Abstract
PARG [poly(ADP-ribose) glycohydrolase] catalyses the hydrolysis of α(1″→2′) or α(1‴→2″) O-glycosidic linkages of ADP-ribose polymers to produce free ADP-ribose. We investigated possible mechanistic similarities between PARG and glycosidases, which also cleave O-glycosidic linkages. Glycosidases typically utilize two acidic residues for catalysis, thus we targeted acidic residues within a conserved region of bovine PARG that has been shown to contain an inhibitor-binding site. The targeted glutamate and aspartate residues were changed to asparagine in order to minimize structural alterations. Mutants were purified and assayed for catalytic activity, as well as binding, to an immobilized PARG inhibitor to determine ability to recognize substrate. Our investigation revealed residues essential for PARG catalytic activity. Two adjacent glutamic acid residues are found in the conserved sequence Gln755-Glu-Glu757, and a third residue found in the conserved sequence Val737-Asp-Phe-Ala-Asn741. Our functional characterization of PARG residues, along with recent identification of an inhibitor-binding residue Tyr796 and a glycine-rich region Gly745-Gly-Gly747 important for PARG function, allowed us to define a PARG ‘signature sequence’ [vDFA-X3-GGg-X6–8-vQEEIRF-X3-PE-X14-E-X12-YTGYa], which we used to identify putative PARG sequences across a range of organisms. Sequence alignments, along with our mapping of PARG functional residues, suggest the presence of a conserved catalytic domain of approx. 185 residues which spans residues 610–795 in bovine PARG.
Keywords: acidic mutation, glycohydrolase, poly(ADP-ribose) glycohydrolase (PARG), poly(ADP-ribosylation)
Abbreviations: ADP-HPD, adenosine diphosphate (hydroxymethyl)pyrrolidinediol; 8-AH-ADP-HPD, 8-(6-aminohexyl)amino-ADP-HPD; PARG, poly(ADP-ribose) glycohydrolase; PARP, poly(ADP-ribose) polymerase; rPARG-CF, recombinant PARG catalytic fragment; SPR, surface plasmon resonance
INTRODUCTION
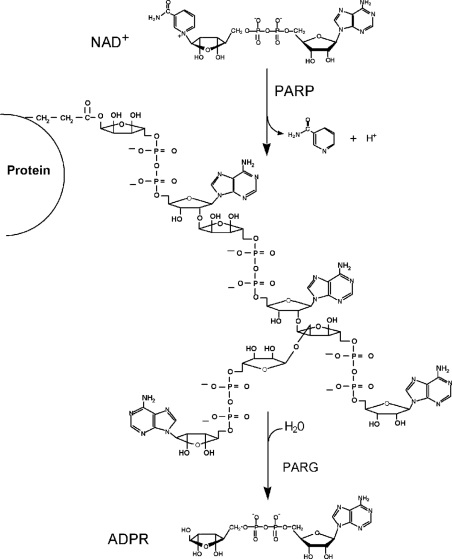
The modification of proteins by ADP-ribose polymers is catalysed by a family of PARPs [poly(ADP-ribose) polymerases] involved in the maintenance of genomic integrity [1–3] (Figure 1). The most characterized member of this family, PARP-1 [4,5], is activated in response to genotoxic stress [6], leading to the modification of nuclear proteins that include histones, p53 and PARP-1 itself. The rapid increase in ADP-ribose polymers in response to DNA damage involves both PARP-1 and PARP-2 [7], whereas polymer degradation is under the control of a single enzyme PARG [poly(ADP-ribose) glycohydrolase] [8,9] (Figure 1). The majority of the studies on ADP-ribose polymer metabolism have focused on PARPs, and include studies of knock-out mice [5,10–12], the utilization of chemical inhibitors [13–12] and antisense RNA techniques [18]. Reduction in PARP-1 or PARP-2 activity leads to genomic instability, and PARP-1 knock-out mice show increased sensitivity to some types of DNA damage, but remarkable resistance to massive DNA damage that occurs in non-proliferating cells following ischaemia and reperfusion stroke and some types of neurotoxic agents [13,19–21].
Figure 1. ADP-ribose polymer cycles catalysed by PARPs and PARG.
The structure and functional relationships of PARG are poorly understood. Bovine PARG cDNA [9] codes for a 111-kDa protein containing a C-terminal catalytic region and a putative N-terminal regulatory region. PARG and PARP-1 are cleaved in cells undergoing apoptosis, and caspase-3 has been implicated in the cleavage [22,23]. Immunofluorescence analysis of Cos-7 cells over-expressing PARG has found it to be mostly cytoplasmic [24], and recently a splice variant containing a nuclear localization signal has been identified that targets PARG to the nucleus [25]. Furthermore, three recent knock-out experiments of PARG in Drosophila [26] and in mice [27,28] show that depletion of PARG plays an important role in neurodegeneration and sensitivity to DNA-damage-induced cell death, revealing a potential new target for anticancer and anti-inflammatory drugs.
