Abstract
Breast cancer is a malignant tumor that poses a serious threat to women's health, and vasculogenic mimicry (VM) is strongly associated with bad prognosis in breast cancer. However, the relationship between VM and immune infiltration in breast cancer and the underlying mechanisms have not been fully studied. On the basis of the Cancer Genome Atlas (TCGA), Fudan University Shanghai Cancer Center (FUSCC) database, GSCALite database, and gene set enrichment analysis (GSEA) datasets, we investigated the potential involvement of VM-related genes in the development and progression of breast cancer. We analyzed the differential expression, mutation status, methylation status, drug sensitivity, tumor mutation burden (TMB), microsatellite instability (MSI), immune checkpoints, tumor microenvironment (TME), and immune cell infiltration levels associated with VM-related genes in breast cancer. We created two VM subclusters out of breast cancer patients using consensus clustering, and discovered that patients in Cluster 1 had better survival outcomes compared to those in Cluster 2. The infiltration levels of T cells CD4 memory resting and T cells CD8 were higher in Cluster 1, indicating an immune-active state in this cluster. Additionally, we selected three prognostic genes (LAMC2, PIK3CA, and TFPI2) using Lasso, univariate, and multivariate Cox regression and constructed a risk model, which was validated in an external dataset. The prognosis of patients is strongly correlated with aberrant expression of VM-related genes, which advances our knowledge of the tumor immune milieu and enables us to identify previously unidentified breast cancer subtypes. This could direct more potent immunotherapy approaches.
Keywords: Vasculogenic mimicry, Immune microenvironment, Consensus clustering, LAMC2, PIK3CA, TFPI2
1. Introduction
Breast cancer is the most common malignant tumor in women and a severe threat to their physical and mental health, accounting for the majority of all female cancers [1]. Despite improvements in treatment options, breast cancer still often spreads and returns. A key factor in the growth and spread of malignancies is angiogenesis [2]. In contrast to traditional tumor angiogenesis, Vasculogenic Mimicry (VM) is a unique pattern of perfusion created by the tumor cells themselves. The notion of VM was introduced by Maniotis et al., in 1999 within the context of their investigation of invasive uveal melanoma [3]. Further studies have revealed the presence of VM in several malignant neoplasms, including hepatocellular carcinoma [4], colorectal cancer [5], lung cancer [6,7], prostate cancer [8], and breast cancer [9]. VM not only provides the necessary nutrients for tumor growth but is also closely related to the tumor immune microenvironment. It has the ability to regulate the immune infiltration status of the tumor microenvironment, although its underlying mechanisms are still unclear [[10], [11], [12]].
In breast cancer, VM formation is related to epithelial-to-mesenchymal transition (EMT) markers and stemness [13]. Muhammad et al. conducted a study that revealed the promotion of VM in breast cancer cells by IL-1β through the activation of the p38/MAPK and PI3K/Akt signaling pathways [14]. Many malignancies have a poor prognosis because VM might foster an environment that is conducive to tumor growth and spread [10,15]. Targeting VM may provide a novel strategy for the treatment of breast cancer. The inhibition of vasculogenic mimicry in breast cancer cells generated by 3D Matrigel is observed when the endoplasmic reticulum stress. This inhibition occurs through the involvement of the TGF-β3/Smad1/2 and β-catenin signaling pathways, resulting in the suppression of tumor development [16]. It has been reported that paclitaxel and matrine, natural compounds with anti-tumor properties, can inhibit VM. Paclitaxel inhibits VM via boosting the proteasome-mediated degradation of EGFR protein, which decreases its level [17]. By suppressing the expression of EphA2, MMP-9, and MMP-2, marrine decreases the production of VM in MDA-MB-231 cells [18]. Since there aren't any specific medications available that target VM in breast cancer, research into the connection between VM and tumor immunity could progress immunotherapy and targeted therapy for the disease.
In this study, we conducted a systematic analysis of VM-related genes, revealing their crucial clinical significance. Furthermore, we utilized VM-related genes to identify unknown subtypes in TCGA data and analyzed the differences in immune checkpoints and immune infiltration among different VM clusters. By employing LASSO and Cox regression algorithms, we screened for prognosis-related genes associated with VM and constructed a risk model. Our research findings reveal the potential correlation between VM and the breast cancer immune microenvironment, providing new evidence for identifying novel prognostic biomarkers for breast cancer immunotherapy. This provides a new perspective on potential treatment targets and strategies for breast cancer.
2. Materials and methods
2.1. Data sets and preparation of VM-related genes
The Cancer Genome Atlas (TCGA) RNA-Seq data, clinical data, and survival data of 1104 patients with breast invasive carcinoma (BRCA) used in this study were downloaded from the UCSC Xena database. The validation set used RNA-Seq data and survival data from 359 triple-negative breast cancer patients obtained from the Fudan University Shanghai Cancer Center (FUSCC) database [19]. The mutation data were obtained from the cBioPortal database (https://www.cbioportal.org/). The genes related with VM were obtained from several sources, including the GSEA gene set, KEGG and relevant review articles.
2.2. Gene set cancer analysis
GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/) is a website for cancer genome analysis for dynamic analysis and visualization of cancer gene sets and drug susceptibility correlations. We used the GSCALite database to analyze genomic features of the VM gene set, the link between gene expression and drug sensitivity, correlation of gene expression with drug sensitivity, and methylation status.
2.3. Survival analysis
We conducted Kaplan-Meier analysis to assess survival differences between different groups using the “survival” and “survminer” R packages, and utilized Cox regression analysis to select genes significantly associated with patient survival.
2.4. Consensus clustering of VM-related model genes
VM-associated genes were used as model genes for consensus clustering (CC) using the “ConsensusClusterPlus” package to categorize TCGA-BRCA cases into 2 subclusters. In order to get the best clustering effect, the parameter “maxK (the maximum K value for cluster analysis)" of the CC algorithm was set to a value of 10 (k = 2–10), the clustering algorithm “pam” was used, and the distance measurement method between data points was set to “spearman".
2.5. Tumor immune microenvironment analysis
The R package “estimate” was utilized to compute ImmuneScore, StromalScore, and ESTIMATEScore, which provide an indication of the relative abundance of immune cells and stromal cells inside the tumor microenvironment. ESTIMATEScore is derived from the summation of ImmuneScore and StromalScore.
CIBERSORT is an algorithm based on gene expression profiles, used to infer the relative abundance of different types of immune cells in tumor samples. It estimates the relative content of different immune cell subtypes by analyzing the correlation between gene expression patterns in the sample and predefined immune cell gene signature profiles. We used the CIBERSORT algorithm to calculate the relative proportions of 22 immune cell types in breast cancer samples from the TCGA database. We also compared the expression differences of the immune checkpoint genes CD274, CTLA4, PDCD1LG2, PDCD1, HAVCR2, LAG3, SIGLEC15 and TIGIT between the two subgroups.
