Abstract
Calpains 1 and 2 are heterodimeric proteases in which large (relative molecular mass Mr 80000) and small (Mr 28000) subunits are linked through their respective PEF (penta-EF-hand) domains. The skeletal muscle-specific calpain 3 is believed not to form a heterodimer with the small subunit but might homodimerize through its PEF domain. Size-exclusion chromatography and analytical ultracentrifugation of the recombinant PEF domain of calpain 3 show that it forms a stable homodimer that does not dissociate on dilution. Molecular modelling suggests that there would be no barriers to the dimerization of the whole enzyme through the PEF domains. This orientation would place the catalytic centres at opposite ends of the dimer.
Keywords: calpain 3, homodimerization, molecular modelling, muscular dystrophy, p94, penta-EF-hand domain (PEF domain)
Abbreviations: DIV, domain IV; DTT, dithiothreitol; IS, insertion sequence; Ni-NTA, Ni2+-nitrilotriacetate–agarose; NS, N-terminal sequence; PEF, penta-EF-hand
INTRODUCTION
Calpains form a widely distributed superfamily of Ca2+-dependent cytosolic cysteine proteases. Numerous calpain isoforms have been identified in vertebrates [1–3], invertebrates [1,3–6], plants [7,8] and microorganisms [1,3,6]. These enzymes are supposed to play a role in many intracellular processes linked to calcium signalling, including cell motility, apoptosis, cell differentiation and cell-cycle regulation [9–11]. They function by modulating the biological activities of their substrates through limited proteolysis [12]. Abnormal changes in calpain activity, including those due to defects in calcium homoeostasis or mutations in calpain genes, contribute to pathologies such as ischaemic injury, Alzheimer's disease and other neurological disorders, muscular dystrophy, cancer and Type II diabetes [3,13].
The two abundant ubiquitous mammalian calpains 1 and 2 (μ- and m- respectively) are the best-characterized members of the family. They are heterodimers of a large (relative molecular mass Mr 80000) and a small (Mr 28000) subunit. The Mr 80000 catalytic subunit has four structural domains (I–IV) [14,15]. The first two domains (I and II) make up the papain-like catalytic core common to all calpains, whereas domains III and IV are the C2-like and PEF (penta-EF-hand) domains respectively [1]. The small subunit contains two domains (V and VI). Its PEF domain (VI) makes extensive contact with the homologous PEF DIV (domain IV) of the large subunit through pairing of the fifth EF hand [16]. Both isoforms are catalytically inactive in the absence of Ca2+. They undergo a series of conformational changes on Ca2+ binding to several domains [17–19]. In addition to these widely distributed μ- and m-isoforms, a dozen more distinctive isoforms have been identified in humans. Although the proteolytic core domains (I and II) are conserved in all isoforms of the calpain family, the other domains can be quite varied. Some homologues seem to lack the small subunit (calpains 3, 5, 6, 7 and 10), suggesting that they may not form heterodimers.
Calpain 3 (p94), the first identified ‘tissue-specific’ isoform, is produced predominantly in skeletal muscles [2]. p94 is implicated in myofibrillogenesis and sarcomere remodelling. Mutations in p94 are genetically linked to limb girdle muscular dystrophy type 2A, signifying a crucial role for p94 in muscle physiology [20,21]. It has a domain organization similar to that of the μ- and m-calpain large subunits, and its amino acid sequence is 54 and 51% identical with their respective large subunits [2]. p94 is Mr 14000 larger than the μ- and m-calpain large subunits. This is due to the presence of three additional unique sequences; an N-terminal extension sequence of 47 amino acids (referred to as NS) that replaces the anchor helix of m-calpain, and two insertion sequences of 48 and 77 amino acids (referred to as IS1 and IS2 respectively). IS1 is located in domain II as an insert that flanks the active site cleft and acts as an internal propeptide [22]; IS2 is located between domains III and IV and binds titin [23].
Attempts to isolate p94 from muscles or to produce it using recombinant systems have been hampered by its rapid autolysis [24–26]. Owing to this instability, the physiological role of p94 and the structure–function aspects of its different domains and additional unique sequences remain obscure. When inactive [C129S (Cys129→Ser)] rat p94 was expressed in COS cells, it was stable, and during size-exclusion chromatography, it eluted with an Mr∼180000, as though it were a dimer [27]. However, there have been no definitive studies performed on the issue of the oligomeric state of p94 in vitro or in vivo. Also, we note that PEF proteins do not naturally occur as monomers [28], suggesting that the PEF domain DIV could be the logical site for dimerization. In the present study, we describe experiments that establish the intrinsic capability of the p94 PEF domain to homodimerize, and we show by molecular modelling that there are no obvious barriers to dimerization of the whole enzyme through this interaction.