The human PARG gene has recently been characterized [25], and genomic sequence searches using a catalytic fragment of PARG (Bos taurus amino acids 380–977) has so far identified only a single mammalian PARG sequence. This raises the possibility that a single gene codes for PARG, which completes the ADP-ribose polymer cycles initiated by many different PARPs. A photolabelling approach with a potent PARG inhibitor has been used to identify an amino acid residue involved in inhibitor binding at a site that presumably is also involved in substrate binding [29]. This approach along with the present investigation has enabled the identification of PARG residues critical for catalytic activity and the elucidation of highly conserved amino acids that constitute a PARG signature sequence.
EXPERIMENTAL
Mutagenesis
The PCR reaction mixtures comprised 50 ng of template DNA, 125 ng of each primer, 1 mM dNTPs and Stratagene reaction buffer (20 mM MgSO4). High-fidelity PfuTurbo DNA polymerase (Stratagene) and 16 PCR cycles were used to reduce the chances of random mutations [30]. The methylated template DNA was digested using DpnI (Stratagene). After digestion, 5 μl of the PCR reaction was used to transform Escherichia coli XL-1 Blue (Stratagene) competent cells. Clones were sequenced with upstream (5′-GGAAACGGCCAGGGCATGCTAC-3′) and downstream (3′-CTGCTGACCGTCTCCGCGTGC-5′) primers to verify the mutation. The cloned plasmid containing the mutation was transformed into E. coli NM522 cells for expression. Primers were made following the guidelines in the QuikChange® site-directed mutagenesis kit (Stratagene). In general, the primers had approx. 10–15 bp on each side of the mutation, the GC content was above 40%, and they terminated in G or C bases. The sequences of primers used for the various acidic mutants are given in Supplementary Table 1 (http://www.BiochemJ.org/bj/388/bj3880493add.htm).
Expression and purification of rPARG-CF (recombinant PARG catalytic fragment) and mutants
rPARG-CF and all mutants we generated in the present study were purified as previously reported [9], with some modifications (see Supplementary Figure 1, http://www.BiochemJ.org/bj/388/bj3880493add.htm). Briefly, transformed E. coli NM522 cells containing the pGEX-2T-MNDV vector encoding glutathione S-transferase and 59-kDa rPARG-CF fusion protein, containing a protease site sensitive to thrombin, were incubated at 37 °C in 1 litre of LB (Luria–Bertani) medium containing 150 μg/ml ampicillin, and were grown to a D600 of approx. 0.6 (approx. 2 h). Expression of rPARG-CF mutant was induced using 0.1 mM IPTG (isopropyl β-D-thiogalactoside) for 4 h, cells were collected by centrifugation (12000 g for 15 min at 4 °C), and the crude extract was prepared by sonication in a buffer containing 20 ml of PBS, 10 mM EDTA and 5% bacterial protease inhibitor cocktail per litre of cell suspension. The bacterial protease inhibitor cocktail contained AEBSF [4-(2-aminoethyl)benzenesulphonyl fluoride], pepstatin A, E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane], bestatin and sodium EDTA in freeze-dried form (Sigma). The volume was adjusted to 80 ml using PBS, and Triton X-100 and dithiothreitol were added to final concentrations of 2% and 4 mM respectively, and the resulting extract was shaken at 4 °C for 30 min using a Clay Adams Brand Nutator Model 421105. The crude extract was then clarified by centrifugation (12000 g for 15 min at 4 °C) and GSH–Sepharose 4B was applied to the cleared lysate to a final concentration of 5% (v/v) for 4 h at 4 °C with constant agitation using the Nutator. The resin was recovered by centrifugation (500 g), washed twice with PBS, and washed once with PBS plus 300 mM NaCl. The resin was resuspended in PBS containing 1 mM CaCl2, 300 mM NaCl and 50 units of thrombin protease. The resulting suspension was agitated at room temperature overnight. The released rPARG-CF mutant was then isolated by centrifugation (500 g) of the GSH–Sepharose 4B suspension and the supernatant was concentrated using an Amicon Centriprep-10 apparatus. The final products were stored at −80 °C in a buffer containing PBS, 300 mM KCl, 10% glycerol, 0.1% Triton X-100, 10 mM 2-mercaptoethanol and protease inhibitors (10 mM EDTA, 1 mM PMSF, 2 μg/ml aprotinin and 100 μM antipain). Protein levels were analysed by SDS/PAGE (12% gel), assayed for PARG activity and the total protein yield was quantified by Bradford protein assay.
