Abstract
Background
Intermediate cells are present in the early stages of human prostate development and adenocarcinoma. While primary cells isolated from benign human prostate tissues or tumors exhibit an intermediate phenotype in vitro, they cannot form tumors in vivo unless genetically modified. It is unclear about the stem cell properties and tumorigenicity of intermediate cells.
Methods
We developed a customized medium to culture primary human intermediate prostate cells, which were transplanted into male immunodeficient NCG mice to examine tumorigenicity in vivo. We treated the cells with different concentrations of dihydrotestosterone (DHT) and enzalutamide in vitro and surgically castrated the mice after cell transplantation in vivo. Immunostaining, qRT-PCR, RNA sequencing, and western blotting were performed to characterize the cells in tissues and 2D and 3D cultures.
Results
We found intermediate cells expressing AR+PSA+CK8+CK5+ in the luminal compartment of human prostate adenocarcinoma by immunostaining. We cultured the primary intermediate cells in vitro, which expressed luminal (AR+PSA+CK8+CK18+), basal (CK5+P63+), intermediate (IVL+), and stem cell (CK4+CK13+PSCA+SOX2+) markers. These cells resisted castration in vitro by upregulating the expression of AR, PSA, and proliferation markers KI67 and PCNA. The intermediate cells had high tumorigenicity in vivo, forming tumors in immunodeficient NCG mice in a month without any genetic modification or co-transplantation with embryonic urogenital sinus mesenchyme (UGSM) cells. We named these cells human castration-resistant intermediate prostate cancer stem cells or CriPCSCs and defined the xenograft model as patient primary cell-derived xenograft (PrDX). Human CriPCSCs resisted castration in vitro and in vivo by upregulating AR expression. Furthermore, human CriPCSCs differentiated into amplifying adenocarcinoma cells of luminal phenotype in PrDX tumors in vivo, which can dedifferentiate into CriPCSCs in vitro.
Conclusions
Our study identified and established methods for culturing human CriPCSCs, which had high tumorigenicity in vivo without any genetic modification or UGSM co-transplantation. Human CriPCSCs differentiated into amplifying adenocarcinoma cells of luminal phenotype in the fast-growing tumors in vivo, which hold the potential to dedifferentiate into intermediate stem cells. These cells resisted castration by upregulating AR expression. The human CriPCSC and PrDX methods hold significant potential for advancing prostate cancer research and precision medicine.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-03917-8.
Keywords: Intermediate cell, Cancer stem cell, Prostate adenocarcinoma, Castration-resistant prostate cancer, Enzalutamide
Background
Prostate cancer is the most commonly diagnosed cancer in men in the US, and it is also the fastest-growing cancer in men in China [1–3]. The majority of prostate cancers are initially diagnosed as adenocarcinomas, which have a luminal phenotype and express androgen receptor (AR), prostate-specific antigen (PSA), keratin 8 (CK8), and CK18 [4]. The androgen signaling pathway is a key regulator in prostate development and adenocarcinoma [5]. Androgen deprivation therapy, also known as castration, has been the standard treatment for advanced and metastatic prostate adenocarcinomas [5]. However, most patients will eventually develop into castration-resistant prostate cancer (CRPC) [5–8]. Prostate stem cells (PSCs) are crucial in the development of prostate cancer and resistance to castration [9, 10]. Studies using lineage-tracing and pulse-chase transgenic mouse models have identified rare castration-resistant PSCs in luminal and basal cell populations that can give rise to prostate cancers upon genetic manipulations [9, 11–15]. Research on human prostate stem cells has significant implications in translational medicine. This is due to the differences between human and mouse prostates in terms of anatomy, histology, and cell biology [16]. Dean Tang’s group identified a subpopulation of cancer stem cells within the LAPC9 prostate cancer cell line, which exhibited a high tumor-initiating capacity [17]. Recent single-cell sequencing studies have identified potential stem cell populations in human prostate normal tissues and tumors [18–22].
The in vitro culture of prostate stem cells or cancer stem cells offers valuable tools for prostate cancer research and precision medicine. Dihydrotestosterone (DHT), a potent endogenous androgen, was typically added to the culture media to support luminal PSC growth [23, 24]. However, primary human prostate stem cells cultured in previous studies exhibited low tumorigenicity. These cells were often genetically engineered to overexpress oncogenes or knock out tumor suppressor genes to promote prostate cancer development [25–29]. Co-transplantation with embryonic urogenital sinus mesenchyme (UGSM) cells was required for these engineered cells to effectively form tumors in vivo [25–29]. However, genetic modifications might hinder the accurate replication of original tumors in vitro, which is crucial for personalized precision medicine. Therefore, it is essential to develop new methods to isolate and culture primary human prostate cancer stem cells with high tumorigenicity.
The prostate epithelial compartment contains multiple cell types, including luminal, basal, neuroendocrine, and intermediate cells [7]. Intermediate cells, characterized by the expression of both luminal and basal markers, have been discovered in mouse and human prostates for several decades [30, 31]. However, it is still unclear whether they represent a distinct cell type or a transitional phenotype [9, 32]. These cells are prevalent in the early stages of human prostate development but are rarely detected in normal adult prostate tissues [33, 34]. Interestingly, some studies have reported the presence of intermediate cells in human prostate tumors, suggesting a potential reactivation of the embryonic program [34–36]. In vitro studies have shown that primary human prostate epithelial cells gained the intermediate phenotype, expressing both luminal (CK8 or CK18) and basal (CK5 or CK14) markers [36–39]. When co-transplanted with rat or mouse UGSM cells, these cells usually form benign prostate glands in vivo [36–41]. However, there have been few reports of the culture of primary prostate stem cells or intermediate cells that have high tumorigenicity in vivo without genetic modifications.
In our previous study, we developed a method to isolate and culture castration-resistant intermediate prostate stem cells from mice [42]. We found these cells expressed luminal and basal cell markers and could differentiate into prostate glands in vivo without co-transplantation with the UGSM cells [42]. In this study, we identified intermediate cells in the luminal compartment in the early stages of human prostate adenocarcinoma and successfully cultured primary human castration-resistant intermediate prostate cancer stem cells (CriPCSCs). We characterized their marker expression, tumorigenicity, and drug resistance. Furthermore, our patient primary cell-derived xenograft (PrDX) model provides a valuable platform for human prostate cancer research and precision medicine.
Methods
This work has been reported in line with the ARRIVE guidelines 2.0.
Primary cell isolation and culture
Two prostate cancer needle biopsies were obtained from a single patient after obtaining informed consent. The procedure was conducted in accordance with the Ministry of Science and Technology guidelines and was approved by the Ethics Committee of the Medical College of Qingdao University, under approval document number QDU-HEC-2022177. Primary cell isolation and culture were performed according to our previous work with minor modifications [42]. The samples were digested in the DMEM containing 2 mg/ml collagenase I and 5 µM Y27632 at 37 °C. The cell suspension was collected every 15 min, centrifuged at 1000 rpm for 5 min, and the cell pellets were suspended in the culture medium. The prostate culture medium included DMEM/F12 (Invitrogen, Cat#12,400,024, USA), 2% fetal bovine serum (FBS, Gibco, Cat#10,091,148, USA), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco, Cat#15,140,122, USA), 1xN2 (Gibco, Cat#17502048), 1xB-27 (Gibco, Cat#A3582801, USA), 10 ng/ml epidermal growth factor (EGF) (Peprotech, Cat#AF-100-15), 10 nM dihydrotestosterone (DHT) (Selleck, Cat#S4757, China), 2 µM Chir99021 (Selleck, Cat#S2924, China), 0.2 µM A83-01 (Tocris, Cat#2939, UK), and 5 µM Y27632 (Selleck, Cat#S1049, China). Accutase (Gibco, Cat#A1110501, USA) was used to passage the cells. The frozen solution was 90% FBS plus 10% DMSO (Sigma, Cat#D2650, USA).
To induce castration of primary cells in vitro, the FBS in the culture medium was replaced with Charcoal-Stripped FBS (HYCEZMBIO, CSFBS2021, China), which is devoid of androgens and is commonly used in prostate cancer research to mimic castration conditions in vitro [17]. There were 4 groups. The cells were cultured in the media containing 10, 1, 0.1, and 0 nM DHT for a week before harvest for qRT-PCR and western blotting. Each group had 3 samples.