2.6. Tumor mutation burden (TMB) and microsatellite instability (MSI)
The TMB, or tumor mutational burden, is a quantitative biomarker that quantifies the amount of mutations present in tumor cells. It is now employed as a contemporary marker for assessing the efficacy of PD-1 antibody therapy. Microsatellite instability (MSI) arises due to functional abnormalities in the DNA mismatch repair system, and it serves as a significant tumor marker in clinical practice. The TMB and MSI data of TCGA-BRCA cases in this study were acquired from a Chinese website ("https://www.aclbi.com/static/index.html#/") and subsequently visualized using the R package “ggplot2” for data visualization purposes.
2.7. Construction and validation of the VMscore
Through least absolute shrinkage and selection operator (Lasso) analysis, 12 genes that significantly affect OS were identified from the VM-related gene set, and 3 genes were finally identified by univariate and multivariate Cox regression analysis for the construction of VMscore. The VMscore for each sample is calculated by multiplying the expression value of the gene by the weights in the multivariate Cox model and then adding them together. Univariate and multivariate Cox regression analyses were then used to verify whether VMscore was independent of other prognostic factors (age, tumor stage and other relevant clinical variables). We used ROC curve and AUC value as indicators to test the performance of VMscore model, and KM curve to show the change of survival probability of high-risk group and low-risk group over time.
2.8. Functional enrichment analysis
The “clusterProfiler” R software was employed to ascertain biological pathways associated with differential gene enrichment in VM gene sets, as well as in high and low risk index groups.
We used the “limma” software with criteria |log2(FC)| > 1 and p-value <0.05 to find DEGs in various VM subgroups as well as high and low risk index groups.
GSEA analysis was used to explore biosignaling pathways in different VM subgroups.
2.9. Single-cell analysis
We performed single-cell analyses based on the Tumor Immunology Single Cell Center (TISCH) databases GSE114727 and GSE138536 to explore cell types in which LAMC2, PIK3CA, and TFPI2 may be expressed.
3. Results
3.1. VM-related gene expression levels and functional enrichment
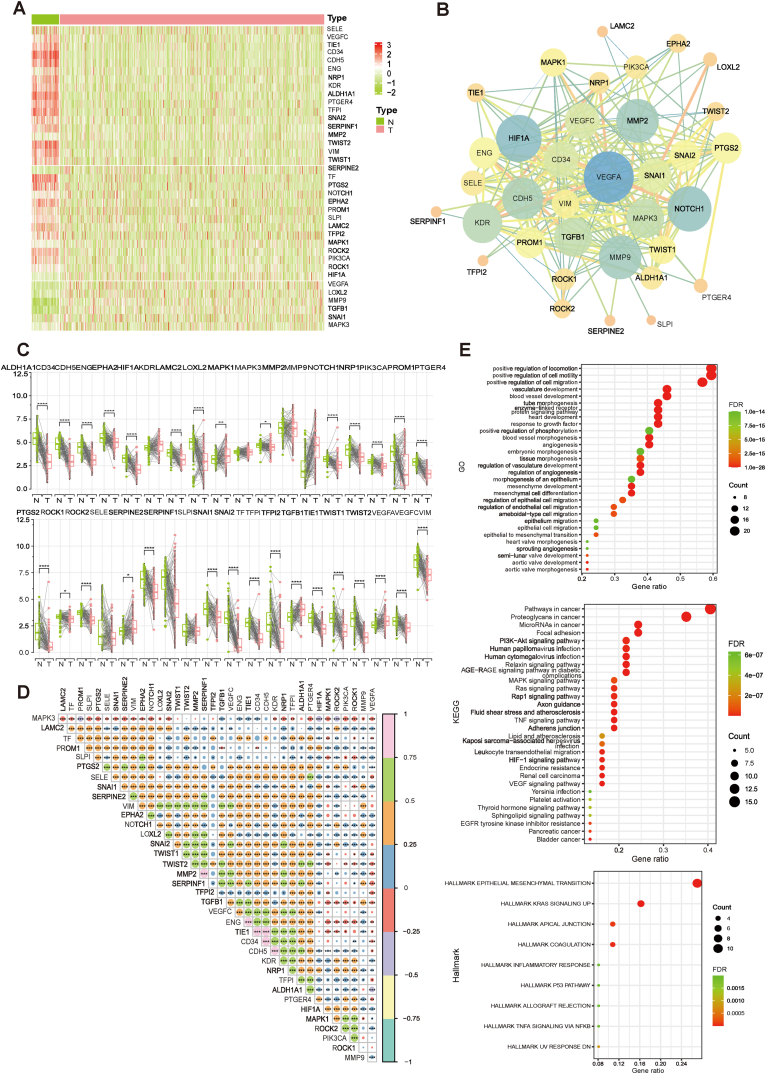
The research flowchart is shown in Fig. S1A. First, we analyzed the expression levels of 37 VM related genes in tumor and normal samples. Heatmaps were drawn and VM genes were found to be generally low-expressed in breast cancer (Fig. 1A). Protein-protein interaction (PPI) analysis was performed by the STRING tool to reveal the interaction of VM genes, and the results indicated that VEGFA, NOTCH1, MMP2, MMP9, HIF1A, FDR, and CDH5 were hub genes (Fig. 1B).
Fig. 1.
Expression of the VM genes in breast cancer. (A) Heatmap of expression levels of VM-related genes between breast cancer and normal tissue. (B) PPI network of VM genes. (C) Differential expression of VM genes between breast cancer and paired normal tissues. (D) Correlation between mRNA expression levels of VM genes (p < 0.05 *; p < 0.01 **; p < 0.001 ***). (E) GO, KEGG and Hallmark enrichment analysis based on VM genes.
Thirty genes were determined to be significantly differently expressed in BRCA versus normal tissues based on the mRNA data of paired tumor and normal samples contained in TCGA-BRCA (Fig. 1C).The correlation between the expression levels of VM genes is shown in Fig. 1D, which showed that most of the VM genes had a significant co-expression relationship (P < 0.05). In addition, kyoto encyclopedia of genes and genomes (KEGG), Gene Ontology (GO), and hallmaker enrichment analyses indicated that VM genes were involved in multiple biological pathways, such as blood vessel development, blood vessel morphogenesis, PI3K-Akt, MAPK, and TNF pathways (Fig. 1E).