PARG assay
This assay utilizes the conditions previously reported [31]. Briefly, a typical reaction mixture was prepared in a 1.5-ml microfuge tube by adding 10 μl of 3×glyco assay buffer (150 mM potassium phosphate buffer, pH 7.5, 150 mM KCl, 0.3 mg/ml BSA and 30 mM 2-mercaptoethanol), 5 μl of 60 μM [α-32P]ADP-ribose polymers (approx. 5000 cpm/μl) and 10 μl of deionized water. The reaction was initiated by the addition of 5 μl of PARG stock or PARG extract, and the reaction mixture was incubated at 37 °C for 3–10 min. The reaction was then stopped by the addition of 2 μl of 3% SDS. Next, 3 μl of the reaction mixture was loaded on to a 2×10 cm poly(ethyleneimine)–cellulose F TLC plate, pre-spotted with 2 μl of 25 mM ADP-ribose. The spot was dried and the plate was placed into a TLC tank with 100 ml of methanol and developed until the solvent front reached the top of the plate. Next, the plate was transferred into a second tank containing 100 ml of a solution comprising 0.9 M acetic acid and 0.3 M LiCl. After developing the plate as before, it was air-dried and the ADP-ribose spot was visualized by using a hand-held short-wave UV lamp. The origin and ADP-ribose spot were excised, placed into 20-ml scintillation vials with 10 ml of EcoLume added, and the radioactivity was counted in a Beckman liquid scintillation counter. One unit of PARG was defined as the amount of enzyme required to convert 1 nmol of ADP-ribose polymer into free ADP-ribose per min under the conditions described above.
SPR (surface plasmon resonance) spectroscopy
A BIAcore X biosensor (Biacore, Uppsala, Sweden) that measures SPR was utilized to determine the interaction of wild-type rPARG-CF and mutants (analytes) with an immobilized inhibitor analogue (ligand) of ADP-HPD [adenosine diphosphate (hydroxymethyl)-pyrrolidinediol], 8-AH-ADP-HPD [8-(6-aminohexyl)amino-ADP-HPD], which has also characterized as a rPARG inhibitor [32]. A standard amine coupling reaction was used to attach the 8-AH-ADP-HPD to a research grade CM5 chip (BIAcore). The buffer used was 10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20 (also called HBS-EP buffer, from BIAcore). The immobilization was carried out by first activating the CM5 chip with a 2-min pulse of NHS/EDC [N-hydroxysuccinimide/N-ethyl-N′-(dimethylaminopropyl)carbodi-imide]. Following the activation, the amine coupling was performed with a 7-min pulse of a 160 nM/μl of solution of 8-AH-ADP-HPD. The chip was then inactivated by a 7-min ethanolamine wash. The immobilization procedure theoretically results in a maximum of 16% coupling to the carboxyl groups of the sensor chip.
For all SPR measurements, two flow cells were simultaneously utilized; one flow cell containing the 8-AH-ADP-HPD modified sensor surface, whereas the second flow cell contained the control sensor surface with no inhibitor coupling in order to obtain parallel measurements of unspecific binding. The result was a real-time reference subtraction, which effectively quantified the analyte–ligand interaction. Protein solutions containing wild-type or mutant enzyme (buffer containing 50 mM Tris/HCl, pH 7.5, 300 mM KCl, 10 mM 2-mercaptoethanol and 0.1% Triton X-100) were diluted in HBS-EP buffer (as described above) to final concentrations ranging from 10–500 nM for analysis. Each experiment began with a 30 s pulse containing HBS-EP buffer in order to establish a baseline, followed by a 90 s sample injection where the association phase is observed. Subsequent to the association phase, HBS-EP buffer was injected for 300 s in order to observe the dissociation phase. At the end of each experiment the CM5 chip was regenerated utilizing a 70–100 μl injection of 6 M urea and 0.1 M boric acid, pH 8.9, and 0.5 M NaCl. For each rPARG-CF sample, a set of sensorgrams was recorded at several different protein concentrations.
PARG sequence analysis
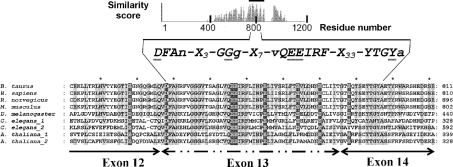
The identification of PARG sequences was performed using PSI-BLAST program against the non-redundant protein database maintained at the National Center for Biotechnology Information (Bethesda, MD, U.S.A.). We used bovine PARG sequence as the query in our BLAST searches. The BLOSUM62 matrix was used for scoring, and 0.01 or 0.001 was used as E-value thresholds for inclusion of sequences in the profile calculation. The sequences were then aligned using CLUSTAL. The aligned sequences are shown below in Figure 3. The conserved acidic residues identified in the alignment were targeted for mutagenesis.
Figure 3. Sequence alignment of a portion of PARG catalytic fragment with highest sequence similarity score that encompasses the essential catalytic residues.