MATERIALS AND METHODS
Cloning of p94DIV
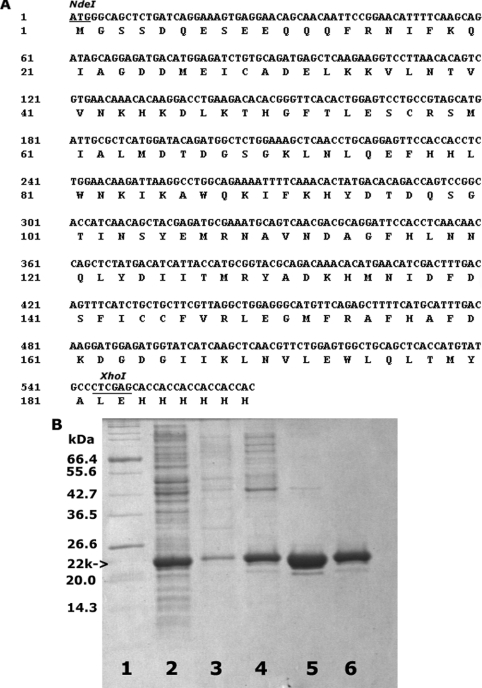
PCR primers (5′-TATACATATGGGCAGCTCTGATCAGGAAAGTG-3′ and 5′-CACCTCGAGGGCATACATGGTGAGCTGCAGCCACTC-3′) were designed to generate the cDNA fragment encoding the DIV region of p94 by PCR amplification of reverse transcripts (reverse transcriptase–PCR) of total RNA from human skeletal muscles (Stratagene, La Jolla, CA, U.S.A.) using an RT-Thermoscript kit (Invitrogen, Carlsbad, CA, U.S.A.) and Expand high-fidelity DNA polymerase (Roche, Indianapolis, IN, U.S.A.). The amplified product was cloned into the pCR 2.1-TOPO vector (Invitrogen). DNA sequences of positive clones were verified. The verified cDNA sequence was ligated into the NdeI and XhoI sites of the pET24a expression vector (Novagen, Madison, WI, U.S.A.) to incorporate an N-terminal Met codon and a C-terminal histidine tag into the p94DIV construct (Figure 1A).
Figure 1. Human calpain p94 domain IV and its purification.
(A) The cDNA sequence of domain IV of human calpain p94 is shown above the encoded amino acid sequence. (B) SDS/PAGE of fractions obtained during purification. Lane 1 shows the molecular-mass standards indicated at the side of the gel. Lanes 2 and 3 were loaded with an aliquot of E. coli supernatant and pellet respectively. Lanes 4–6 represent pooled fractions from purifications steps on DEAE-Sepharose, Ni-NTA and Sephadex G-75 columns respectively. The amount of protein in lane 5 is twice that loaded in lane 6.
Protein expression and purification
The plasmid encoding p94DIV was transfected into Escherichia coli BL21(DE3) (Novagen) grown in LB (Luria–Bertani) medium under kanamycin selection to an absorbance A600 of 0.8–1.0 at 37 °C. Protein expression was induced with 0.4 mM isopropyl β-D-thiogalactoside for 3 h. Cells harvested from the culture by centrifugation were resuspended in lysis buffer (25 mM Tris/HCl, pH 7.6, 5 mM EDTA, 5%, v/v, glycerol, 10 mM 2-mercaptoethanol and 0.1 mM PMSF) and lysed by sonication. The lysate was centrifuged at 27000 g for 45 min and the supernatant was applied to a DEAE-Sepharose column. Protein was eluted by a 0–0.75 M NaCl salt gradient in lysis buffer, and fractions containing p94DIV were identified by the presence of a prominent Mr 22000 band on SDS/PAGE. These fractions were pooled and applied to a 10 ml NTA (Ni2+-nitrilotriacetate–agarose) column (Qiagen, Chatsworth, CA, U.S.A.). The column was washed with N-buffer (50 mM Tris/HCl, pH 7.6, 100 mM NaCl, 5 mM imidazole and 0.01% sodium azide) and eluted with N-buffer and 250 mM imidazole. The fractions containing p94DIV were further purified by gel-permeation chromatography.
Size-exclusion chromatography
A Sephadex G-75 column (2.6 cm×100 cm; Amersham Biosciences) was equilibrated overnight with 25 mM Tris/HCl (pH 7.6), 150 mM NaCl, 5 mM EDTA, 2% glycerol and 0.05% sodium azide at a flow rate of 0.5 ml/min. The column was then calibrated with the following protein standards: Blue Dextran (Mr 2000000), BSA (Mr 66000), ovalbumin (Mr 44000), carbonic anhydrase (Mr 29000), myoglobin (Mr 17000) and cytochrome c (Mr 12400), with inorganic phosphate to mark the total volume. The protein sample containing p94DIV was loaded on to the column and eluted at 0.5 ml/min. All chromatographic runs were performed at 4 °C. Peak volumes are standardized to a KD (diffusion constant) value [30] given by KD=(Ve−Vo)/(Vt−Vo), where Ve is the elution volume of the centre of a protein peak, Vo the void volume (determined by elution of Blue Dextran) and Vt the total elution volume (determined by elution of inorganic phosphate). The absorbance of each fraction was measured at 280 nm, and those in which Blue Dextran and myoglobin eluted were measured at 256 and 415 nm respectively. The presence of phosphate in the eluate was detected by a colorimetric assay [31].
Purified p94DIV was concentrated using a Centricon Mr 10000 concentrator (Millipore, Bedford, MA, U.S.A.) to a final protein concentration of 25 mg/ml in 10 mM Hepes (pH 7.5) and 10 mM DTT (dithiothreitol), then divided into 400 μl aliquots and flash-frozen in liquid nitrogen for storage.