3.2. Genome-wide analysis of VM genes
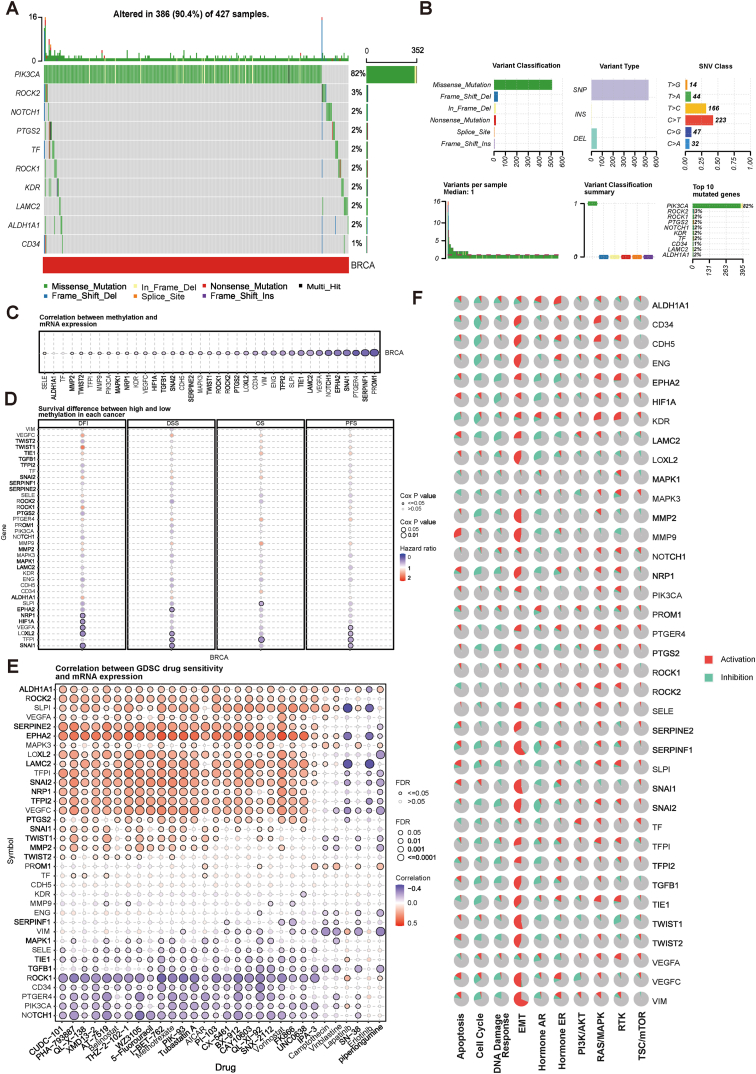
VM genes were analyzed in the GSCALite database, and the results of the genome-wide analysis showed that 10 out of 37 VM genes had mutations, including PIK3CA, ROCK2, NOTCH1, PTGS2, TF, ROCK1, KDR, LAMC2, ALDH1A1, and CD34, of which PIK3CA had the highest mutation frequency and its mutation type was dominated by Missense_Mutation, followed by ROCK1, and CD34 had the lowest mutation frequency (Fig. 2A and B). PIK3CA had the highest mutation frequency and its mutation type was dominated by Missense_Mutation, followed by ROCK1, while CD34 had the lowest mutation frequency (Fig. 2A and B). Methylation of VM genes and mRNA expression all showed negative correlation (Fig. 2C). The results showed that the methylation status of SLPI, EPHA2, NRP1, HIF1A, VEGFA, LOXL2, TFPI, and SNAI1 was negatively correlated with survival (Fig. 2D).
Fig. 2.
Genetic mutation mapping of the VM genes. (A-B) Mutation frequency of the VM genes. (C) Correlation between VM genes methylation and mRNA expression. (D) Survival differences between hypermethylation and hypomethylation of the VM genes in breast cancer. (E) Correlation between GDSC drug sensitivity and mRNA expression of the VM genes. (F) Activation or inhibition of 10 major tumor-related pathways by VM gene expression levels.
We analyzed the correlation between drug sensitivity and mRNA expression in the GDSC database and showed that ALDH1A1, ROCK2, SLPI, VEGFA, SERPINE2, EPHA2, MAPK3, LOXL2, LAMC2, TFPI, SNAI2, NRP1, TFPI2, and VEGFC were positively correlated with most drugs and ROCK1, CD34, PTGER4, PIK3CA and NOTCH1 were negatively associated with most drugs (Fig. 2E). Tumor-associated pathway activity analysis revealed that VM genes were significantly associated with EMT pathway activation, especially SERPINF1, SNAI1, TWIST1, and VIM showed more than 50 % activation (Fig. 2F).
3.3. Identification and analysis of VM gene clusters in breast cancer
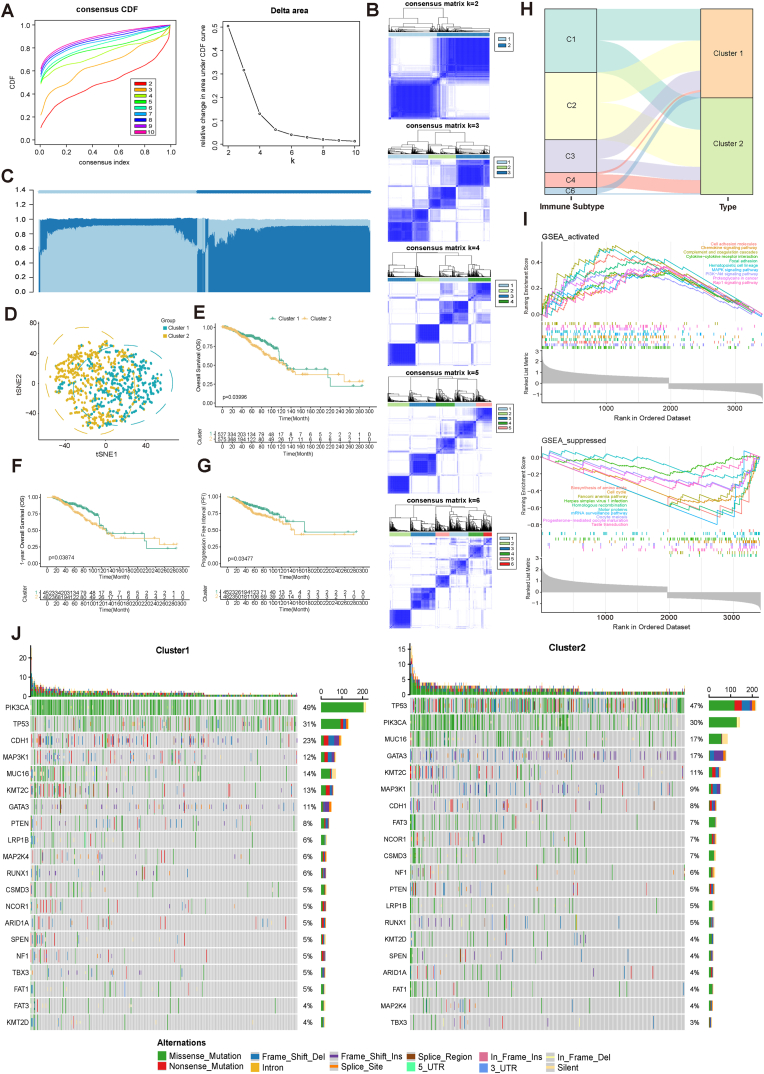
We performed consensus clustering analysis using VM genes as model genes to explore unknown subtypes of breast cancer (Fig. 3A). Based on the “ConsensusClusterPlus” R package, all TCGA-BRCA cases were categorized into K subclusters (K = 2–10). Based on the analysis results, it was finally determined that K = 2, which indicated a more significant difference between the two subclusters (Cluster 1 and Cluster 2) of 1104 breast cancer patients (Fig. 3B and C and Table S1). t-SNE downscaling classification results showed that this classification could effectively differentiate between the two subclusters of BRCA patients (Fig. 3D).
