1 mM EDTA and 1 mM dithiothreitol containing 50 mM NaCl. Protein concentrations were determined using the Bio-Rad Protein Assay kit.
Purification of activators
Azotobacter vinelandii NifA and E.coli PspFΔHTH were prepared as N-terminal His-tagged proteins as previously described (23,38)
In vitro assays of σ54 function
These were conducted as described previously (12,14) using the S.meliloti or K.pneumoniae nifH promoters and E.coli core RNA polymerase (Epicentre Technologies). The heteroduplex DNA sequences used and transcription vectors pMKC28 (for S.meliloti nifH) and pNH8 (for K.pneumoniae nifH) have been described (22,39). For reproducibility of data experiments were repeated at least four times. Gel shift assays yielded replicate data within 5% of each experiment.
RESULTS
Mutagenesis of rpoN
Initially we explored the use of a high copy number vector carrying K.pneumoniae rpoN in combination with E.coli rpoN mutants to establish a screen for non-functional plasmid borne rpoN alleles. This procedure proved unreliable because of toxicity associated with the high copy number leading to a loss of expression of σ54 through spontaneous formation of stop codons in the early translated sequences (data not shown). The most reliable combination for screening defects in rpoN proved to be the low copy rpoN plasmid pMM83 in K.pneumoniae UNF2792 (26) rather than in E.coli rpoN mutants. Utilisation of arginine as a nitrogen source (aut phenotype) in K.pneumoniae (as in E.coli) requires a functional σ54 (40). This phenotype was easily discernible by growth on minimal medium agar plates inoculated from liquid cultures (see Materials and Methods and Fig. 1A). Random mutations were introduced into pMM83 (see Materials and Methods). Amongst 10 000 transformants screened for arginine utilisation, 151 were identified that were defective after retransformation and retesting for arginine utilisation. A final immunoblot screen was used to eliminate from our study mutations that resulted in the production of truncated or unstable σ54, possibly arising through gross losses of structural integrity. Of the 151 candidates, five were found to produce a full-length σ54 protein in immunoblots (Fig. 1A). The remainder did not produce any detectable protein or produced only a truncated protein (data not shown). Only one mutant (later characterised as I23N) had a steady-state level equal to the wild-type, suggesting that for the other mutants reduced levels of σ54 could contribute to the lower in vivo activities measured. However, reductions in σ54 levels do not always correlate with reduced in vivo activities (33).
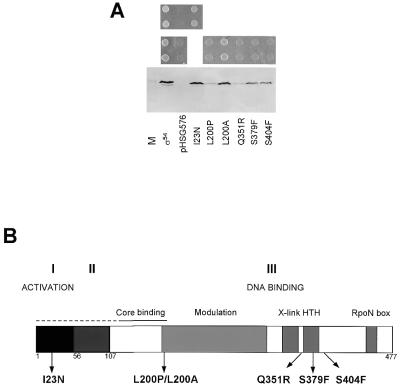
Figure 1.
(A) Growth of K.pneumoniae UNF 2792 on arginine with plasmids containing wild-type σ54 (pMM83) or mutant rpoN genes or with vector (pHSG576). The top panel compares growth of I23N with wild-type and vector. The middle panel compares growth of L200P, L200A, Q351R, S379F and S404F with wild-type and vector. The bottom panel shows the corresponding immunoblot. M, marker (purified σ54 protein). (B) Domain structure of σ54 and its three regions (I–III). The position of each of the random mutants is indicated. X-link, cross linking to promoter DNA; HTH, helix–turn–helix motif; Modulation, domain influencing DNA binding.
For each of the five defective σ mutants which produced full-length protein the entire rpoN and associated natural promoter region that drives expression of rpoN in pMM83 were sequenced. Each mutant harboured base changes that resulted in a single amino acid substitution in σ54 and DNA sequencing confirmed that the σ54 protein detected by immunoblotting was full-length. The five clones selected exhibited the following changes: ATT→AAT (I23N), CTG→CCG (L200P), CAG→CGG (Q351R), TCC→TTC (S379F) and TCC→TTC (S404F). Mutations at these five positions have not been described previously or have not been extensively characterised (26,27). Locations of the individual amino acid substitutions for each mutant are distributed throughout σ54 (Fig. 1B; see also 41). The mutations map to regions associated with activation, core RNA polymerase binding and DNA binding. This confirms that the strategy was random and hence useful in determination of important novel residues. All substitutions were at residues displaying a high degree of conservation, indicating that important sites had been mutated. Secondary structure predictions strongly indicate that L200 lies in an α-helical secondary structure element. Because of the potentially structurally damaging effects of L200P, a L200A substitution was made for strict comparison.