CD spectroscopy
CD spectra were recorded using an Olis RSM 1000 CD spectrophotometer (Bogart, GA, U.S.A.). The CD spectra spanning wavelengths 260–180 nm were collected using a 0.1 mm path length quartz cuvette at 4 °C. p94DIV was dialysed into 5 mM Tris/HCl (pH 7.6), 0.1 mM DTT and 0.2 mM EDTA. The protein concentration was 2.2 mg/ml. Five scans were recorded, averaged and corrected for buffer background signal. Deconvolution of the data was performed using CDNN software [32]. Results are expressed in terms of mean residue ellipticity [θ] (deg·cm2·dmol−1).
Intrinsic tryptophan fluorescence studies
Intrinsic tryptophan fluorescence measurements were performed using a PerkinElmer LS50B fluorescence spectrophotometer equipped with a stirrer-adapted 4 ml cuvette (Hellma, Concord, Ontario, Canada) at a constant temperature of 24 °C. Excitation and emission wavelengths were set at 280 and 340 nm respectively. The reaction buffer was 50 mM Tris/HCl (pH 7.6) containing 100 mM NaCl, 1 mM EDTA and 1 mM DTT. p94DIV (2 μM) was added to the assay buffer and intrinsic fluorescence was recorded for 10 min. At this point, 50 mM CaCl2 dissolved in the assay buffer was pumped continuously into the cuvette at 4 μl/min using an injector pump (Harvard Apparatus pump 22). Similar experiments were performed under the same conditions but using MgCl2 instead of CaCl2. In a control experiment, intrinsic tryptophan fluorescence changes were monitored during CaCl2 addition to purified rat μI-II (1 μM) [18] by the method described above for p94DIV.
Analytical ultracentrifugation
Ultracentrifugation experiments were performed on p94DIV at 20 °C in a Beckman (Palo Alto, CA, U.S.A.) Optima XL-I analytical ultracentrifuge equipped with automated scanning absorption optics (at 280 nm) by the method of Laue and Stafford [29]. Protein solutions were exhaustively dialysed against 20 mM Pipes (pH 6.5), containing 150 mM NaCl, 0.1 mM DTT and either 1 mM EDTA or 2 mM CaCl2. The rotor used was a Beckman An60-Ti. Sedimentation velocity runs were performed at two different speeds, 30000 and 42000 rev./min, for 7 and 4 h duration respectively and a final protein concentration of 0.25 mg/ml both in the absence and presence of Ca2+ (2 mM). The results were analysed by SEDFIT software [33]. Sedimentation equilibrium experiments were performed at three different speeds, 12000, 16000 and 20000 rev./min, and at six different protein concentrations (0.05, 0.1, 0.2, 0.3, 0.4 and 0.5 mg/ml) both in the presence and absence of Ca2+ (2 mM). Each speed was maintained until there was no significant difference in r2/2 versus absorbance. After 24 h of equilibration at each speed, a scan was taken at each hour for the next 5 h (total five scans). The results were analysed using Origin4.1 software (Beckman).
Homology modelling of the p94DIV dimer
Modelling of the p94DIV dimer was performed using PyMOL, MODELLER and GROMACS [34–37]. A total of 100 models of p94DIV was created with the program MODELLER by mapping DIV (Glu649-Ala821) on to the crystal structure of the rat calpain small subunit DVI homodimer (Glu1-Ser173) [38] to verify that there was sufficient sequence identity to form a dimer. The structures were examined with PyMOL to detect clashes at the interface, and the complementarity of the models was measured with the program sc from the CCP4 program suite [39]. The model with the highest complementarity was subjected to energy minimization for 10000 steps or until convergence (tolerance, 1000 kJ·mol−1·nm−1) and molecular dynamics (3 ns or 30000 steps, at 300 K, with a constant temperature bath) using GROMACS. Co-ordinates for the p94DIV dimer have been deposited in the Protein Data Bank (PDB ID 1Y9V). A second model was generated to examine the possibility of heterodimerization of p94DIV with DVI. In this case, the sequence of p94DIV was mapped on to one subunit of the rat calpain DIV homodimer, whereas the sequence of the human DVI was mapped on to the other. As a control, the process of energy minimization and molecular dynamics was repeated for the crystal structure of the rat DVI homodimer (PDB ID 1DVI) and for the DIV–DVI heterodimer (PDB ID 1KFU) from the Ca2+-free human calpain crystal structure. The trajectories from the molecular dynamics simulations were analysed for energetic stability and complementarity between the subunits.
Graphic model of the m-calpain large-subunit dimer
A homodimer of two m-calpain large subunits was generated by superimposing DIV of one m-calpain heterodimer on to the small subunit (DVI not shown) of a second m-calpain heterodimer (Figure 6). This computer-graphics model of a full-length m-calpain large-subunit dimer associated through their PEF domains was constructed using PyMOL. The structure was visually examined for structural clashes but no other manipulations were performed.
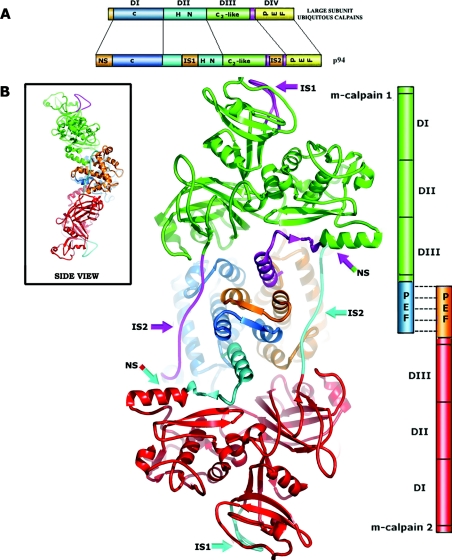
Figure 6. Graphic model of the large subunit dimer.