Fig. 3.
VM subtypes and characterization of the two subclusters. (A) Cumulative distribution function (CDF) curves in consensus cluster analysis. CDF curves are shown for consensus scores for different numbers of subtypes (k = 2–10). The relative change in the area under the CDF curve for k = 2–10 is also shown. (B) Consensus score matrix for all samples when k = 2–6. (C) Sample distribution of the consensus matrix for k = 2. (D) The t-SNE analysis showed differences in the transcriptome between the two subclusters. (E-G) OS, OS (1-year), and PFI survival analysis of two subclusters. (H) Distribution of different immune subtypes in two subclusters. (I) GSEA enrichment analysis of two subclusters differential genes. (J) Genomic characterization of high-frequency mutant genes in two breast cancer subgroups.
We performed Kaplan-Meier survival analysis of patients in both subclusters and found that Cluster 2 patients had worse OS, PFI relative to Cluster 1 patients (Fig. 3E–G).
Thorsson et al. [20] conducted an extensive immunogenomic analysis of 33 different cancer types using data collected by TCGA. Six immune subtypes were ultimately identified, including: wound healing, IFN-γ dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGF-β dominant. We analyzed these six subtypes in Cluster 1 and Cluster 2, and it can be seen that the lymphocyte depleted type is more distributed in Cluster 2, the TGF-β dominant type is more in Cluster 1, and the rest of the subtypes are evenly distributed in both groups (Fig. 3H).
We explored the characterization of the differential genes between Cluster 1 and Cluster 2 and their associated signaling pathways, and we observed that the differential genes of both subclusters showed activation in Chemokine, MAPK, PI3K-Akt, and Rap1 signaling pathways, but inhibition in Biosynthesis of amino acids and Cell cycle showed inhibition (Fig. 3I). The 20 genes with the highest mutation frequencies in BRCA were subjected to analysis, and subsequently, the mutation frequencies of these 20 genes were compared between two subclusters (Fig. 3J). It was observed that FAT3 had a higher frequency of mutations in Cluster2 compared to Cluster1 (Fig. 3J).
3.4. Impact of VM genes on the tumor immune microenvironment
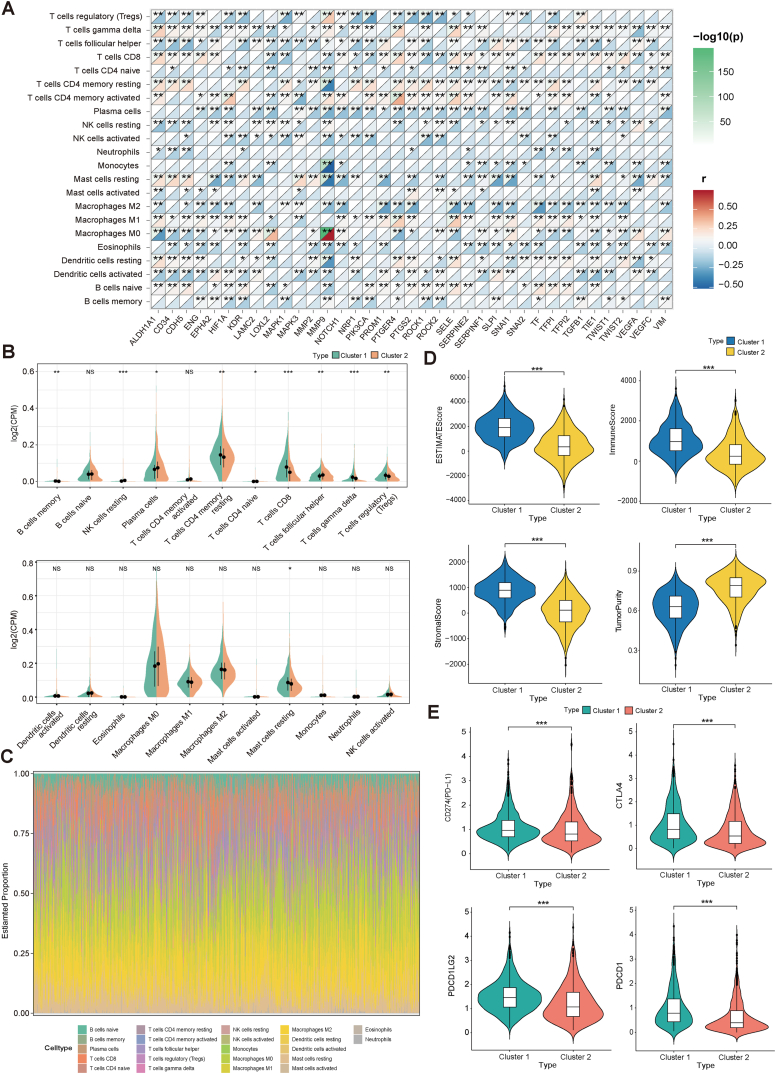
The CIBERSORT algorithm was used to evaluate the extent of immune cell infiltration in the TCGA-BRCA samples (Table S2). Fig. 4A illustrates the relationship between the expression level of VM genes and the quantity of immune cells. Notably, MMP9 expression exhibited a statistically significant positive correlation with Macrophages M0, while displaying a robust negative correlation with T cells CD4 memory resting, Monocytes, and Mast cells activated.
Fig. 4.
The immune microenvironment of two subclusters. (A) Correlation between VM genes expression levels and the percentage of different immune cells in breast cancer. (B) Differences in the extent of immune cell infiltration among the two subpopulations. (C) Immune cell infiltration in breast cancer. (D) TumorPurity, StromalScore, ImmuneScore and ESTIMATEScore for two subclusters. (E) Differential expression of immune checkpoints in two subclusters.