Promoter activation in vivo
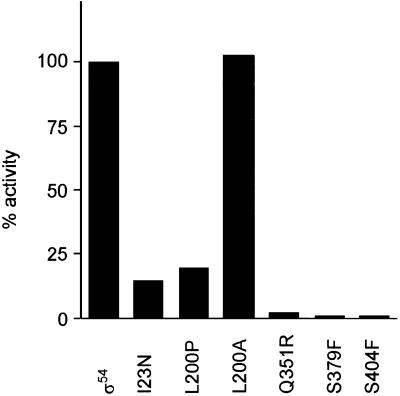
To extend characterisation of the in vivo phenotype and provide a preliminary quantitative characterisation, the five clones directing the synthesis of full-length σ54 that failed to support the aut phenotype were assessed in a promoter activation assay. Results in Figure 2 show that each mutant gave reduced activation of the K.pneumoniae nifH promoter. They were not rescued by the use of the high affinity σ54 mutant nifH promoter present in pWVC88049 (data not shown; 36). This suggests that several of the mutants did not have simple defects in promoter occupancy, but could be defective in post-promoter binding steps. None of the clones tested elevated the unactivated expression level (assayed under nitrogen replete growth conditions; data not shown) showing that they were all activator-dependent mutants in vivo. A range of activity was identified in the in vivo activation assays. Taking into account the different levels of σ54 (Fig. 1A), it seems that I23N, S379F and S404F have clearly reduced activities, suggesting that different types of mutants were represented. The activity of the I23N mutant was clearly substantially reduced at the K.pneumoniae nifH promoter. This contrasts with the modest defect in the arginine growth test (Fig. 1A) and suggests that certain combinations of activator and promoter can reveal a range of transcription deficiencies for any one mutant σ54. Subsequently, each mutant σ54 was purified as a histidine-tagged protein, together with the wild-type, and characterised by a range of in vitro activity assays. In some cases mutant σ54 proteins were found in inclusion bodies and refolded, together with a refolded wild-type σ54 for strict comparison (see Materials and Methods).
Figure 2.
Activity of K.pneumoniae UNF 2792 (pMB1) (nifH::lacZ) carrying pMM83 (σ54) or mutant derivatives. β-Galactosidase activities were calculated as a percentage of the wild-type. Means of a minimum of 10 assays are shown, with a relative variation of not more than 0.15 of the net value.
Core RNA polymerase binding
We used a native gel mobility shift assay to measure binding of the five mutant σ proteins to core polymerase (10). In this assay core polymerase runs as a slow species, the wild-type holoenzyme as a faster single band and the free σ closest to the gel front. Among the mutants, only L200P and S379F behaved differently to the wild-type and displayed a modestly reduced affinity for core polymerase (data not shown). However, the S379F mutant failed to run cleanly into the native gel, suggesting a changed conformation and possibly a reduced availability for binding to core polymerase in vitro. It seems that transcription defects with S379F and L200P might include a contribution from altered core interactions.
Activation of transcription in vitro
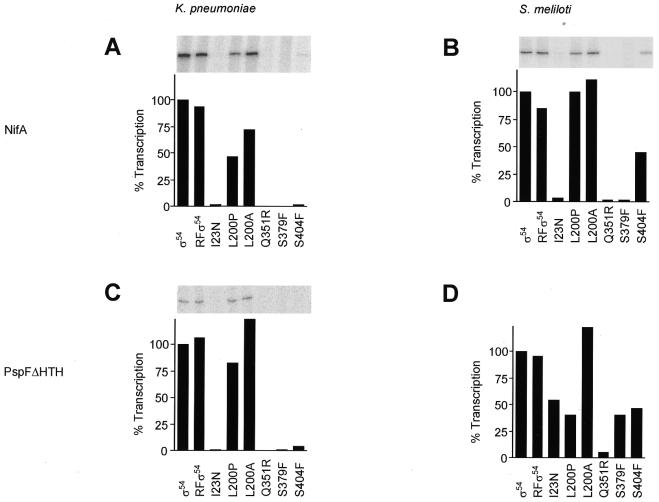
We next determined the in vitro transcription activities of the purified proteins to learn if the purified proteins had activities comparable to those determined in vivo. The ability of the wild-type and mutant holoenzymes to form transcriptionally active open complexes at the K.pneumoniae and S.meliloti nifH promoters was measured using supercoiled template DNA. The S.meliloti promoter was used as it provides the basis of many other assays of σ54 function (22,42–44). Two activators, A.vinelandii NifA and E.coli PspFΔHTH, were used to gauge activator-dependent transcription and to relate to the in vivo assays using NifA. Since PspFΔHTH lacks DNA binding activity, its use simplifies DNA gel shift assays (13). In transcription assays dGTP was added as the hydrolysable nucleotide used by the activator, to limit the reaction to formation of an open complex. Subsequently, heparin was added to destroy residual closed complexes and unstable open complexes, together with the four rNTPs to allow initiation and transcript elongation from the preformed stable open complex. Results of these single round transcription assays are shown in Figure 3. Refolded wild-type σ54 and L200A were able to perform activator-dependent transcription at the K.pneumoniae and S.meliloti nifH promoters, using either NifA or PspFΔHTH as activator. They produced transcripts to a level comparable to that of wild-type σ54. Mutant Q351R was defective for transcription in all the conditions assayed. Mutants I23N and S379F were unable to transcribe from the K.pneumoniae nifH promoter or from the S.meliloti nifH promoter using NifA as activator. Their activities recovered to 50% of the wild-type activity using S.meliloti nifH as template and PspFΔHTH as activator (Fig. 3D). In contrast, mutant S404F was capable of NifA-dependent transcription at the S.meliloti promoter (Fig. 3B). Mutant L200P showed promoter-specific behaviour with some decreased activity in certain transcription assays (compare Fig. 3B and D). Although some mutants showed low levels of in vivo transcription (Fig. 2), the combination of PspFΔHTH and the S.meliloti promoter allowed I23N, Q351R and S379F some increase in transcription levels in vitro (Fig. 3). Overall it appears that the purified mutant proteins have diminished activities in accord with the in vivo activities and protein levels. The results also show that the activities of the mutant σ54 proteins in the transcription assays varied with promoter and activator used, demonstrating that the magnitude of the defects in transcription measured can be specific to the combination of promoter and activator used.