To conduct castration and enzalutamide (Selleck, Cat#S1250, China) experiments on PrDX-derived cells in vitro, the cells were isolated from the first generation of PrDX tumors, which were digested in DMEM containing 2 mg/ml collagenase I and 5 µM Y27632 at 37 °C. The FBS in the culture medium was replaced by Charcoal-Stripped FBS (HYCEZMBIO, CSFBS2021, China). There were 6 groups and each group had 3 samples. The cells were cultured in the media containing 10 nM DHT, 1 nM DHT, 0.1 nM DHT, 0 nM DHT, 0.1 nM DHT with 10 µM enzalutamide, and 0 nM DHT with 10 µM enzalutamide for a week before harvest for western blotting. The enzalutamide concentration was set at 10 µM, based on previous studies [42, 43]. For 2D culture, the cells were cultured in the culture dishes coated with diluted Matrigel (0.1 mg/ml). For 3D organoid culture, the cell suspension was mixed with Matrigel at a concentration of 2 × 106 cells/ml and incubated in a 37 °C incubator for 30 min before adding the culture medium.
Culture of cell lines
The cell lines were purchased from Wuhan Pricella Biotechnology Co., Ltd. (China). PC-3 (Cat# CL-0185) and VCaP (Cat# CL-0241) were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. 22Rv1 (Cat# CL-0004) and LNCaP (Cat# CL-0143) were cultured in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Animal procedures
All the animal procedures were performed according to the Ministry of Science and Technology guide and approved by the Ethics Committee of the Medical College of Qingdao University, with the approval document number QDU-AEC-2022305. The male NCG mice (Strain NO. T001475) of 4–5 weeks were purchased from Gempharmatech Co., Ltd. in China. The animals were housed in individually ventilated cages in a specific-pathogen free animal facility with a light cycle of 12-h light and 12-h dark. There were a total of 7 animals used in this study.
For the primary PrDX, the primary cells were digested with Accutase, centrifuged at 1000 rpm for 5 min, and mixed with Matrigel (Corning, Cat#354234) before subcutaneous injection into a male NCG mouse. The animal was anesthetized with 2% isoflurane (Cat#: R510-22-10, RWD Life Science Co., Ltd, China) delivered by mask. Toe pinch was performed to confirm deep anesthesia. Due to the limited number of primary cells, 1 NCG mouse was used for the first generation of PrDX tumors, which were harvested after 4 weeks.
For animal castration experiments, the cells isolated from the first generation of PrDX tumors were transplanted into NCG mice for the generation of secondary PrDX tumors. There were a total of 6 animals that were randomly divided into 2 groups: 3 in the control group and 3 in the castration group. After a week of cell transplantation, the mice in the castration group underwent surgical castration while the control group underwent a sham operation with a skin wound but no castration. The animals were anesthetized by 2% isoflurane followed by confirmation of deep anesthesia by toe pinch. Aseptic surgeries were performed. The PrDX tumors were harvested after another 4 weeks. The animals were euthanized by an overdose of isoflurane before tissue harvest.
Different investigators were involved in the experiments. JM performed animal surgery. Another investigator RL labeled the animals and was the only person who knew the group allocation. DW and YZ collected tissues. JM performed the qRT-PCR analysis and was unaware of the animal treatment. The animals that died or were inactive before the end of the experiments should be excluded. All the animals were healthy, and no exclusions were made from this study.
Histology and immunostaining
The tumor tissues were fixed in 4% paraformaldehyde (PFA) overnight, followed by dehydration in 15% and 30% sucrose before being embedded in OCT for cryosection. The cultured cells were fixed in 4% PFA for 15 min. For immunostaining, the cryosections and fixed cells were washed with PBS, permeabilized by 0.5% Triton X-100 for 10 min, blocked in 5% NDS for 1 h, incubated in primary antibodies at 4 °C overnight, followed by secondary antibodies for 1 h. The antibodies were listed in the supplemental materials. DAPI (Sigma, Cat#D9542) was used for nuclear staining. Confocal microscopy was performed on a Leica SP8 confocal microscope.
RNA sequencing
The tumor tissues with and without castration treatment were snap frozen in liquid nitrogen and stored at -80oC before RNA sequencing. The mRNA library preparation and sequencing were performed on the Illumina Novaseq 6000 platform of Majorbio (China). The volcano plot was analyzed on Majorbio Cloud. The heatmap was plotted by https://www.bioinformatics.com.cn, an online platform for data analysis and visualization.
Quantitative RT-PCR analysis
The tumor tissues and cells were collected in Trizol (Invitrogen, Cat#15596026, USA). The total RNA was extracted for cDNA preparation by the Evo M-MLV RT Mix Kit (Accurate Biotechnology, Cat#AG11728, China) according to the instructions. The primers are listed in the supplemental materials. Quantitative RT-PCR was performed in a QuantStudio 3 Real-Time PCR System (ABI, USA). The relative mRNA expression levels were calculated using the 2^-ΔΔCt method. GAPDH was the reference gene.
Western blotting
The cells cultured in 2D were washed with cold PBS three times and lysed with 1% SDS lysis buffer (10 mM Tris, 1% SDS, 1 mM Na3VO4, and protease inhibitor). The 3D organoids were digested with 1 mg/ml collagenase II for cell dissociation, followed by centrifugation at 10,000 rpm for 5 min to obtain cell precipitate, which was then lysed with 1% SDS lysis buffer. Protein concentration was determined by a BCA protein assay kit (Sangon Biotech, Cat#C503051-0500, China). Equal amounts of protein samples were loaded for SDS-PAGE and transferred to the PVDF membrane (Millipore, Cat# IPVH00010, USA). The membranes were blocked with 5% BSA (Biosharp, Cat#BS114, China), followed by incubation with primary antibodies, including AR (1:2000, Abcam, Cat#ab108341, USA), Actin (1:2000, Sangon Biotech, Cat#D191048, China), PSA (1:2000, Proteintech, Cat#10679-1-AP, China) at 4℃ overnight. Secondary antibodies (1:10000, Sangon Biotech, Cat#D110011, China) were incubated at room temperature for 1 h. ECL detection reagent (Beyotime Biotechnology, Cat#P0018FS, China) was prepared according to the kit instructions, and the bands were visualized using an enhanced chemiluminescence detection system (Vilber Bio Imaging, Vilber solo 4s, France).
Statistical analysis
All statistical analyses were conducted using GraphPad Prism 8 software. Data were presented as mean ± standard deviation. The significant differences between multiple groups were analyzed using one-way ANOVA followed by Bonferroni post hoc tests. The significant differences between the two groups were analyzed by t-test. Statistical significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Luminal location of intermediate cells in human prostate adenocarcinoma
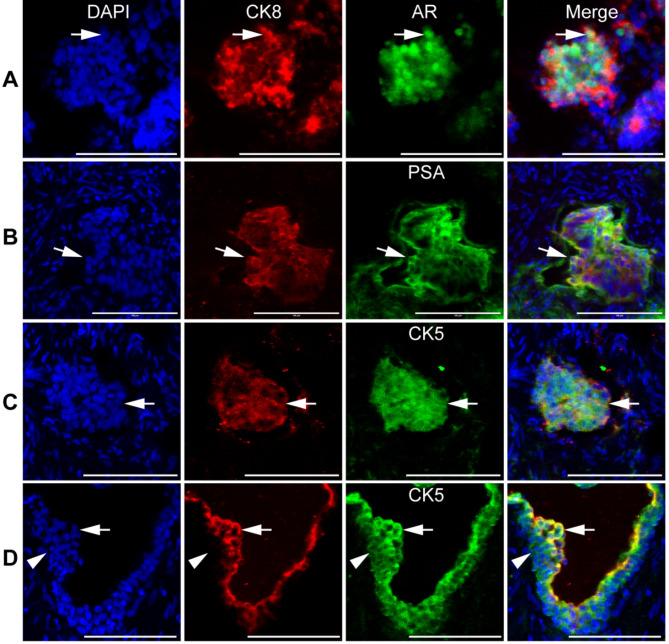
We conducted immunostaining on cryosections of human prostate adenocarcinoma biopsies and found intermediate cells expressing both luminal (AR, PSA, CK8) and basal (CK5) markers (Fig. 1). Most prostate adenocarcinoma cells expressed AR, PSA, CK8, and CK5 (Fig. 1, A-C, arrows). In some areas, we observed intermediate CK8+CK5+ cells in the luminal compartment (Fig. 1, D, arrows), with the underlying basal cells expressing CK8−CK5+ (Fig. 1, D, arrowheads). This suggests that luminal cells are a potential source of intermediate cells in human prostate adenocarcinoma.
Fig. 1.