The differences in immune cell infiltration levels between Cluster 1 and Cluster 2 are illustrated in Fig. 4B and C. B cells memory, T cells CD4 memory resting, T cells CD8, T cells gamma delta, T cells regulatory(Tregs) and Mast cells resting have higher infiltration levels in Cluster 1 than in Cluster 2, while NK cells resting, Plasma cells, T cells CD4 naive and T cells follicular helper have higher infiltration levels in Cluster 2 than in Cluster 1. The other immune cells B cells naive, T cells CD4 memory activated, Dendritic cells activated, Dendritic cells resting, Eosinophils, Macrophages M0, Macrophages M1, Macrophages M2, Mast cells activated, Monocytes, Neutrophils and NK cells activated show no significant difference in infiltration levels between the two subclusters.
TMB is used to reflect the total mutation level in tumor cells, and MSI refers to the phenomenon that the mismatch repair mechanism fails during DNA replication, resulting in an increase or decrease in the number of repetitive fragments that cannot be repaired by the body, causing changes in the length of microsatellite bodies. The MSI scores in Cluster 1 and Cluster 2 were not significantly different, while the TMB scores of samples in Cluster 1 were significantly higher than those in Cluster 2 (Fig. S1B).
The “estimate” R package was used to calculate the immune and stromal scores for each sample in TCGA-BRCA, and it was further observed that the entire TCGA dataset with StromalScore, ImmuneScore, ESTIMATEScore, and TumorPurity were significantly different in Cluster 1 and Cluster 2 samples. The StromalScore, ImmuneScore, and ESTIMATEScore of Cluster 1 samples were higher than those of Cluster 2 samples, while TumorPurity of Cluster 2 samples was higher than that of Cluster 1 (Fig. 4D).
We further analyzed the difference in the expression of immune checkpoints in Cluster 1 and Cluster 2, and the expression of CD274, CTLA4, PDCD1LG2, PDCD1, HAVCR2, LAG3, SIGLEC15, and TIGIT was significantly different in both subclusters (Fig. 4E and Fig. S1B).
3.5. Construction and validation of risk prediction model for VM genes
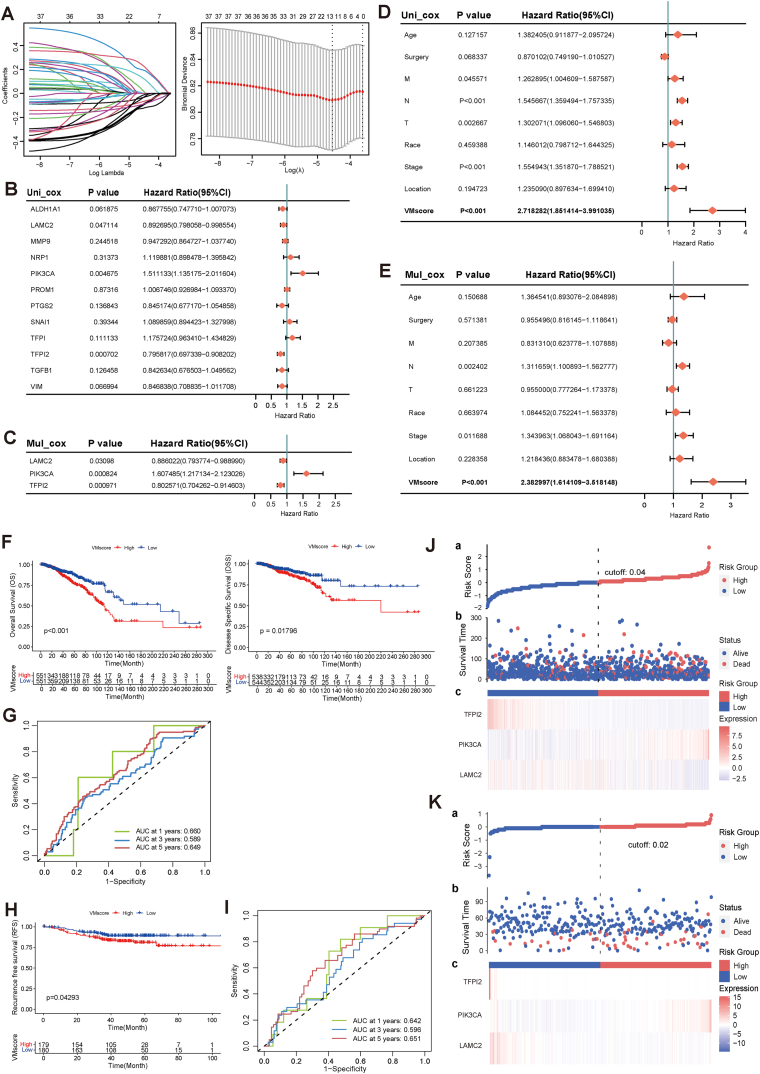
Twelve genes were screened by LASSO, and then univariate and multivariate Cox regression analyses were performed to finally obtain three prognostically significantly associated genes including LAMC2, PIK3CA, and TFPI2 for the construction of the risk model (Fig. 5A–C). Our VM index (VMscore) was calculated by the following formula VMscore = (−0.12101358)*LAMC2 expression + 0.47467087*PIK3CA expression + (−0.21993498)*TFPI2 expression. Univariate Cox regression analysis showed that TMN staging, Stage and VMscore were significantly associated with BRCA prognosis. Multifactorial Cox regression analysis confirmed that VMscore was an independent prognostic factor after adjusting for other clinicopathologic factors (Fig. 5D–E). Using the median VMscore as the threshold in the TCGA-BRCA dataset, OS (P < 0.001) and DSS (P = 0.018) were better in patients with low VMscore than in patients with high VMscore (Fig. 5F, G and 5J). Then, the role of VMscore was verified by the Fudan dataset. As shown in Fig. 5H, I, and 5K, the prognosis of patients in the high VMscore subgroup was significantly worse compared to the low VMscore subgroup, consistent with the results of the TCGA dataset. Subsequently, ROC analysis and AUC were used to evaluate the performance of the model at different follow-up times. The AUC values of OS and RFS are better in the TCGA dataset and test dataset for 1 and 5 years, both above 60 % (Fig. 5G and I).
Fig. 5.
Construction and validation of VM risk models. (A) Trajectories for each independent variable and confidence intervals with different lambda values. (B–C) Univariate and multivariate cox analysis of VM prognostic genes. (D–E) Univariate and multivariate cox analysis of VM risk scores and clinical factors. (F) Kaplan-Meier curves of OS and DSS for TCGA-BRCA high and low VMscore subgroups. (G) ROC curves, OS sensitivity and specificity based on VMscore prediction of the training sets for 1, 3 and 5 years. (H) Kaplan-Meier curves of the RFS for the high VMscore and low VMscore subgroups of the validation sets. (I) ROC curves, RFS sensitivity and specificity based on VMscore prediction of the validation sets for 1, 3 and 5 years. (J–K) Risk score for training and validation sets.