Figure 3.
In vitro transcription activity of wild-type and mutant σ54 holoenzymes. Assays were repeated at least eight times to enhance reproducibility and activities were within 15% of the average shown. (A) At the K.pneumoniae nifH promoter in the presence of activator protein NifA at 100 nM. (B) At the S.meliloti nifH promoter in the presence of NifA at 100 nM. (C) At the K.pneumoniae nifH promoter in the presence of PspFΔHTH at 100 nM. (D) At the S.meliloti nifH promoter in the presence of PspFΔHTH at 100 nM.
DNA binding assays
We used a gel shift assay with S.meliloti nifH promoter DNA to measure σ and holoenzyme promoter binding. In order to determine the defects associated with each mutation, different templates were used to measure binding of σ and holoenzyme to DNA, as occurs on initial closed complex formation, in the closed complex and in the open complex.
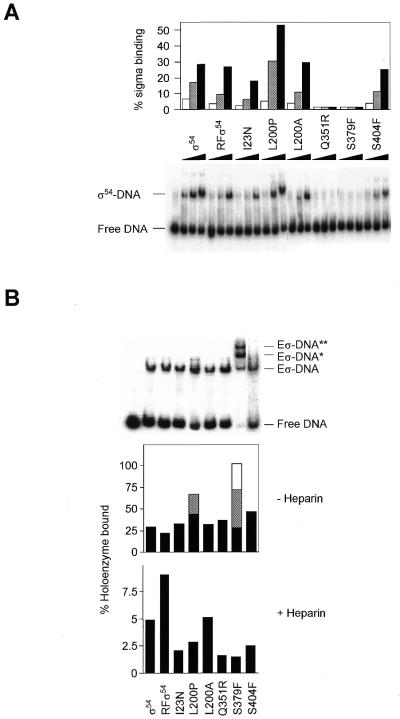
Sigma binding to double-stranded promoter DNA. Binding to the S.meliloti nifH promoter by the wild-type and mutant σ proteins was measured using double-stranded homoduplex DNA (12) to estimate their abilities to support normal closed complex formation (Fig. 4A). Refolded wild-type, L200A and S404F behaved like wild-type. I23N showed a modestly reduced ability to bind homoduplex DNA, but Q351R and S379F were greatly impaired for binding. With L200P binding appeared to be improved compared to wild-type. These results indicate that mutants I23N, Q351R, S379F and L200P have altered interactions with promoter DNA.
Figure 4.
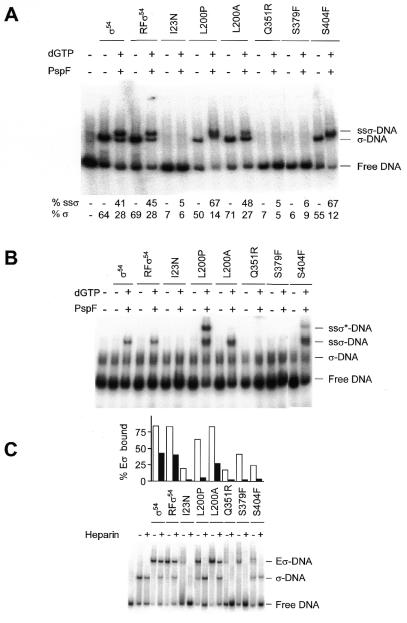
Binding of wild-type and mutant σ54 proteins and their holoenzymes to S.meliloti homoduplex promoter DNA. (A) A constant amount of homoduplex DNA (16 nM) was incubated with increasing amounts of σ54 proteins (0.5, 1 and 2 µM). The percentage of holoenzyme-bound DNA was quantified by phosphorimager analysis and plotted. (B) Gel mobility shift assay showing binding of the wild-type and mutant σ54 holoenzymes (Eσ 100 nM) to homoduplex DNA (16 nM) in the presence of PspFΔHTH (4 µM) and dGTP (4 mM) in the absence of heparin (top). The histograms below indicate the number of initial bound complexes (– Heparin) and those surviving a 5 min heparin challenge (+ Heparin, gel not shown). Black, grey and empty bars are additive and represent the Eσ–DNA, Eσ–DNA* and Eσ–DNA** complexes, respectively.
Holoenzyme binding to double-stranded promoter DNA. Amongst the mutants, only L200P and S379F behaved differently to the wild-type in their ability to form closed complexes at the double-stranded S.meliloti nifH promoter (Fig. 4B). In addition to the standard holoenzyme–DNA conformer, both mutants formed additional conformers with reduced mobilities. It seems that core RNA polymerase can recover some of the lost DNA binding activity seen in the σ–DNA binding assays, particularly for Q351R. In all cases the holoenzyme complexes were disrupted by addition of heparin, which is known to dissociate the σ54 holoenzyme closed complexes (data not shown; see also 10). Only in the presence of activator and a hydrolysable nucleoside triphosphate (dGTP) did heparin-stable complexes form (Fig. 4B). Under these conditions wild-type σ54 holoenzyme produced the most heparin-stable complexes. I23N, L200P, Q351R, S379F and S404F consistently formed fewer complexes stable to the heparin challenge. The pattern of reduced stable complex formation by mutants is similar to the activity patterns observed in in vitro transcription assays.