Intermediate prostate cells in human prostate adenocarcinoma. The cryosections of human prostate cancer biopsies were immunostained by the antibodies against CK8, AR (A), PSA (B), and CK5 (C and D). Arrows pointed to double positive cells. Arrowheads pointed to the CK8−CK5+ basal cells. DAPI stained nuclei. Scale bars, 100 μm
Expansion of primary human intermediate prostate cells in vitro
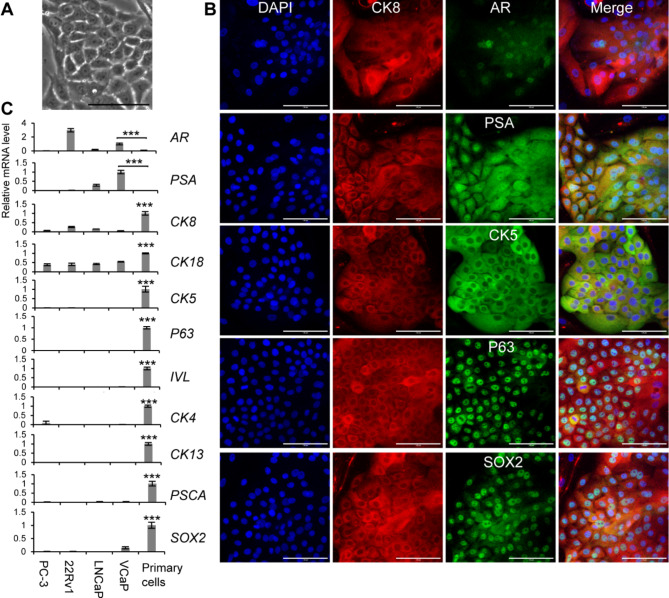
Using a custom-made medium based on our previous study [42], we were able to successfully culture primary human prostate cells in vitro (Fig. 2, A). Immunostaining revealed that the primary human prostate cells expressed luminal markers (CK8, AR, and PSA), basal markers (CK5 and P63), and stem cell marker SRY-box transcription factor 2 (SOX2) (Fig. 2, B). Several prostate cancer cell lines (PC-3, 22Rv1, LNCaP, and VCaP) have been widely used in prostate cancer research. One concern regarding the use of cell lines is that they may lose critical features of prostate cancer. Therefore, we performed qRT-PCR to compare the primary cells from this study with the prostate cancer cell lines. The qRT-PCR results showed that the primary cells had lower AR and PSA expression but higher levels of other genes compared to the prostate cancer cell lines. These genes included luminal markers (CK8 and CK18), basal markers (CK5 and P63), the intermediate cell marker (IVL), and stem cell markers (CK4, CK13, prostate stem cell antigen / PSCA, and SOX2) (Fig. 2, C). The marker expression profile of the primary human prostate epithelial cells in vitro indicates that they may be potential intermediate stem cells.
Fig. 2.
Marker expression profile of primary human intermediate prostate cells. A, The phase contrast image of primary human prostate cells. B, Immunostaining of primary human prostate cells by the antibodies against CK8, AR, PSA, CK5, P63, and SOX2. DAPI stained nuclei. Scale bars, 100 μm. C, qRT-PCR analysis of primary human prostate cells and prostate cancer cell lines, including PC-3, 22Rv1, LNCaP, and VCaP. The reference group was “VCaP” for AR and PSA, and “Primary cells” for the other genes. Data were presented as mean ± SD. N = 3. One-way ANOVA was performed on the data, followed by Bonferroni post hoc tests. ***p < 0.001
Castration resistance of primary human intermediate prostate cells in vitro
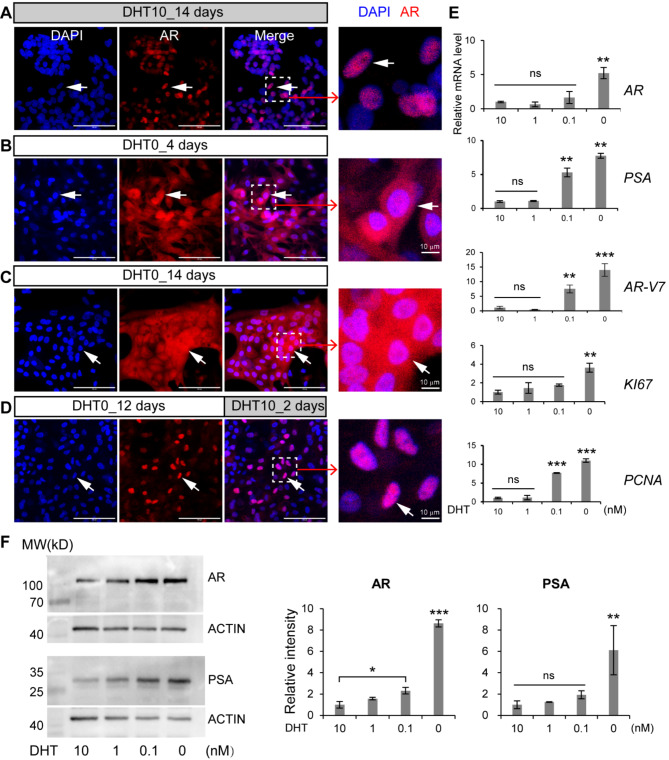
We next examined how primary human intermediate prostate cells respond to castration. We designed 4 experimental groups and cultured the cells in the media with (1) dihydrotestosterone (DHT) for 14 days, (2) no DHT for 4 days, (3) no DHT for 14 days, and (4) no DHT for 12 days followed by 2 days of 10 nM DHT, respectively. Our immunostaining results showed that some AR proteins were translocated from the nucleus to the cytoplasm upon castration (Fig. 3, A-C). However, when we restored DHT concentration in the medium, the AR proteins moved back into the nucleus (Fig. 3, D). We also conducted qRT-PCR and western blotting experiments to measure the expression of AR, PSA, and proliferation markers KI67 and proliferating cell nuclear antigen (PCNA) in response to different concentrations of DHT. We found that lower concentrations of DHT (0.1 and 0 nM) increased AR and PSA mRNA and protein levels, as well as the expression of KI67 and PCNA (Fig. 3, E and F). However, there were no significant differences between the 10 and 1 nM DHT groups (Fig. 3, E and F). Complete deprivation of DHT from the medium (0 nM) significantly elevated AR and PSA mRNA and protein levels and the expression of KI67 and PCNA mRNA (Fig. 3, E and F). It is unclear whether there are such regions in vivo that are absent of androgens. Some in vitro studies used 0.1 nM DHT to mimic low androgen concentrations in vivo for patients undergoing androgen deprivation therapy [44]. Our study showed that there were some variations between mRNA and protein levels of AR and PSA in the primary human intermediate prostate cells treated with 0.1 nM DHT. The AR protein level in the 0.1 nM DHT group was higher than that in the 10 nM group, although the difference in mRNA was insignificant (Fig. 3, E and F). PSA mRNA expression in the 0.1 nM group was higher than in the 10 and 1 nM groups. However, their differences in protein levels were insignificant (Fig. 3, E and F). The proliferation gene PCNA expression in the 0.1 nM group was higher than in the 10 and 1 nM groups, although another proliferation gene, KI67, showed no significant differences (Fig. 3, E). These results showed that the primary human intermediate prostate cells were able to resist castration treatment by increasing the expression of their AR, PSA, and proliferation markers.
Fig. 3.