3.6. Functional identification of differential genes in high and low VM subgroups and single-cell level analysis of three prognostic genes
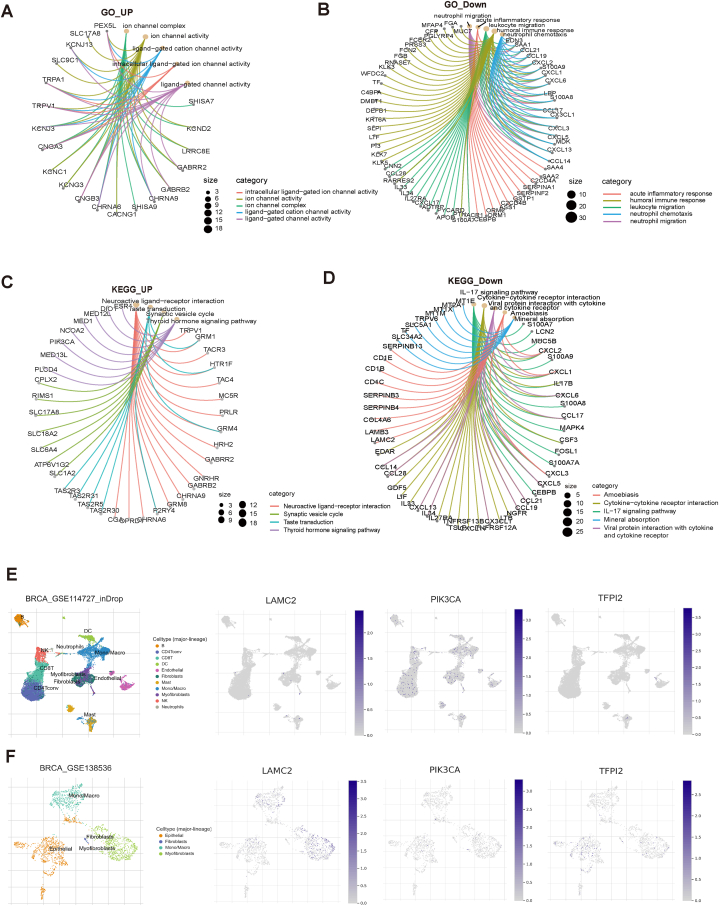
Differential genes were identified using a comparison of gene expression levels between the high and low VMscore groups. These genes were then subjected to functional enrichment analysis, with up-regulated genes and down-regulated genes being separately examined. The findings of the GO analysis indicated that the genes that were up-regulated exhibited a significant enrichment in intracellular ligand-gated ion channel activity, ion channel activity, and ligand-gated channel activity, as depicted in Fig. 6A. Conversely, the down-regulated genes were primarily enriched in immune-related pathways such as acute inflammatory response, leukocyte migration, and neutrophil chemotaxis, as illustrated in Fig. 6B. The results of the KEGG analysis revealed that the up-regulated genes exhibited a significant enrichment in the Neuroactive ligand-receptor interaction pathway (Fig. 6C). On the other hand, the down-regulated genes were predominantly enriched in the Cytokine-cytokine receptor interaction, IL-17 signaling pathway, and Viral signaling pathway, all of which are pathways relevant to immunity (Fig. 6D). Additionally, the down-regulated genes were also found to be enriched in the Viral protein interaction with cytokine and cytokine receptor pathway.
Fig. 6.
Functional enrichment and single-cell analysis. (A) High and low VMscore subgroups up-regulate differential gene GO enrichment network map. (B) High and low VMscore subgroups down-regulate differential gene GO enrichment network map. (C) High and low VMscore subgroups up-regulate differential gene KEGG enrichment network map. (D) High and low VMscore subgroups down-regulate differential gene KEGG enrichment network map. (E–F) Single-cell analysis of LAMC2, PIK3CA, and TFPI2.
We used two GEO datasets, GSE114727 and GSE138536 to localize LAMC2, PIK3CA, and TFPI2 at the single-cell level to examine their potential functions. It can be observed that PIK3CA is commonly expressed in immune cells, LAMC2 is expressed in Myofibroblasts cells, while TFPI2 is expressed in small amounts in Myofibroblasts and Epithelial (Fig. 6E and F).
4. Discussion
Considering the important role of VM, this study thoroughly assessed the differential expression of 37 genes associated with VM at the transcriptome level. Our study revealed that a majority of these genes had a downregulation in tumor tissues when compared to normal tissues. Furthermore, it was observed that these genes played a key role in activating the epithelial-mesenchymal transition (EMT) pathway. Several studies have demonstrated that EMT promotes the formation of VM and upregulates EMT related transcription factors in tumor cells with VM formation [21]. According to Xing et al. inhibition of Tenascin-C suppresses ERK-triggered EMT, which in turn prevents vasculogenic mimicry in gastric cancer [22]. Huangfuchuan protein A suppresses CXCL12-mediated EMT in HCC, hence inhibiting vasculogenic mimicry [23]. By inhibiting Claudin15 expression, the EMT regulator Twist1 speeds up tumor vasculogenic mimicry in triple-negative breast cancer [24].
Then, we divided TCGA-BRCA cases into two clusters based on the expression levels of all VM genes through consensus clustering. The survival analysis demonstrated that individuals belonging to Cluster 2 exhibited poorer OS and PFI outcomes in comparison to those in Cluster 1. Furthermore, we observed significant differences in immune-related parts between the two clusters. Our analysis revealed that the infiltration levels of CD8 T cells were significantly higher in Cluster 1 as compared to Cluster 2. This observation suggests that Cluster 1 exhibits a more prominent immunological activation characteristic in comparison to Cluster 2. It is noteworthy to acknowledge that the responsiveness of malignancies to targeted immune checkpoint inhibitor (ICI) therapy appears to be contingent upon the extent and composition of immune cell infiltration inside the tumor microenvironment. The absence of T lymphocytes within tumor microenvironments has been identified as a contributing factor to the development of resistance towards immunotherapeutic interventions [25]. CD8 T cells has the ability to identify and eliminate cancer cells by means of discharging cytotoxic chemicals and cytokines. Therefore, the infiltration level of CD8 T cells directly determines the anti-tumor killing capacity, which is closely associated with extended survival, improved immunotherapy efficacy, and enhanced response to ICI therapy in cancer patients [[26], [27], [28]]. Therefore, it is hypothesized that Cluster 1 may demonstrate a more favorable response to ICI therapy compared to Cluster 2.
In addition, we have identified genes that exhibit significant changes in expression across the two groups and conducted enrichment analysis. The results showed that the MAPK and PI3K-Akt signaling pathway were significantly activated in Cluster 2. During the process of tumor vasculogenic mimicry, vascular endothelial cells and tumor cells can secrete various growth factors and chemokines. These bioactive molecules activate a series of signal transduction pathways by binding to corresponding receptors, promoting cell proliferation and migration, and forming new vascular networks. In previous studies, IL-1β was reported to promote angiogenesis in breast cancer cells through the p38/MAPK and PI3K/Akt signaling pathways [14]. The PI3K/Akt/mTOR signaling pathway has been documented to facilitate the development of tumor VM by activating downstream signaling molecules such as E-cadherin, MMP2, MMP9, and other related factors [29]. Therefore, it can be inferred that the activation of the MAPK and PI3K-Akt signaling pathways in Cluster 2 may promote the formation of VM, thereby influencing tumor metastasis and progression in patients.