Sigma binding to early melted DNA. In closed promoter complexes a local σ54-dependent DNA opening occurs next to the GC promoter element (19). We judged the ability of the mutants to productively interact with DNA fragments containing this early melted DNA. Binding of wild-type and mutant σ54 proteins to an early melted S.meliloti nifH promoter was measured using heteroduplex DNA mismatched at –12/–11, next to the –14/–13 GC element (13,14). Refolded wild-type, L200A, L200P and S404F behaved like wild-type σ54 but I23N, Q351R and S379F were unable to bind (Fig. 5A). This result shows that I23N is distinct amongst the mutants in that it binds double-stranded DNA well (see Fig. 4 above) but fails to bind early melted DNA. In contrast, Q351R and S379F have defects with both template types. They may therefore have defects in a major overall DNA binding determinant.
Figure 5.
(A) Binding of wild-type and mutant σ54 proteins to S.meliloti –12/–11 heteroduplex promoter DNA and formation of supershifted complexes. Gel shift assays were conducted with 1 µM protein and 16 nM DNA, in the absence and presence of PspFΔHTH (4 µM) and dGTP (4 mM). The percentage of σ–DNA complex and the supershifted complex (ssσ–DNA) was quantified by phosphorimager analysis. (B) Binding of wild-type and mutant σ54 proteins to K.pneumoniae –12/–11 heteroduplex promoter DNA and formation of supershifted complexes (ssσ–DNA). Assays were conducted as in (A). (C) Holoenzyme mobility shift assays of the wild-type and mutant σ54 holoenzymes (Eσ, 100 nM) with S.meliloti –12/–11 heteroduplex promoter DNA (16 nM), represented by the white bars. Holoenzyme–DNA complexes were challenged with heparin (100 µg/ml) for 5 min prior to loading, represented by the grey bars.
Isomerisation of σ bound to early melted DNA. A stage in open complex formation can be studied without core subunits using DNA pre-opened next to the promoter –12 GC element, so called early melted DNA, σ54 and activator (13,14). σ54 bound to early melted DNA responds to activator in a nucleotide hydrolysis-dependent reaction to form a supershifted DNA complex (ssσ–DNA) in which the DNA has melted and the σ isomerised (13,14). In this assay refolded wild-type and L200A showed similar affinity for early melted DNA and similar efficiency of conversion to the isomerised ssσ–DNA complex using the activator protein PspFΔHTH and hydrolysable nucleotide dGTP (Fig. 5A). I23N, Q351R and S379F failed to bind the early melted DNA and ssσ–DNA complex was not detected after addition of activator and hydrolysable nucleotide (Fig. 5A). Although mutants L200P and S404F showed a similar affinity for early melted DNA to wild-type, the amount of ssσ–DNA complex formed was significantly greater. This same relationship was evident over a range of PspFΔHTH concentrations (data not shown).
A second set of assays using early melted K.pneumoniae nifH promoter, the activator protein PspFΔHTH and hydrolysable nucleotide dGTP confirmed and extended the results obtained with S.meliloti early melted DNA (Fig. 5B). The initial σ54–DNA complex with early melted K.pneumoniae nifH promoter DNA was difficult to detect, consistent with the known low binding of σ54 and its holoenzyme to this promoter (42). With σ54 at 10 µM an initial complex was evident (data not shown). However, under activating conditions with 1 µM σ54 the supershifted complex was evident, suggesting that conversion of the initial complex to the isomerised one is rapid. The mutants I23N, Q351R and S379F all failed to give any detectable supershifted complex (Fig. 5B), probably because of poor initial binding (Figs 4 and 5A). The L200P and S404F mutants behave differently on the K.pneumoniae DNA, compared to the S.meliloti early melted DNA (Fig. 5A): in addition to the standard ssσ–DNA complex seen with wild-type σ54 and L200A, L200P and S404F formed an additional conformer with reduced mobility (ssσ*–DNA). The slower moving activator-dependant complex formed with the L200P and S404F mutants and early melted K.pneumoniae nifH DNA could result from either a new σ conformation or from activator recruitment. To explore these possibilities assays were done using 32P-end-labelled PspFΔHTH and unlabelled early melted DNA plus the L200P and S404F mutants. No gel mobility shift of the activator was observed, suggesting that the L200P and S404F mutations result in a new activator-dependent change in complex formation rather than in stable recruitment of activator to the σ–DNA complex (data not shown).
Overall, the results with early melted DNA indicate a specific early melted DNA interaction defect in I23N and altered activator responses in L200P and S404F and confirm a strong DNA binding defect in Q351R and S379F.
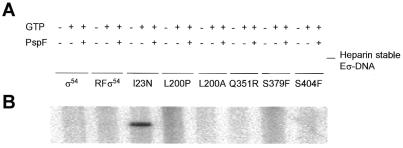
Holoenzyme binding to early melted DNA. To determine whether the mutant proteins would support formation of a closed complex on early melted DNA we next measured binding of holoenzymes to early melted DNA (Fig. 5C). As anticipated from the poor binding of I23N, Q351R and S379F, their holoenzymes bound poorly. Surprisingly, so did the holoenzyme formed with S404F, which alone bound early melted DNA well (Fig. 5A). Holoenzymes formed with L200P, L200A and wild-type σ54 all bound >60% of the DNA.