Castration resistance of the primary human intermediate prostate cells. A-D, The cells were cultured in the media with 10 nM DHT (DHT10) for 14 days (A), no DHT (DHT0) for 4 days (B), no DHT for 14 days (C), no DHT for 12 days followed by 2 days of 10 nM DHT (D), before immunostaining by the antibody against AR. DAPI stained nuclei. Arrows pointed to AR+ cells. Scale bars, 100 μm. The cells were cultured in the media supplemented with different concentrations of DHT (10, 1, 0.1, and 0 nM) for 7 days before analysis by qRT-PCR (E) and western blotting (F). The 10 nM DHT group served as the reference. Data were presented as mean ± SD. N = 3. One-way ANOVA was performed on the data followed by Bonferroni post hoc tests. “ns”, not significant. *p < 0.05. **p < 0.01. ***p < 0.001. Full-length blots are presented in supplemental Figure S1
Tumorigenicity and castration resistance of primary human intermediate prostate cells in vivo
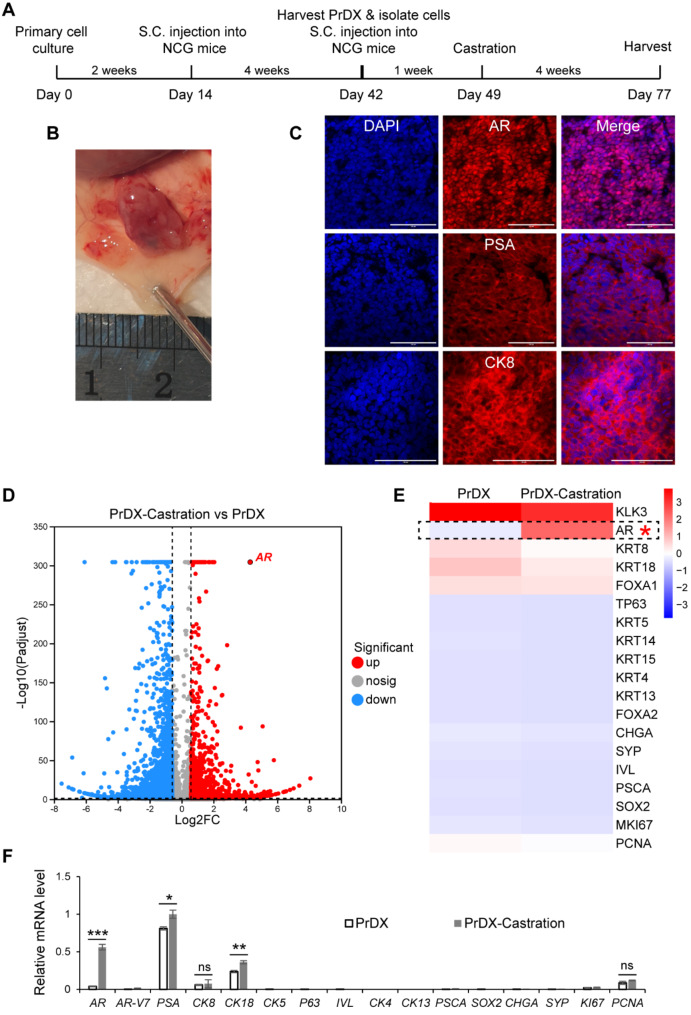
We cultured the primary human intermediate prostate epithelial cells based on the method that we had previously established for mouse prostate stem cells [42]. However, it was more challenging to expand primary human prostate cells, which lost proliferation capacity after a month of culture in vitro. To test their tumorigenicity in vivo, we thawed the first passage of cells, cultured them for about 2 weeks in vitro, and transplanted them into immunodeficient NCG mice subcutaneously within Matrigel (Fig. 4, A). We were able to obtain prostate tumors after 4 weeks (Fig. 4, B). Immunostaining revealed that these prostate tumors had strong and uniform staining signals of luminal markers such as AR, PSA, and CK8 (Fig. 4, C). We isolated the tumor cells from the first generation of PrDX tissues, injected them into NCG mice subcutaneously, and surgically castrated the mice after a week (Fig. 4, A). After 4 weeks, we obtained tumors, indicating their resistance to castration. We performed mRNA sequencing on the tumor tissues before and after castration and found that AR expression was significantly elevated upon castration in vivo (Fig. 4, D). The heatmap of the key prostate lineage biomarkers confirmed that castration did not change the lineage characteristic of the luminal adenocarcinoma, with high levels of luminal markers and low or no basal, intermediate or stem cell markers. This was also confirmed by qRT-PCR (Fig. 4, D and F). We found that compared to AR-V7, full-length AR expression was significantly increased (fold change = 14.5) upon surgical castration (Fig. 4, D-F). This might be the main reason underlying castration resistance.
Fig. 4.
Tumorigenicity and castration resistance of CriPCSCs in vivo. A, Workflow of cell and animal experiments. B, The PrDX tumor after 4 weeks in vivo. C, Immunostaining of the PrDX tumor cryosections by the antibodies against AR, PSA, and CK8. DAPI stained cell nuclei. Scale bars, 100 μm. D, The volcano map of differentially expressed genes after castration. E, The heatmap of selected genes. F, qRT-PCR analysis of the PrDX tumor samples before and after castration. The PSA level in the PrDX-Castration group served as the reference. Data were presented as mean ± SD. N = 3. The significance of the difference between the samples before and after castration was analyzed by t-test. “ns”, not significant. *p < 0.05, **p < 0.01, ***p < 0.001. The mRNA sequencing data for D and E are available in the supplemental materials
We cultured primary cells derived from prostate tumors for a short period of time in vitro before xenografting in vivo. This method differs from the classical patient-derived xenograft (PDX) model, where patient tumor tissues are directly transplanted into immunodeficient mice [45]. We thus named this method the patient primary cell-derived xenograft or PrDX. Based on the marker expression profile, tumorigenicity in vivo, and castration resistance in vitro and in vivo, we defined these cells as human castration-resistant intermediate prostate cancer stem cells or human CriPCSCs. The thriving culture of primary human CriPCSCs may be critical for successful PrDX.
Differentiation and dedifferentiation of human CriPCSCs
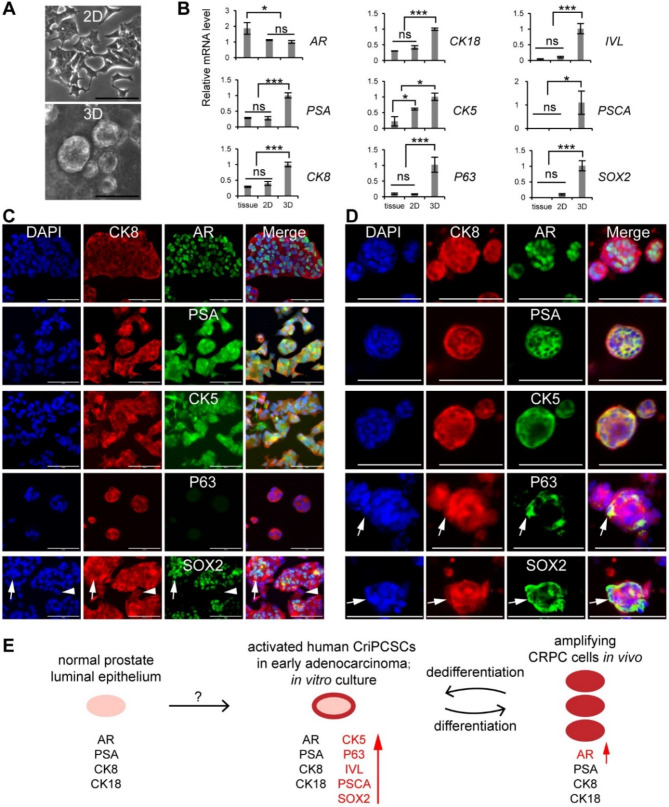
In this study, we observed that human CriPCSCs lost their intermediate cell markers and differentiated into luminal adenocarcinoma cells in fast-growing tumors in vivo (Fig. 4). We speculated that there could be some stem cells present among the luminal adenocarcinoma cells in vivo. To investigate their growth and gene expression patterns, we isolated cells from PrDX tumors and cultured them in vitro. The cells derived from PrDX tumors grew as monolayers in 2D culture and formed organoids in 3D Matrigel (Fig. 5, A). Gene expression analysis revealed that AR expression was downregulated in vitro, while PSA, CK8, and CK18 were elevated in 3D culture (Fig. 5, B). Immunostaining confirmed the presence of luminal markers (CK8, AR, and PSA) in both 2D and 3D cultures in vitro (Fig. 5, C and D). Additionally, the basal marker CK5 expression was elevated in both 2D and 3D cultures, as confirmed by qRT-PCR and immunostaining (Fig. 5, B-D). Further qRT-PCR analysis revealed that P63, IVL, PSCA, and SOX2 were elevated in 3D culture (Fig. 5, B). Immunostaining results showed few P63+ cells in 2D culture (Fig. 5, C) and only a few P63+ cells in 3D culture (Fig. 5, D), whereas many SOX2+ cells were present in both 2D and 3D cultures (Fig. 5, C and D, arrows). The study also found some CK8+SOX2− cells, which could be terminally differentiated adenocarcinoma cells (Fig. 5, C and D, arrowheads). These findings suggest that some luminal adenocarcinoma cells in PrDX tumors have the potential to dedifferentiate into stem cells.
Fig. 5.