(A) Schematic representation of the large subunit domain structures of m-calpain and p94. The domain organization of the two proteins is similar, but p94 has three additional unique sequences; an N-terminal (NS) extension sequence and two insertion sequences (IS1 and IS2). (B) Homodimer of two m-calpain large subunits generated by superimposing domain VI of one m-calpain heterodimer on to domain IV of the other (domain VI not shown). They associate with each other through the PEF domain IV. The locations of IS1 and IS2 and the NS sequence in p94 (coloured purple and cyan respectively) are indicated by thick arrows. The inset to (B) shows a side view (rotated 90° about the vertical axis).
RESULTS
Expression and purification of recombinant human p94 domain IV
The DNA sequence encoding p94DIV is shown in Figure 1(A) above the amino acid sequence. The predicted protein has 189 residues, including the His6 C-terminal tag, with a theoretical pI of 5.92 and a calculated Mr of 22181. The amino acid sequence is approx. 52% identical with DIV of μ-calpain and approx. 47% identical with DIV of m-calpain. p94DIV was produced as the major soluble product in E. coli supernatant (Figure 1B, lane 2) and was purified using a combination of three column separations, DEAE-Sepharose, Ni-NTA and Sephadex G-75. The pooled fractions from the DEAE-Sepharose column contained several higher Mr contaminating E. coli proteins, which were largely removed in the subsequent Ni-NTA affinity chromatography step. The final purification product from Sephadex G-75 was estimated to be more than 95% pure, as indicated by the SDS/PAGE (Figure 1B, lane 6). Soluble p94DIV was stable throughout the purification and concentration steps. A final yield of approx. 250 mg of protein was obtained from a 4 litre culture.
CD spectra and intrinsic tryptophan fluorescence studies
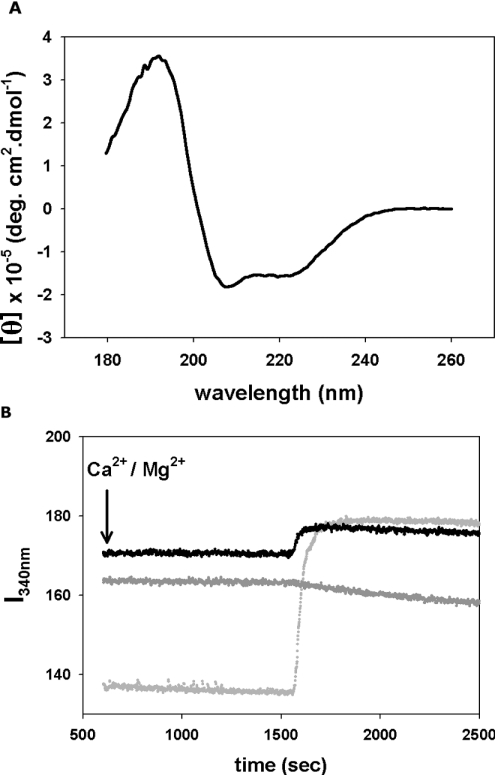
To assess that recombinant p94DIV is indeed properly folded, we determined its secondary structure by CD (Figure 2A). The CD spectra of p94DIV are those of a predominantly α-helical protein. Deconvolution of the CD spectra showed a high α-helical content (∼59%) and a very low percentage of random coil. A comparison of the secondary structure profile of p94DIV with that of the homologous rat small subunit determined from its crystal structure (PDB ID 1DVI) shows that the two proteins share high structural similarity (Table 1). There are only a few percentage points difference in the abundance of each secondary structure type. The same is true for the model of p94DIV. These results strongly suggest that p94DIV was produced as a well-folded protein in E. coli.
Figure 2. CD spectra and intrinsic tryptophan studies on p94DIV.
(A) Plot of the molar ellipticity θ as a function of wavelength. Spectra of p94DIV were collected over the far- and near-UV range (180–260 nm). (B) Ca2+-induced conformational change in p94DIV (black) and μI-II (light grey). Intrinsic fluorescence intensity was monitored at 340 nm (I340) by exciting p94DIV and μI-II at 280 nm while continuously adding CaCl2. Under similar conditions, no change in protein conformation was observed when MgCl2 was added to p94DIV (dark grey).
Table 1. Comparison of secondary structure content of p94DIV, based on CD spectra or modelling, with that of the rat Mr 21000 small-subunit dimer obtained from its crystallographic structure (PDB ID 1DVI).
| Rat small subunit (%) | p94DIV (%) | p94DIV model (%) | |
|---|---|---|---|
| Helix | 63 | 59 | 64 |
| Random coil | 17 | 16 | 17 |
| β-Strand | 5 | 5 | 5 |
| β-Turn | 14 | 13 | 14 |
| Total | 99 | 93 | 100 |
To study the effect of Ca2+ on the conformation of p94DIV, we monitored its intrinsic tryptophan fluorescence in the presence and absence of this ion. The p94DIV monomer has three tryptophan residues and five putative EF-hand Ca2+-binding sites. p94DIV displayed a significant shift in fluorescence intensity on the addition of Ca2+ (Figure 2B), which is indicative of a conformational change in the protein caused by binding Ca2+. Under the same conditions, Mg2+ did not produce a change in intrinsic tryptophan fluorescence. For a positive control, we used the proteolytic core of μ-calpain (μ I–II), which undergoes a large conformational change when it binds Ca2+ (Figure 2B, light grey trace) [18].