We also measured the stability of the holoenzyme–early melted DNA complexes in a heparin challenge assay (Fig. 5C). Wild-type σ54 holoenzyme binds early melted DNA to give a significant proportion of stable complexes after a 5 min challenge with heparin (14). I23N, L200P, Q351R, S379F and S404F formed holoenzyme complexes with early melted DNA that were significantly less resistant to a heparin challenge (Fig. 5C). In the case of I23N, Q351R and S379F formation of heparin-sensitive complexes correlates with poor binding of the mutant σ to early melted DNA (Fig. 5A). Mutants L200P and S404F are exceptions to this as both of these mutant σ54 proteins bind early melted DNA with similar efficiency to the wild-type. Note that in the heparin challenge a σ–DNA complex forms with the L200P and S404F mutants but not the I23N, Q351R or S379F mutants (Fig. 5C). The presence of a fainter σ–DNA band in the minus heparin lanes for these same two mutants and its virtual absence from the wild-type and L200A lanes further suggests a weaker interaction of S404F and L200P with core RNA polymerase in the assay.
Overall, the results with early melted DNA show that the amount of holoenzyme complex and its stability is reduced in mutants I23N, L200P, Q351R, S379F and S404F. The basis for the reductions seems different in that L200P and S404F are not themselves defective in binding early melted DNA whereas the other mutants are.
Holoenzyme and σ binding to late melted DNA. Binding of the wild-type and mutant σ proteins (without core) to the –10 to –1 opened DNA followed a pattern very similar to that found on double-stranded DNA (Fig. 4), except for S404F at 2 µM, which showed a 7-fold decrease in affinity for late melted DNA.
In open complexes formed with the σ54 holoenzyme, DNA is opened in the region from –10 to –1 (5,12). Mutants of σ54 deregulated for transcription can bind heteroduplex DNA templates opened from –10 to –1 independently of activator and nucleotide hydrolysis because the holoenzyme has isomerised (12,22,33). The holoenzyme formed with wild-type σ54 still requires activator and nucleotide hydrolysis to stably bind such templates (12). To determine whether any of the mutant σ54 proteins were deregulated or changed in their interactions with late melted DNA, the ability of the wild-type and mutant holoenzymes to bind a fully melted S.meliloti nifH promoter was measured using heteroduplex DNA with the top strand mismatched from –10 to –1 (12). All mutant holoenzymes bound late melted DNA with similar efficiency as did wild-type holoenzyme and this binding was largely unaffected by addition of activator and GTP (data not shown). However, differences were revealed with a heparin challenge to measure stability (Fig. 6A). As expected, activator and nucleotide were needed for the wild-type holoenzyme to make the greatest number of stable complexes. L200A and S379F behaved similarly. In contrast, I23N and, to a lesser extent, L200P, Q351R and S404F very weakly formed heparin-resistant holoenzymes on late melted DNA in the presence of GTP (the S.meliloti nifH mRNA starts 5′-GGG; see 22) but absence of activator. This result suggests that some activator-independent isomerisation of the holoenzyme occurred and was stabilised by GTP.
Figure 6.
Bypass activity of σ mutants. (A) Heparin-stable holoenzyme complexes. Binding of the wild-type and mutant σ54 holoenzymes (Eσ, 100 nM) to S.meliloti –10 to –1 heteroduplex DNA (16 nM) in the presence of PspFΔHTH (4 µM) and GTP (4 mM). Heparin (100 µg/ml) was added for 5 min before gel loading. (B) Activator-independent in vitro transcription assay of wild-type and mutant σ54 holoenzymes (100 nM) on the S.meliloti nifH promoter (pMKC28, 10 nM).
It seems that S404F has a specific defect in binding to late melted DNA which is corrected by core RNA polymerase. Overall, the results suggest that the conformation of the holoenzymes formed with the I23N, L200P and Q351R mutants are different to the wild-type holoenzyme and allow activator-independent interactions with DNA opened for –10 to –1.
Activator bypass transcription
We next explored whether the mutant σ proteins would support activator-independent transcription from a supercoiled DNA template. With this assay we are able to identify mutants that allow holoenzyme isomerisation without activator. The variation in the standard transcription activation assay employs GTP as the initiating nucleotide and omits activator. Heparin is added after initiation has occurred so as to disrupt unstable complexes, trap stable initiated complexes and limit transcription to a single round. The subsequent addition of the remaining three NTPs thus allows an estimate of the number of stable initiated complexes that have formed from activator-independent open complexes (Fig. 6B). The DNA binding assays with –10 to –1 opened DNA indicated that the I23N, L200P and Q351R mutants were candidates for producing activator-independent transcripts, however, only the I23N holoenzyme produced them. Judged by the lower level of detection, all other mutants and the wild-type holoenzyme formed no more than 50-fold fewer transcripts. The failure of Q351R to support activator-independent transcription may reflect formation of fewer closed complexes or the inability of the holoenzyme to stabilise open DNA unless activated. The failure of L200P may relate to defects in σ interactions with core RNA polymerase.
DISCUSSION
We have identified five new single amino acid substitutions important for σ54 activity. Our in vitro assays clearly show that several of the mutants alter steps after the initial closed complex forms. Previously, the role of σ54 in the activator-dependent initiation process has been explored using directed mutagenesis of motifs or through random mutagenesis of a restricted set of segments of rpoN (15–18,26–31). In many cases multiple substitutions were required to observe a phenotype and conserved residues proved to be surprisingly tolerant to mutation. Further, inferences that certain residues were important for function were drawn from the apparent non-mutability of particular sequences using a statistical analysis of mutation data (27). In our work altered σ54 proteins defective in supporting growth on arginine and in transcription from the K.pneumoniae nifH promoter were obtained by random mutagenesis of rpoN. The amino acid substitutions leading to defects in σ54 functioning fell in conserved Regions I and III of σ54. Since Region II is largely dispensable for σ54 function and deletions and insertions in Region II lead to modest defects in functionality (45,46), it is likely that there are very few Region II positions where disruptive single amino acid substitutions can occur. Differences in activities we have not directly measured in vitro, such as promoter escape, σ release and rebinding, may also contribute to the lower levels of transcription seen with the mutants in vivo. Some reduction in σ54 holoenzyme levels through reduced σ54 levels are also possible (Fig. 1A), but we note that reduced levels of σ54 are tolerated in in vivo promoter activation assays. Below we discuss the activities of the mutants we obtained in Regions I and III.