Dedifferentiation of PrDX-derived cells to CriPCSCs in vitro. A, Phase contrast images of PrDX-derived cells in 2D and 3D culture. B, qRT-PCR analysis of the PrDX tissue, PrDX-derived cells in 2D and 3D culture. The 3D culture group served as the reference. Data were presented as mean ± SD. N = 3. One-way ANOVA was performed on the data followed by Bonferroni post hoc tests. “ns”, not significant. *p < 0.05, ***p < 0.001. C and D, Immunostaining of PrDX-derived cells cultured in 2D (C) and 3D (D). The antibodies were against CK8, AR, PSA, CK5, P63, and SOX2. Scale bars, 100 μm. E, Illustration of CriPCSCs activation, differentiation, and dedifferentiation
Castration and enzalutamide resistance of human CriPCSCs in vitro
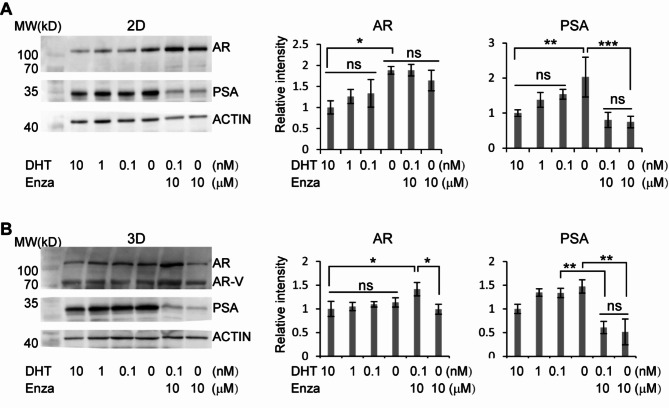
Human CriPCSCs isolated from PrDX tumors are a promising source of cells for drug screening in vitro. In our study, we treated these cells with varying concentrations of DHT, with or without enzalutamide (Enza), in 2D or 3D culture. Our results showed that the PrDX-derived cells sustained AR and PSA protein levels under low DHT conditions (Fig. 6) and even upregulated AR and PSA levels in the medium without DHT (0 nM) in 2D culture (Fig. 6, A), similar to primary cells isolated from clinical biopsies (Fig. 3). Our study further revealed that enzalutamide treatment significantly reduced PSA levels of the cells, consistent with its clinical performance, but failed to downregulate AR in both 2D and 3D cultures (Fig. 6). Interestingly, we even observed an increase of AR protein in the cells cultured in 0.1 nM DHT and 10 μm enzalutamide in 3D (Fig. 6, B). Additionally, western blotting of the cells in 3D culture showed bands of AR variants (Fig. 6, B), suggesting that the microenvironment in 2D and 3D cultures can influence AR variant expression. Based on our other results from in vivo (Figs. 4) and 2D culture in vitro (Fig. 3), we believe that the full-length AR may play a major role in the castration resistance of prostate adenocarcinoma. These findings indicate that human CriPCSCs may be an effective tool in the development of new treatments for prostate adenocarcinoma.
Fig. 6.
Castration and enzalutamide resistance of PrDX-derived CriPCSCs. 2D (A) and 3D (B) culture of PrDX-derived CriPCSCs in the media supplemented with different concentrations of DHT (10, 1, 0.1, and 0 nM) and enzalutamide (Enza, 10 µM) for a week before harvest for western blotting of AR and PSA. ACTIN was the internal control. The 10 nM group served as the reference. Data were presented as mean ± SD. N = 3. One-way ANOVA was performed on the data, followed by Bonferroni post hoc tests. *p < 0.05, **p < 0.01, ***p < 0.001. Full-length blots are presented in supplemental Figure S2
Discussion
Prostate cancer incidence has been increasing dramatically in recent years affecting millions of people worldwide [1, 2]. Understanding the cellular origins of prostate cancers is crucial for developing effective treatments. Recent studies supported luminal stem cells as the major origins of prostate adenocarcinoma [46, 47]. Our study suggests that some subpopulations among luminal stem cells give rise to intermediate stem cells contributing to prostate adenocarcinoma.
During the early stages of prostate development, intermediate prostate cells are the most dominant cell population [33, 34]. However, the number of these cells decreases significantly in the normal adult prostate [33]. Wang et al. found a low number of CK5+CK18+ intermediate cells in normal mouse prostate tissues (1.6%) and genetically engineered mouse prostate tumors (3%) [30]. Goldstein et al. found some intermediate cells in the human prostate by flow cytometry [26]. Recent single-cell sequencing studies identified intermediate cell populations in mouse prostate and human prostate tumors [31, 48]. Several lines of evidences support the luminal origin of intermediate cells during the development of prostate adenocarcinoma. Wang et al. used genetic lineage tracing mouse models and found that luminal origin tumors had more CK5+CK18+ intermediate cells than basal origin tumors of mice [30]. Chua et al. reported that luminal progenitor cells in mouse and human prostates can generate prostate organoids in vitro and can differentiate into basal cells and intermediate cells [49]. It was interesting that the human organoids seemed to have more intermediate cells than mouse organoids [49]. Our study demonstrated that CK5+CK8+ intermediate cells were located in the luminal compartment of prostate adenocarcinoma at the early stages (Fig. 1). Furthermore, the luminal adenocarcinoma cells derived from the PrDX tissues could dedifferentiate into intermediate cancer stem cells in vitro (Fig. 5). These findings further support the concept that luminal cells are an important source of prostate cancer stem cells. The differentiation and dedifferentiation mechanisms of human CriPCSCs in prostate adenocarcinoma require further study.
In terms of cell localization, previous immunohistological studies showed that the intermediate cells were mainly located in the luminal compartment in the early stage of normal prostate development and prostate tumors [34, 35]. Our study has confirmed this by double staining of CK5 and CK8 in clinical biopsies (Fig. 1). However, there have been conflicting reports on the location of these intermediate cells. Henry et al. reported that there were CK5+CK18+ cells underlying the CK5−CK18+ luminal cells in human prostate cryosections [50], suggesting the basal origin of intermediate cells. On the other hand, Shibata et al. used lineage-tracing mouse models and defined the CK5+CK8+ cells as the “transient bipotent luminal progenitor” during the early stage of mouse prostate development [32]. It is possible that certain subpopulations within luminal and basal cells may differentiate or dedifferentiate into intermediate cells under some unknown conditions. Further studies are required to fully understand the heterogeneity and origins of intermediate or bipotent prostate cells.
The emergence of intermediate cells in prostate tumors was believed to be the reactivation of the embryonic intermediate program [34–36]. While some studies have shown that intermediate cells can be cultured in vitro, few of them have demonstrated high tumorigenicity in vivo, except through genetic modification. Several groups have reported that when normal human prostate cells were isolated and cultured in vitro, most of them acquired the intermediate phenotype, expressing both luminal (CK8 or CK18) and basal (CK5 or CK14) markers [36–39]. In one study, Dean G. Tang’s group isolated primary cells from human prostate tissues and found that these cells expressed CK5 and CK18 without AR or PSA. However, these cells lost their proliferative capacity and the expression of CK5 and CK18 after a few passages in vitro [37]. The researchers then genetically immortalized the primary intermediate cells by p16 inhibition and hTERT overexpression to obtain prostate stem cells with the basal phenotype. Upon co-transplantation in vivo with rat UGSM [40] for three months, the immortalized human prostate stem cells differentiated into well-organized prostate glands containing luminal, basal, and neuroendocrine cells [37]. Another study by Jiang et al. expanded primary human prostate cells (BHPrE1) (PSA+CK14+P63+) by spontaneous immortalization in a custom-made culture medium [41]. These cells differentiated into benign prostate glands in vivo by tissue recombination xenografting with rat UGSM [41]. Additionally, Goldstein et al. sorted CD49floTrop2hi luminal cells and CD49fhiTrop2hi basal cells from human benign prostate tissues by flow cytometry [26]. They then infected the primary prostate cells with the lentivirus carrying the oncogenes AKT and ERG and transplanted them with mouse UGSM into NSG mice. In vivo tumor formation occurred only in the CD49fhiTrop2hi basal cells, which actually had an intermediate phenotype due to CK18 expression [26]. Studies from our group and others suggest that the intermediate program can be easily activated in vitro. However, the intermediate phenotype may not guarantee tumor formation in vivo. One possible reason for the successful tumor formation in our study may be the short-term culture of primary cells, which still express some unknown genes critical for tumorigenicity. Further studies are needed to understand the mechanisms regulating the intermediate phenotype and tumorigenicity.