P94DIV is a stable dimer in solution
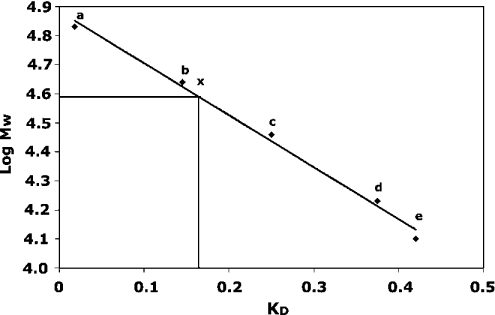
The fraction obtained after Ni-NTA purification was chromatographed on Sephadex G-75, both as a purification step and to determine its Mr. A standard curve was generated by plotting the KD values for proteins of known molecular masses against the logarithm of their molecular masses (Figure 3). Recombinant p94DIV eluted immediately after ovalbumin as a single macromolecular species with an Mr∼40000±5000, estimated by interpolation of its KD value on to the standard curve. This is twice the expected molecular mass (Mr 22181) of p94DIV. These results suggest that p94DIV forms a homodimer in solution. To confirm this and obtain quantitative information on the strength of dimerization, ultracentrifugation analysis was performed.
Figure 3. Size-exclusion (Sephadex G-75) chromatography standard curve.
KD values obtained from peak volumes of different protein standards are plotted against log Mr. The points a–e correspond to the protein standards used: (a) BSA (Mr 66000), (b) ovalbumin (Mr 44000), (c) carbonic anhydrase (Mr 29000), (d) myoglobin (Mr 17000) and (e) cytochrome c (Mr 12400). Point x represents the intersection of the KD value of p94IV obtained from its elution volume with the standard curve giving a log Mr value. The Mr was then calculated to be approx. 40000 with an error value of ±5000.
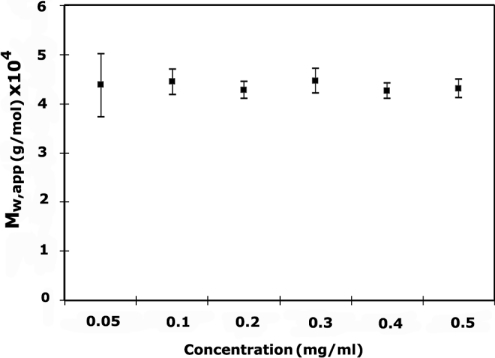
Sedimentation velocity analysis of recombinant p94DIV showed that the sample solution was homogeneous both in the presence and absence of 2 mM CaCl2. A value of s020,w∼3.41 was obtained, which in this case corresponds to an Mr of approx. 44000, assuming the protein to be globular in shape. From sedimentation equilibrium studies, an Mr of 43580 was obtained, which is consistent with the expected theoretical value for the p94DIV dimer (44360). The dependence of Mr on the ultracentrifuge cell-loading concentration c is shown in Figure 4 for the entire range of protein concentrations used (approx. 0.05–0.6 mg/ml). Mr is constant as a function of c, indicating that the sample behaves as an ideal single component. No evidence for dissociation of the homodimer (at concentrations of 0.05, 0.1 and 0.2 mg/ml) was observed either in the presence or absence of Ca2+, even at the lowest concentrations tested, suggesting strong dimer interaction.
Figure 4. Sedimentation equilibrium distribution for p94IV.
The Mr is plotted against the cell loading concentration c (mg/ml). The graph shows that the Mr of p94DIV is constant as a function of concentration (c), indicating an ideal single component in sample. Graphical data are consistent with the expected molecular mass range for the p94DIV domain dimer (∼43500) throughout the concentration range in the absence of Ca2+. Similar values were obtained in the presence of 2 mM Ca2+.
Homology modelling of p94DIV
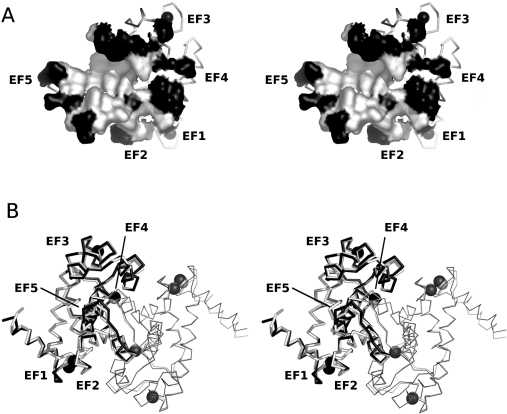
Mapping of the sequence of p94DIV on to the rat calpain DVI homodimer showed that the dimer interface is well conserved (Figure 5). Energy minimization and molecular dynamics did not show any evidence that the p94DVI homodimer is unstable. Calculation of the average complementarity from every 100th structure in the dynamics trajectories for the human m-calpain DIV–DVI heterodimer, the rat DVI homodimer and the p94DIV homodimer models gave values of 0.76, 0.74 and 0.73 respectively. Average complementarity of the p94DIV homodimer lies within the minimum and maximum values calculated for the trajectory for the human m-calpain DIV–DVI heterodimer (0.70–0.80 respectively). This suggests that the p94DIV is capable of dimerization. When a model of the p94DIV-DVI heterodimer was generated, there were no obvious steric clashes or charge repulsions that would prevent heterodimerization and the average complementarity value 0.72 was comparable with the others.