Region I mutant I23N
A number of mutagenesis studies have shown that sequences between residues 15 and 47 in Region I are important for the full function of σ54, but few single substitution mutants have been obtained or characterised and, where they have, these are clustered between residues 25 and 37 (15–17,31,33). Our results show that the I23N mutation results in diminished transcription in response to activator and the acquisition of an activator-independent transcription activity. This result agrees with a patch mutagenesis in which a mutant containing three contiguous alanine substitutions at positions 21–23 showed similar properties (14,33). Mutant I23N is specifically defective in binding to a DNA structure representing the early DNA opening found in closed complexes. This and other evidence leads to the view that the conformational stability of the closed complex is diminished, allowing the I23N holoenzyme to isomerise and engage with melted DNA so as to transcribe without activator and to form stable complexes on heteroduplex templates where DNA is opened from –10 to –1. The defects in activator-dependent transcription can be traced to a reduced activity in the σ isomerisation assay. It seems that I23N contributes both to limiting isomerisation in the absence of activator and to activator responsiveness.
Region III mutant L200P
The L200P mutation may be structurally damaging since the L200A mutation was indistinguishable from the wild-type in all assays applied to it. Probably, L200P disrupts only a local structure since it retained many of the usual activities of σ54. Assays of core polymerase binding by L200P indicated disruption of a structure that contributes to binding to the core polymerase. We note that L200 is within the 120–215 fragment of σ54 that binds core polymerase with an affinity comparable to that of intact σ54 and that an Fe-BABE form of σ54 derivatised at position 198 is active for cleaving core polymerase (10,24). Many of the defects of the L200P holoenzyme can be rationalised in terms of an altered binding of σ54 to core RNA polymerase. However, L200P also showed altered behaviour in the σ isomerisation assays (evident in formation of a new conformer and increased formation of the isomerised product), suggesting a role for the core binding surfaces of σ54 at steps other than initial formation of the holoenzyme. The defects in open complex formation of the L200P holoenzyme may reflect altered core polymerase to σ contacts important for progression to the open complex where changing interactions between σ54 and core polymerase are envisaged. A mutant L199P of Salmonella typhimiuruim σ54 (31) showed a related set of defects and also led to the conclusion that the sequence in σ54 contributed to a set of interactions after closed complex formation. The results with σ54 are fully consistent with the emerging view that the interface of σ with core polymerase is extensive and functionally specialised (10,23). We note that L200P lies within a domain of σ54 that influences DNA binding (9). Altered properties of L200P evident in DNA binding assays are consistent with this.
Region III mutant Q351R
The Q351R mutant showed a clear strong and specific defect in overall DNA binding, but core polymerase binding appeared unaffected. Q351 clearly falls within the minimal functional DNA-binding domain of σ54 (residues 329–477; see also 8). Although previously suggested to be unimportant for DNA binding (27), it appears that Q351 makes an important contribution to the activity of the DNA-binding domain of σ54. The properties of the Q351R holoenzyme are readily understood in terms of diminished DNA binding by mutant σ54 restricting levels of closed complex formation. In experiments where the solvent accessibility of σ54 was probed using hydroxyl radical cleavage several protected regions were identified in closed complexes. Strongest DNA-dependent protection from cleavage was at residues 362–386 and 397–432, less strong at residues 305–315 and 325–335 (21). It seems that Q351R could interfere with DNA binding through changing the activity of these or other DNA interacting sequences rather than establishing a direct DNA contact itself. We note that Q350 is implicated in DNA binding by statistical analysis of mutant data (27). Interestingly, Q351R holoenzyme was able to form some heparin-stable complexes independently of activator if the DNA was opened from –10 to –1. This suggests that holoenzyme isomerisation was partly accomplished. Reduced promoter occupancy and/or dissociation of the closed complex on a time scale faster than holoenzyme isomerisation could further explain the transcription defects in Q351R.
Region III mutant S379F
Two defects were evident in the S379F mutant: one appeared to relate to aggregation, preventing the mutant protein from running into native gels, the other to a marked defect in overall DNA binding. Protein footprints showed that in the closed complex σ54 was protected from hydroxyl radical cleavage at positions 362–386, consistent with a DNA contact being made in the 379 region (21). S379 falls within the helix–turn–helix motif of σ54 where certain mutations are known to reduce promoter occupancy (26,28). Hydrophobic substitutions at position 379 led to diminished transcription in vivo, but the effects of substitution by Phe were not reported. S379F holoenzyme showed reduced promoter DNA binding and did not allow activator-independent transcription. It seems that properties of the S379F mutation are in accord with a role for 379 in setting promoter occupancy rather than restricting polymerase isomerisation in the absence of activator. As discussed for Q351R above, reduced DNA binding by S379F holoenzyme could also contribute to reduced levels of transcripts.