The culture medium is essential for expanding stem cells in vitro. Several studies have developed custom-made culture media for expanding prostate stem cells. For instance, Zhang et al. formulated a culture medium (WIT) to expand primary luminal and basal stem cells from human benign prostate tissues [25]. They observed that culture medium can up or downregulate luminal or basal gene expression [25]. Similarly, Chan et al. discovered that in vitro culture and drug treatment could promote the intermediate cell phenotype in organoids from genetically engineered mouse prostate cancer cells and human prostate cancer cells [51]. Our previous studies formulated culture media for expanding adult stem cells, including mouse prostate stem cells [42, 52–54]. This study takes it a step further with a custom-made medium mainly supplemented with the Wnt signaling agonist Chir99021, TGFβ signaling inhibitor A83-01, and ROCK inhibitor Y27632, to culture human CriPCSCs. The human CriPCSCs expressed luminal (CK8, CK18, AR, PSA), basal (CK5, P63), intermediate (IVL), and stem cell markers (CK4, CK13, PSCA, SOX2), similar to the mouse prostate stem cells in our previous study [42]. Most importantly, these cells had high tumorigenicity in vivo, forming prostate tumors in just one month without the need for genetic modification or UGSM co-transplantation (Fig. 4). This is especially significant because establishing PDX from primary human prostate cancer biopsies can be challenging [9]. By expanding primary stem cells in vitro before xenografting in vivo, our PrDX method offers an alternative approach to successful PDX models.
Multiple mechanisms contribute to CRPC, such as AR amplification, AR alternative splicing, the loss of AR, lineage plasticity, aberrant metabolism, and genetic and epigenetic alterations [4–6, 51, 55–66]. In this study, AR amplification was found to be the primary cause of castration resistance, while AR variants may have played a minor role. However, the exact roles of the basal (CK5 and P63), intermediate (IVL), and stem cell markers (CK4, CK13, PSCA, and SOX2) in human CriPCSCs are still unclear, which warrants further studies. SOX2 upregulation has been linked to luminal to neuroendocrine transition [67, 68]. Our results showed that SOX2 expression was reversible in human CriPCSCs, which lost SOX2 in vivo. Although our experiments did not find significant neuroendocrine gene expression during in vitro culture and in vivo tumor formation, it is important to investigate whether long-term castration can induce SOX2 and neuroendocrine marker expression in the future.
Due to the limited number of tumor cells isolated from the clinical biopsies, PrDX-derived cells offer a rich source of cells for in vitro drug screening. The responses of human CriPCSCs to castration and enzalutamide were different from those of the mouse cells in our previous study [42], highlighting the value of human cells in translational medicine. The primary human CriPCSCs isolated from patient biopsies and the PrDX-derived cells were resistant to castration in 2D and 3D organoid culture by upregulating AR and PSA expression. This makes them the ideal model for testing anti-AR therapies. Upon treatment with the AR antagonist enzalutamide, PSA levels significantly dropped, consistent with clinical observations [69]. However, AR levels remained high even with enzalutamide treatment. In 3D culture, there was even a small increase in AR proteins in the group receiving 0.1 nM DHT and 10 µM enzalutamide. The 2D and 3D cultures showed similar trends in AR and PSA upon castration and enzalutamide treatment, except that there were bands of AR variants in 3D culture groups. In vivo data showed that full-length AR played a major role in castration resistance, but the role of AR variants in drug resistance needs clarification in the future. Although enzalutamide promoted patient survival and reduced PSA levels, most patients inevitably become resistant [70–74]. Further studies are required to investigate the mechanisms underlying enzalutamide resistance.
This study has some limitations that need to be considered. Firstly, it was challenging to culture the human CriPCSCs in vitro for an extended period of time. Although they grew well in the first few weeks, they eventually underwent senescence and lost their proliferation capacity after about four weeks in vitro. Therefore, primary cells were cultured for approximately two weeks in vitro before they were xenografted into immunodeficient mice. Secondly, the PrDX-derived cells showed an upregulation of basal and stem cell markers in vitro, which is different from the luminal adenocarcinoma cells in vivo. This means that in vitro drug tests on these cells may not fully replicate the cell in vivo. In prostate cancers and other cancer types, patient-derived organoids have been widely used for drug screening [44, 75–83]. It is worth noting that the phenotype change of the organoids may be a factor to consider when interpreting results.
Conclusions
Our study revealed the presence of intermediate cells in the luminal compartment of human prostate adenocarcinoma. We have successfully cultivated human castration-resistant intermediate prostate cancer stem cells (CriPCSCs) and established patient primary cell-derived xenograft (PrDX), which can serve as a valuable platform for further research. The differentiation and dedifferentiation of human CriPCSCs may play an essential role in the development of prostate adenocarcinoma and drug resistance. We believe that further investigation into the mechanisms regulating human CriPCSCs will bring new insights into prostate adenocarcinoma and lead to the development of more effective therapies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Thanks to Zhishang Chang, Qian Wen, Xuxia Song, and Bing Wang at the Laboratory of Biomedical Center, Qingdao University, for their technical help. Thanks to Lulu Song and Guangdong Mei for their help in the animal facility of Qingdao University. We apologize to the scientists whose work was not cited due to space constraint.
Abbreviations
- CRPC
Castration-resistant prostate cancer
- CriPCSC
Castration-resistant intermediate prostate cancer stem cell
- PrDX
Patient primary cell-derived xenograft
- AR
Androgen receptor
- PSA
Prostate-specific antigen
- PSCA
Prostate stem cell antigen
- UGSM
Urogenital sinus mesenchyme
- DHT
Dihydrotestosterone
Author contributions
D.W., H.Z., J.L., and A.L. conceived and designed the study. J.M., R.L., Y.Z., and Y.L. performed cell culture. J.M., R.L., M.Z., W.M., and M.C. performed animal experiments. J.M. performed qRT-PCR and western blotting. J.M. and R.L. performed immunostaining and J.M. and D.W. performed confocal imaging. D.W., H.Z., J.L., A.L., and J.M. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the Natural Science Foundation of Shandong Province (No. ZR2023MH327 and ZR2019LZL001), the National Nature Science Foundation of China (No. 32101020), the Natural Science Foundation of Qingdao (No. 23-2-1-193-zyyd-jch), and the “Medicine+” Program of Medical College of Qingdao University.
Data availability
The mRNA sequencing data for Fig. 4, panels D and E, are available in the supplemental materials. The antibodies and primers are listed in Tables S1 and S2, respectively, in the supplemental materials. The uncropped blots of Figs. 3 and 6 are presented in Figures S1 and S2, respectively, in the supplemental materials.
Declarations
Ethics approval and consent to participate
The patient prostate cancer biopsies were obtained and animal procedures were performed according to the Ministry of Science and Technology guide and approved by the Ethics Committee of Medical College of Qingdao University. The patient provided written informed consent for participation in the study and the use of samples. The ethics document for the human study was approved on September 26, 2022, with the document number QDU-HEC-2022177. The ethics document for the animal study was approved on April 29, 2022, with the document number QDU-AEC-2022305.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aihua Liu, Email: liuah@qdu.edu.cn.
Jing Li, Email: jingli@qdu.edu.cn.
Hai Zhu, Email: shijingzhou@163.com.
Dong Wang, Email: dongwang@qdu.edu.cn.