Using sequence conservation and the identification of catalytic acidic residues, we have developed a sequence fingerprint indicated in this Figure. The catalytic fragment of PARG is the region of highest sequence similarity. The residues of highest sequence identity are highlighted in light grey, the acidic residues targeted for investigation are shown in white on dark grey boxes (Glu728, Asp738, Glu756, Glu757, Glu774, Glu780 and Glu788). Also indicated in the sequence alignment are the exon boundaries derived from the human PARG gene [25]. In addition to the acidic mutations, Tyr796, which has been previously identified as an active site residue that interacts with ADP-HPD, is indicated [29]. Accession numbers for sequences are: B. taurus (GI27806491), Homo sapiens (GI4505609), Mus musculus (GI6754986), Rattus norvegicus (GI23618924), Drosophila melanogaster (GI17137486), A. thaliana_1 (GI42571009), A. thaliana_2 (GI14701908), C. elegans_1 (GI32566022), C. elegans_2 (GI32566024).
RESULTS
Acidic residue screening reveals three essential catalytic residues in PARG
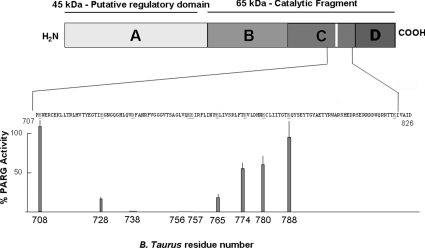
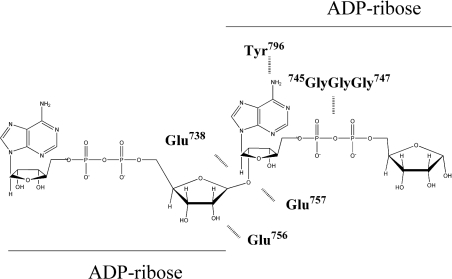
Previously, we identified an inhibitor-binding residue of bovine PARG, Tyr796, that maps to a highly conserved region of PARG [29]. PARG is an O-glycosidase, which, in general, are enzymes that contain two acidic residues essential for activity surrounding the glycosidic bond to be hydrolysed. Thus we used site-directed mutagenesis to screen conserved acidic residues of PARG in the region of Tyr796 for their effect on PARG activity. Our strategy was to convert all conserved glutamic acid or aspartic acid residues into asparagine. Asparagine was chosen because it removes the negative charge associated with the acidic residue at physiological pH, without any significant changes in the volume of amino acid side chain for aspartate residues and a side chain one carbon atom shorter in the case of glutamate residues. The latter alteration has the rationale of preventing any possible adaptation by the enzyme in case hydrogen bonding is playing a role in catalysis. Bovine PARG contains a series of conserved acidic amino acids between residues 700 to 800 (708, 728, 738, 756, 757, 765, 774, 780 and 788). Following generation of mutants and their expression, all of the mutants were purified and analysed by SDS/PAGE prior to analysis (Supplementary Figure 1, http://www.BiochemJ.org/bj/388/bj3880493add.htm). This analysis allowed confirmation of expression and purity of each protein sample used in the analysis. The results of the activity assays are shown in Table 1 and Figure 2. As shown in Figure 2, mutagenesis had no effect on the catalytic activity of Glu708, but progressing towards the C-terminus the activity decreased to a minimum where mutation of three acidic residues, Asp738, Glu756 and Glu757, resulted in no detectable activity, followed by a progressive increase in activity to Glu788 where, again, no effect on activity was observed.
Table 1. Effect of site-directed mutagenesis of glutamate and aspartate residues in rPARG on PARG activity and inhibitor binding (KD).
n, number of different experiments in which enzyme specific activity was measured, and each experiment was carried out in triplicate; ND, no detectable activity with a limit of detection of 1% of wild-type specific activity (units/mg); NE, binding not examined, those values reported have φ2<10.6.
| PARG analysed | PARG specific activity (% of wild-type) | n | KD for 8-AH-ADP-HPD (nM) |
|---|---|---|---|
| Wild-type | 100 | 10 | 8.2 |
| E708N | 119±7.3 | 4 | 3.3 |
| E728N | 18±1.9 | 4 | 3.2 |
| D738N | ND | 4 | 11.6 |
| E756N | ND | 6 | 1.03 |
| E757N | ND | 2 | 7.7 |
| E756N/E757N | ND | 2 | NE |
| E765N | 29±4.0 | 2 | NE |
| E774N | 61±8.0 | 2 | NE |
| E780N | 57±12.0 | 2 | NE |
| E788N | 100±22 | 2 | NE |
Figure 2. Catalytic activity of conserved acidic residues of PARG catalytic fragment of bovine rPARG-CF.
The specific activity of each mutant was measured and compared with wild-type rPARG-CF. The graph shows the relative percentage of specific activity of mutant rPARG-CF compared with wild-type rPARG-CF as a function of residue number within the catalytic fragment.