Figure 5. Homology model of p94DIV.
(A) Molecular surface of the ‘A’ chain at the dimer interface between the two monomers of the rat DVI dimer. Residues that are identical between rat DVI and p94DIV are coloured white, while those that differ are coloured black. (B) Alpha-carbon trace of the superimposed structures of rat DVI dimer (black) and the p94DIV dimer (light grey) created with the modeller program. The view in this Figure is rotated 90° about the vertical axis from that in (A). The ‘A’ chain of each dimer is drawn in thick tubes, while the ‘B’ chain is drawn in thin tubes. The spheres represent Ca2+ ions.
DISCUSSION
The premise for our study of p94DIV dimerization stems from the circumstantial evidence that p94 might associate with itself rather than with the small subunit (Mr 28000) found in calpains 1 and 2. This small subunit has not been seen in preparations of p94 from skeletal muscles [27], even though muscle cells do produce the small-subunit-containing calpains [40]. Also, yeast two-hybrid assays on p94 (both inactive and active) failed to show binding of the small subunit to p94 [23,41]. Evidence for homodimerization comes mainly from reports that an active-site mutant of p94(C129S) elutes as an Mr 180000 protein during size-exclusion chromatography [27]. This is consistent with homodimer formation, although an asymmetrical p94 monomer could also produce the same result. In a comparable system, the homologous large subunits of chicken calpain (Mr 80000) have been reported to form homodimers when expressed alone in SF9 cells, but form heterodimers when co-expressed with the Mr 28000 small subunit [27,42]. Most PEF proteins are known to form either heterodimers or homodimers. This has been established by biochemical analyses, co-immunoprecipitation and yeast two-hybrid assays on PEF proteins such as calpain, ALG-2, grancalcin, sorcin and peflin [28]. This observation that PEF domains are typically paired as heterodimers [3,14,15,43] or homodimers [44–46] argues for p94 homodimerization and points to the PEF domain as being the probable homodimerization interface.
p94DIV was massively produced in E. coli and gave yields that were comparable with those obtained with the homodimer of the rat calpain small subunit [46]. It accumulated in the soluble fraction and behaved during column chromatography as a single entity with a sharp elution profile, consistent with it being a stable, well-folded protein. This was confirmed by analytical ultracentrifugation and CD spectroscopy studies. On Ca2+ binding, it underwent a conformational change that was detected by an increase in intrinsic tryptophan fluorescence. Evidence for p94DIV being a homodimer comes from both size-exclusion chromatography and analytical ultracentrifugation studies. The Mr values of the p94DIV domain deduced from size-exclusion chromatography (40000±5000) and analytical ultracentrifugation studies (43500) are consistent with a calculated theoretical dimer molecular mass of 44362. The value determined by size-exclusion chromatography is also deemed to be reliable because crystallography of calpain PEF domain homo- and heterodimers shows them to be roughly globular in shape (47 Å×48 Å×54 Å; 1 Å=10−10 m) both with and without Ca2+ present [38,47]. The dimer molecular mass calculated from analytical ultracentrifugation (Mr 43500) is constant over a 10-fold concentration range, suggesting that the dimer is very stable.
To investigate further the implications of our biophysical studies described above, we performed some modelling studies. Initially, we mapped the p94DIV sequence on to the rat small subunit (DIV) and observed that the dimer interface is well conserved. We generated a homology model of p94DIV and performed energy minimization and molecular dynamics studies. These showed that p94DIV is capable of forming a homodimer through the same interface as the rat DVI homodimer and the human m-calpain heterodimer (Figure 5).
Given the known propensity of PEF domain proteins to form dimers, we explored the possibility that full-length p94 could dimerize through the pairing of its PEF domain. In the absence of a p94 structure, we made a computer-graphics model of an m-calpain large-subunit homodimer (Figure 6). The m-calpain large subunit has a domain composition similar to that of p94 but lacks the three unique additional sequences (NS1, IS1 and IS2). The regions of the human m-calpain that are replaced by these additional sequences (NS, IS1 and IS2) in p94 are highlighted in Figure 6. Since these additional sequences have no known structural homologues, we were not able to model their structures with confidence. Nevertheless, we can draw some conclusions from this mock-up. The IS1 sequence is remote from the dimerization contact and is unlikely to be affected by dimer formation. The NS sequence of p94 replaces the anchor helix of m-calpain. Although it appears to be in close proximity to the DIV homodimer, its sequence is rich in proline and glycine residues and shows no homology to the anchor helix of m-calpain. Therefore we predict that it will be intrinsically disordered and will not occupy the same space as the m-calpain anchor helix. Thus it is unlikely to interfere with dimer association. The IS2 sequence is present in the m-calpain ‘transducer’ region, which lies away from the dimer interface. Although we have no structural information about IS2, there is nothing in our model that suggests that it should interfere with homodimerization of p94. Also, since IS2 is the binding site for titin, we assume that it would project outwards to favour titin association. In our model, the catalytic cores are orientated in opposite directions (at the two extreme ends), and they are unlikely to autoproteolyse each other. Dimerization removes the fifth EF-hand of DIV as a potential binding site for the interactions of p94 with other proteins. However, dimerization means that all other possible contact sites are duplicated, which could result in stronger binding or simultaneous binding to two ligands (e.g. binding sites on titin) [23].