Region III mutant S404F
The defects in transcription seen with the S404F mutant were clearly due to a step(s) after initial DNA binding since the S404F mutant bound double-stranded DNA and core polymerase well. However, the stability of the S404F holoenzyme on early melted DNA was compromised, but binding of S404F σ54 to the same DNA was not. Moreover, activator-dependent isomerisation of S404F bound to early melted DNA occurred with an efficiency at least as great as that of the wild-type σ54. It seems that S404 is important for an early step in the activator-dependent initiation pathway, which is associated with the interaction the holoenzyme makes with early melted DNA. It seems likely that S404 contributes to holoenzyme isomerisation through being part of the interface between σ54 and core polymerase that is operational when early melted DNA forms. Consistent with this, an altered behaviour of S404F in the σ isomerisation assay was evident, with formation of a new conformer and increased formation of the isomerised product on early melted DNA templates, as also seen with L200P (Fig. 5B).
S404 is within a conserved part of the DNA-binding domain, where either Ser or Thr are found adjacent to two invariant Phe residues at positions 402 and 403. Prior directed mutagenesis showed that F402 and F403 were important for holoenzyme function and led to the suggestion that residues 402 and 403 contributed to productive interactions with early melted DNA (47). Results obtained with S404F extend this view by showing that the DNA binding activity to a number of different DNA templates, including early melted DNA, is intact, but that holoenzyme stability is changed on early melted DNA. The changed stability does not lead to activator-independent open complex formation or engagement with melted DNA, suggesting that the inhibitory properties of Region I and R336 that prevent activator bypass activities are largely intact in the S404F mutant (22,47). Although S404F holoenzyme produced a low level of activator-independent, heparin-stable complex on heteroduplex DNA opened from –10 to –1, bypass transcription was not evident (Fig. 6). It seems that S404F might not be able to sufficiently overcome the restriction of isomerisation imposed by its efficient binding to the early melted DNA that forms in closed complexes. Poor binding to early melted DNA, as occurs with I23N, seems necessary to allow sufficient isomerisation for a productive interaction with the transiently melted DNA forming on the supercoiled template (Figs 5 and 6; see also 14). However, S404F alone bound late melted DNA poorly. This suggests that S404F may also be defective in interactions closely associated with late DNA opening, needed for unregulated open complex formation. Protein footprints of σ54 implicate residues between positions 397 and 432 in interactions with core polymerase and with promoter DNA that also involve regulatory Region I (21,23). The close relationship between core-dependent protections seen in the absence of Region I and in the presence of Region I plus promoter DNA suggests that some activities of Region I and the 402–404 patch are linked. The phenotypes of holoenzymes assembled with σ54 mutant at position 402, 403 or 404 are consistent with a defect at an early step in initiation that requires the functionality of Region I (47; this work). This functionality seems to be Region I-directed binding of σ54 to early melted DNA that allows the holoenzyme to assume a heparin-stable complex on such DNA structures (13,14,20).
Holoenzyme functionality
Overall, our results help identify single amino acids important for σ54 function. In several of the mutants more than one functional defect was evident and some of these were shared by other mutants, arguing that several sequences in σ54 interact for its full function. Probably a large fraction of holoenzyme functioning involves σ54 exchanging information about promoter structure with the core subunits via an extensive set of core–σ54 and σ54–DNA interfaces. The L200P and S404F mutants have altered σ isomerisation properties and some altered holoenzyme properties, suggesting that they could have changes in a communication pathway that links changes in σ to interactions with core polymerase. Current models for activation of the σ54 holoenzyme suggest that the activator engages σ54 bound to early melted DNA as created in closed complexes by the σ54 holoenzyme (13,14,19). A network of interactions involving Region I, core polymerase and DNA contacts is then changed to allow isomerisation of σ54 and holoenzyme and binding to melted DNA. Mutants in σ54 with defects in activator contact could have the phenotype of binding to early melted DNA but failing in isomerisation. No such mutants were found in our screen. Further, none of our mutants were differentially rescued by increasing amounts of activator in the isomerisation assay. Although the mutagenesis was clearly not saturating, it is possible that residues in σ54 that contact the activator have several roles, therefore complicating their identification. Activators of the σ54 holoenzyme lack motifs found in helicases, indicating that activation is via a contact to σ54 rather than through creation of a favourable DNA structure for σ54 binding.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by funding from the BBSRC to M.B. M.T.G. received a CEC TMR fellowship. We thank M.J. Merrick for pMM83 and UNF2972, D. Studholme, M. Chaney and J. Shumacher for their comments on the manuscript and W. Cannon for oligonucleotides.