PARG mutants are able to bind a known inhibitor, 8-AH-ADP-HPD
Although we identified acidic residues which when mutated eliminated enzymatic activity, it was possible that these alterations resulted in a loss of activity by effects on protein folding that affected the ability of the mutant proteins to bind to substrate. In order to examine this possibility, we utilized SPR spectroscopy where an inhibitor of PARG, 8-AH-ADP-HPD, was immobilized on a dextran surface and used to compare binding of the wild-type and mutant protein to the inhibitor. Previous studies have shown that the wild-type protein binds to 8-AH-ADP-HPD with high affinity [29]. The binding constants (KD) of the mutants (Glu708, Glu728, Asp738, Glu756 and Glu757) to 8-AH-ADP-HPD are given in Table 1. The binding constants clearly show that acidic mutations were able to bind to the inhibitor at the nanomolar level, suggesting that the mutations are not having any structural effect, leading to a misfolded protein or even more subtle domain effects that interfere with the ability to recognize substrate-like moieties. When one considers the physical chemical properties of the inhibitor ADP-HPD, which contains a negatively charged phosphate group [much like the substrate poly(ADP-ribose)], it is reasonable to rationalize that mutations of the type Glu→Asn will significantly reduce charge repulsion, while retaining polarity, as well as the steric characteristics, of the native side chain. In the worst possible case, the PARG mutants (Glu→Asn) are expected to display little to no effect on inhibitor binding, and may, in the best case, improve inhibitor binding.
Signature sequence of PARG function
The three essential acidic residues identified within the PARG catalytic fragment are localized in a region of PARG coded by exons 13 and 14 of the human PARG gene [25], which also contains an inhibitor-binding site residue identified through photolabelling of Tyr796 [29] and a glycine-rich region Gly745-Gly-Gly747 previously shown to be involved with control of circadian rhythms [33]. It can be seen in Figure 3 that this region contains blocks of amino acid residues that are highly conserved in PARG sequences across a wide range of organisms. Taken together, these data allow us to define a ‘PARG signature’ sequence that is also shown in Figure 3.
DISCUSSION
There are several different enzymes involved in ADP-ribose polymer synthesis with apparently specialized roles, but there remains only a single ADP-ribose-polymer-degrading enzyme in mammals, PARG. Recently, we used a photoprobe variant of a PARG inhibitor (8-N3-ADP-HPD) to identify the inhibitor-binding site [29]. Peptide mapping of the photoderivatized enzyme led to the identification of a highly conserved region of PARG (Figure 3). This region of PARG contained a series of conserved acidic residues. A large class of O-glycosidases utilize acidic residues for catalysis, and a recent analysis of a vast majority of enzyme active sites reveals the preponderance of catalytic residues to be either histidine (∼18%) or acidic (aspartic acid, ∼15%; glutamic acid, ∼11%) [34]. Therefore we decided to use site-directed mutagenesis to identify essential acidic residues.
Our investigation led to the identification of three essential acidic residues, two of which are vicinal, Glu756 and Glu757, and a third Asp738 (Figures 2 and 3), and all three proved to be indispensable for catalytic activity. Usually, acidic residues are involved in metal binding, but there is no known metal requirement for PARG activity. The requirement for two catalytic vicinal acidic residues has also been found in the mammalian and bacterial ADP-ribosylarginine hydrolase [35]. In the family GH11 glycosidases we also found another case of an enzyme with two neighboring catalytic acidic residues (dispersed in sequence) that provide a general base assistance by activating a water molecule for direct nucleophilic attack [36]. A third case has been shown in α-glucuronidase (glycosidase family GH67), where two active site acidic residues (Glu392 and Asp364) trap a nucleophilic water molecule which attacks the anomeric carbon of the substrate [37]. Another interesting case of two structurally neighbouring acidic residues is found in the enzyme enoyl-CoA hydratase where, again, a water molecule is found trapped between the acidic residues and functionally adds to a double bond [38]. In the latter case, a mutation of either glutamic acid residue to glutamine reduces the kcat/Km value by three orders of magnitude, with little or no effect in Km [39]. The alterations we have introduced in PARG were sufficient to reduce activity below levels of detection for the two vicinal glutamic acid residues, Glu756 and Glu757, and Asp738.
In order to rationalize the mechanistic importance of these residues for PARG activity we have drawn from the large body of data available from structural and functional characterization of glycosidases [40,41]. Although both PARG and glycosidases cleave O-glycosidic bonds, they are designed to recognize distinct sugars, the former binding furanose polymers whereas the latter bind polymers of pyranose rings. From a mechanistic point of view, glycosidases act via acid/base catalysis, requiring two critical acidic residues located at opposite faces of the labile O-glycosidic bond [42,43]. Glycosidases have two distinct mechanisms, one with inversion of configuration and another with retention of configuration about the anomeric carbon. The first design of PARG inhibitors [44,45] capitalized on mechanistic analogies between PARG and glycosidases with the synthesis of ADP-HPD, a nitrogen-in-the-ring analogue of ADP-ribose [32]. ADP-HPD mimics the positive charge development of a potential oxocarbenium ion-like intermediate that may occur in PARG catalysis. The impermeability of ADP-HPD makes it unsuitable for biological characterization of PARG inhibition, but useful in mapping PARG substrate recognition sites. Taking all of the above into consideration, it seems likely that we have identified acidic residues which may be mechanistically similar to catalytic acidic residues found in glycosidases.