References
- 1.Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2016. J Natl Cancer Cent. 2022;2(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. Cancer J Clin. 2022;72(1):7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 3.Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, Wang H, Yu Y, Yang C, Gao X, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580(7801):93–9. 10.1038/s41586-020-2135-x [DOI] [PubMed] [Google Scholar]
- 4.Tang DG. Understanding and targeting prostate cancer cell heterogeneity and plasticity. Sem Cancer Biol. 2022;82:68–93. 10.1016/j.semcancer.2021.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai C, Heemers H, Sharifi N. Androgen signaling in prostate Cancer. Cold Spring Harbor Perspect Med 2017, 7(9). [DOI] [PMC free article] [PubMed]
- 6.Tang F, Xu D, Wang S, Wong CK, Martinez-Fundichely A, Lee CJ, Cohen S, Park J, Hill CE, Eng K, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science. 2022;376(6596):eabe1505. 10.1126/science.abe1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebello RJ, Oing C, Knudsen KE, Loeb S, Johnson DC, Reiter RE, Gillessen S, Van der Kwast T, Bristow RG. Prostate cancer. Nat Reviews Disease Primers. 2021;7(1):9. 10.1038/s41572-020-00243-0 [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Wang C, Huang H, Yang Y, Dai L, Han S, Xing N, Ren S. SEMA3A-mediated crosstalk between prostate cancer cells and tumor-associated macrophages promotes androgen deprivation therapy resistance. Cell Mol Immunol. 2021;18(3):752–4. 10.1038/s41423-021-00637-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JJ, Shen MM. Prostate stem cells and Cancer Stem cells. Cold Spring Harbor Perspect Med 2019, 9(6). [DOI] [PMC free article] [PubMed]
- 10.Skvortsov S, Skvortsova II, Tang DG, Dubrovska A. Concise review: prostate Cancer stem cells: current understanding. Stem Cells. 2018;36(10):1457–74. 10.1002/stem.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZA, Toivanen R, Bergren SK, Chambon P, Shen MM. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8(5):1339–46. 10.1016/j.celrep.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Jeter C, Gong S, Tracz A, Lu Y, Shen J, Tang DG. Histone 2B-GFP label-retaining prostate luminal cells possess Progenitor Cell properties and are intrinsically resistant to Castration. Stem cell Rep. 2018;10(1):228–42. 10.1016/j.stemcr.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu S, Dong Y, Kumar R, Jeter C, Tang DG. Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis. Sem Cancer Biol. 2022;78:90–103. 10.1016/j.semcancer.2021.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Shen MM. Prostate cancer cell heterogeneity and plasticity: insights from studies of genetically-engineered mouse models. Sem Cancer Biol. 2022;82:60–7. 10.1016/j.semcancer.2021.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo YA, Vatapalli R, Lysy B, Mok H, Desouki MM, Abdulkadir SA. The role of castration-resistant Bmi1 + Sox2 + cells in driving recurrence in prostate Cancer. J Natl Cancer Inst. 2019;111(3):311–21. 10.1093/jnci/djy142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ittmann M. Anatomy and Histology of the Human and Murine Prostate. Cold Spring Harbor perspectives in medicine 2018, 8(5). [DOI] [PMC free article] [PubMed]
- 17.Chen X, Li Q, Liu X, Liu C, Liu R, Rycaj K, Zhang D, Liu B, Jeter C, Calhoun-Davis T, et al. Defining a Population of stem-like human prostate Cancer cells that can generate and propagate castration-resistant prostate Cancer. Clin cancer Research: Official J Am Association Cancer Res. 2016;22(17):4505–16. 10.1158/1078-0432.CCR-15-2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Q, Butler W, Zhou Y, Zhang H, Tang L, Perkinson K, Chen X, Jiang XS, McCall SJ, Inman BA, et al. Pre-existing castration-resistant prostate Cancer-like cells in primary prostate Cancer Promote Resistance to Hormonal Therapy. Eur Urol. 2022;81(5):446–55. 10.1016/j.eururo.2021.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, Carver B, Gopalan A, Abida W, Laudone V, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368(6490):497–505. 10.1126/science.aay0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Li L, He J, Liu Z, Han M, Li F, Xia X, Zhang X, Zhu Y, Wei Y, et al. Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips. Nat Genet. 2020;52(9):908–18. 10.1038/s41588-020-0642-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, et al. Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate. Nat Commun. 2020;11(1):706. 10.1038/s41467-020-14296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R et al. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife 2020, 9. [DOI] [PMC free article] [PubMed]
- 23.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5(4):702–13. 10.1038/nprot.2010.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25(11):2760–9. 10.1634/stemcells.2007-0355 [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Lin K, Lu Y, Rycaj K, Zhong Y, Chao HP, Calhoun-Davis T, Shen J, Tang DG. Developing a Novel two-Dimensional Culture System to enrich human prostate luminal progenitors that can function as a cell of origin for prostate Cancer. Stem Cells Translational Med. 2017;6(3):748–60. 10.5966/sctm.2016-0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate Cancer. Science. 2010;329(5991):568–71. 10.1126/science.1189992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362(6410):91–5. 10.1126/science.aat5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, Reiter RE, Huang J, Witte ON. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. 2011;6(5):656–67. 10.1038/nprot.2011.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunha GR, Cao M, Derpinghaus A, Baskin LS. Human urogenital sinus mesenchyme is an inducer of prostatic epithelial development. Am J Clin Experimental Urol. 2021;9(4):329–36. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15(3):274–83. 10.1038/ncb2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Zhu G, Yang Y, Wang F, Xiao YT, Zhang N, Bian X, Zhu Y, Yu Y, Liu F, et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nat Cell Biol. 2021;23(1):87–98. 10.1038/s41556-020-00613-6 [DOI] [PubMed] [Google Scholar]
- 32.Shibata M, Epsi NJ, Xuan S, Mitrofanova A, Shen MM. Bipotent progenitors do not require androgen receptor for Luminal specification during prostate organogenesis. Stem cell Rep. 2020;15(5):1026–36. 10.1016/j.stemcr.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Hayward S, Cao M, Thayer K, Cunha G. Cell differentiation lineage in the prostate. Differ Res Biol Divers. 2001;68(4–5):270–9. [DOI] [PubMed] [Google Scholar]
- 34.Xue Y, Smedts F, Debruyne FM, de la Rosette JJ, Schalken JA. Identification of intermediate cell types by keratin expression in the developing human prostate. Prostate. 1998;34(4):292–301. [DOI] [PubMed] [Google Scholar]
- 35.Verhagen AP, Ramaekers FC, Aalders TW, Schaafsma HE, Debruyne FM, Schalken JA. Colocalization of basal and luminal cell-type cytokeratins in human prostate cancer. Cancer Res. 1992;52(22):6182–7. [PubMed] [Google Scholar]
- 36.van Leenders G, Dijkman H, Hulsbergen-van de Kaa C, Ruiter D, Schalken J. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab Invest. 2000;80(8):1251–8. 10.1038/labinvest.3780133 [DOI] [PubMed] [Google Scholar]
- 37.Bhatia B, Jiang M, Suraneni M, Patrawala L, Badeaux M, Schneider-Broussard R, Multani AS, Jeter CR, Calhoun-Davis T, Hu L, et al. Critical and distinct roles of p16 and telomerase in regulating the proliferative life span of normal human prostate epithelial progenitor cells. J Biol Chem. 2008;283(41):27957–72. 10.1074/jbc.M803467200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garraway LA, Lin D, Signoretti S, Waltregny D, Dilks J, Bhattacharya N, Loda M. Intermediate basal cells of the prostate: in vitro and in vivo characterization. Prostate. 2003;55(3):206–18. 10.1002/pros.10244 [DOI] [PubMed] [Google Scholar]
- 39.Tokar EJ, Ancrile BB, Cunha GR, Webber MM. Stem/progenitor and intermediate cell types and the origin of human prostate cancer. Differ Res Biol Divers. 2005;73(9–10):463–73. [DOI] [PubMed] [Google Scholar]
- 40.Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR. Interactions between adult human prostatic epithelium and rat urogenital sinus mesenchyme in a tissue recombination model. Differ Res Biol Divers. 1998;63(3):131–40. [DOI] [PubMed] [Google Scholar]