REFERENCES
- 1.Gross C.A., Chan,C., Dombroski,A., Gruber,T., Sharp,M., Tupy,J. and Young,B. (1998) Cold Spring Harbor Symp. Quant. Biol., 63, 141–155. [DOI] [PubMed] [Google Scholar]
- 2.Lonetto M., Gribskov,M. and Gross,C.A. (1992) J. Bacteriol., 174, 3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrick M.J. (1993) Mol. Microbiol., 10, 903–909. [DOI] [PubMed] [Google Scholar]
- 4.Popham D.L., Szeto,D., Keener,J. and Kustu,S. (1989) Science, 243, 629–635. [DOI] [PubMed] [Google Scholar]
- 5.Wedel A. and Kustu,S. (1995) Genes Dev., 9, 2042–2052. [DOI] [PubMed] [Google Scholar]
- 6.Sasse-Dwight S. and Gralla,J.D. (1990) Cell, 62, 945–954. [DOI] [PubMed] [Google Scholar]
- 7.Studholme D.J. and Buck,M. (2000) FEMS Microbiol. Lett., 186, 1–9. [DOI] [PubMed] [Google Scholar]
- 8.Cannon W., Missailidis,S., Smith,C., Cottier,A., Austin,S., Moore,M. and Buck,M. (1995) J. Mol. Biol., 248, 781–803. [DOI] [PubMed] [Google Scholar]
- 9.Cannon W.V., Chaney,M.K., Wang,X. and Buck,M. (1997) Proc. Natl Acad. Sci. USA, 94, 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallegos M.T. and Buck,M. (1999) J. Mol. Biol., 288, 539–553. [DOI] [PubMed] [Google Scholar]
- 11.Wong C., Tintut,Y. and Gralla,J.D. (1994). J. Mol. Biol., 236, 81–90. [DOI] [PubMed] [Google Scholar]
- 12.Cannon W., Gallegos,M.T., Casaz,P. and Buck,M. (1999) Genes Dev., 13, 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon W., Gallegos,M.T. and Buck,M. (2000) Nature Struct. Biol., 7, 594–601. [DOI] [PubMed] [Google Scholar]
- 14.Gallegos M.T. and Buck,M. (2000) J. Mol. Biol., 297, 849–859. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh M. and Gralla,J.D. (1994) J. Mol. Biol., 239, 15–24. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh M., Tintut,Y. and Gralla,J.D. (1994) J. Biol. Chem., 269, 373–378. [PubMed] [Google Scholar]
- 17.Syed A. and Gralla,J.D. (1998) J. Bacteriol., 180, 5619–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J.T., Syed,A. and Gralla,J.D. (1997) Proc. Natl Acad. Sci. USA, 94, 9538–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris L., Cannon,W., Claverie-Martin,F., Austin,S. and Buck,M. (1994) J. Biol. Chem., 269, 11563–11571. [PubMed] [Google Scholar]
- 20.Guo Y., Wang,L. and Gralla,J.D. (1999) EMBO J., 18, 3736–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casaz P. and Buck,M. (1999) J. Mol. Biol., 285, 507–514. [DOI] [PubMed] [Google Scholar]
- 22.Chaney M. and Buck,M. (1999) Mol. Microbiol., 33, 1200–1209. [DOI] [PubMed] [Google Scholar]
- 23.Casaz P. and Buck,M. (1997) Proc. Natl Acad. Sci. USA, 94, 12145–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigneshweraraj S.R., Fujita,N., Ishihama,A. and Buck,M. (2000) EMBO J., 19, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp M.M., Chan,C.L., Lu,C.Z., Marr,M.T., Nechaev,S., Merritt,W., Severinov,K., Roberts,J.W. and Gross,C.A. (1999) Genes Dev., 13, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coppard J.R. and Merrick,M.J. (1991) Mol. Microbiol., 5, 1309–1317. [DOI] [PubMed] [Google Scholar]
- 27.Guo Y. and Gralla,J.D. (1997) J. Bacteriol., 179, 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrick M.J. and Chambers,S. (1992) J. Bacteriol., 174, 7221–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tintut Y., Wong,C., Jiang,Y., Hsieh,M. and Gralla,J.D. (1994) Proc. Natl Acad. Sci. USA, 91, 2120–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tintut Y. and Gralla,J.D. (1995) J. Bacteriol., 177, 5818–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly M.T. and Hoover,T.R. (1999) J. Bacteriol., 181, 3351–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Casaz P., Gallegos,M. and Buck,M. (1999) J. Mol. Biol., 292, 229–239. [DOI] [PubMed] [Google Scholar]
- 34.Jishage M., Iwata,A., Ueda,S. and Ishihama,A. (1996) J. Bacteriol., 178, 5447–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck M., Khan,H. and Dixon,R. (1985) Nucleic Acids Res., 13, 7621–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck M. and Cannon,W. (1989) Nucleic Acids Res., 17, 2597–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon R., Eady,R.R., Espin,G., Hill,S., Iaccarino,M., Kahn,D. and Merrick,M. (1980) Nature, 286, 128–132. [DOI] [PubMed] [Google Scholar]
- 38.Wassem R., de Sousa,E.M., Yates,M.G., de Oliviera Pedrosa,F. and Buck,M. (2000) Mol. Microbiol., 35, 756–764. [DOI] [PubMed] [Google Scholar]
- 39.Austin S., Buck,M., Cannon,W., Eydmann,T. and Dixon,R. (1994) J. Bacteriol., 176, 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrick M.J. and Edwards,R.A. (1995) Microbiol. Rev., 59, 604–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buck M., Gallegos,M., Studholme,D.J., Guo,Y. and Gralla,J. (2000) J. Bacteriol., 182, 4129–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck M. and Cannon,W. (1992) Nature, 358, 422–424. [DOI] [PubMed] [Google Scholar]
- 43.Gallegos M.T., Cannon,W. and Buck,M. (1999) J. Biol. Chem., 274, 25285–25290. [DOI] [PubMed] [Google Scholar]
- 44.Oguiza J.A. and Buck,M. (1997). Mol. Microbiol., 26, 655–664. [DOI] [PubMed] [Google Scholar]
- 45.Wong C. and Gralla,J.D. (1992) J. Biol. Chem., 267, 24762–24768. [PubMed] [Google Scholar]
- 46.Cannon W., Chaney,M. and Buck,M. (1999) Nucleic Acids Res., 27, 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oguiza J.A., Gallegos,M.T., Chaney,M.K., Cannon,W.V. and Buck,M. (1999) Mol. Microbiol., 33, 873–885. [DOI] [PubMed] [Google Scholar]