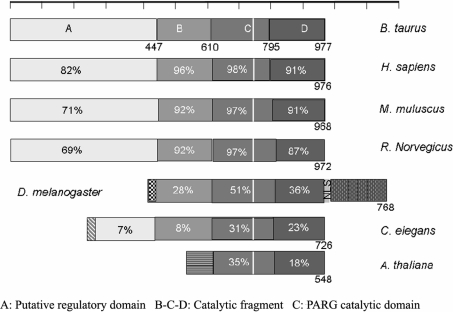
The identification of three essential acidic residues, along with the inhibitor-binding site and the recent functional characterization of a glycine-rich region [33], also within this highly conserved region of PARG, has allowed us to define a functional fingerprint for PARG activity (Figure 3). We have used this sequence fingerprint along with PHI-BLAST [46] to search for distantly related PARG sequences with sequence identity below 20–30%, sometimes referred to as the ‘twilight zone’ [47]. Our sequence searches have only been able to identify putative PARG sequences in plants, invertebrates and vertebrates, all of which have sequence identity above the ‘twilight zone’ (Figure 4). A potential functional redundancy in PARG sequences, which may mimic that observed in PARP sequences, has only been found in Caenorhabditis elegans and Arabidopsis thaliana where there are two genes with a conserved rPARG-CF motif, but not all have been functionally characterized. The vertebrate PARG sequences have essentially four conserved sequence blocks or domains based on the domain classification of ProDom [48]. The PFAM classification identifies a single region spanning ProDom C and D (PFAM 0528.5). We labelled the ProDom domains A (PD156514), B (PD251687), C (PD147216) and D (PD025592). The catalytic fragment of PARG spans ProDom B, C and D. This organization does not work when comparing vertebrate and invertebrate sequences and plants. In the latter, we see greater divergence in sequence outside of the conserved PARG catalytic fragment, suggesting different domain composition (Figure 4). However, the comparison of distantly related PARGs reveals that domains C and D are the ones most conserved among PARG sequences. A secondary sequence prediction for domains C and D reveals a mostly β-sheet domain (domain C, Figure 4), followed by a sharp boundary (around residue 800 of bovine PARG), leading to a mostly α-helical domain (domain D, Figure 4) [49]. All of the catalytic mutants of PARG map to domain C, thus, taken together, all the data suggest domain C to be the PARG catalytic domain.
Figure 4. Domain organization of the various PARG sequences identified using iterative PSI-BLAST.
Notice that the domain organization of PARGs changes dramatically when comparing vertebrates, invertebrates and plants. The various grey shading refers to conserved regions within a sequence, for example, the regulatory domain found in invertebrates is highly conserved with identity >25% at the amino acid level. The percentage conservation of the catalytic fragment is indicated in each block with respect to the bovine PARG. Accession numbers for sequences are as described in Figure 3.
In summary we are now able to offer the first prediction of a PARG active site (Figure 5). We have found residues involved in the recognition of the adenine ring [29], as well as the potential residues that are playing roles in catalysis either nucleophile or general acid. Finally, our sequence analysis of the PARG catalytic fragment suggests that domain C is the minimal catalytic fragment which contains a mostly β-sheet structure [49].
Figure 5. Residues that comprise the PARG catalytic site, along with their respective functional roles with respect to glycosidic α(1″→2′) bond cleavage.
One of the two vicinal acidic residues may be the nucleophile or may also be trapping a water molecule that may act as a nucleophile. Previous work has identified the cluster of glycine residues as vital for catalytic activity, which in many proteins are found to interact with phosphate groups [33]. Finally, in previous work we have identified Tyr796 [29] as forming a potential stacking interaction with an adenine ring of the PARG inhibitor ADP-HPD.
Online data
Acknowledgments
This work was supported by research grants from the American Cancer Society (85-001-13-IRG), University of Kentucky Research support grant and Kentucky Lung Cancer Research Grant to M.A.O. and the National Institutes of Health (CA 43894) to M.K.J.