By using LASSO and Cox regression algorithms to assess the effect of genes on survival time, we identified three VM prognostic genes, including LAMC2, PIK3CA, and TFPI2. Laminin subunit gamma 2 (LAMC2) belongs to the family of extracellular matrix glycoproteins and is a key element in ECM receptor signaling. It is involved in various biological processes, including cell adhesion, differentiation, migration, signal transduction, neural outgrowth, and metastasis [30]. ECM remodeling and ECM signal transduction function are crucial for the formation of VM [10]. LAMC2 in ECM may activate receptors on tumor cells, thereby initiating signal transduction pathways and promoting the development of vascular mimicry.
PIK3CA is one of the most frequently mutated genes in recurrent breast cancer and has been found to play an important role in many other cancer types. PIK3CA directly participates in the signal transduction of the PI3K/AKT pathway by encoding the catalytic subunit of PI3K. PIK3CA mutations can lead to abnormal activation of this signaling pathway, thereby promoting tumor cell growth, survival, and anti apoptosis [31,32]. PI3K/AKT signaling pathway is also closely associated with the progression of VM. Activation of the PI3K/AKT pathway leads to overexpression of EMT related factors E-cadherin, vimentin, and Twist, as well as VM formation related signal transduction proteins VE cadherin, VEGF-A/VEGFR2, EphA2, and MMPS, thereby promoting VM [29].
Tissue factor pathway inhibitor 2 (TFPI2) belongs to the superfamily of serine protease inhibitors. TFPI2 has been reported to be downregulated in breast cancer tissue, and its low expression is significantly associated with tumor size, metastasis, pathological staging, and poor prognosis in breast cancer patients [29]. Downregulation of TFPI2-mediated TWIST1 reduces integrin α5, thereby inhibiting the progression of breast cancer [33]. According to reports, TFPI2 can upregulate the expression of TMPRSS4 and play an important regulatory role in EMT [34]. TFPI2 is considered a key factor that can inhibit ECM degradation by inhibiting the activation of MMP1 and pro MMP13, and ECM degradation is crucial for VM formation [35].
There are still some limitations to this study. The correlation between VM and tumor immune microenvironment deserves further exploration in order to achieve better clinical applications. In addition, we constructed a prognostic model based on public databases and validated it, which should be further validated in prospective real-world studies in the future.
5. Conclusion
In conclusion, our research provides a new perspective for understanding VM in breast cancer. VM in breast cancer shows heterogeneity, and its functional role in tumor progression may be closely related to the infiltration of immune cells. Different VM subtypes in breast cancer have different prognosis and immune invasion patterns. By identifying different subtypes of VM, more precise classification can be provided for clinical practice, and more personalized treatment plans can be provided for patients. We also developed a VM risk scoring model based on LAMC2, PIK3CA and TFPI2, which can accurately and conveniently predict the prognosis of breast cancer patients and provide a promising molecular target for VM immunotherapy. The biomarkers we screen can serve as important tools for disease diagnosis, prognosis, and treatment response evaluation.
Ethics approval
Not applicable.
Data availability statement
The public data set of this study can be found in the TCGA database (https://portal.gdc.cancer.gov/).
Funding
The study were supported by the Natural Science Foundation of Shaanxi Province, China (No.2024JC-YBQN-0932); The National Natural Science Foundation of China (No.82002803); The Key R&D projects in Shaanxi Province, China (No.2020SF-029).
CRediT authorship contribution statement
Xiao Liang: Writing – original draft, Visualization, Methodology, Data curation, Conceptualization. Xinyue Ma: Writing – review & editing, Methodology, Formal analysis. Feiyang Luan: Methodology, Formal analysis. Jin Gong: Methodology. Shidi Zhao: Data curation. Yiwen Pan: Methodology. Yijia Liu: Visualization. Lijuan Liu: Writing – original draft, Conceptualization. Jing Huang: Writing – original draft. Yiyang An: Investigation. Sirui Hu: Methodology, Data curation. Jin Yang: Writing – review & editing. Danfeng Dong: Funding acquisition, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to Fudan University Shanghai Cancer Center (FUSCC) for providing RNA-seq data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36565.
Contributor Information
Jin Yang, Email: yangjin@mail.xjtu.edu.cn.
Danfeng Dong, Email: qiwudanfeng@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zeng X., Liu C., Yao J., Wan H., Wan G., Li Y., Chen N. Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol. Res. 2021;163 doi: 10.1016/j.phrs.2020.105320. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Zhen F., Sun Y., Han B., Wang H., Zhang Y., Zhang H., Hu J. Single-cell RNA sequencing reveals small extracellular vesicles derived from malignant cells that contribute to angiogenesis in human breast cancers. J. Transl. Med. 2023;21:570. doi: 10.1186/s12967-023-04438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniotis A.J., Folberg R., Hess A., Seftor E.A., Gardner L.M., Pe'er J., Trent J.M., Meltzer P.S., Hendrix M.J. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am. J. Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng N., Zhang S., Wu W., Zhang N., Wang J. Regulatory mechanisms and therapeutic targeting of vasculogenic mimicry in hepatocellular carcinoma. Pharmacol. Res. 2021;166 doi: 10.1016/j.phrs.2021.105507. [DOI] [PubMed] [Google Scholar]