- 41.Jiang M, Strand DW, Fernandez S, He Y, Yi Y, Birbach A, Qiu Q, Schmid J, Tang DG, Hayward SW. Functional remodeling of benign human prostatic tissues in vivo by spontaneously immortalized progenitor and intermediate cells. Stem Cells. 2010;28(2):344–56. 10.1002/stem.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Mu J, Zhou Z, Leng Y, Yu Y, Song X, Liu A, Zhu H, Li J, Wang D. Expansion of mouse castration-resistant intermediate prostate stem cells in vitro. Stem Cell Res Ther. 2022;13(1):299. 10.1186/s13287-022-02978-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rycaj K, Cho EJ, Liu X, Chao HP, Liu B, Li Q, Devkota AK, Zhang D, Chen X, Moore J, et al. Longitudinal tracking of subpopulation dynamics and molecular changes during LNCaP cell castration and identification of inhibitors that could target the PSA-/lo castration-resistant cells. Oncotarget. 2016;7(12):14220–40. 10.18632/oncotarget.7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. 10.1016/j.cell.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karkampouna S, La Manna F, Benjak A, Kiener M, De Menna M, Zoni E, Grosjean J, Klima I, Garofoli A, Bolis M, et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat Commun. 2021;12(1):1117. 10.1038/s41467-021-21300-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crowell PD, Fox JJ, Hashimoto T, Diaz JA, Navarro HI, Henry GH, Feldmar BA, Lowe MG, Garcia AJ, Wu YE, et al. Expansion of Luminal Progenitor cells in the Aging mouse and human prostate. Cell Rep. 2019;28(6):1499–e15101496. 10.1016/j.celrep.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, et al. Low CD38 identifies progenitor-like inflammation-Associated Luminal cells that can initiate human prostate Cancer and predict poor outcome. Cell Rep. 2016;17(10):2596–606. 10.1016/j.celrep.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MT, Malladi VS, et al. Urethral luminal epithelia are castration-insensitive cells of the proximal prostate. Prostate. 2020;80(11):872–84. 10.1002/pros.24020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16(10):951–61. 10.1038/ncb3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, et al. A Cellular anatomy of the normal adult human prostate and Prostatic Urethra. Cell Rep. 2018;25(12):3530–e35423535. 10.1016/j.celrep.2018.11.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan JM, Zaidi S, Love JR, Zhao JL, Setty M, Wadosky KM, Gopalan A, Choo Z-N, Persad S, Choi J, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 2022;377(6611):1180–91. 10.1126/science.abn0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Wang E, Liu K, Xia CH, Li S, Gong X. Roles of TGFbeta and FGF signals during growth and differentiation of mouse lens epithelial cell in vitro. Sci Rep. 2017;7(1):7274. 10.1038/s41598-017-07619-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, Wu F, Yuan H, Wang A, Kang GJ, Truong T, Chen L, McCallion AS, Gong X, Li S. Sox10(+) cells contribute to Vascular Development in multiple organs-brief report. Arterioscler Thromb Vasc Biol. 2017;37(9):1727–31. 10.1161/ATVBAHA.117.309774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D, Wang A, Wu F, Qiu X, Li Y, Chu J, Huang WC, Xu K, Gong X, Li S. Sox10(+) adult stem cells contribute to biomaterial encapsulation and microvascularization. Sci Rep. 2017;7:40295. 10.1038/srep40295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirk JS, Wang J, Tracz A, Long M, Rosario SR, Ji Y, Kumar R, Liu X, Singh PK, Puzanov I et al. Integrated single-cell analysis defines the epigenetic basis of castration-resistant prostate luminal cells. bioRxiv: the preprint server for biology 2023. [DOI] [PMC free article] [PubMed]
- 56.Zhang SY, Zeng Y. Research progress of m(6)a methylation in prostate cancer. Asian J Androl. 2023;25(2):166–70. 10.4103/aja202265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Li WJ, Puzanov I, Goodrich DW, Chatta G, Tang DG. Prostate cancer as a dedifferentiated organ: androgen receptor, cancer stem cells, and cancer stemness. Essays Biochem. 2022;66(4):291–303. 10.1042/EBC20220003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Chen J, Gao WQ, Yang R. METTL14 promotes prostate tumorigenesis by inhibiting THBS1 via an m6A-YTHDF2-dependent mechanism. Cell Death Discovery. 2022;8(1):143. 10.1038/s41420-022-00939-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng S, Hu P, Xiao YT, Lu W, Guo D, Hu S, Xie J, Wang M, Yu W, Yang J, et al. Single-cell analysis reveals EP4 as a target for restoring T-Cell infiltration and sensitizing prostate Cancer to Immunotherapy. Clin cancer Research: Official J Am Association Cancer Res. 2022;28(3):552–67. 10.1158/1078-0432.CCR-21-0299 [DOI] [PubMed] [Google Scholar]
- 60.Han M, Li F, Zhang Y, Dai P, He J, Li Y, Zhu Y, Zheng J, Huang H, Bai F, et al. FOXA2 drives lineage plasticity and KIT pathway activation in neuroendocrine prostate cancer. Cancer Cell. 2022;40(11):1306–e13231308. 10.1016/j.ccell.2022.10.011 [DOI] [PubMed] [Google Scholar]
- 61.Ahmed M, Soares F, Xia JH, Yang Y, Li J, Guo H, Su P, Tian Y, Lee HJ, Wang M, et al. CRISPRi screens reveal a DNA methylation-mediated 3D genome dependent causal mechanism in prostate cancer. Nat Commun. 2021;12(1):1781. 10.1038/s41467-021-21867-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu B, Liu Y, Luo H, Fu J, Li Y, Shao C. Androgen receptor splicing variant 7 (ARV7) inhibits docetaxel sensitivity by inactivating the spindle assembly checkpoint. J Biol Chem. 2021;296:100276. 10.1016/j.jbc.2021.100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong B, Miao J, Wang Y, Luo W, Ji Z, Lai H, Zhang M, Cheng X, Wang J, Fang Y, et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biology. 2020;3(1):778. 10.1038/s42003-020-01476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q, Deng Q, Chao HP, Liu X, Lu Y, Lin K, Liu B, Tang GW, Zhang D, Tracz A, et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat Commun. 2018;9(1):3600. 10.1038/s41467-018-06067-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Y, Lu J, Ye Z, Hao S, Wang L, Kohli M, Tindall DJ, Li B, Zhu R, Huang H. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018;46(4):1895–911. 10.1093/nar/gkx1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abudurexiti M, Zhu W, Wang Y, Wang J, Xu W, Huang Y, Zhu Y, Shi G, Zhang H, Shen Y, et al. Targeting CPT1B as a potential therapeutic strategy in castration-resistant and enzalutamide-resistant prostate cancer. Prostate. 2020;80(12):950–61. 10.1002/pros.24027 [DOI] [PubMed] [Google Scholar]
- 67.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–8. 10.1126/science.aah4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwon OJ, Zhang L, Jia D, Zhou Z, Li Z, Haffner M, Lee JK, True L, Morrissey C, Xin L. De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc. Oncogene. 2020;39(48):7142–51. 10.1038/s41388-020-01487-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merseburger AS, Haas GP, von Klot CA. An update on enzalutamide in the treatment of prostate cancer. Ther Adv Urol. 2015;7(1):9–21. 10.1177/1756287214555336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Chu Y, Shi G, Wang X, Ye W, Shan C, Wang D, Zhang D, He W, Jiang J, et al. A novel inhibitor of ARfl and ARv7 induces protein degradation to overcome enzalutamide resistance in advanced prostate cancer. Acta Pharm Sinica B. 2022;12(11):4165–79. 10.1016/j.apsb.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng H, Fazli L, Jeffrey Karnes R, Jimenez R, Wang L, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun. 2021;12(1):1521. 10.1038/s41467-021-21860-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoshi S, Meguro S, Imai H, Matsuoka Y, Yoshida Y, Onagi A, Tanji R, Honda-Takinami R, Matsuoka K, Koguchi T, et al. Upregulation of glucocorticoid receptor-mediated glucose transporter 4 in enzalutamide-resistant prostate cancer. Cancer Sci. 2021;112(5):1899–910. 10.1111/cas.14865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Peng Y, Dong S, Hou D, Li N, Li H, Li T, Zhang Z, Wang H. A comprehensive characterization of the transcriptome in enzalutamide resistance prostate cancer. Annals Translational Med. 2021;9(24):1782. 10.21037/atm-21-6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, Aggarwal R, Cetnar J, Ryan CJ, Tabatabaei S, et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci USA. 2020;117(22):12315–23. 10.1073/pnas.1922207117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richards Z, McCray T, Marsili J, Zenner ML, Manlucu JT, Garcia J, Kajdacsy-Balla A, Murray M, Voisine C, Murphy AB, et al. Prostate stroma increases the viability and maintains the branching phenotype of human prostate organoids. iScience. 2019;12:304–17. 10.1016/j.isci.2019.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M, et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun. 2018;9(1):2404. 10.1038/s41467-018-04495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, Jansson KH, Yang Q, McGowen KM, Yin J, et al. A PDX/Organoid Biobank of Advanced Prostate Cancers captures genomic and phenotypic heterogeneity for Disease modeling and therapeutic screening. Clin cancer Research: Official J Am Association Cancer Res. 2018;24(17):4332–45. 10.1158/1078-0432.CCR-18-0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S, Gao D, Chen Y. The potential of organoids in urological cancer research. Nat Reviews Urol. 2017;14(7):401–14. 10.1038/nrurol.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11(2):347–58. 10.1038/nprot.2016.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fong EL, Wan X, Yang J, Morgado M, Mikos AG, Harrington DA, Navone NM, Farach-Carson MC. A 3D in vitro model of patient-derived prostate cancer xenograft for controlled interrogation of in vivo tumor-stromal interactions. Biomaterials. 2016;77:164–72. 10.1016/j.biomaterials.2015.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–75. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pamarthy S, Sabaawy HE. Patient derived organoids in prostate cancer: improving therapeutic efficacy in precision medicine. Mol Cancer. 2021;20(1):125. 10.1186/s12943-021-01426-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Servant R, Garioni M, Vlajnic T, Blind M, Pueschel H, Muller DC, Zellweger T, Templeton AJ, Garofoli A, Maletti S, et al. Prostate cancer patient-derived organoids: detailed outcome from a prospective cohort of 81 clinical specimens. J Pathol. 2021;254(5):543–55. 10.1002/path.5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mRNA sequencing data for Fig. 4, panels D and E, are available in the supplemental materials. The antibodies and primers are listed in Tables S1 and S2, respectively, in the supplemental materials. The uncropped blots of Figs. 3 and 6 are presented in Figures S1 and S2, respectively, in the supplemental materials.