References
- 1.Smulson M. E., Simbulan-Rosenthal C. M., Boulares A. H., Yakovlev A., Stoica B., Iyer S., Luo R., Haddad B., Wang Z. Q., Pang T., et al. Roles of poly(ADP-ribosyl)ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv. Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson M. K., Jacobson E. L. Discovering new ADP-ribose polymer cycles: protecting the genome and more. Trends Biochem. Sci. 1999;24:415–417. doi: 10.1016/s0968-0004(99)01481-4. [DOI] [PubMed] [Google Scholar]
- 3.Smith S. The world according to PARP. Trends Biochem. Sci. 2001;26:174–179. doi: 10.1016/s0968-0004(00)01780-1. [DOI] [PubMed] [Google Scholar]
- 4.Shieh W. M., Ame J. C., Wilson M. V., Wang Z. Q., Koh D. W., Jacobson M. K., Jacobson E. L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 5.Shall S., de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T., Satoh M. S., Poirier G. G., Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber V., Ame J. C., Dolle P., Schultz I., Rinaldi B., Fraulob V., Menissier-de Murcia J., de Murcia G. PARP-2 is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 8.Davidovic L., Vodenicharov M., Affar E. B., Poirier G. G. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 9.Lin W., Ame J. C., Aboul-Ela N., Jacobson E. L., Jacobson M. K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 10.Trucco C., Oliver F. J., de Murcia G., Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pieper A. A., Brat D. J., Krug D. K., Watkins C. C., Gupta A., Blackshaw S., Verma A., Wang Z. Q., Snyder S. H. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid G., Wang Z. Q., Wesierska-Gadek J. Compensatory expression of p73 in PARP-deficient mouse fibroblasts as response to a reduced level of regularly spliced wild-type p53 protein. Biochem. Biophys. Res. Commun. 1999;255:399–405. doi: 10.1006/bbrc.1999.0191. [DOI] [PubMed] [Google Scholar]
- 13.Tentori L., Portarena I., Graziani G. Potential clinical applications of poly(ADP-ribose) polymerase (PARP) inhibitors. Pharmacol. Res. 2002;45:73–85. doi: 10.1006/phrs.2001.0935. [DOI] [PubMed] [Google Scholar]
- 14.White A. W., Almassy R., Calvert A. H., Curtin N. J., Griffin R. J., Hostomsky Z., Maegley K., Newell D. R., Srinivasan S., Golding B. T. Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase. J. Med. Chem. 2000;43:4084–4097. doi: 10.1021/jm000950v. [DOI] [PubMed] [Google Scholar]
- 15.Martin D. R., Lewington A. J., Hammerman M. R., Padanilam B. J. Inhibition of poly(ADP-ribose) polymerase attenuates ischemic renal injury in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000;279:R1834–R1840. doi: 10.1152/ajpregu.2000.279.5.R1834. [DOI] [PubMed] [Google Scholar]
- 16.Shiokawa D., Maruta H., Tanuma S. Inhibitors of poly(ADP-ribose) polymerase suppress nuclear fragmentation and apoptotic-body formation during apoptosis in HL-60 cells. FEBS Lett. 1997;413:99–103. doi: 10.1016/s0014-5793(97)00887-9. [DOI] [PubMed] [Google Scholar]
- 17.Griffin R. J., Curtin N. J., Newell D. R., Golding B. T., Durkacz B. W., Calvert A. H. The role of inhibitors of poly(ADP-ribose) polymerase as resistance-modifying agents in cancer therapy. Biochimie. 1995;77:408–422. doi: 10.1016/0300-9084(96)88154-5. [DOI] [PubMed] [Google Scholar]
- 18.Simbulan-Rosenthal C. M., Rosenthal D. S., Ding R., Jackman J., Smulson M. E. Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influence on genomic stability, chromatin organization, DNA repair, and DNA replication. Prog. Nucleic Acid Res. Mol. Biol. 1996;55:135–156. doi: 10.1016/s0079-6603(08)60192-0. [DOI] [PubMed] [Google Scholar]
- 19.Pieper A. A., Verma A., Zhang J., Snyder S. H. Poly(ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol. Sci. 1999;20:171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- 20.Eliasson M. J., Sampei K., Mandir A. S., Hurn P. D., Traystman R. J., Bao J., Pieper A., Wang Z. Q., Dawson T. M., Snyder S. H., Dawson V. L. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 21.Mandir A. S., Przedborski S., Jackson-Lewis V., Wang Z. Q., Simbulan-Rosenthal C. M., Smulson M. E., Hoffman B. E., Guastella D. B., Dawson V. L., Dawson T. M. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Affar E. B., Germain M., Winstall E., Vodenicharov M. D., Shah R. G., Salvesen G. S., Poirier G. G. Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase during apoptosis. J. Biol. Chem. 2000;276:2935–2942. doi: 10.1074/jbc.M007269200. [DOI] [PubMed] [Google Scholar]
- 23.Boulares A. H., Yakovlev A. G., Ivanova V., Stoica B. A., Wang G., Iyer S., Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 24.Winstall E., Affar E. B., Shah R., Bourassa S., Scovassi I. A., Poirier G. G. Preferential perinuclear localization of poly(ADP-ribose) glycohydrolase. Exp. Cell Res. 1999;251:372–378. doi: 10.1006/excr.1999.4594. [DOI] [PubMed] [Google Scholar]