Homodimerization in this manner might apply to all of the single-chain calpains that have a PEF domain, such as the Drosophila calpains A–C. Furthermore, it should not be assumed that other PEF-containing mammalian calpains (e.g. 8, 11, 12 and 13) will necessarily form a heterodimer with the small subunit in preference to a homodimer, until this is proven by experimentation as with calpains 1, 2 and 9 [48].
Acknowledgments
This work was supported by the Heart and Stroke Foundation of Ontario. We are grateful to the Protein Function Discovery (PFD) Facility of Queen's University and K. Munro for assistance with analytical ultracentrifugation and CD analysis and to S. Gauthier for technical assistance. R. R. is supported by a tuition bursary and a QGA award from Queen's University. P. L. D. holds a Canada Research Chair in Protein Engineering.
References
- 1.Sorimachi H., Suzuki K. The structure of calpain. J. Biochem. (Tokyo) 2001;129:653–664. doi: 10.1093/oxfordjournals.jbchem.a002903. [DOI] [PubMed] [Google Scholar]
- 2.Sorimachi H., Imajoh-Ohmi S., Emori Y., Kawasaki H., Ohno S., Minami Y., Suzuki K. Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- μ-types. Specific expression of the mRNA in skeletal muscle. J. Biol. Chem. 1989;264:20106–20111. [PubMed] [Google Scholar]
- 3.Goll D., Thompson V., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 4.Pinter M., Stierandova A., Friedrich P. Purification and characterization of a Ca2+-activated thiol protease from Drosophila melanogaster. Biochemistry. 1992;31:8201–8206. doi: 10.1021/bi00150a012. [DOI] [PubMed] [Google Scholar]
- 5.Barnes T. M., Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996;15:4477–4484. [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X., Czerwinski E., Mellgren R. Purification and properties of the Dictyostelium calpain-like protein, Cpl. Biochemistry. 2003;42:1789–1795. doi: 10.1021/bi026461+. [DOI] [PubMed] [Google Scholar]
- 7.Lid S. E., Gruis D., Jung R., Lorentzen J. A., Ananiev E., Chamberlin M., Niu X., Meeley R., Nichols S., Olsen O. A. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5460–5465. doi: 10.1073/pnas.042098799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margis R., Margis-Pinheiro M. Phytocalpains: orthologous calcium-dependent cysteine proteinases. Trends Plant Sci. 2003;8:58–62. doi: 10.1016/S1360-1385(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 9.Carafoli E., Molinari M. Calpain: a protease in search of a function? Biochem. Biophys. Res. Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- 10.Santella L. The role of calcium in the cell cycle: facts and hypotheses. Biochem. Biophys. Res. Commun. 1998;244:317–324. doi: 10.1006/bbrc.1998.8086. [DOI] [PubMed] [Google Scholar]
- 11.Wang K. K. Calpain and caspase: can you tell the difference? Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- 12.Glading A., Lauffenburger D. A., Wells A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol. 2002;12:46–54. doi: 10.1016/s0962-8924(01)02179-1. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y., Wang K. K. The calpain family and human disease. Trends Mol. Med. 2001;7:355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- 14.Hosfield C. M., Elce J. S., Davies P. L., Jia Z. Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999;18:6880–6889. doi: 10.1093/emboj/18.24.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strobl S., Fernandez-Catalan C., Braun M., Huber R., Masumoto H., Nakagawa K., Irie A., Sorimachi H., Bourenkow G., Bartunik H., et al. The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc. Natl. Acad. Sci. U.S.A. 2000;97:588–592. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kretsinger R. H. EF-hands embrace. Nat. Struct. Biol. 1997;4:514–516. doi: 10.1038/nsb0797-514. [DOI] [PubMed] [Google Scholar]
- 17.Tompa P., Emori Y., Sorimachi H., Suzuki K., Friedrich P. Domain III of calpain is a Ca2+-regulated phospholipid-binding domain. Biochem. Biophys. Res. Commun. 2001;280:1333–1339. doi: 10.1006/bbrc.2001.4279. [DOI] [PubMed] [Google Scholar]
- 18.Moldoveanu T., Hosfield C. M., Lim D., Elce J. S., Jia Z., Davies P. L. A Ca2+ switch aligns the active site of calpain. Cell (Cambridge, Mass.) 2002;108:649–660. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 19.Reverter D., Strobl S., Fernandez-Catalan C., Sorimachi H., Suzuki K., Bode W. Structural basis for possible calcium-induced activation mechanisms of calpains. Biol. Chem. 2001;382:753–766. doi: 10.1515/BC.2001.091. [DOI] [PubMed] [Google Scholar]
- 20.Kramerova I., Kudryashova E., Tidball J. G., Spencer M. J. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 21.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell (Cambridge, Mass.) 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 22.Diaz B. G., Moldoveanu T., Kuiper M. J., Campbell R. L., Davies P. L. Insertion sequence 1 of muscle-specific calpain, p94, acts as an internal propeptide. J. Biol. Chem. 2004;279:27656–27666. doi: 10.1074/jbc.M313290200. [DOI] [PubMed] [Google Scholar]