- 5.Liu X., He H., Zhang F., Hu X., Bi F., Li K., Yu H., Zhao Y., Teng X., Li J., et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13:483. doi: 10.1038/s41419-022-04950-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu R., Du W., Ding Z., Wang Y., Li Y., Zhu J., Zeng Y., Zheng Y., Liu Z., Huang J.A. HIF-1α promoted vasculogenic mimicry formation in lung adenocarcinoma through NRP1 upregulation in the hypoxic tumor microenvironment. Cell Death Dis. 2021;12:394. doi: 10.1038/s41419-021-03682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi B., Li H., Cai H., Lou X., Yu M., Li Z. LOXL1-AS1 communicating with TIAR modulates vasculogenic mimicry in glioma via regulation of the miR-374b-5p/MMP14 axis. J. Cell Mol. Med. 2022;26:475–490. doi: 10.1111/jcmm.17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y., Yang Z., Yu Y., Zhang P. HIF1α lactylation enhances KIAA1199 transcription to promote angiogenesis and vasculogenic mimicry in prostate cancer. Int. J. Biol. Macromol. 2022;222:2225–2243. doi: 10.1016/j.ijbiomac.2022.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Morales-Guadarrama G., García-Becerra R., Méndez-Pérez E.A., García-Quiroz J., Avila E., Díaz L. Vasculogenic mimicry in breast cancer: clinical relevance and drivers. Cells. 2021;10 doi: 10.3390/cells10071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei X., Chen Y., Jiang X., Peng M., Liu Y., Mo Y., Ren D., Hua Y., Yu B., Zhou Y., et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol. Cancer. 2021;20:7. doi: 10.1186/s12943-020-01288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng S., Guo G., Yang Z., Lu Y., Lu K., Fu W., Huang Q. Vasculogenic mimicry regulates immune infiltration and mutational status of the tumor microenvironment in breast cancer to influence tumor prognosis. Environ. Toxicol. 2024;39:2948–2960. doi: 10.1002/tox.24165. [DOI] [PubMed] [Google Scholar]
- 12.Gu Y., Huang Q., Wang Y., Wang H., Xiang Z., Xu Y., Wang X., Liu W., Wang A. The vasculogenic mimicry related signature predicts the prognosis and immunotherapy response in renal clear cell carcinoma. BMC Cancer. 2024;24:420. doi: 10.1186/s12885-024-12107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andonegui-Elguera M.A., Alfaro-Mora Y., Cáceres-Gutiérrez R., Caro-Sánchez C.H.S., Herrera L.A., Díaz-Chávez J. An overview of vasculogenic mimicry in breast cancer. Front. Oncol. 2020;10:220. doi: 10.3389/fonc.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nisar M.A., Zheng Q., Saleem M.Z., Ahmmed B., Ramzan M.N., Ud Din S.R., Tahir N., Liu S., Yan Q. IL-1β promotes vasculogenic mimicry of breast cancer cells through p38/MAPK and PI3K/akt signaling pathways. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.618839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treps L., Faure S., Clere N. Vasculogenic mimicry, a complex and devious process favoring tumorigenesis - interest in making it a therapeutic target. Pharmacol. Ther. 2021;223 doi: 10.1016/j.pharmthera.2021.107805. [DOI] [PubMed] [Google Scholar]
- 16.Liu H., Wang H., Chen D., Gu C., Huang J., Mi K. Endoplasmic reticulum stress inhibits 3D Matrigel-induced vasculogenic mimicry of breast cancer cells via TGF-β1/Smad2/3 and β-catenin signaling. FEBS Open Bio. 2021;11:2607–2618. doi: 10.1002/2211-5463.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu D.G., Yu Y., Lee C.H., Kuo Y.L., Lu Y.C., Tu C.W., Chang W.W. Hinokitiol inhibits vasculogenic mimicry activity of breast cancer stem/progenitor cells through proteasome-mediated degradation of epidermal growth factor receptor. Oncol. Lett. 2016;11:2934–2940. doi: 10.3892/ol.2016.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M.R., Wei P.F., Suo M.Z., Hu Y., Ding W., Su L., Zhu Y.D., Song W.J., Tang G.H., Zhang M., et al. Brucine suppresses vasculogenic mimicry in human triple-negative breast cancer cell line MDA-MB-231. BioMed. Res. Int. 2019;2019 doi: 10.1155/2019/6543230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y.Z., Ma D., Suo C., Shi J., Xue M., Hu X., Xiao Y., Yu K.D., Liu Y.R., Yu Y., et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–440.e425. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.-H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q., Qiao L., Liang N., Xie J., Zhang J., Deng G., Luo H., Zhang J. The relationship between vasculogenic mimicry and epithelial-mesenchymal transitions. J. Cell Mol. Med. 2016;20:1761–1769. doi: 10.1111/jcmm.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang X., Xu E., Wang X., Qian L., Yang Z., Yu H., Wang C., Ren C., Wang Y., Lu X., et al. Tenascin-c knockdown suppresses vasculogenic mimicry of gastric cancer by inhibiting ERK- triggered EMT. Cell Death Dis. 2021;12:890. doi: 10.1038/s41419-021-04153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao T., Bao J., Tian J., Lin R., Zhang Z., Zhu Y., He Y., Gao D., Sun R., Zhang F., et al. Flavokawain A suppresses the vasculogenic mimicry of HCC by inhibiting CXCL12 mediated EMT. Phytomedicine. 2023;112 doi: 10.1016/j.phymed.2023.154687. [DOI] [PubMed] [Google Scholar]
- 24.Zhang D., Sun B., Zhao X., Sun H., An J., Lin X., Zhu D., Zhao X., Wang X., Liu F., et al. Twist1 accelerates tumour vasculogenic mimicry by inhibiting Claudin15 expression in triple-negative breast cancer. J. Cell Mol. Med. 2020;24:7163–7174. doi: 10.1111/jcmm.15167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruni D., Angell H.K., Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 27.Jansen C.S., Prokhnevska N., Master V.A., Sanda M.G., Carlisle J.W., Bilen M.A., Cardenas M., Wilkinson S., Lake R., Sowalsky A.G., et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 2019;576:465–470. doi: 10.1038/s41586-019-1836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y.T., Sun Z.J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics. 2021;11:5365–5386. doi: 10.7150/thno.58390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J., Wang C., Hou Y., Tian Y., Li Y., Zhang H., Zhang L., Li W. Molecular mechanisms of Thrombospondin-2 modulates tumor vasculogenic mimicry by PI3K/AKT/mTOR signaling pathway. Biomed. Pharmacother. 2023;167 doi: 10.1016/j.biopha.2023.115455. [DOI] [PubMed] [Google Scholar]
- 30.Fu T., Liu J.X., Xie J., Gao Z., Yang Z. LAMC2 as a prognostic biomarker in human cancer: a systematic review and meta-analysis. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2022-063682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelman J.A., Luo J., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 32.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao D., Qiao J., He H., Song J., Zhao S., Yu J. TFPI2 suppresses breast cancer progression through inhibiting TWIST-integrin α5 pathway. Mol. Med. 2020;26:27. doi: 10.1186/s10020-020-00158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Xu Y., Wang Q., Xie C., Liu Y., Tu Z. Tissue factor pathway inhibitor-2 induced hepatocellular carcinoma cell differentiation. Saudi J. Biol. Sci. 2017;24:95–102. doi: 10.1016/j.sjbs.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang L., Dunterman M., Guo S., Khan F., Liu Y., Taefi E., Bahrami A., Geula C., Hsu W.H., Horbinski C., et al. Kunitz-type protease inhibitor TFPI2 remodels stemness and immunosuppressive tumor microenvironment in glioblastoma. Nat. Immunol. 2023;24:1654–1670. doi: 10.1038/s41590-023-01605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public data set of this study can be found in the TCGA database (https://portal.gdc.cancer.gov/).