- 25.Meyer R. G., Meyer-Ficca M. L., Jacobson E. L., Jacobson M. K. Human poly(ADP-ribose) glycohydrolase (PARG) gene and the common promoter sequence it shares with inner mitochondrial membrane translocase 23 (TIM23) Gene. 2003;314:181–190. doi: 10.1016/s0378-1119(03)00738-8. [DOI] [PubMed] [Google Scholar]
- 26.Hanai S., Kanai M., Ohashi S., Okamoto K., Yamada M., Takahashi H., Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes U., Tong W. M., Coyle D. L., Meyer-Ficca M. L., Meyer R. G., Petrilli V., Herceg Z., Jacobson E. L., Jacobson M. K., Wang Z. Q. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koh D. W., Lawler A. M., Poitras M. F., Sasaki M., Wattler S., Nehls M. C., Stoger T., Poirier G. G., Dawson V. L., Dawson T. M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh D. W., Patel C. N., Ramsinghani S., Slama J. T., Oliveira M. A., Jacobson M. K. Identification of an inhibitor binding site of poly(ADP-ribose) glycohydrolase. Biochemistry. 2003;42:4855–4863. doi: 10.1021/bi0272048. [DOI] [PubMed] [Google Scholar]
- 30.Cline J., Braman J. C., Hogrefe H. H. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menard L., Poirier G. G. Rapid assay of poly(ADP-ribose) glycohydrolase. Biochem. Cell Biol. 1987;65:668–673. doi: 10.1139/o87-088. [DOI] [PubMed] [Google Scholar]
- 32.Koh D. W., Coyle D. L., Mehta N., Ramsinghani S., Kim H., Slama J. T., Jacobson M. K. SAR analysis of adenosine diphosphate (hydroxymethyl)pyrrolidinediol inhibition of poly(ADP-ribose) glycohydrolase. J. Med. Chem. 2003;46:4322–4332. doi: 10.1021/jm020541u. [DOI] [PubMed] [Google Scholar]
- 33.Panda S., Poirier G. G., Kay S. A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev. Cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 34.Barlett G. j., Porter C. T., Borkakoti N., Thornton J. M. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 35.Konczalik P., Moss J. Identification of critical, conserved vicinal aspartate residues in mammalian and bacterial ADP-ribosylarginine hydrolases. J. Biol. Chem. 1999;274:16736–16740. doi: 10.1074/jbc.274.24.16736. [DOI] [PubMed] [Google Scholar]
- 36.Joshi M. D., Sidhu G., Pot I., Brayer G. D., Withers S. G., McIntosh L. P. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 2000;299:255–279. doi: 10.1006/jmbi.2000.3722. [DOI] [PubMed] [Google Scholar]
- 37.Nurizzo D., Nagy T., Gilbert H. J., Davies G. J. The structural basis for catalysis and specificity of the Pseudomonas cellulosa α-glucuronidase, GlcA67A. Structure. 2002;10:547–556. doi: 10.1016/s0969-2126(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 38.Bahnson B. J., Anderson V. E., Petsko G. A. Structural mechanism of enoyl-CoA hydratase: three atoms from a single water are added in either an E1cb stepwise or concerted fashion. Biochemistry. 2002;41:2621–2629. doi: 10.1021/bi015844p. [DOI] [PubMed] [Google Scholar]
- 39.Hofstein H. A., Feng Y., Anderson V. E., Tonge P. J. Role of glutamate 144 and glutamate 164 in the catalytic mechanism of enoyl-CoA hydratase. Biochemistry. 1999;38:9508–9516. doi: 10.1021/bi990506y. [DOI] [PubMed] [Google Scholar]
- 40.Zechel D. L., Withers S. G. Dissection of nucleophilic and acid–base catalysis in glycosidases. Curr. Opin. Chem. Biol. 2001;5:643–649. doi: 10.1016/s1367-5931(01)00260-5. [DOI] [PubMed] [Google Scholar]
- 41.Vasella A., Davies G. J., Bohm M. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 2002;6:619–629. doi: 10.1016/s1367-5931(02)00380-0. [DOI] [PubMed] [Google Scholar]
- 42.Davies G. J., Mackenzie L., Varrot A., Dauter M., Brzozowski A. M., Schulein M., Withers S. G. Snapshots along an enzymatic reaction coordinate: analysis of a retaining β-glycoside hydrolase. Biochemistry. 1998;37:11707–11713. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 43.Koshland D. E. Stereochemistry and mechanism of enzymatic reactions. Biol. Rev. 1953;28:416–438. [Google Scholar]
- 44.Slama J. T., Aboul-Ela N., Goli D. M., Cheesman B. V., Simmons A. M., Jacobson M. K. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J. Med. Chem. 1995;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- 45.Slama J. T., Aboul-Ela N., Jacobson M. K. Mechanism of inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J. Med. Chem. 1995;38:4332–4336. doi: 10.1021/jm00021a023. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z., Schaffer A. A., Miller W., Madden T. L., Lipman D. J., Koonin E. V., Altschul S. F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doolittle R. F. Stein and Moore Award address. Reconstructing history with amino acid sequences. Protein Sci. 1992;1:191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servant F., Bru C., Carrere S., Courcelle E., Gouzy J., Peyruc D., Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 49.Oliveira M. A., Koh D. W., Patel C. N., Jacobson M. K. Structure-based characterization of a novel anticancer target poly(ADC-ribose) glycohydrolase (PARG): Evidence for the presence of an ADP-ribosyltransferase (ADPRT) fold. Proc. Am. Assoc. Cancer Res. 2001;42:832. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.