- 23.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., et al. Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J. Biol. Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 24.Sorimachi H., Ishiura S., Suzuki K. Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 1993;268:10593–10605. [PubMed] [Google Scholar]
- 25.Federici C., Eshdat Y., Richard I., Bertin B., Guillaume J. L., Hattab M., Beckmann J. S., Strosberg A. D., Camoin L. Purification and identification of two putative autolytic sites in human calpain 3 (p94) expressed in heterologous systems. Arch. Biochem. Biophys. 1999;363:237–245. doi: 10.1006/abbi.1998.1091. [DOI] [PubMed] [Google Scholar]
- 26.Branca D., Gugliucci A., Bano D., Brini M., Carafoli E. Expression, partial purification and functional properties of the muscle-specific calpain isoform p94. Eur. J. Biochem. 1999;265:839–846. doi: 10.1046/j.1432-1327.1999.00817.x. [DOI] [PubMed] [Google Scholar]
- 27.Kinbara K., Ishiura S., Tomioka S., Sorimachi H., Jeong S. Y., Amano S., Kawasaki H., Kolmerer B., Kimura S., Labeit S., et al. Purification of native p94, a muscle-specific calpain, and characterization of its autolysis. Biochem. J. 1998;335:589–596. doi: 10.1042/bj3350589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maki M., Kitaura Y., Satoh H., Ohkouchi S., Shibata H. Structures, functions and molecular evolution of the penta-EF-hand Ca2+-binding proteins. Biochim. Biophys. Acta. 2002;1600:51–60. doi: 10.1016/s1570-9639(02)00444-2. [DOI] [PubMed] [Google Scholar]
- 29.Laue T. M., Stafford W. F., III Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 1999;28:75–100. doi: 10.1146/annurev.biophys.28.1.75. [DOI] [PubMed] [Google Scholar]
- 30.Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem. J. 1964;91:222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackereth F. J. H. Ambleside: Freshwater Biological Association; 1978. Water Analysis: Some Revised Methods for Limnologists. [Google Scholar]
- 32.Bohm G., Muhr R., Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 33.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano W. L. San Carlos, CA: DeLano Scientific; 2003. The PyMOL molecular graphics system. http://www.pymol.org. [Google Scholar]
- 35.Sali A., Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl E., Hess B., van der Spoel D. A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 37.Berendsen H. J. C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comp. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 38.Blanchard H., Grochulski P., Li Y., Arthur J. S., Davies P. L., Elce J. S., Cygler M. Structure of a calpain Ca2+-binding domain reveals a novel EF-hand and Ca2+-induced conformational changes. Nat. Struct. Biol. 1997;4:532–538. doi: 10.1038/nsb0797-532. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Farkas A., Tompa P., Friedrich P. Revisiting ubiquity and tissue specificity of human calpains. Biol. Chem. 2003;384:945–949. doi: 10.1515/BC.2003.106. [DOI] [PubMed] [Google Scholar]
- 41.Herasse M., Ono Y., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., et al. Expression and functional characteristics of calpain 3 isoforms generated through tissue-specific transcriptional and posttranscriptional events. Mol. Cell. Biol. 1999;19:4047–4055. doi: 10.1128/mcb.19.6.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinbara K., Sorimachi H., Ishiura S., Suzuki K. Skeletal muscle-specific calpain, p49: structure and physiological function. Biochem. Pharmacol. 1998;56:415–420. doi: 10.1016/s0006-2952(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 43.Kitaura Y., Matsumoto S., Satoh H., Hitomi K., Maki M. Peflin and ALG-2, members of the penta-EF-hand protein family, form a heterodimer that dissociates in a Ca2+-dependent manner. J. Biol. Chem. 2001;276:14053–14058. doi: 10.1074/jbc.M008649200. [DOI] [PubMed] [Google Scholar]
- 44.Blanchard H., Li Y., Cygler M., Kay C. M., Simon J., Arthur C., Davies P. L., Elce J. S. Ca2+-binding domain VI of rat calpain is a homodimer in solution: hydrodynamic, crystallization and preliminary X-ray diffraction studies. Protein Sci. 1996;5:535–537. doi: 10.1002/pro.5560050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamada H., Okochi E., Oh-hara T., Tsuruo T. Purification of the Mr 22000 calcium-binding protein (sorcin) associated with multidrug resistance and its detection with monoclonal antibodies. Cancer Res. 1988;48:3173–3178. [PubMed] [Google Scholar]
- 46.Graham-Siegenthaler K., Gauthier S., Davies P. L., Elce J. S. Active recombinant rat calpain II. Bacterially produced large and small subunits associate both in vivo and in vitro. J. Biol. Chem. 1994;269:30457–30460. [PubMed] [Google Scholar]
- 47.Lin G. D., Chattopadhyay D., Maki M., Wang K. K., Carson M., Jin L., Yuen P. W., Takano E., Hatanaka M., DeLucas L. J., et al. Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding. Nat. Struct. Biol. 1997;4:539–547. doi: 10.1038/nsb0797-539. [DOI] [PubMed] [Google Scholar]
- 48.Lee H. J., Tomioka S., Kinbara K., Masumoto H., Jeong S. Y., Sorimachi H., Ishiura S., Suzuki K. Characterization of a human digestive tract-specific calpain, nCL-4, expressed in the baculovirus system. Arch. Biochem. Biophys. 1999;362:22–31. doi: 10.1006/abbi.1998.1021. [DOI] [PubMed] [Google Scholar]